Abstract
Patients with advanced hepatocellular carcinoma (HCC) face a dismal prognosis due to a lack of any effective therapies. To address this situation, we conducted a preclinical investigation of the therapeutic efficacy of oligonucleotides directed against the oncogenic microRNA miR-221 which has been implicated in HCC. Of 9 chemistries evaluated, we determined that a 2′-O-methyl phosphorothioate-modified anti-miR-221 oligonucleotide was most effective at reducing proliferation in vitro. A cholesterol-modified isoform of anti-miR-221 (chol-anti-miR-221) exhibited improved pharmacokinetics and liver tissue distribution compared to unmodified oligonucleotide. Chol-anti-miR-221 significantly reduced miR-221 levels in liver within a week of intravenous administration and in situ hybridization studies confirmed accumulation of the oligonucleotide in tumor cells in vivo. Within the same period, chol-anti-miR-221 reduced tumor cell proliferation and increased markers of apoptosis and cell cycle arrest, elevating the tumor doubling time and increasing mouse survival. Taken together, our findings offer a preclinical proof of efficacy for chol-anti-miR-221 in a valid orthotopic mouse model of HCC, suggesting that this targeted agent could benefit treatment of advanced HCC patients.
Keywords: microRNA, antisense, antagomiR, HCC, liver
Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of cancer death worldwide. Accounting for an estimated 549,000 deaths per year, it represents 10% of all deaths from cancer (1). At its earliest stages, HCC is treatable by resection or transplantation. Percutaneous ablation is an option in patients who are afflicted with early HCC and who are not candidates for resection or transplantation (2). Transarterial chemoembolization has been effective in patients with intermediate stage HCC (3). HCC patients that are diagnosed with advanced disease or whose cancer recurs following regional therapy have a dismal prognosis. Doxorubicin chemotherapy produces response rates of 10 to 15% with no survival advantage (4). Combination therapy with cisplatin/interferon/doxorubicin/5-fluorouracil produced somewhat higher response rates than single agent doxorubicin, however at the expense of significantly higher toxicity (5, 6). New therapies for HCC include EGFR inhibitors (7) and antiangiogenic compounds such as bevacizumab (8, 9) and sunitinib (10). In a phase III trial, advanced HCC patients treated with the molecular targeted agent sorafenib, reported an increase in survival of ∼ 3 months (11). Clearly, new agents must be developed to treat advanced HCC.
Extensive profiling studies over the past several years have demonstrated that various miRNAs are differentially expressed in HCC and other types of cancers (12-20). miRNAs with increased expression in the tumor often target tumor suppressors. Differentially expressed miRNAs in HCC include miR-221, miR-21 and miR-18 (increased expression in HCC) and miR-122a, miR-199a* and miR-200 (reduced expression in HCC) (reviewed in (21, 22)). In the present study, we focused our attention on miR-221 as a therapeutic target. miR-221 expression was increased in the HCC tumors compared to non-diseased and adjacent benign liver (12-14, 18, 20). In highly aggressive HCCs, miR-221/-222 was among the most up-regulated of all miRNAs studied (18). miR-221 targets a number of key tumor suppressors including p27Kip1(23-25), p57Kip2(26, 27), PTEN (28), an tissue inhibitor of metalloproteinase-3 (TIMP3) (28) and the DNA damage-inducible transcript 4 (DDIT4), a modulator of the mTOR pathway (18). miR-221 antisense oligonucleotides reduced in vitro cell proliferation in HCC (18) and pancreatic cancer (29).
Antisense oligonucleotides targeted to miRNA are effective in mice (30, 31) and primates (32, 33). A human clinical trial using anti-miR-122 to treat hepatitis C infection has recently been completed (Santaris Pharma, ClinicalTrials.gov). An issue that has impeded the use of antisense oligonucleotides is the delivery and cellular accumulation to the target organ. It is well known that antisense and other oligonucleotides preferentially accumulate in highly perfused organs such as the liver. Therefore targeting the liver with anti-miRNA oligonucleotides presents an ideal opportunity. Injection of 2′ MOE antisense oligonucleotides to the liver specific miR-122 was effective at miR-122 degradation, target gene up-regulation and cholesterol lowering in mice (30). LNA antisense oligonucleotides to miR-122 administered to monkeys resulted in depletion of mature miR-122 and reduced plasma cholesterol levels (32). Primates dosed with LNA antisense to miR-122 had reduced HCV viral load without evidence of viral resistance or side effects (33).
The purpose of this study was to evaluate anti-miR-221 oligonucleotides as a potential therapeutic for HCC in mice. Our goal was to demonstrate that anti-miR-221 oligonucleotides could be delivered to liver tumors and be effective in modulating miRNA expression and produce a functional effect on validated targets in orthotopic human HCC xenografts. We purposed that anti-miR-221 oligonucleotides would accumulate in the HCC tumors, reduce endogenous miR-221 oligonucleotides, modulate miR-221-related protein levels, and enhance the survival of tumor bearing mice. Our results demonstrate the successful accomplishment of these goals.
Materials and Methods
Cell lines
The PLC/PRF/5 cell line was purchased from American Type Tissue Collection prior to 2005 (Manassas, VA). The PLC/PRF/5 cells were authenticated using short tandem repeat analysis in March, 2011 by Johns Hopkins University. Our analysis demonstrated that the cells were successfully authenticated. Cells were cultured in RPMI 1640 medium with 10% fetal bovine serum using standard conditions. PLC/PRF/5 cells were stably transfected with luciferase (luc) expressing construct to generate PLC/PRF/5-luc cells.
Oligonucleotides
miR-221 antisense and scrambled control oligonucleotides were synthesized from ThermoFisher (Lafayette, CO). Oligonucleotides contained the natural nucleotides and phosphodiester bond or were chemically modified to contain a phosphorothioate (PS) linkage, 2′ OMe, 2′ F, LNA, 5′ cholesterol or a 3′ inverted deoxythymidine (idT) (Supplemental Table S1). The oligonucleotides for the mouse studies were processed for in vivo work by the manufacturer and include sterilization and testing for endotoxin.
