Abstract
N-ethyl-N-nitrosourea (ENU) mutagenesis is presented as a powerful approach to developing models for human disease. The efforts of three NIH Mutagenesis Centers established for the detection of neuroscience-related phenotypes are described. Each center has developed an extensive panel of phenotype screens that assess nervous system structure and function. In particular, these screens focus on complex behavioral traits from drug and alcohol responses to circadian rhythms to epilepsy. Each of these centers has developed a bioinformatics infrastructure to track the extensive number of transactions that are inherent in these large-scale projects. Over 100 new mouse mutant lines have been defined through the efforts of these three mutagenesis centers and are presented to the research community via the centralized Web presence of the Neuromice.org consortium (http://www.neuromice.org). This community resource provides visitors with the ability to search for specific mutant phenotypes, to view the genetic and phenotypic details of mutant mouse lines, and to order these mice for use in their own research program.
Keywords: N-ethyl-N-nitrosourea (ENU), Functional genomics, Mouse mutants behavioral phenotype, Neurogenetics
1. Introduction
The publication of the human genome sequence was a cause for celebration and sober assessment. While sequence was presented for the whole genome [17,18], it was also clear that over 40% of the genes did not readily fit into a gene ontology category, and the vast majority of genes had little if any functional annotation. With the goal to understand the function of all the genes in the human genome, and the acknowledgment that the mouse was the best model organism to attain this objective, a variety of complementary strategies in the mouse has emerged as the means to this end [1,10]. To understand the function of genes in the central nervous system is arguably one of the most challenging of these initiatives, as we are still learning how human neurological and behavioral phenotypes manifest themselves in mice. Until recently, neurological phenotypes in mice were detected in a cottage-industry setting at a steady but slow pace (e.g., investigators studying select phenotypes of mice with natural variants or spontaneous mutations or gene knockouts). The wonderful productivity of mutagenesis programs in fly, worm, and fish, and the demonstration by the German (http://www.gsf.de/ieg/groups/enu-mouse.html) and UK (http://www.mgu.har.mrc.ac.uk/mutabase/) groups that these efforts can be scaled to the mouse led NIH to support three mutagenesis centers in the United States to screen for mutations that affected various aspects of nervous system structure and function. The phenotype is the driving force in these efforts at Northwestern University, the Jackson Laboratory, and the Tennessee Mouse Genome Consortium (TMGC). These Centers are all using a forward-genetic strategy of combining chemical mutagenesis with high-throughput phenotypic screening, allowing the altered phenotypes of mutants to lead to their identification. Identification of mutants, in turn, allows for the association of genes with functions, as well as providing model systems for studying the molecular and genetic mechanisms underlying the affected phenotype.
2. ENU mutagenesis approach
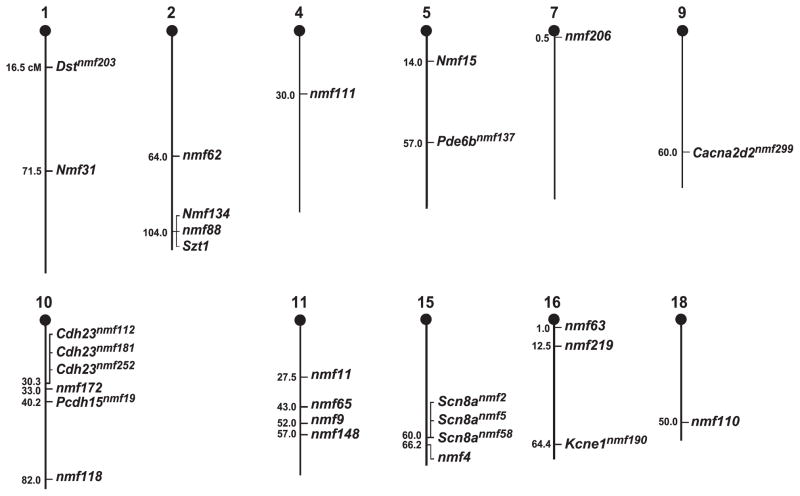
The screens being carried out by the three mutagenesis centers employ the chemical supermutagen N-ethyl-N-nitrosourea (ENU). ENU is an ethylating agent that is both mutagenic and cytotoxic in mouse spermatogonial stem cells [16]. The average per-locus mutation frequencies achievable by ENU treatment, historically measured at visibly marked loci, depend on the dose, gene (target) size, and ability to recognize phenotype, and range from 1/1500 for a single 250 mg/kg dose to ~1/700 for a fractionated 4×100 mg/kg dose [4,16], but the rate can vary substantially among loci [13,16]. ENU is particularly valuable for inducing allelic series of mutations since it causes primarily base-pair substitutions [11] (for recent NMF results see Fig. 3). Such base-pair substitutions can result in a wide range of mutation outcomes, including generation of amorphs (nulls), hypomorphs (observed as less-severe or leaky mutations), neomorphs (proteins with novel functions or binding activities, often recognized as dominant mutations), or antimorph mutations (e.g., dominant-negative mutations where one mutant member of a protein homo-or heterocomplex reduces or eliminates the function of the complex). Importantly, about 70% of the 38,000 human mutations in over 1500 genes that have been identified are of the single-base pair variety (http://archive.uwcm.ac.uk/uwcm/mg/docs/hahaha.html). Thus, the ENU-induced mutations resulting from this experimental approach in mice mimic the predominant cause of human genetic disease.
