Abstract
Arsenic, a toxic metalloid widely existing in the environment, causes a variety of health problems. The ars operon encoded by Escherichia coli plasmid R773 has arsD and arsA genes, where ArsA is an ATPase that is the catalytic subunit of the ArsAB As(III) extrusion pump, and ArsD is an arsenic chaperone for ArsA. ArsD transfers As(III) to ArsA and increases the affinity of ArsA for As(III), allowing resistance to environmental concentrations of arsenic. Cys12, Cys13 and Cys18 in ArsD form a three sulfur-coordinated As(III) binding site that is essential for metallochaperone activity. ATP hydrolysis by ArsA is required for transfer of As(III) from ArsD to ArsA, suggesting that transfer occurs with a conformation of ArsA that transiently forms during the catalytic cycle. The 1.4 Å x-ray crystal structure of ArsD shows a core of four β-strands flanked by four α-helices in a thioredoxin fold. Docking of ArsD with ArsA was modeled in silico. Independently ArsD mutants exhibiting either weaker or stronger interaction with ArsA were selected. The locations of the mutations mapped on the surface of ArsD are consistent with the docking model. The results suggest that the interface with ArsA involves one surface of α1 helix and metalloid binding site of ArsD.
Keywords: Arsenic, ArsD, Metallochaperone, ArsA, ATP-driven efflux pump
Introduction
Arsenic ranks first on the superfund list of hazardous chemicals http://www.atsdr.cdc.gov/cercla/07list.html, in part because it is the most ubiquitous of all environmental toxic compounds. The metalloid is a carcinogen and is considered a causative agent of a number of other diseases, including cardiovascular and neurological disorders. Both prokaryotes and eukaryotes have arsenic detoxifying systems, frequently involving extrusion from cytosol.
In bacteria, arsenic resistance genes are organized as ars operons. The majority have three genes, arsRBC. ArsR is an As(III)-responsive transcriptional repressor (Xu and Rosen 1999), ArsB is a As(OH)3/H+ antiporter that extrudes As(III), conferring resistance (Meng et al. 2004), and ArsC is an arsenate reductase that converts As(V) to As(III), the substrate of ArsB, extending the range of resistance to include As(V) (Mukhopadhyay and Rosen 2002). Some have two more genes, arsD and arsA, such as the arsRDABC operon in E. coli plasmid R773. ArsA and ArsB forms the ArsAB complex, a pump utilizing the energy of ATP hydrolysis for As(III) or Sb(III) extrusion (Dey et al. 1994a, b). Cells expressing the five genes arsRDABC are more resistant to As(V) and As(III) than those expressing only the arsRBC genes.
ArsA is a 583-amino acid ATPase with two pseudo-symmetric halves, A1 and A2, connected by a short linker. A1 and A2 have consensus nucleotide-binding domains (NBDs) at their interface (Zhou et al. 2000). A high affinity metalloid binding domain (MBD) about 20 Å from the NBDs binds As(III) or Sb(III) (Zhou et al. 2001, 2000) using conserved residues Cys113, Cys72 and Cys422 (Ruan et al. 2006). Nucleotide binding at the NBDs stimulates metalloid binding, and, reciprocally, metalloid binding brings A1 and A2 together, stimulating ATP hydrolysis.
ArsD is a metallochaperone that transfers As(III) or Sb(III) to ArsA (Lin et al. 2006). ArsD is a homodimer of two 120-residue subunits (Chen and Rosen 1997). Although ArsD exhibits weak repressor activity, its primary function is as an arsenic metal-lochaperone (Chen and Rosen 1997; Lin et al. 2006; Wu and Rosen 1993). ArsD has three conserved cysteine residues, Cys12, Cys13 and Cys18 (Lin et al. 2007a). These form a high affinity As(III) binding site required for ArsD to deliver As(III) to the ArsA ATPase, increasing the affinity of ArsA for As(III), producing increased efflux and resistance at environmental concentrations of arsenic (Lin et al. 2007b; Yang et al. 2010).
It is not known how these two proteins interact and how the metalloid is transferred. The transfer of copper from Atx1-like chaperones to the N-terminal domains of copper efflux pumps involves sequential transfer of metal between cysteine thiolates (Boal and Rosenzweig 2009, 2002). We have proposed a similar sequential transfer mechanism from the cysteine thiolates of ArsD to the cysteine thiolates of ArsA (Yang et al. 2010). According to this model, metalloid binding sites of ArsA and ArsD come close to each other during interaction. Crystal structures of ArsA and ArsD have been solved. Based on the structure of ArsA and ArsD, ArsA-ArsD complex structure was modeled by in silico docking (Ye et al. 2010). The results of recent genetic analysis are consistent with the docking model (Yang 2011).