Antisense Oligonucleotide Transfection
The cells (1,200 cells/well) were plated 1 day before transfection and the oligonucleotides were transfected using Lipofectamine 2000 and Opti-MEM medium (Invitrogen, Carlsbad, CA) following the manufacturer's protocol. Transfection efficiency was measured using a DY547 fluorescently-labeled miR-16 oligonucleotide.
Cell Proliferation Assay
Cell proliferation was performed using the reagent WST-1 (Roche, Indianapolis, IN). Cells were seeded into 96-well plates at 1,200 cells per well. On the following day, the cells were transfected with anti-miRNA oligonucleotides and allowed to incubate for predetermined time (e.g., 48 hours). Fifteen μl of WST-1 was then added to the cell culture medium and incubated for 2 hours. Sample absorbance was analyzed using a microplate enzyme-linked immunosorbent assay reader at 490 nm. All experiments were performed at least in triplicate.
qPCR of microRNA
The mature miRNA was quantified by qPCR using TaqMan® microRNA Assays (Applied Biosystems Foster City, CA). The references gene was 18S rRNA. qPCR (10 μl reaction) was performed using 1 μl of a 1:50 dilution of cDNA. Duplicate PCRs were performed for each miRNA. The mean CT was determined from the duplicate PCRs and the data were presented using the comparative CT method.
Pilot pharmacokinetic studies
All animal experiments were carried out under protocols approved by the Ohio State University Institutional Laboratory Animal Care and Use Committee. Female C57BL/6 mice were obtained from Harlan Sprague Dawley and maintained in a clean room with a 12 h light-dark cycle and provided food and water ad libitum. A cholesterol-modified isoform of anti-miR-221 (chol-anti-miR-221) and unmodified anti-miR-221 (non-chol-anti-miR-221) were freshly formulated with phosphate buffered saline and administrated to mice through tail vein injection at the dose of 7.5 mg/kg. After administration, mice were euthanized at various time points between 5 minutes and 36 hours with carbon dioxide followed immediately by exsanguination via cardiac puncture. Blood samples were transferred to lithium heparin tubes and centrifuged at 6000×g for 2 min. Liver tissue was flash frozen. Liver and plasma were stored at -80°C until analysis.
qPCR of anti-miR-221 for pharmacokinetic analysis
To perform the pharmacokinetic studies, we developed a novel qPCR method to assay anti-miR-221 in plasma and tissue (additional details provided in the legend to Supplemental Fig. S1). Since the 3′ cholesterol interfered with the reverse transcription (data not shown), the cholesterol label was moved to the 5′ end of the oligo and in its place a 3′ inverted idT was added to inhibit 3′ exonuclease activity. This compound is referred to here as chol-anti-miR-221 and is essentially identical to the antagomiR chemistry (31) with the exception of the aforementioned changes to the 5′ and 3′ ends of the oligo. To quantitatively assay the amount of anti-miR-221 in mouse plasma and tissue homogenate samples, duplicate standard curves were produced in control plasma and liver homogenate using 1 to 109 copies of anti-miR-221 oligo per PCR (representative curves were shown in Supplemental Fig. S1A and B). Two μl of samples from standard curves and mouse blood and liver homogenates was used to synthesize the first strand cDNA as described (34) using primers and probes specific to the anti-miR-221 oligo. Accuracy and precision of the PCR assay were determined for both intra- and inter-runs. CV (coefficients of variation) is an indication of precision. The percentage relative error at three concentrations of oligonucleotide, representing the entire range of samples analyzed, was summarized in Supplemental Table S2.
Pharmacokinetic analysis
Pharmacokinetic data analysis and modeling were performed using WinNonlin v.5.2 (Pharsight, Mountain View, CA). Non-compartmental analyses were completed with uniform weighting and linear trapezoidal AUC calculations. For compartmental analysis, 2- and 3- compartment models were utilized for non-chol-anti-miR-221 and chol-anti-miR-221, respectively.
Western blot analysis
Protein was harvested using CellLytic™ MT (Sigma–Aldrich, St. Louis, MO) and 1× protease and phosphatase inhibitor (Pierce, Rockford, IL) using standard techniques. Protein concentration was measured using the BCA Protein Assay Kit (Pierce). Thirty micrograms of total protein extract was separated on a 10% SDS–PAGE gel. Blotting was performed for p27kip1, p57kip2, and PTEN (Santa Cruz Biotechnology, Santa Cruz, CA). β-actin (Abcam, Cambridge, MA) was used as the loading control. Secondary horseradish peroxidase antibody was detected using ECL Western Blotting Analysis System (Amersham Biosciences, Piscataway, NJ).
Northern blotting
Total RNA was isolated from the frozen mouse tissues using Trizol reagent (Life Technologies, Carslbad, CA) or RNeasy® Plus Mini kit (Qiagen, Germany) following the manufacturer's protocol. Northern blotting was performed as previously described (35). LNA probes specific to the mature miR-221 were purchased from Exiqon (Denmark).
Establishment of murine orthotopic HCC xenografts
Orthotopic tumors were established by the direct intrahepatic injection of PLC/PRF/5-luc cells (1,000,000 cells suspended in matrigel) into the left hepatic lobe. Starting 10-14 days after orthotopic implantation, and every week thereafter, tumor burden was determined by bioluminescence imaging using the IVIS200 imaging system (Xenogen Corp., Alameda, CA), 10 minutes after intraperitoneal administration of 150 mg/kg body weight D-luciferin (Gold Biotechnology, St. Louis, MO). Once bioluminescence exceeded 1 × 106 photons/sec, mice were randomized to receive either 60 mg/kg chol-anti-miR-221(n=7) or chol-labeled scrambled control antisense oligonucleotide (n=5) three days a week, for two weeks, intravenously. Body weight was measured and bioluminescence imaging was performed weekly for the first four weeks. Tumor doubling time was calculated by modeling the data to the standard equation for exponential growth (At = A0ekt). Mice were followed for 10 weeks, and survival curves analyzed using the Kaplan-Meier method.