Figure 3.
Chromosomal Mapping of NMF Mutants (see text for description).
ENU mutagenesis is complementary to other types of mutagenesis (e.g., gene-trap insertional mutations or specific-gene knockouts achieved by homologous recombination in ES cells). Thus, when screening for new mutant phenotypes, the range of mutational types induced by ENU increases the potential for discovery of novel neurodevelopmental or behavioral phenotypes, in cases where early embryonic lethality might be the only phenotype ascertained from a complete null at a particular locus. Furthermore, the knockout allele may have a very different phenotype than other less “invasive” perturbations of the gene, such as with ENU. A recent illustration of this outcome comes from the knockout phenotype of the Af4 gene. This gene is associated with lymphoblastic leukemia in humans and its elimination by knockout led to deficits in B and T cell development [5]. However, a recent report from the UK mutagenesis effort found that an ENU-mutation in the Af4 gene led to a progressive degeneration of cerebellar Purkinje cells and attendant functional abnormalities such as ataxia [6], with no notable alterations in immune function.
3. Genome-wide and regional approaches to mutagenesis
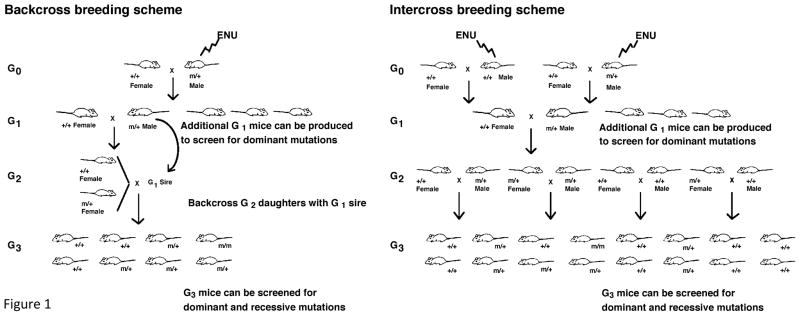
Two general approaches can be used with ENU mutagenesis. The more widely used approach is to target the whole genome (Fig. 1). The projects at Northwestern and the Jackson Laboratory are using ENU to produce mutations throughout the genome, followed by three generations of breeding to produce homozygous mutants. By screening third-generation (G3) progeny of mutagen-treated mice, dominant, semidominant, and recessive mutations may all be recovered. In this approach, the target is broad but the ascertainment of mutants is dependent upon the number of mice that are screened. Thus, in the schema presented in Fig. 1 (left), only 1/8th of the tested mice will carry the homozygous mutation and the cohort size for screening needs to take into account these odds (typically, 12–16 mice are screened).
Figure 1.
Whole genome mutagenesis strategies. See text for description.
3.1. The Neurogenomics Project at Northwestern University
3.1.1. Overview
The NIH Neurogenomics Project at the Northwestern University Center for Functional Genomics is focused upon identifying genes involved in a number of neuroscience-relevant systems and behaviors. Two different breeding schemes for ENU mutagenesis have been used for the screening of either dominant or recessive mutations (shown in Fig. 1). These are a backcross scheme (at left) and an intercross scheme (at right). In the case of a first-generation screen for dominant mutations, each G1 mouse represents one mutagenized gamete. In the case of a screen for recessive mutations, each male G1 mouse may be used to found a three-generation pedigree in which the G2 females are backcrossed to the G1 male in order to isolate G3 progeny that are homozygous for the mutagenized gamete (Fig. 1, left). In the case of an intercross breeding scheme, two G1 mice are used to found the pedigree, and G2 progeny are intercrossed with siblings (Fig. 1, right), so that two mutagenized gametes are represented in the same pedigree.
In both schemes, G1 mice are produced as follows: ENU (90–100 mg/kg body weight) is injected intraperitoneally into wild-type C57BL/6J (JAX # 000664) male mice once a week for 3 weeks, starting at 6–8 weeks of age. On average, the G0 male mice recover fertility 12–14 weeks post-injection, but only approximately 50% of all ENU-treated mice ever regain fertility to sire G1 offspring. Each G1 mouse produced from the ENU-treated mice represents one mutagenized G0 gamete. After this point, the two breeding schemes diverge.
In the backcross scheme, male G1 mice are mated to wild type C57BL/6J females to produce G2 offspring. The G2 females are then backcrossed to their G1 fathers to produce G3 progeny. In the intercross scheme, two G1 mice are bred together so that two mutagenized G0 gametes are represented in each kindred. G2 sibs are then intercrossed to produce G3 progeny. The target rate of production and screening is 10,000 G3 mice/year. To date, the Northwestern group has screened N16,000 mice with production and screening ongoing.