Genomics of arsenic detoxification
As a consequence of the prevalence of arsenic in the environment, bacteria are continuously exposed to arsenic and have evolved arsenic resistance systems encoded by ars operon that have been found in nearly every sequenced bacterial genome. To date more than 400 bacterial and archaeal ars operons containing arsA and arsD genes have been identified, and the arsA and arsD are nearly always adjacent to each other, suggesting 1) that they co-evolved and later moved as a unit into an ars operon, and 2) that they have linked biochemical functions in arsenic detoxification.
Cell biology of ArsD function
Since ArsA is the catalytic subunit of an arsenic efflux pump, it was considered likely that a function of ArsD would be to augment the ability of ArsA to extrude arsenite, enhancing arsenic detoxification. The resistance of cells expressing different ars genes was compared. Cells carrying arsDAB gene were slightly more resistant to high concentrations of As(III) compared to cells carrying only arsAB, indicating that ArsD works together with ArsA to increase the efficiency of the ArsAB pump (Lin et al. 2006).
However, the concentration of arsenic in such laboratory growth experiments is usually in the millimolar range, which might be occasionally found in extreme environments such as volcanic areas in Yellowstone National Park but rarely in human habitats. The advantage conferred by the arsD gene at concentrations of arsenic commonly found in the environment was demonstrated by a competitive growth assay of mixed cultures (Lin et al. 2006). E. coli strain AW3110, in which the chromosomal ars operon had been deleted, was transformed with plasmids with arsAB genes or arsDAB genes. Equivalent numbers of cells carrying either arsAB or arsDAB genes were mixed and grown with 10 μM sodium arsenite, a subtoxic concentration present in many deep tube wells in Bangladesh and elsewhere in the world. After a little more than a week, the cells with arsDAB took over the population, demonstrating that ArsD provides a competitive growth advantage in soil or water with moderate amounts of arsenic contamination.
The advantage conferred by ArsD was supported by the ability of ArsD to lower As(III) accumulation in intact cells (Lin et al. 2006). Cells expressing only arsB were able to lower the intracellular level of As(III) compared to a strain with no ars genes since ArsB catalyzes As(OH)3/H+ exchange (Meng et al. 2004). Expression of arsA increased the efficiency of As(III) extrusion when co-expressed with arsB by forming the ArsAB pump that catalyzes ATP-driven As(III) efflux (Dey and Rosen 1995). When arsD was co-expressed with arsAB, As(III) accumulation in the cells was further reduced (Lin et al. 2006). These lines of evidence support the hypothesis that ArsD increases the efficiency of the ArsAB pump.
Interaction between ArsD and ArsA
The association of ArsD and ArsA in arsenic resistance was strongly supported by the demonstration of interaction between ArsD and ArsA in yeast two-hybrid analysis (Lin et al. 2006). Interactions among the four soluble proteins encoded by the R773 arsRDABC operon were examined. ArsD was found to interact with ArsA but not with the ArsR repressor or the ArsC arsenate reductase; ArsD also interacted with itself, which would be expected since ArsD is a homodimer (Chen and Rosen 1997). ArsA only interact with ArsD but not with ArsR or ArsC. Since there is no arsenic or antimony in yeast cells during analysis, ArsD and ArsA must interact to some extent in the absence of metalloid. But for arsenic transfer from ArsD to ArsA (described in more detail below), the arsenic-bound ArsD should interact more strongly with ArsA. Whether arsenic increases interaction between ArsD and ArsA is currently under investigation.
Interaction between ArsD and ArsA is also evidenced by chemical crosslinking with dibromobimane (dBBr) (Kosower et al. 1980). dBBr is a homobifunctional thiol-specific crosslinker, reacting with cysteine residues within 3 to 6 Å of each other. An ArsD-ArsA complex was trapped by dBBr crosslinking, as shown by SDS–PAGE analysis and blotting with anti-ArsA and ArsD antibodies. This complex had an apparent mass of approximately 90 kDa, close to the predicted mass of a monomer of ArsA crosslinked with either an ArsD dimer or monomer. ArsD-ArsA crosslinking efficiency was increased by addition of MgATP but not by antimonite. These results suggest 1) that cysteine-rich metalloid binding sites of ArsD and ArsA come close to each other during interaction, 2) that ArsD interacts with nucleotide-bound form of ArsA and 3) that apo-ArsD is able to interact with ArsA (although it is not known whether the interaction of apoArsD or As(III)-ArsD with ArsA occurs with equal affinity).