In situ detection of miRNA and immunohistochemistry
LNA probes that hybridized to endogenous miR-221 and to the miR-221 antisense oligonucleotide and 5′ end labeled with digoxigenin were purchased from Exiqon (Denmark). In situ hybridization was performed on the FFPE sections of tumor tissue as described (36) and were blinded to the experimental conditions. Nuclear fast red was used as the counterstain. p27Kip1, Ki-67, and cleaved caspase-3 (Santa Cruz Biotechnology) were detected using standard immunohistochemistry techniques. Co-localization analysis was performed with the Nuance system from Cambridge Research Institute (Woburn, MA) according to their specifications.
Results
Evaluation of chemically modified anti-miR-221
The natural oligonucleotide chemistry (2′ OH and phosphodiester backbone) as well as eight chemical modifications of the anti-miR-221 and scrambled control oligonucleotides (Supplemental Table S1) were evaluated. The anti-proliferative activity of the modified anti-miR-221 or scrambled control oligos was studied in PLC/PRF/5 cells following lipofectamine transfection. The most effective chemical modification at reducing cell proliferation was the 2′ OMe, PS; reducing cell proliferation by 25% at both the 48 and 72 h time points (Supplemental Fig. S2A and B). Since previous work showed that a cholesterol end label was effective at increasing targeting and uptake to the liver in vivo (31), all of the subsequent in vivo work was performed using a cholesterol labeled 2′ OMe, PS anti-miR-221 (chol-anti-miR-221) and cholesterol labeled scrambled control oligos (Supplemental Table S1).
Pharmacokinetics of cholesterol and non-cholesterol labeled anti-miR-221
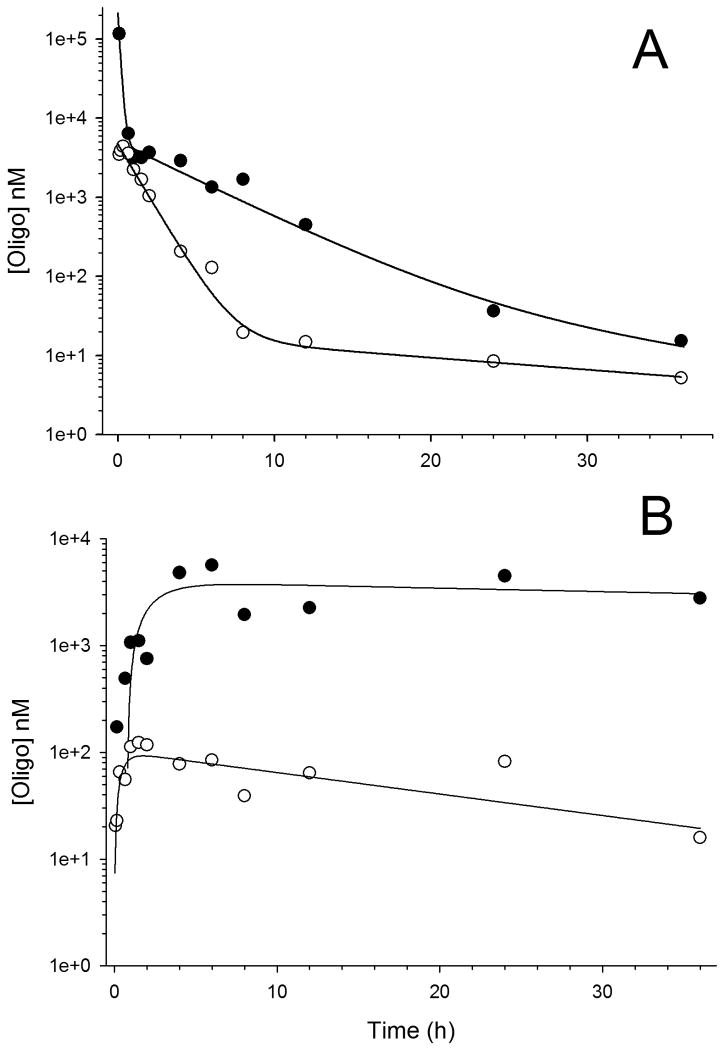
Pilot studies were completed to evaluate plasma pharmacokinetics and liver distribution of both chol-anti-miR-221 and non-chol-anti-miR-221 in C57Bl/6 mice. Fig. 1 displays concentration-time profiles and model fits for the plasma and liver data. Estimated non-compartmental pharmacokinetic parameters are provided in Table 1. A comparison of the chol-anti-miR-221 and non-chol-anti-miR-221 data indicates the cholesterol modification significantly alters anti-miR-221 oligonucleotide pharmacokinetics. Fig. 1A indicates a prolonged distribution phase of chol-anti-miR-221, resulting in significantly increased plasma area under the curve (AUClast 74.4 vs. 7.3 hr*μg/ml for chol-anti-miR-221 and non-chol-anti-miR-221, respectively). This altered pharmacokinetic behavior is reflected in the liver data (Fig. 1B), which indicates approximately 50-fold higher maximum concentrations (Cmax) and AUCs of chol-anti-miR-221 compared to non-chol-anti-miR-221. The modified pharmacokinetic properties of chol-anti-miR-221 demonstrate the cholesterol modification achieves the desired effects of increasing concentrations and prolonging exposures of the anti-miR-221 in liver tissue.
Figure 1. Plasma and liver pharmacokinetics of anti-miR-221 oligonucleotides.
Plasma (A) and liver (B) concentration-time profiles of chol-anti-miR-221 (solid circles) and non-chol-anti-miR-221 (open circles) after single i.v. injections of oligo (7.5 mg/kg) into C57Bl/6 mice. Lines represents one- (liver data) and two- (plasma data) compartment model fits (WinNonlin v. 5.2).