3.1.2. Phenotypic screens
The Northwestern group has selected five phenotypic domains on which to focus their screens. These screens were selected not only because of their central interest to neuroscience, but because their potential interrelationships would allow the tests to be mutually reinforcing. In addition, a set of “preliminary assessment” tests was conducted to provide supporting information to aid interpretation of other assays. All mice go through all the tests (unless there are concerns over the animal’s ability to perform a test, or it is to be bred); the sequence of tests is designed to minimize carry-over influences of one test to the next. The specific assay procedures or conditions have been adjusted in many cases so that both low-and high-scoring outliers can be detected, or so that variance within a normal population is minimized.
Preliminary Assessment
The Preliminary Assessment consists of recording the animal’s body weight at two ages (6 and 10 weeks), a hearing screen (IHR Clickbox, Preyer reflex), and two emotional behavioral assays: the elevated plus maze and the open-field behavior.
Neuroendocrine Response to Stress
A standardized, acute stress (10 min restraint) is presented. The tip of the tail is cut and blood samples are collected at the beginning and end of the stress using capillary tubes. Serum samples are radioimmunoassayed for Corticosterone and for Thyroid Stimulating Hormone (TSH) to assess the responsiveness of the hypothalamic–pituitary–thyroid and hypothalamic–pituitary–adrenal axes to stress. Putative mutants with both high and low, basal and/or stress corticosterone values are being tested for heritability of the trait, as well as putative mutants with both high and low TSH values.
Learning and Memory
Fear conditioning is used as the screening test for learning and memory. Mice are trained to fear the test chamber with a series of four 0.75-mA footshocks, 1 min apart. Twenty-four hours after training, the mice are returned to the same test chamber for 5 min and fearful behavior (freezing) is recorded as a measure of context-dependent learning or memory. An “overtraining” protocol is used so that mice are subject to a second training session of four footshocks following the 24-h test, and a second test session is conducted the next day. Putative mutants with both high and low freezing scores are being tested for heritability of the trait.
Response to Psychostimulants
The hyperlocomotion response to administration of 20 mg/kg cocaine is recorded. Drug-naïve wild-type C57BL/6J mice exhibit a robust induction of activity in response to this dose; hyper-or hyporesponsive mice are selected. Test-crosses to assess heritability are underway for both hyper-and hyporesponsive putative mutants.
Circadian Rhythmicity
The circadian rhythm of wheel-running activity is monitored. Animals are maintained first in a light–dark cycle, then constant darkness (to remove time cues). The period and persistence of a free-running rhythm, and phase angle of entrainment are studied. Two heritable mutant lines are presently available, part-time, an autosomal recessive mutation with shortened circadian period, and Overtime, an autosomal dominant mutation with lengthened period.
3.1.3. Vision
Two assays of the visual system are used. The fundus of the eye is examined and photographs are taken of all mice. These are reviewed by the ophthalmology team at the University of Iowa. Dark-adapted electroretinograms also are recorded in response to a series of light flashes of different irradiances. An autosomal dominant mutation, Noerg1, has been identified and is available. This mutation produces lesions in the fundus as well as no electroretinogram response to light of any irradiance.
3.2. The Neuroscience Mutagenesis Facility at the Jackson Laboratory
3.2.1. Overview
The majority of mutagenized mice at the Neuroscience Mutagenesis Facility of The Jackson Laboratory (NMF) is of C57BL/6J or A.B6-Tyr origin, although some mutants are produced on a mixed (e.g. C57BL/6J and BALB/cByJ) background. Applying the above described approach of genome-wide phenotypic screening of third generation families, the NMF has so far screened over 28000 G3 mice, which represent over 1600 families or distinct mutagenized genomes. These efforts resulted in the detection of more than 40 neurological mutant mouse lines, which have been characterized and made available to the public; in addition, over 28 phenotypic deviants with abnormalities that are (due to their non-neurological characteristics and/or commonness) of lesser interest to the NMF have been identified and made available to the public.