Biochemistry of ArsD function
As a metallochaperone, ArsD can transfer As(III) (Yang et al. 2010) or Sb(III) (Lin et al. 2006) to ArsA. To assay transfer between purified proteins, a maltose binding protein (MBP) fused ArsD with As(III) in the metalloid binding site was bound to amylose column, following which ArsA and different nucleotides were passed through the column, and, finally, ArsD was eluted with maltose (Yang et al. 2010). The elution positions of the two proteins and As(III) was determined. Transfer was estimated from the amount of As(III) that eluted with ArsA. Transfer of As(III) from ArsD to ArsA was compared under catalytic and noncatalytic conditions. At room temperature in the presence of MgATP, nearly all of the As(III) was transferred from ArsD to ArsA. At 4°C, where ArsA hydrolyzes ATP very slowly, almost no As(III) was transferred. No As(III) was transferred to ArsA with the product MgADP or with the nonhydrolyzable nucleotides MgATP-γ-S or MgAMP-PNP. These results demonstrate that ATP hydrolysis is required for As(III) transfer from ArsD to ArsA. We interpret this result as As(III) transfer occurring with a conformation of ArsA that transiently forms during the catalytic cycle. This is also supported by the fact that MgATP increases the efficiency of ArsA–ArsD crosslinking by dBBr (Lin et al. 2006).
How is As(III) transferred from ArsD to ArsA? It could be a thermodynamic dissociation and re-association process. Or it could be mediated by direct interaction of ArsD and ArsA that allows for step-wise As(III) transfer directly from the thiols of ArsD to the thiols of ArsA (Lin et al. 2007a), similar to the mechanism proposed for copper transfer from the yeast Atx1p chaperone to the Ccc2p copper efflux pump (Pufahl et al. 1997). The As(III) chelator dimercaptosuccinic acid (DMSA) was used to differentiate between channeling and a dissociation/re-association mechanism. Direct transfer of As(III) from ArsD to ArsA would be expected to be insensitive to DMSA. In contrast, if As(III) dissociates from ArsD before binding to ArsA, an excess of DMSA should prevent transfer. The results showed that the As(III) transfer from ArsD to ArsA was insensitive to DMSA (Yang et al. 2010), consistent with channeling of metalloid from one protein to the other, suggesting that ArsD and ArsA form an interface at their metal binding sites.
ArsD not only delivers As(III) to ArsA (Yang et al. 2010) but also enhances its ATPase activity at low As(III) concentrations by increasing the affinity of ArsA for As(III) (Lin et al. 2006). In the presence of ArsD, the apparent affinity of ArsA for As(III) is increased from the millimolar range to around 10 μM, but the Vmax for ATP hydrolysis remains unchanged. Since ArsA ATPase activity is activated by As(III), ArsD makes ArsA more effective at environmentally appropriate concentrations of As(III) by increasing its affinity for As(III). As a metallochaperone, ArsD recycles to keep loading ArsA with As(III) during cycle of catalysis, effectively increasing the affinity of ArsA for As(III).
Although ArsD transfer of metalloid to ArsA involves specific ArsD-ArsA interaction, the driving force for the metalloid transfer is not clear. ArsD has an affinity of approximately 2 μM for As(III), as determined by tryptophan fluorescence quenching (Yang et al. 2010). ArsA has an apparent affinity of about 0.5 to 1 mM for As(III), as determined from the stimulation of ATP hydrolysis as a function of As(III) concentration (Bhattacharjee et al. 1995; Lin et al. 2006; Yang et al. 2010). Thus As(III) transfer from ArsD to ArsA seems to be thermodynamically unfavorable. However, ATP hydrolysis is required for the metalloid transfer. This may provide the energy to make this process favorable. It is possible that ATP hydrolysis and protein–protein interaction could lead to increased affinity of ArsA or decreased affinity of ArsD for metalloid. Finally, when the metalloid bound to the ArsAB pump is pumped out of the cell, mass action will provide the directionality of the reaction. Thus the driving force could be either thermodynamic or kinetic factors or a combination of the two.
Metalloid binding sites in ArsD
Native R773 ArsD has 120 residues with three vicinal cysteine pairs, Cys12–Cys13, Cys112–Cys113 and Cys119–Cys120. Phenylarsine oxide (PAO) affinity chromatography was used to examine the metal binding ability of ArsD mutants or truncations. Only a single vicinal cysteine pair was sufficient to bind to the PAO column, suggesting that each cysteine pair forms an independent metalloid binding site (Li et al. 2001). A series of cysteine mutants and truncations of R773 ArsD were purified and assayed directly for metalloid binding (Li et al. 2001, 2007a). The native R773 ArsD bound three Sb(III) per monomer, but variants with only a single vicinal pair bound only one Sb(III) per monomer, showing that each cysteine pair forms an independent metalloid binding site with similar affinity for Sb(III) of approximately 1 μM (Lin et al. 2007a).