Table 1. Non-compartment pharmacokinetic parameters for non-chol-anti-miR-221 and chol-anti-miR-221.
| Mouse plasma | ||
| Non-chol-anti-miR-221 | Chol-anti-miR-221 | |
| T1/2 (hr) | 15.8 | 4.1 |
| AUClast (hr*mg/ml) | 7.3 | 74.4 |
| Cl (ml/hr/kg) | 1,013 | 101 |
|
| ||
| Mouse liver | ||
| Non-chol-anti-miR-221 | Chol-anti-miR-221 | |
| T1/2 (hr) | 1.5 | 6.0 |
| Cmax (ng/ml) | 123 | 5,654 |
| AUClast (hr*mg/ml) | 2.3 | 118 |
Note. T1/2 = half life; AUClast = area under the concentration-time curve from time zero to last time with the quantifiable concentration; Cl = clearance; Cmax = maximum observed concentration of an oligo after intravenous administration.
Hepatic accumulation and activity of chol-anti-miR-221 in normal and tumor bearing mice
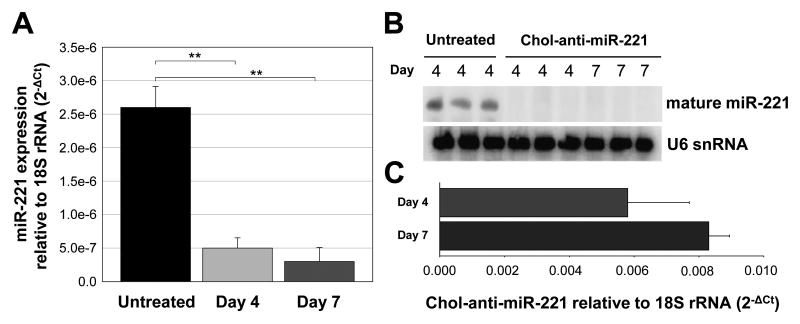
C57Bl/6 mice were injected via the tail vein with 7.5 mg/kg of chol-anti-miR-221 three times over a 10 day period; mice were then sacrificed at days 11, 14 and 17. The amount of endogenous miR-221 in the whole liver was reduced by about 2-fold at day 11, but the effect was short lived with no significant change in miR-221 levels at days 14 and 17 (data not shown). For this reason we increased the dose to 30 mg/kg. C57Bl/6 mice injected with 3 daily doses of 30 mg/kg via the tail vein were sacrificed on days 4 and 7 and the amount of chol-anti-miR-221 and endogenous miR-221 was assayed in whole liver. Both qPCR and Northern blotting demonstrated that chol-anti-miR-221 significantly reduced the levels of endogenous miR-221 (Fig. 2A and B). The reduction in endogenous miR-221 correlated with increased accumulation of the chol-anti-miR-221 in the liver (Fig. 2A and C). Endogenous miR-221 was reduced by 80 and 90% at days 4 and 7, respectively (Fig. 2A). The expression of 4 control miRNAs was not altered by the chol-anti-miR-221 treatment (Supplemental Fig. S3).
Figure 2. Liver accumulation and activity of chol-anti-miR-221 in normal mice.
C57BL/6 mice were injected via the tail vein with 3 daily doses of 30 mg/kg of chol-anti-miR-221. Mice were sacrificed on days 4 and 7 and the endogenous miR-221 in the liver was determined by both qPCR (A) and Northern blotting (B). Hepatic chol-anti-miR-221 levels were also determined in the liver by qPCR (C). ** P < 0.01, Student's t-test.
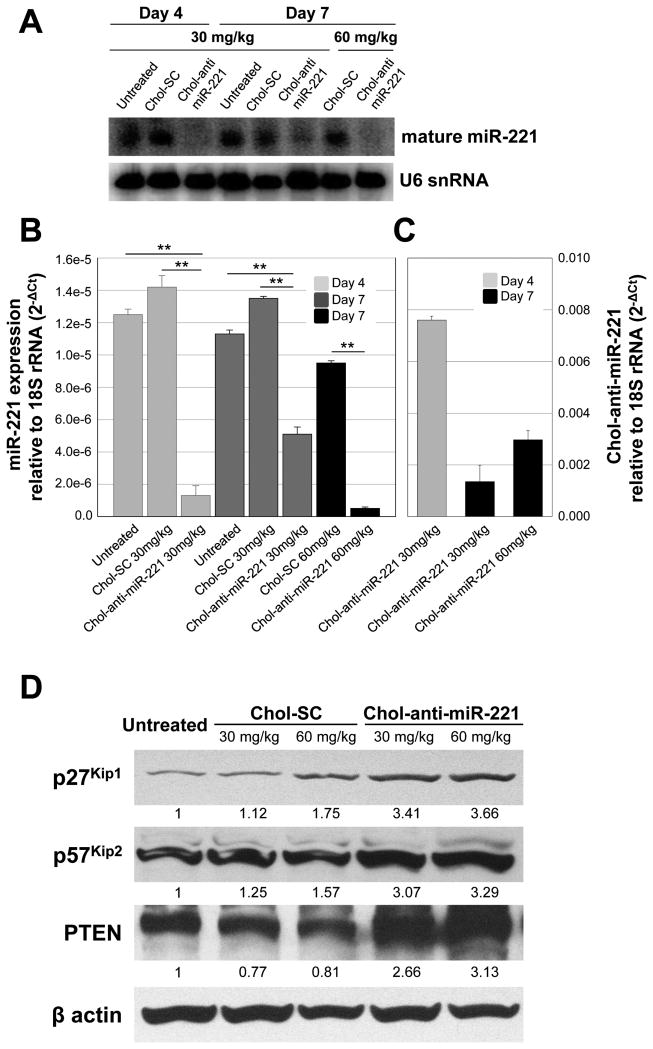
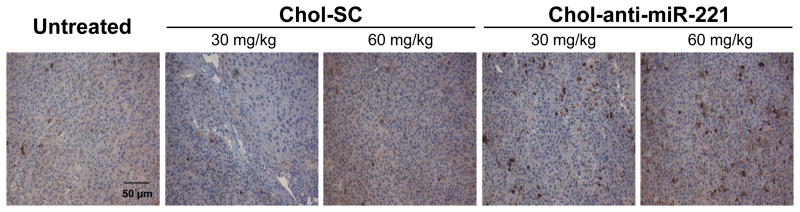
Mice bearing xenograft orthotopic tumors were injected via the tail vein with 30 or 60 mg/kg of the chol-anti-miR-221 or cholesterol labeled scrambled control oligo for 3 consecutive daily doses. Mice were sacrificed at days 4 and 7 and the amount of endogenous miR-221 and chol-anti-miR-221 was assayed in the tumor tissue. Similar to the effect in normal liver, chol-anti-miR-221 produced a significant reduction in the endogenous miR-221 levels in the tumor (Fig. 3A and B). The miR-221 levels in the cholesterol labeled scrambled control oligo treatment did not differ from that of the untreated