3.2.2. Phenotypic screens
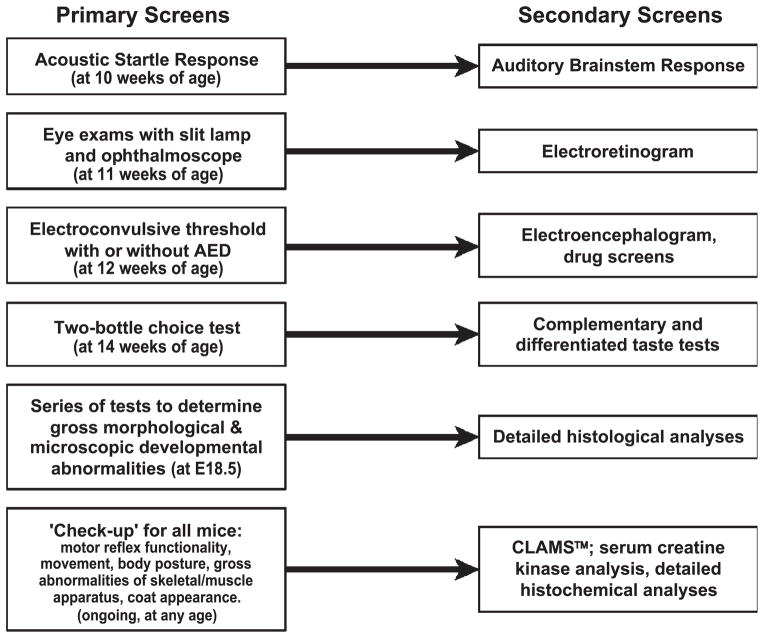
The NMF uses a series of high-throughput screens to detect mutations with phenotypes affecting visual, auditory, vestibular, gustatory, and metabolic functions, seizure thresholds, and the progression of growth and development. They include overt screens, such as the routine inspection of the animals for signs of motor, behavioral or morphological abnormalities, examination of the cornea and fundus, a determination of the electroconvulsive threshold, etc., and also more complex screens, like the Comprehensive Lab Animal Monitoring System (CLAMS™), which allows the monitoring of several physiological and behavioral parameters over time (i.e., 3 days), including activity levels, volume of food and liquid consumption, temperature, and metabolism (Fig. 2).
Figure 2.
Phenotype Screens at the NMF (see text for description).
For a quantitative evaluation of phenotypes, scores which deviate two or more standard deviation from the mean of B6 mice or represent 1% or less of the normal B6 population are regarded provisionally as abnormal. In general, mice with abnormal scores in at least two consecutive test runs are regarded as phenotypic deviants. For a qualitative evaluation of phenotypes, phenotypic deviants are defined by overt behavioral abnormalities, such as head bobbing, paralysis of limbs, hyperactivity, etc., or other phenotypes that are not normally observed in B6 mice. G3 families that show phenotypic deviants are selectively mated and, once heritability of the observed abnormal behavior has been confirmed by vertical transmission, the affected mice are regarded as mutants. They then undergo further testing with phenotype-dependent secondary screens, such as electroretinograms, electroencephalograms, auditory brainstem responses, etc., (Fig. 2), to characterize the abnormality in more detail. These mostly behavioral observations are complemented through the results of pathology work-ups and histochemical and/or immunohistochemical procedures which can provide information about anatomical and/or neurochemical correlates of the observed behavioral abnormalities.
To support workflow management for the colony managers, phenotyping personnel, and associated NMF scientists, we have developed a laboratory information system, called MutaJAX. MutaJAX has a client–server architecture, which is ideal for information systems that serve multiple, distributed users. The database has been implemented as a relational database in Oracle 9i running on a Unix (SunOS) platform. The primary data entry interface for MutaJAX is implemented as a JAVA application. The query and report interfaces are implemented as Web-based forms that return results in either html format or as Excel spreadsheets.
The NMF has established, to date, 43 mutant mouse lines, and heritability testing of approximately 90 phenotypic deviants is in progress. Established mutant lines show abnormal functions of motor and neuromuscular (n=24), visual (n=7), auditory and/or vestibular systems (n=7), metabolism (n=1), and/or abnormal seizure thresholds (n=4) and are characterized by various abnormalities such as hindlimb spasms and paralysis, unsteady gait, reduced grip strength, head nodding, circling behavior, deafness, obesity, and/or thin or mottled retinae.
The majority of the observed abnormalities is recessive (N80%), and a chromosomal location has been determined for over 60% of the observed mutations (see Fig. 3) by either determining a chromosomal region or the specific location of the mutation, or through the results of complementation tests. In some cases the identified mutations appear to link to new genes; in other cases they link to alleles of known genes, i.e., of Pcdh15 (nmf19), Pde6b (nmf137), Kcne1 (nmf190), Dst (nmf203), and Cacna2d2 (nmf299). In two cases three alleles of known genes have been identified, i.e., nmf2, nmf5 and nmf58 as alleles of Scn8a, and nmf112, nmf181 and nmf252 as alleles of Cdh23; of course other (pending) gene identifications may have been made by scientists who have received NMF mice.
Our results confirm the usefulness of the ENU-induced mutagenesis to detect mutations genome wide and provide effective tools for the investigations of genetic mutations that may lead to an enhanced understanding of neurological diseases. Detailed information about NMF mutant lines, e.g., heritability, mapping results, screens, videos, photo documentation of anatomical or histological abnormalities that were observed, and information about how to obtain mutant mice, is available at the NMF Web site (http://nmf.jax.org and http://www.neuromice.org).