However, only the Cys12–Cys13 pair and an additional cysteine, Cys18, are conserved in homologues. For example, Alkaliphilus metalliredigens QYMF has two ars operons, each encoding an ArsD of only 99 residues and having only Cys12, Cys13 and Cys18 (Fu et al. 2010). If any of the three cysteines residues was mutated, high affinity metalloid binding was eliminated, suggesting that each of the three conserved cysteine residues, Cys12, Cys13 and Cys18, form a 3-coordinate thiolate metalloid binding site (Lin et al. 2007a). The nature of this metalloid binding site was confirmed by extended X-ray absorption fine structure spectroscopy (EXAFS) (Yang et al. 2010). The As(III) binding kinetics and dissociate constant of this site was further characterized using protein fluorescence (Yang et al. 2010). Two single tryptophan derivatives of ArsD, T15 W and V17 W, exhibited quenching of intrinsic protein fluorescence upon binding of As(III) or Sb(III). Interestingly, in the absence of added thiols, As(III) quenches fluorescence quite slowly, requiring more than 2 h to attain a steady-state level of fluorescence when 1 μM protein was mixed with equimolar As(III). This allowed estimation of the rates of binding and affinities for metalloid. The two single tryptophan derivatives, T15 W and V17 W, reported similar affinity for As(III) of approximately 2 μM. Substitution of Cys12, Cys13 or Cys18 decreased the affinity for As(III) more than tenfold. When the glutathione conjugate, As(GS)3 was used in place of As(OH)3 (the solution form of arsenite (Ramírez-Solis et al. 2004)), the rate of quenching was too fast to measure by standard fluorescence instrumentation but was at least 1000-fold faster. Given that intracellular GSH is in the millimolar range, nearly all intracellular arsenite would be in the form of As(GS)3, so it should not be surprising that this is the source of As(III) for ArsD.
Although R773 ArsD has three metalloid binding sites, but only the N-terminal conserved site formed by Cys12, Cys13 and Cys18 is required and sufficient to transfer arsenic to ArsA and activate ArsA ATPase (Lin et al. 2007a; Yang et al. 2010). A construct truncated at residue 109 (denoted as ArsD109) lacking the last two metalloid binding sites interacted with ArsA by yeast-two-hybrid analysis, and the purified ArsD109 stimulated ArsA ATPase activity as well as the native ArsD (Lin et al. 2007a). In contrast, an ArsD derivative lacking this site but retaining the last two was unable to interact with ArsA in yeast-two-hybrid assay and did not stimulate ArsA activity. The effect of these constructs on arsenic accumulation was examined. Cells expressing an ArsD with only the first site reduced the intracellular accumulation of arsenic (reflecting active extrusion) as well as wild type ArsD when co-expressed with ArsAB. Any construct lacking the first site did not increase the ability of the cells to extrude arsenite. These in vivo results strongly support the hypothesis that the Cys12-Cys13-Cys18 metalloid binding site is involved in metallochaperone function.
Crystal structure of ArsD
A chitin binding domain fusion of ArsD109 (ArsD109-CDB) was constructed for purification purposes. After the intein tag and chitin binding domain were removed, both ArsD109 and ArsD109/C12A/C13A/C39S proteins crystallized in space group P21. ArsD109 crystals diffracted to 2.1 Å, and ArsD109/C12A/C13A/C39S diffracted to 1.4 Å. The ArsD109 structure could be superimposed with the ArsD109/C12A/C13A/C39S structure with an RMS deviation 0.17 Å, so the higher resolution mutant structure was used for subsequent analysis. There are two ArsD molecules in the asymmetric unit. They form a dimer of approximate 50 × 35 × 30 Å3. The two monomers are nearly identical, with a RMSD of 0.52 Å. The termini, especially the C-terminus, are more divergent. The ArsD monomer has a core of four β-strands flanked by four α-helices. Helices α1 and α4 are on one side of the β-strands; α2 and α3 are on the other side. α2 is almost perpendicular to α3, with only one residue between them. β1 is between β2 and 3, parallel to β2 and anti-parallel to β3. β3 and 4 form a β-hairpin. Residues 70 to 72 form a short 310 helix. On molecule B, residues 102 to 104 also form a short 310 helix.