control (Fig. 3A and B). There was a trend between the accumulation of chol-anti-miR-221 (30 mg/kg dose) and endogenous miR-221 levels in the tumor between days 4 and 7; comparing the 30 and 60 mg/kg doses (day 7), endogenous miR-221 decreased with increasing accumulation of chol-anti-miR-221 (Fig. 3C). To determine if the chol-anti-miR-221 modulated three miR-221 target proteins, we assayed p27Kip1, p57Kip2 and PTEN levels by western blotting. Following 7 days of treatment, the 60 mg dose of chol-anti-miR-221 increased the p27Kip1, p57Kip2 and PTEN protein levels by 3-fold (Fig. 3D). There was a slight increase in the p27Kip1 and p57Kip2 protein in the cholesterol labeled scrambled control oligo treated tumors (60 mg/kg) compared to the untreated control (Fig. 3D). To determine the degree of cycling cells, we performed Ki-67 staining on these tissues. Ki-67 staining was significantly reduced in the tumors of the mice treated with chol-anti-miR-221 compared to those mice treated with cholesterol labeled scrambled control (Fig. 4A, B, and Supplemental Fig. S4).
Figure 3. Tumor accumulation and activity of chol-anti-miR-221 in orthotopic tumor xenograft mice.
Tumor bearing mice were generated by injecting 1 × 106 luciferase expressing PLC/PRF/5 cells directly into the left hepatic lobe of an immunodeficient mouse. Mice that displayed sufficient bioluminescence of tumor cells (> 1 × 106 photon/sec) were dosed via tail vein injection with 3 daily doses of 30 or 60 mg/kg of chol-anti-miR-221 or cholesterol labeled scrambled control (chol-SC). Mice were sacrificed on days 4 and 7 and the endogenous miR-221 in the tumor was determined by both Northern blotting (A) and qPCR (B). (C) Tumor levels of chol-anti-miR-221 as determined by qPCR. (D) p27Kip1, p57Kip2, PTEN and β actin protein levels in the tumors of mice treated with cholesterol-labeled scrambled control oligo (chol-SC) or chol-anti-miR-221 was determined by western blotting. ** P < 0.01, Student's t-test.
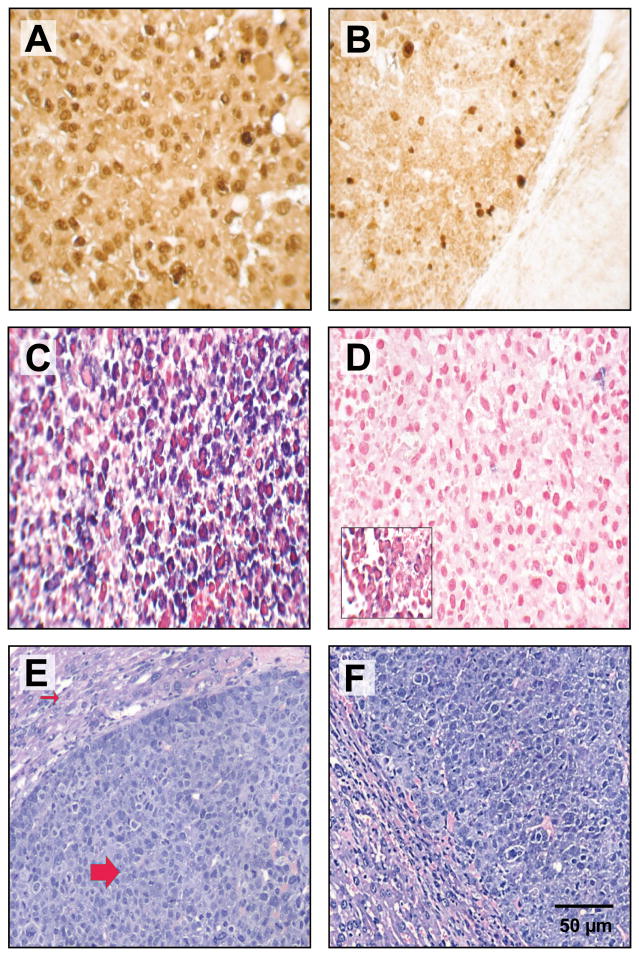
Figure 4. Chol-anti-miR-221 cellular activity and localization in orthotopically implanted hepatocellular carcinoma.
Mice orthotopically implanted with hepatocellular carcinoma were injected via the tail vein with 3 daily doses of 60 mg/kg chol-anti-miR-221 or cholesterol-labeled scrambled control oligonucleotide (chol-SC). Mice were sacrificed 7 days after receiving the initial dose. Ki-67 staining in the tumors of mice treated with 60 mg/kg chol-SC oligo (A) or chol-anti-miR-221 (B). In situ hybridization using probes to anti-miR-221 (C) or endogenous miR-221 (D) was performed in the tumors of the mice treated with chol-anti-miR-221 (C and D) or chol-SC oligo (D, insert). Sections were counterstained with nuclear fast red dye. H&E staining in the tumors of mice treated with 60 mg/kg chol-SC oligo (E) or chol-anti-miR-221 (F). The H&E shows the highly anaplastic tumor cells (large arrow) invading the adjacent liver (small arrow).