3.3. The regional mutagenesis approach of the TMGC
3.3.1. Overview
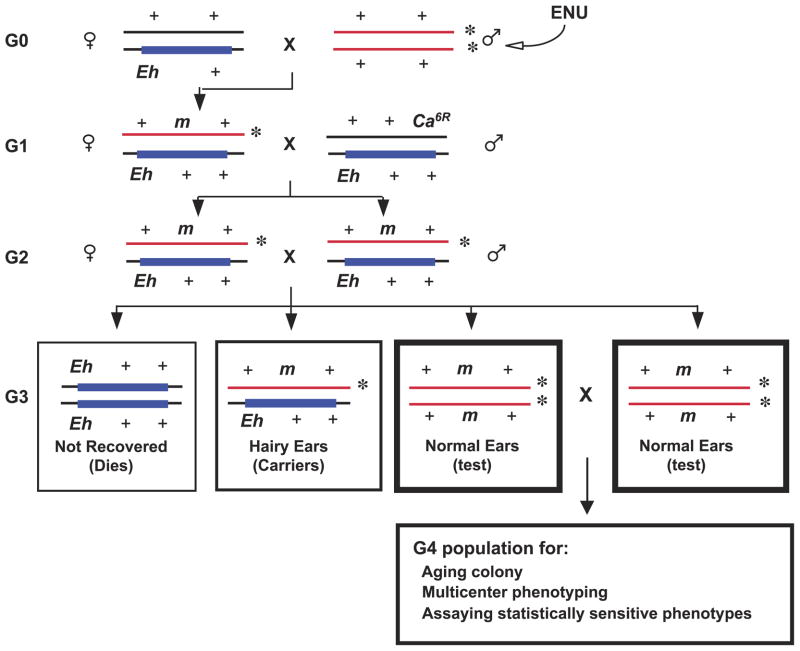
The Tennessee Mouse Genome Consortium uses a regional approach. Here, a genetic marker (ideally, a simple-to-identify visible marker) is used to follow a specific mutagenized chromosome through generations of the breeding protocol. Usually, chromosomal rearrangements, such as deletions and inversions, are used to facilitate these crosses (Fig. 2; reviewed in Ref. [14]). Because of the use of visible (or molecular) markers in the design of a regional screen, a visibly identifiable class of animals that should express a recessive phenotype if a mutation occurs in the region under study is present in each pedigree. Thus, the trade-off for the regional approach is studying confirmed “test class” mice but for only a limited part of the genome. The TMGC’s mutagenesis efforts are directed toward recovering new recessive mutations, affecting neurodevelopment and behavior, in several genomic regions. In two-generation screens, G2 test-class animals are hemizygous for a mutagenized chromosome region brought about by placing mutagenized chromosomes opposite a deletion [12] or opposite the Y chromosome in male test-class animals (e.g., see Ref. [9]). In three-generation screens, the G3 test-class animals are homozygous for a segment of the mutagenized chromosome [7,14,15]. See Fig. 4 for an example of the TMGC regional screen.
Figure 4.
An inversion-mediated screen of the distal portion of Chr 15. The shaded rectangular box on a particular Chr 15 indicates the In (15)Eh 2Rl inversion (denoted as ‘Eh’, which has a hairy-ear phenotype when heterozygous and is lethal when homozygous). This inversion suppresses the recovery of crossover products in the distal part of Chr 15. Ca6R is a radiation-induced, recessive-lethal allele of the Caracul locus, which maps within the region spanned by the In (15) 2Rl (Eh) inversion (L.B. Russell and E.M. Rinchik, unpublished data). Ca6R/+ animals have a mild phenotype of wavy fur and whiskers that can be easily observed whether or not the In (15) 2Rl/+ hairy ears phenotype is present. The normal-eared G3 test class will be homozygous for a particular mutagenized Chr 15 (identified by the asterisk). Absence of G3 mice with normal ears at weaning in any pedigree indicates the presence of a prenatal-, perinatal-, or juvenile-lethal mutation, which can be easily propagated from marked G3 inversion-heterozygotes. If these G3 test-class mice are recovered (i.e., if the pedigree is not segregating a distal Chr-15 lethal mutation), they are bred to generate a larger G4 population that is homozygous for the same mutagenized Chr. These larger G4 populations are important for multicenter phenotyping, for establishment of an aging colony for ascertainment of progressive or late-onset recessive disorders, and for assaying statistically sensitive mutant phenotypes.
In regional screens, the entire genome is mutagenized in the G0 male, but newly induced recessive mutations are readily recovered only if they are located in particular regions (corresponding to the visible markers used in the cross). Consequently, since one is ignoring the majority of the genome, cursory evaluation of such regional screens might indicate that they are less efficient and/or far less “deep” in coverage than whole-genome screens. We argue, however, that, in practice, the opposite is true for reasons that pertain to the screen itself as well as to future maintenance, use, and analysis of the resulting mutant stocks. The regional approach has numerous advantages that include: (1) Test-class animals can be visibly or molecularly identified with minimal effort, (2) genetically identical test-class mice can be sent out for multisite testing in statistically relevant numbers, (3) test-class mice can be set aside for analysis of aging phenotypes, (4) embryonic lethal phenotypes can be ascertained, (5) the identified phenotypes are virtually pre-mapped for the targeted chromosome.