The overall structure of the ArsD monomer has a thioredoxin fold, with a core of four beta-strands flanked by four α-helices (PDB code 3MWH). The region of ArsD with the cysteines that form the arsenic binding site was not visible in that structure, but a structure of an oxidized form of an ArsD homologue, showing the location of the cysteines in disulfide bonds, has been reported (PDB code 3KTB) (vida infra). A Dali search using ArsD structure returned thioredoxins as one of the closest structural homologues of ArsD, with the highest Z-score of 6.5 for the E. coli thioredoxin TrxA (PDB ID 2TRX). The root mean square deviation (RMSD) between the Cα atoms of the 72 aligned residues is 2.5 Å. TrxA has an N-terminal sequence not present in ArsD, and ArsD α3 is absent in TrxA. The TrxA molecule has a four residue loop connecting β1 and α1. The TrxA active site sequence C32PGC35 is at the beginning of a long α-helix that extends from residue 32 to residue 49, which renders Cys35 solvent inaccessible. Neither of the active site cysteine residues is congruent with Cys12, Cys13 or Cys18 in ArsD. Interestingly, a CACA mutant of TrxA, in which the active site was changed to C32AC34A, has the end of the extended helix from residues 35–40 unraveled, exposing Cys34 to solvent, facilitating its participation in catalysis. In this structure the loop (residues 29 to 39) is of similar length to the putative loop in ArsD (residues 12–22).
NMR solution structure of ArsD
NMR characterization of ArsD provides a basis for understanding the structural aspects of how the protein functions. A solution structure of ArsD would allow comparison with the static crystal structure and, even more important, identification of amino acid-specific chemical environmental changes in ArsD coupled with binding of arsenite and antimonite and/or during interaction with ArsA.
For those reasons an initial NMR characterization was performed (Ye et al. 2010). Dispersion in the high-resolution 15N TROSY HSQC spectrum (Pervushin et al. 2000) indicates the protein is well folded, with the number of peaks consistent with the protein existing as a symmetric homodimer. Assignments for the 118-residue construct were completed for the majority of native protein residues. Chemical shift analysis of the backbone atom resonances (Wishart et al. 1991) was applied to predict the secondary structure for the protein in solution. Prediction analysis suggested that the protein is flanked by a 7 residue N-terminal β-strand. In the absence of As(III), amide chemical shifts for the three conserved cysteines (Cys12, Cys13 and Cys18) occur in the unfolded region of the spectrum, indicating these residues exist in the unstructured region between β-strand 1 and α-helix 1. In addition, the protein contains an extensive C-terminal unstructured tail ranging from residues 100 to 118.
A comparison of predicted secondary structural elements for ArsD in solution with the crystal structure results indicates a reasonable agreement between the structures, with a few differences. The solution structure is less ordered than the crystal structure, especially in the length and location of α-helix 2 and β-strand 3. By both methods, the conserved cysteines (Cys12, Cys13 and Cys18) exist in an unstructured region between β-strand 1 and α-helix 1, as earlier indicated for the predicted solution structure. It is only when the cysteine residues are artifactually oxidized to form disulfide bonds that they could be resolved by crystallography (vida infra). These results suggest this region of apoArsD is normally very flexible and folds around As(III), allowing the three cysteine residues to form the binding site. Therefore, these NMR data provide the basis for future experiments directed at monitoring structural changes coupled with binding of As(III) and/or ArsA. Using the crystal structure as the global structural template, it should be possible using NMR spectroscopy to map the binding surfaces between ArsD and ArsA and also to monitor structural changes during As(III) binding and delivery. These results will provide significant insight in the molecular mechanism of ArsD metallochaperone function.
Modeling the metalloid binding site
The 2.1 Å structure of a homologue from B. vulgatus ATCC 8482 was deposited in the PDB data base (PDB code: 3KTB; Kim, Y., Tesar, C., Feldmann, B., Joachimiak, A., Midwest Center for Structural Genomics (MCSG), unpublished). 3KTB and R773 ArsD109 can be superimposed with an RMSD of 1.0 Å. In contrast to native ArsD109, where the cysteines are completely reduced, all of the cysteines residues of 3KTB are oxidized in intra- and intermolecular disulfide bonds, likely an artifactual result of oxidation during purification. The 11 residues not visible in the R773 ArsD109 structure are contained within a 14-residue loop in B. vulgatus ArsD, perhaps because disulfide bond formation rigidified the loop. A structure-based alignment of the two proteins shows that the loops in both are of identical length, and 10 of the 14 loop residues are identical and 3 are similar between the two proteins. The close homology between the two loops allows the residues not visible in the R773 ArsD109 structure to be modeled with confidence. Based on this information, a model of the metallated form of the R773 ArsD109 was constructed taking into account the bond distances of 2.24 Å between the sulfur atoms of Cys12, Cys13 and Cys18 and the central arsenic atom determined by EXAFS (Yang et al. 2010). We postulate that when the unbound form is presented with As(III), it forms an intermediate in which As(III) is weakly bound to Cys13 and Cys18 thiolates. In the B. vulgatus homologue, the Cys12 thiolate is oriented away from other two cysteines, but in the EXAFS structure, all three sulfur atoms become equidistant to the arsenic atom, suggesting that Cys12 could reorient to form the third ligand, forming the high affinity binding site. Further experimental results will be necessary to test this proposed order of binding.