To visualize the hepatic accumulation and activity of chol-anti-miR-221, in situ hybridization was performed in the liver and tumors of orthotopic mice treated with both chol-anti-miR-221 and cholesterol labeled scrambled control oligo. Significant accumulation of anti-miR-221 occurred in the tumors (Fig. 4C) of mice 7 days after the initiation of therapy. The chol-anti-miR-221 was predominately localized to the cytoplasm in the tumor (Fig. 4C). Probing the tumors for endogenous miR-221 showed a complete lack of miR-221 expression in the mice treated with chol-anti-miR-221 (Fig. 4D) but not cholesterol labeled scrambled control oligo (Fig. 4D, insert). There is no evidence of hepatocellular toxicity in the chol-anti-miR-221, cholesterol labeled scrambled oligo (Fig. 4E and F) or saline control treated mice (not shown). Co-localization studies were performed to determine the spatial expression of both chol-anti-miR-221 and p27Kip1 protein. As described above, chol-anti-miR-221 was abundant in the tumors of the treated mice (Supplemental Fig. S5A) as was expression of the p27Kip1 (Supplemental Fig. S5B). The intense yellow color in the merged figure (Supplemental Fig. S5C) demonstrates that p27Kip1 protein is increased in areas that contain high levels of chol-anti-miR-221. There was a dose-dependent increase in the active, cleaved-form of caspase-3 in the chol-anti-miR-221 treated tumors compared to cholesterol labeled control oligo (Fig. 5 and Supplemental Fig. S6).
Figure 5. Activated caspase-3 expression in orthotopically implanted hepatocellular carcinoma.
Mice were dosed via tail vein injection with 3 daily doses of 30 or 60 mg/kg of chol-anti-miR-221 or cholesterol labeled scrambled control (chol-SC). Mice were sacrificed 7 days after receiving the initial dose. Immunohistochemistry was performed to examine caspase-3 activity with anti-cleaved caspase-3 antibody. Sections were counterstained with H&E.
Anti-miR-221 treatment in orthotopic tumor xenograft mice
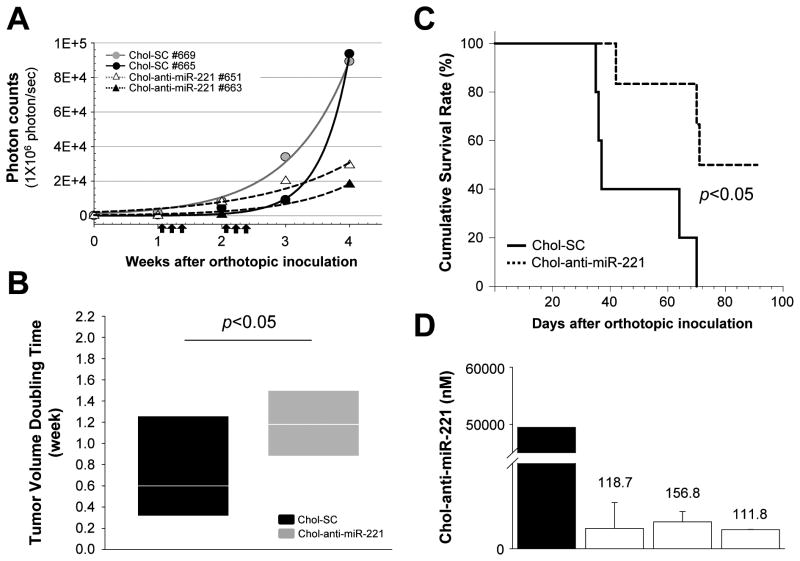
In order to further study the efficacy of chol-anti-miR-221, survival studies were initiated. Nude mice implanted with PLC/PRF/5-luc cells were examined for bioluminescence prior to the beginning of the study. Mice bearing detectable bioluminescence (> 1 × 106 photon/sec) were randomized and then injected 3 times per week with 60 mg/kg of the chol-anti-miR-221 or cholesterol labeled scrambled control oligo for 2 weeks. There was no discernable difference in either bioluminescence or body weight among the treated and control groups at the beginning of the study (Supplemental Fig. S7). Mice were imaged weekly for 4 weeks. Chol-anti-miR-221 oligo increased the tumor doubling time by 1.6-fold (P < 0.05, Fig. 6A and B). Chol-anti-miR-221 treatment improved survival of the orthotopic tumor xenograft mice compared to the cholesterol labeled scrambled control (Fig. 6C, P = 0.0470).
Figure 6. Chol-anti-miR-221 treatment in orthotopic tumor xenograft mice.
Tumor bearing mice were generated by injecting 1 × 106 cells of luciferase expressing PLC/PRF/5 cells directly into the left hepatic lobe of an immunodeficient mouse. Bioluminescence imaging was performed weekly to monitor tumor growth. Once the bioluminescence of tumor cells exceeded 1 × 106 photon/sec, mice were randomized into 2 groups that received treatment with chol-anti-miR-221 or cholesterol labeled scrambled control oligo (chol-SC) at 60 mg/kg intravenously, 3 days/week for 2 weeks. (A) Kinetics of in vivo tumor growth of representative mice from each group receiving chol-SC or chol-anti-miR-221. Arrows indicate treatment points of oligos. (B) Tumor burden (photon count) and tumor doubling time were calculated by injecting mice intraperitoneally with 150 mg/kg D-luciferin every 7 day following tumor injection and were imaged using the IVIS xenogen system. Treatment of chol-anti-miR-221 increased the in vivo doubling time for PLC/PRF/5 cells from 0.75 ± 0.52 to 1.25 ± 0.4 weeks (mean ± SD). (C) Cumulative survival curve of chol-anti-miR-221 and chol-SC treated mice by the Kaplan-Meier method. Chol-anti-miR-221 treatment improved survival of the orthotopic tumor xenograft mice (P = 0.0470, log-rank test). (D) Chol-anti-miR-221 levels were determined by qPCR in the tumors of 3 mice treated with chol-anti-miR-221 that survived for 10 weeks. A black bar indicates mouse sacrificed 7 days after start of treatment. Open bars denote mice sacrificed following 10 weeks of treatment.