These several advantages more than make up for the smaller apparent target size in the regional approach when compared to whole genome approaches. In other words, for any particular class of phenotypes, total mutation yield in a regional screen may be lower than in a whole-genome screen, but the favorable attributes of regional screens make it likely that a greater number of phenotypes per unit genome length will be ascertained in the long run. Likewise, because these regional strategies nonetheless involve inbreeding crosses (e.g., the G2 and G3 matings in Fig. 4), we can observe nontarget-chromosome recessive phenotypes at variable frequency, although clearly not at the frequency that can be achieved by performing father–daughter backcrosses, which are typically done for whole-genome recessive screens [3,8]. Thus, both visible recessive and dominant mutations on other chromosomes can also be ascertained along with the pre-mapped, more wide-ranging phenotypic spectrum of mutations in the region under study. In addition, depending on the type of phenotype screen employed, concentrating on a single region allows a larger number of gametes to be screened for the same total number of mice bred, which sets the stage for recovering multiple alleles at individual loci, usually a highly desirable result.
3.3.2. The TMGC screen
The TMGC Neuromutagenesis project has employed a regional approach with both visibly identifiable marker strains (with inversions on chromosomes 7, 10, 15, and X) as well as strains with molecularly identifiable markers (using consomic lines with introgressed chromosomes 14 or 19). As briefly described above, this regional-mutagenesis approach provides at the outset a simple way to identify test-class mice, which can be generated in statistically useful numbers. Using this approach then, the TMGC mutagenesis program has identified many mutation-induced overt embryonic lethal mutant pedigrees (n=29), as well as several overt morphological and obvious behavioral phenotypes (n=26). We have taken further advantage of this scheme in our behavioral screens, allowing us to send statistically relevant cohorts of test-class mice to each screening domain, providing greater confidence in assessing potential outlier status for each pedigree (or mutagenized gamete) tested. The regional approach is particularly advantageous for performing the Aging and Neurohistology screens, since we can identify and set aside test-class mice from each pedigree for long-term or terminal analyses. The TMGC has cast a wide phenotypic net to take advantage of this test-class cohort approach by assessing behavioral measures in nine phenotypic domains, including: General Behavioral, Ethanol Responses, Drug Abuse, Social Behavior, Epilepsy, Auditory, Vision and Eye, Neurohistology, and Aging. Detailed descriptions of the screens (and PDF versions of all protocols) for each phenotypic domain can be found by following the links provided at http://www.tnmouse.org/neuromutagenesis/. Integral to this project, we have developed a Web-based information management system (termed MuTrack) that tracks animal husbandry information, phenotype data acquisition, and has a wide range of statistical analysis package that aids PIs in identifying outlier pedigrees for further characterization as mutants. Through these quantitative and histologogical screens, we have identified “subtle” mutant phenotypes in 26 pedigrees of mice.
3.3.2.1. General behavior
The core behavioral screen consists of the assessment of six independent behavioral phenotypes, including: locomotion and exploratory activity, learning and memory, startle and prepulse inhibition, nociception, behavioral despair, and behavioral anxiety. (1) The-open field test is used to assess locomotor activity and anxiety-related behavioral patterns during a 20-min test period in an open-field activity monitor. (2) Learning and memory is assessed using a standard fear-conditioning paradigm, where a series of footshocks are paired with an auditory cue during a training phase. The efficacy of learning is tested 24 h following training where cue and context-dependent associations are tested independently. (3) A startle/prepulse inhibition paradigm are used to screen for neuropsyciatric behavioral phenotypes, where the influence of variable intensity stimuli prepulses are measured in comparison to maximal startle responses. Deficits in this type of paradigm have been associated schizophrenia in humans. (4) The behavioral assessment of sensitivity to nociceptive stimuli is performed with a standard forepaw withdrawal test using a 52°C hotplate. (5) The tail suspension test is used to determine the relative susceptibility to behavioral despair. Individuals or pedigrees prone to despair-like phenotypes exhibit longer times immobile while suspended during a 6-min trial. (6) In order to assess anxiety-related behaviors, we have employed a light/dark box test, where the relative time spent in a darkened compartment compared with the light compartment reflects the general anxiety levels of a particular individual/pedigree. Through these screens, we achieve a general picture of the baseline behavioral phenotype of potentially mutant pedigrees.
The Behavioral core screen has been the most productive of the TMGC screens, identifying eight mutant pedigrees, from Tail Suspension (n=2), Open Field (n=3), Prepulse Inhibition (n=2), and Nociception (n=1).