Modeling the ArsD-ArsA interaction
Metallated ArsD interacts with and transfers As(III) to ArsA during catalysis, when the ATPase cycles between open and closed conformations (Li and Rosen 2000). The X-ray crystal structure of ArsA has been solved in the closed form (Zhou et al. 2000). A Saccharomyces cerevisiae homologue of ArsA termed Arr4p or Get3 is involved in targeting tail-anchored proteins in the endoplasmic reticulum (Auld et al. 2006). Recently crystal structures of Get3 were solved in the open (nucleotide free) and closed (ADP-AlF4−) conformations (Bozkurt et al. 2009; Mateja et al. 2009). ArsA has two homologous halves, ArsA1 and ArsA2, each with a nucleotide binding domain (NBD) connected by a short linker, while Get3 is a dimer of two identical monomers, each homologous to both ArsA1 and ArsA2. In the open Get3 conformation there is a large conformational change, and the two monomers are separated by approximately 37°, with rotation of one subunit to the other relative to their orientation in the closed form, which is more compact. The ADP–AlF4− closed ArsA structure (Zhou et al. 2001) can be superimposed with the Get3 closed structure with an RMSD of 3.4 Å for 420 Cα atoms. ArsA NBD1 (residue 1–296) and NBD2 (305–583) can be superimposed with the two monomers in the Get3 open structure, with an RMSD of 2.3 Å and 3.0 Å, respectively. An open model of the ArsA structure based on the open Get3 structure was generated for use for analysis of ArsA–ArsD interactions and docking studies (Yang et al. 2010).
Views of the open form of ArsA from opposite directions displays a cavity into which ArsD can fit. To examine whether the structures have complementary surfaces, the open ArsA and metallated ArsD models were docked by using the fully automated, web-based program ClusPro Version 2.0 with default parameters. Based on the biochemical results, the one most consistent with the demonstrated proximity of the cysteine residues of ArsD and ArsA had ArsD sulfur atoms of Cys12 and Cys13 within 4.1–7.5 Å of the sulfur atoms of ArsA residues Cys113 and Cys172. Overall, 15 ArsD residues are predicted to be within 3.2 Å of 13 residues of ArsA.
Genetic mapping of the interface between ArsD and ArsA ATPase
As described above, As(III) is transferred from ArsD to an ArsA conformation formed during ATP hydrolysis, and this process is protected from the As(III) chelator dimercaptosuccinic acid (Yang et al. 2010), suggesting these two proteins form an interface at their metal binding sites. This hypothesis is supported by the facts that the metal binding sites of ArsA and ArsD could be crosslinked by dBBr (Lin et al. 2006), and that ArsD almost lost interaction with ArsA when the conserved cysteine residues, Cys12, Cys13 or Cys18 were mutated to alanine or serine (Lin et al. 2007a). A genetic approach was used to test this hypothesis. Random mutagenesis of ArsD was used to create ArsD mutants that exhibit increased or decreased interaction with ArsA, as selected by several variations of the yeast two-hybrid assay (Yang 2011). Nine ArsD mutants (S14R, T20I, D28T, D28V, T31A, Q34R, Q38R and V61A) exhibiting increased interaction with ArsA were isolated by repressed transactivator yeast two-hybrid selection. Five ArsD mutants (V17A, V22A, V27D, Q51H and F55L) exhibiting decreased interaction with ArsA were isolated by reverse yeast two-hybrid selection. Additionally, two mutants in Lys-37 and 62 were identified as being involved in ArsD function by a combination of site-directed mutagenesis and chemical modification. When these two lysine residues are substituted by alanine, ArsD lost the capability to stimulate ArsA ATPase activity and lost interaction with ArsA as measured by yeast two-hybrid analysis. However, substitution at either position with arginine was tolerated, suggesting participation of a positive charge. All the residues were mapped on the surface of the ArsD structure, and their locations are consistent with the structural model generated by in silico docking. Four (Ser14, Val17, Thr20 and Val22) are close to metalloid binding site residues Cys-12, Cys-13 and Cys-18, and seven (Gln24, Val 27, Asp28, Thr31, Gln34, Lys37 and Gln38) are on the surface of helix 1. In addition, substitutions at residues 12, 13 or 18 lost the ability to interact with ArsA (vida supra) (Lin et al. 2007a). The agreement of the genetic, biochemical and in silico data clearly indicates that the interface with ArsA involves one surface of helix 1 and the metalloid binding site of ArsD. We had proposed a cycle with a step-wise transfer of As(III) from ArsD to ArsA (Lin et al. 2007b). The combination of genetic, biochemical and structural analysis described here allows us to hypothesize a molecular description of As(III) binding by ArsD and transfer to ArsA (Fig. 1).