Discussion
In these studies, we demonstrated the feasibility and therapeutic efficacy of selectively targeting a miRNA that is over-expressed in HCC using antisense oligonucleotides. These observations are novel for several reasons. First, a survival benefit was observed using intravenous administration of a therapeutic agent in an orthotopic model of HCC. Useful pre-clinical information is provided using a systemic therapy to directly target intrahepatic tumors, an approach which is highly germane and relevant to human HCC. Next, antisense oligonucleotides were used to selectively target miRNA. Their use for targeting miRNA has distinct advantages compared to their use for targeting mRNA. Finally, these studies demonstrate the therapeutic efficacy of targeting a single aberrantly over-expressed miRNA, which may have multiple cancer-relevant targets. Thus, they provide proof-of-principle for further development of miRNA targeted therapies.
Treatment of tumor-bearing mice with chol-anti-miR-221 resulted in a survival advantage compared to mice exposed to cholesterol-modified control oligonucleotides. These observations support an anti-tumoral effect of targeting miR-221, one of the most consistently over-expressed miRNA in HCC. Our observations show an increase in tumoral expression of p27Kip1, p57Kip2 and PTEN by chol-anti-miR-221. These three cell-cycle regulatory factors/tumor suppressors have been implicated in human HCC. Thus, alterations in these factors support reduced tumor cell cycle progression as a mechanism by which chol-anti-miR-221 enhances survival of tumor bearing mice. Further support for this is provided by the significant reduction of Ki-67, increase in tumor doubling time and increased cleaved caspase-3 in tumors treated with chol-anti-miR-221. Given that a single miRNA may have multiple functional targets, it is possible that other tumoral or systemic effects of chol-anti-miR-221 could also contribute to the improved survival. Further studies to elucidate and identify the mechanisms of increased survival with chol-anti-miR-221 are clearly warranted, for example the effect of chol-anti-miR-221 on the formation of micrometastasis.
The feasibility of targeting miRNA to modulate metabolism in the liver (30), or to achieve antiviral effects (37) has been reported. Anti-tumor effects as a result of miRNA modulation have been shown with the use of adenoviral vectors to over-express miRNA such as miR-26 (38). Recently it was shown that antisense to miR-191 could reduce tumor weight in an orthotopic mouse model of HCC (39). In contrast to these studies, our study is the first to our knowledge that shows a survival advantage by directly targeting intrahepatic tumors with anti-miRNAs. Our study provides useful preclinical information to guide further development of therapeutic strategies using similar approaches in humans. We demonstrate here that systemically administered naked oligonucleotides accumulate in the normal liver and within liver tumors at very high levels. Chol-anti-miR-221 was effective at reducing endogenous miR-221 and altering miR-221 target proteins. Notably, chol-anti-miR-221 was detectable (110-160 nM) in the tumors of all 3 mice treated with chol-anti-miR-221 that survived for ten weeks (Fig. 6D). Our data parallel that of Krutzfeldt, et al., who showed anti-miRNA regulation from a single dose of anti-miRNA oligo 28 weeks following injection (31). The doses used here (i.e. 30, 60 mg/kg) were similar to those used in other in vivo studies for targeting liver (31). It may be possible to reduce the dose by formulating the antisense oligos into nanoparticles (40). Since miR-221 is increased in a number of solid tumors including pancreas (41), non small cell lung cancer (28), glioblastoma (42) and thyroid cancer (25), optimizing the delivery of antisense oligos may be a useful approach to target tumors besides HCC. While the 2′-O-methyl modification used here dramatically reduced non-specific effects (43), the phosphorothioate backbone employed in our study has been shown to have excellent antisense activity but with sequence-independent effects on cellular function (44, 45). For example, interaction of phosphorothioate oligos to cellular proteins such as basic fibroblastic growth factor results in non-specific effects (45). The slight increase in p27Kip1, p57Kip2, and cleaved caspase-3 (Fig. 3D and Supplemental Fig. S6) in cholesterol labeled scrambled control treated tumors might be due to sequence independent activity of the oligo backbone. Lowering the amount of oligo is one possible way to minimize the non specific oligo effects (44).
The choice of antisense oligonucleotides to miRNA as a targeting strategy for liver cancer is attractive for several reasons. Over the past two decades, major efforts have been directed to develop antisense as therapeutic agents by targeting mRNA, but with disappointing results. A drawback of antisense has been inappropriate targeting and poor cellular uptake in the target tissue. Our results corroborate what has been known for many years; oligonucleotides accumulate in highly perfused tissues such as the liver. Liver uptake is enhanced by the presence of the cholesterol label. Unlike with the design of antisense for mRNA, where the optimal antisense is determined by screening dozens of potential oligos, antisense to mature miRNA is simply the reverse compliment of the mature miRNA sequence. While siRNA has many useful applications for gene knockdown, targeting miRNA with siRNA is not practical. The cytoplasmic RISC complex that cleaves the RNA target of siRNA would not be expected to cleave the nuclear pri-miRNA. Targeting the loop region of pre-miRNAs with siRNA required much higher doses of siRNA than what is required for siRNA inhibition of mRNA (46). miRNA sponges represents another option to inhibit mature miRNA levels (47), however these are vector driven approaches. A more clinically relevant scenario to interfere with the function of the mature miRNA is to simply inhibit the mature miRNA with antisense oligos.