3.3.2.2. Ethanol responses
The screens employed by the ethanol domain are focused to identify mutant pedigrees that reflect either alterations in particular physiological and behavioral responses to ethanol or are involved in the predisposition for altered ethanol consumption. An initial series of screens assess the effects of a moderate dose of ethanol (2.25 g/kg) on anxiety-related behavior, locomotor activity, motor coordination, thermoregulation, and alcohol metabolism. Behavioral anxiety is assessed in an elevated plus maze, 10 min following IP injection of ethanol, where the relative time spent in the open arms of the maze during a 5-min test trial reflects the anxiolytic effects of ethanol. The mice are then assessed for the influence of ethanol on locomotor behavior in a computer-monitored activity chamber, where the level of hyper-or hypoactivity is measured over a 10-min session. Ethanol-induced motor incoordination is assessed by measuring performance on an accelerating rotarod after ethanol treatment (the assessment of rotarod training is conducted 1 day prior to ethanol exposure and testing). Sixty minutes following the ethanol injection, the influence on this exposure on core body temperature is measured and then tail vein blood is sampled to determine the blood alcohol concentration. The following day baseline core body temperature is measured to allow a calculation of the influence of ethanol exposure on thermoregulation. Finally, the two-bottle ethanol choice test is used to measure a propensity of a given pedigree to consume ethanol or water.
The Ethanol screen has presently identified three mutant pedigrees: two pedigrees represent altered activity (hyperactivity) in response to ethanol exposure, and a third pedigree was identified as drinking excessive amounts of water in the two-bottle choice test.
3.3.2.3. Drug abuse
The Drug abuse domain employs three independent screens to assess behaviors that are associated with illicit drug abuse in humans, specifically patterns of novelty-seeking or risk-taking behaviors and behavioral responses to cocaine. An initial screen assesses drug-naïve mice for novelty-seeking behavior and food neophobia as measures indicating a propensity for susceptibility to addiction or drug use. Because behavioral response to stressors have also been associated with vulnerability to addiction, a second screen assesses locomotion in a novel environment and locomotion in response to a saline injection as the stressors. Behavioral responses to cocaine are systematically assessed by measuring locomotion elicited by acute and repeated administration of cocaine and conditioned place preference (CCP) in response to cocaine. These assays provide an indication of genetic modifications that may influence the activation of mesocorticolimbic dopamine and/or dopamine dependent or independent elements of mesocorticolimbic system.
The Drug screen has identified one pedigree that demonstrates low responsiveness to the activity-stimulating effects of cocaine.
3.3.2.4. Social behavior
The Social behavior domain uses screens to identifying mutant pedigrees that display abnormal sociosexual responses to either conspecific scent presentation (direct or indirect) or with physical interaction with conspecific control mice. An odor preference screen assesses an individual’s attention to or, interest in, either same-sex or opposite-sex scents in an isolated cage environment. In conjunction with indirect odor preference determination, this screen also assesses habituation and dishabituation of responses to repeatedly presented odors. A T-maze is used to assess behavioral investigation of compartments that have been scent marked by either opposite-sex or same-sex individuals. Lastly, in order to assess the influence of genetic modifications on the aggression in mice, we have implemented an intruder-encounter aggression screen.
The Social Behavior screen has identified two mutant pedigrees that exhibit female specific behaviors involving preference and environmental interactions to conspecific odors.
3.3.2.5. Epilepsy
The Epilepsy domain has developed high-throughput screens to identify pedigrees with altered susceptibility to hypothermia-induced seizures in preweanling pups and drug-induced seizures in adult mice. This febrile seizure screen (hyperthermia-induced) is unique to the mutagenesis realm, but not unique to behavioral neurogenetics, where several febrile seizure-susceptibility genes have been identified in humans. For this screen, postnatal day 14 pups are subjected to whole body hyperthermia by circulated heated air (~50 °C) until a seizure is induced. At seizure onset, the core body temperature is recorded as seizure threshold, and the length and intensity of the seizure are also recorded. Similarly, in adults, methyl-betacarboline 3-carboxylate (h-CCM) is used to determine susceptibility for seizure induction. In conjunction with a complex seizure score, the latency to induce and severity of seizures are measured across test-class pedigrees in order to determine deviation from the population.
This newly added screen has only screened 40 pedigrees and has yet to identify a mutant line.
3.3.2.6. Neurohistology
The Neurohistology domain employs several screens to identify pedigrees that display altered growth, abnormal morphological character, and/or changes in the cytoarchitecture of the brain. As an initial gross measure of brain growth and health, brain weights are taken from freshly isolated tissue, and this tissue is then quick frozen in isopentane. A series of relatively simple histochemical and immunocytochemical stains on a representative brain from each pedigree of test-class mice. These stains assess the cytoarchitectonics, myelinated fiber pathways, terminal fields, astroglia and neuronal populations, activity state, and proliferative populations and neuropathology in brain. A panel of morphometric measures (such as area measures of the cerebral cortex, corpus callosum, and cerebellum) are also taken from the histological material to assess overt regional changes in brain structure and size.
The Neurohistology screen has identified five mutant phenotypes, One quantitative phenotype of low brain weight, and four exhibiting histological dysmorphology of the cerebellum or hippocampus.