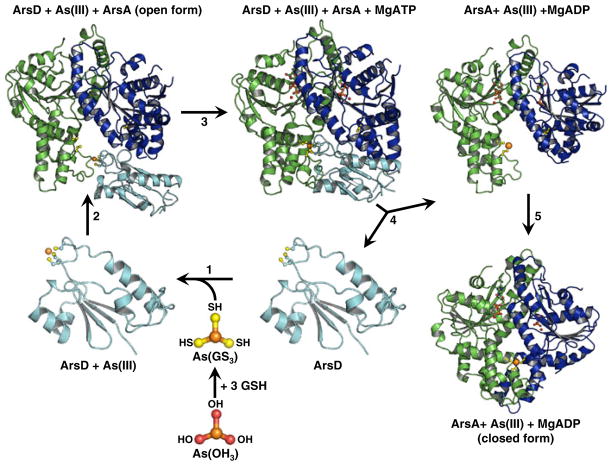
Fig. 1.
Proposed reaction scheme for transfer of As(III) from ArsD to ArsA. In the cytosol As(OH)3, the solution form or arsenite, reacts with 3 GSH to form As(GS)3, the arsenic donor to ArsD (Yang et al. 2010). Step 1 ArsD binds As(III) by exchange of the three thiols of As(GS)3 for the thiols of cysteines residues Cys12, Cys13 and Cys18. Although ArsD is a dimer, the structure of the monomer of apoArsD is shown (Ye et al. 2010). Step 2 As(III)-bound ArsD binds to the open form of ArsA. Step 3 As(III) is transferred in a step-wise exchange from the three thiols of ArsD to the thiols of Cys113, Cys172 and Cys422 of ArsA. Step 4 Transfer of As(III) results in dissociation of the ArsD-ArsA complex. Step 5 As(III)-bound ArsA undergoes a conformational change in ArsA to the closed form concomitant with activation of ATP hydrolysis
Acknowledgments
This work was supported by United States Public Health Service Grants AI43428 and GM55425.
Contributor Information
A. Abdul Ajees, Department of Cellular Biology and Pharmacology, Herbert Wertheim College of Medicine, Florida International University, Miami, FL 33199, USA.
Jianbo Yang, Department of Biochemistry and Molecular Biology, School of Medicine, Wayne State University, Detroit, MI 48201, USA.
Barry P. Rosen, Email: brosen@fiu.edu, Department of Cellular Biology and Pharmacology, Herbert Wertheim College of Medicine, Florida International University, Miami, FL 33199, USA
References
- Auld KL, Hitchcock AL, Doherty HK, Frietze S, Huang LS, Silver PA. The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae. Genetics. 2006;174:215–227. doi: 10.1534/genetics.106.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee H, Li J, Ksenzenko MY, Rosen BP. Role of cysteinyl residues in metalloactivation of the oxyanion-translocating ArsA ATPase. J Biol Chem. 1995;270:11245–11250. doi: 10.1074/jbc.270.19.11245. [DOI] [PubMed] [Google Scholar]
- Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109:4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt G, Stjepanovic G, Vilardi F, Amlacher S, Wild K, Bange G, Favaloro V, Rippe K, Hurt E, Dobberstein B, Sinning I. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci USA. 2009;106:21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Rosen BP. Metalloregulatory properties of the ArsD repressor. J Biol Chem. 1997;272:14257–14262. doi: 10.1074/jbc.272.22.14257. [DOI] [PubMed] [Google Scholar]
- Dey S, Rosen BP. Dual mode of energy coupling by the oxyanion-translocating ArsB protein. J Bacteriol. 1995;177:385–389. doi: 10.1128/jb.177.2.385-389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Dou D, Rosen BP. ATP-dependent arsenite transport in everted membrane vesicles of Escherichia coli. J Biol Chem. 1994a;269:25442–25446. [PubMed] [Google Scholar]
- Dey S, Dou D, Tisa LS, Rosen BP. Interaction of the catalytic and the membrane subunits of an oxyanion-translocating ATPase. Arch Biochem Biophys. 1994b;311:418–424. doi: 10.1006/abbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- Fu HL, Rosen BP, Bhattacharjee H. Biochemical characterization of a novel ArsA ATPase complex from Alkaliphilus metalliredigens QYMF. FEBS Lett. 2010;584:3089–3094. doi: 10.1016/j.febslet.2010.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosower NS, Newton GL, Kosower EM, Ranney HM. Bimane fluorescent labels. Characterization of the bimane labeling of human hemoglobin. Biochim Biophys Acta. 1980;622:201–209. [PubMed] [Google Scholar]
- Li J, Rosen BP. The linker peptide of the ArsA ATPase. Mol Microbiol. 2000;35:361–367. doi: 10.1046/j.1365-2958.2000.01696.x. [DOI] [PubMed] [Google Scholar]
- Li S, Chen Y, Rosen BP. Role of vicinal cysteine pairs in metalloid sensing by the ArsD As(III)-responsive repressor. Mol Microbiol. 2001;41:687–696. doi: 10.1046/j.1365-2958.2001.02546.x. [DOI] [PubMed] [Google Scholar]