Deregulation of miRNAs is consistently observed in human cancers. Therefore targeting aberrantly expressed miRNA is logical and attractive if critical targetable miRNA can be identified. The precise target gene effects of these interventions are not readily elucidated because of the promiscuity of potential miRNA targets. However, this does not preclude modulation of miRNA as a therapeutic strategy when identifiable effects on tumor growth can be demonstrated. Thus, the choice of target miRNA is crucial, and careful validation is needed. While our studies validate miR-221 as a suitable target, future studies to determine the relative efficacy of targeting miR-221 compared to other miRNAs that are aberrantly expressed in HCC may be needed to identify the most optimal target miRNA for further translation into clinical trials.
Supplementary Material
Acknowledgments
This work was supported by the Provost's Targeted Investment in Excellence (TIE) grant from the Ohio State University and grants CA114304 (T.D.S.) and DK069370 (T.P.).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–6. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Lencioni R, Cioni D, Crocetti L, Bartolozzi C. Percutaneous ablation of hepatocellular carcinoma: state-of-the-art. Liver Transpl. 2004;10:S91–7. doi: 10.1002/lt.20043. [DOI] [PubMed] [Google Scholar]
- 3.Raoul JL, Sangro B, Forner A, Mazzaferro V, Piscaglia F, Bolondi L, et al. Evolving strategies for the management of intermediate-stage hepatocellular carcinoma: Available evidence and expert opinion on the use of transarterial chemoembolization. Cancer Treat Rev. 2010;37:212–20. doi: 10.1016/j.ctrv.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Abou-Alfa GK, Huitzil-Melendez FD, O'Reilly EM, Saltz LB. Current management of advanced hepatocellular carcinoma. Gastrointest Cancer Res. 2008;2:64–70. [PMC free article] [PubMed] [Google Scholar]
- 5.Leung TW, Patt YZ, Lau WY, Ho SK, Yu SC, Chan AT, et al. Complete pathological remission is possible with systemic combination chemotherapy for inoperable hepatocellular carcinoma. Clin Cancer Res. 1999;5:1676–81. [PubMed] [Google Scholar]
- 6.Yeo W, Mok TS, Zee B, Leung TW, Lai PB, Lau WY, et al. A randomized phase III study of doxorubicin versus cisplatin/interferon alpha-2b/doxorubicin/fluorouracil (PIAF) combination chemotherapy for unresectable hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:1532–8. doi: 10.1093/jnci/dji315. [DOI] [PubMed] [Google Scholar]
- 7.Philip PA, Mahoney MR, Allmer C, Thomas J, Pitot HC, Kim G, et al. Phase II study of Erlotinib (OSI-774) in patients with advanced hepatocellular cancer. J Clin Oncol. 2005;23:6657–63. doi: 10.1200/JCO.2005.14.696. [DOI] [PubMed] [Google Scholar]
- 8.Thomas MB, Morris JS, Chadha R, Iwasaki M, Kaur H, Lin E, et al. Phase II trial of the combination of bevacizumab and erlotinib in patients who have advanced hepatocellular carcinoma. J Clin Oncol. 2009;27:843–50. doi: 10.1200/JCO.2008.18.3301. [DOI] [PubMed] [Google Scholar]
- 9.Zhu AX, Blaszkowsky LS, Ryan DP, Clark JW, Muzikansky A, Horgan K, et al. Phase II study of gemcitabine and oxaliplatin in combination with bevacizumab in patients with advanced hepatocellular carcinoma. J Clin Oncol. 2006;24:1898–903. doi: 10.1200/JCO.2005.04.9130. [DOI] [PubMed] [Google Scholar]
- 10.Faivre S, Raymond E, Boucher E, Douillard J, Lim HY, Kim JS, et al. Safety and efficacy of sunitinib in patients with advanced hepatocellular carcinoma: an open-label, multicentre, phase II study. Lancet Oncol. 2009;10:794–800. doi: 10.1016/S1470-2045(09)70171-8. [DOI] [PubMed] [Google Scholar]
- 11.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 12.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 13.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 14.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 16.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 18.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2009;107:264–9. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–15. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 21.Braconi C, Patel T. MicroRNA expression profiling: a molecular tool for defining the phenotype of hepatocellular tumors. Hepatology. 2008;47:1807–9. doi: 10.1002/hep.22326. [DOI] [PubMed] [Google Scholar]
- 22.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27kip1. J Biol Chem. 2007;282:23716–24. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 24.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. Embo J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791–8. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 26.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 27.Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–80. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas. 2009;38:e190–9. doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- 30.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 32.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 33.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmittgen TD, Jiang J, Liu Q, Yang L. A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4:107–15. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 38.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elyakim E, Sitbon E, Faerman A, Tabak S, Montia E, Belanis L, et al. hsa-miR-191 Is a Candidate Oncogene Target for Hepatocellular Carcinoma Therapy. Cancer Res. 2010;70:8077–87. doi: 10.1158/0008-5472.CAN-10-1313. [DOI] [PubMed] [Google Scholar]
- 40.Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles Modified With Tumor-targeting scFv Deliver siRNA and miRNA for Cancer Therapy. Mol Ther. 2010;18:1650–6. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 43.Yoo BH, Bochkareva E, Bochkarev A, Mou TC, Gray DM. 2′-O-methyl-modified phosphorothioate antisense oligonucleotides have reduced non-specific effects in vitro. Nucleic Acids Res. 2004;32:2008–16. doi: 10.1093/nar/gkh516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein CA. The experimental use of antisense oligonucleotides: a guide for the perplexed. J Clin Invest. 2001;108:641–4. doi: 10.1172/JCI13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guvakova MA, Yakubov LA, Vlodavsky I, Tonkinson JL, Stein CA. Phosphorothioate oligodeoxynucleotides bind to basic fibroblast growth factor, inhibit its binding to cell surface receptors, and remove it from low affinity binding sites on extracellular matrix. J Biol Chem. 1995;270:2620–7. doi: 10.1074/jbc.270.6.2620. [DOI] [PubMed] [Google Scholar]
- 46.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–41. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 47.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.