- Lin YF, Walmsley AR, Rosen BP. An arsenic metallochaperone for an arsenic detoxification pump. Proc Natl Acad Sci USA. 2006;103:15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YF, Yang J, Rosen BP. ArsD residues Cys12, Cys13, and Cys18 form an As(III)-binding site required for arsenic metallochaperone activity. J Biol Chem. 2007a;282:16783–16791. doi: 10.1074/jbc.M700886200. [DOI] [PubMed] [Google Scholar]
- Lin YF, Yang J, Rosen BP. ArsD: an As(III) metallochaperone for the ArsAB As(III)-translocating ATPase. J Bioenerg Biomembr. 2007b;39:453–458. doi: 10.1007/s10863-007-9113-y. [DOI] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng YL, Liu Z, Rosen BP. As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. J Biol Chem. 2004;279:18334–18341. doi: 10.1074/jbc.M400037200. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP. Arsenate reductases in prokaryotes and eukaryotes. Environ Health Perspect. 2002;110(5):745–748. doi: 10.1289/ehp.02110s5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Braun D, Fernandez C, Wuthrich K. [15 N, 1H]/[13C, 1H]-TROSY for simultaneous detection of backbone 15 N–1H, aromatic 13C–1H and side-chain 15 N–1H2 correlations in large proteins. J Biomol NMR. 2000;17:195–202. doi: 10.1023/a:1008399320576. [DOI] [PubMed] [Google Scholar]
- Pufahl RA, Singer CP, Peariso KL, Lin SJ, Schmidt PJ, Fahrni CJ, Culotta VC, Penner-Hahn JE, O’Halloran TV. Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science. 1997;278:853–856. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- Ramírez-Solis A, Mukopadhyay R, Rosen BP, Stemmler TL. Experimental and theoretical characterization of arsenite in water: insights into the coordination environment of As-O. Inorg Chem. 2004;43:2954–2959. doi: 10.1021/ic0351592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig AC. Metallochaperones: bind and deliver. Chem Biol. 2002;9:673–677. doi: 10.1016/s1074-5521(02)00156-4. [DOI] [PubMed] [Google Scholar]
- Ruan X, Bhattacharjee H, Rosen BP. Cys-113 and Cys-422 form a high affinity metalloid binding site in the ArsA ATPase. J Biol Chem. 2006;281:9925–9934. doi: 10.1074/jbc.M600125200. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Bio. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Wu J, Rosen BP. The arsD gene encodes a second transacting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol Microbiol. 1993;8:615–623. doi: 10.1111/j.1365-2958.1993.tb01605.x. [DOI] [PubMed] [Google Scholar]
- Xu C, Rosen BP. Metalloregulation of soft metal resistance pumps. In: Sarkar B, editor. Metals and genetics. Plenum Press; New York: 1999. pp. 5–19. [Google Scholar]
- Yang J, Rawat S, Stemmler TL, Rosen BP. Arsenic binding and transfer by the ArsD As(III) metallochaperone. Biochemistry. 2010;49:3658–3666. doi: 10.1021/bi100026a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Abdul Ajees A, Rosen BP. Genetic mapping of the interface between the ArsD metallochaperone and the ArsA ATPase. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2010.07494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Ajees AA, Yang J, Rosen BP. The 1.4 A crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase. Biochemistry. 2010a;49:5206–5212. doi: 10.1021/bi100571r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, He Y, Skalicky J, Rosen BP, Stemmler TL. Resonance assignments and secondary structure prediction of the As(III) metallochaperone ArsD in solution. Biomol NMR Assign. 2010b;2:211–219. doi: 10.1007/s12104-010-9279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J. 2000;19:1–8. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Radaev S, Rosen BP, Gatti DL. Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation. J Biol Chem. 2001;276:30414–30422. doi: 10.1074/jbc.M103671200. [DOI] [PubMed] [Google Scholar]