3.3.2.7. Eye
The Eye domain employs three assays to identify pedigrees with alterations in the morphology of the eye and retina. Taking advantage of the regional test-class approach, we have used an initial screen of fresh eye weights, which provides a simple means to identify possible disrupted eye growth or development phenotypes. The indirect ophthalmoscopy and fundus exam screens provide noninvasive measures for altered eye morphology and have proven to be the most successful screen for identifying heritable mutants. Lastly, a simple histological screen of retinal tissues (incorporating several histochemical and immunocytochemical stains) provides an excellent survey of the morphological underpinnings of the fundus phenotype as well general assessment of more subtle changes in retinal cellular makeup.
The Eye screen has identified eight mutant pedigrees: One quantitative phenotype of low eye weight, and seven pedigrees identified initially through the fundus exam, four of which have presently been followed up to show a histological pheneotype of degeneration, hypervascularization, or lamination defects.
3.3.2.8. Auditory
The Auditory domain uses two screens that can identify pedigrees with deficits in auditory processing and hair cell function. The auditory brainstem response (ABR) is used to determine a threshold for normal ABR wave using broadband click stimuli as well as pure-tone stimuli at 8, 16, and 32 kHz. Pedigrees that consistently display high thresholds for any of the stimuli are considered to have wide-range to high-frequency hearing loss. As a secondary screen for mutants, a distortion otoacoustic emissions (DPOAE) analysis is used to determine whether the auditory deficits measured with ABR result from altered hair cell function.
The hearing screen has identified one mutant pedigree that exhibits high-frequency hearing loss.
3.3.2.9. Aging
The aging domain takes full advantage of the test-class cohort approach that is provided by the TMGC regional mutagenesis screen. For this screen, eight genetically identical (within the targeted chromosomal region) mice (4 male/4 female) are set aside to age for 18–28 months. Over the course of this time frame, body weights are measured at 4, 6, and 8 weeks, and then monthly until death or 28 months. The morbidity and mortality of these aging mice are monitored for signs of decreased or increased longevity. In addition, at 18 months of age, four individuals are tested in the General Behavior screen (described above) to determine whether there are any aging-related differences in these behaviors. At this age, one male and one female are sacrificed and organs, brain, and eyes are collected and assessed for pathology using standard histological staining (as indicated in the Neurohistology and Eye domains). Lastly, one 18-month old individual per pedigree undergoes a micro-CT analysis, that identifies any major alterations in skeletal and soft tissue. All measures produced in the aging screens are compared to similar aging pedigrees as well as to measures from young mice from that same pedigree. In this way, this screen will more precisely determine differences in the aging process and identify mutation-induced accelerated decrepitude or enhanced longevity.
3.3.2.10. Summary
Through this regional screen, we have identified 81 different mouse lines that exhibit heritable mutant phenotypes. Inherent in this approach is the ability to identify embryonic lethal pedigrees, of which we have identified 29 lines linked to three chromosomes (13 on Chr7, 8 on Chr10, and 8 on Chr15).
4. Bioinformatic tools
The amount of information that is needed to be assembled and processed is quite substantial in such an animal-and screen-intensive program. To make each of the mutagenesis programs run successively, it has been critical to establish Web-based bioinformatic tools to manage the multitude of transactions that start from the time of mutagenesis, subsequent breeding of several generations, phenotype screens, identification of outliers, and then pushing the whole process through verification and confirmation of the original outlying pedigree. These issues have been addressed for our three projects [2] and a summary of the bioinformatics infrastructure to enable each of our mutagenesis programs is provided in Table 1.
Table 1.
Bioinformatic infrastructure supporting neuroscience mutagenesis centers
| Center | Northwestern University Center for Neurogenomics | The Jackson Lababoratory Neuroscience Mutagenesis Facility | Tennessee Mouse Genome Consortium Neuromutagensis Project |
|---|---|---|---|
| Center home page | http://genome.northwestern.edu/neuro/ | http://nmf.jax.org | http://www.tnmouse.org/neuromutagenesis/ |
| Database management system/operating system | Oracle 9i on Windows 2003 | Oracle 9i on Solaris | Oracle 9i on Solaris |
| Architecture | Multi-tier: database server, application (middleware) server, Web clients | Multi-tier: database server, application server (middleware), Web and JAVA clients | Multi-tier, Client–server (Web clients—Apache server) |
| Data entry interface | ColdFusion | Java | PHP |
| Data query/report interfaces | Oracle, ColdFusion, Perl, Java | JSP, ASP, Oracle | Oracle, SAS, PHP |
| Special features | Store mouse behavioral videos —about 14 TB of videos are currently on line | Underlying data model and applications support extension of the information system to a wide range of phenotyping workflows. | Acquires phenotype data and automates processes across multiple, widely dispersed labs and institutions. |
| Report graphics | Java servlets, flash | Crystal Reports, Oracle | GD, SAS, PHP |
| System information | http://genome.northwestern.edu/neuro/mousedb.html | http://nmf.jax.org | http://www.tnmouse.org/mutrack.html |
| Accessibility | Internal use only | Primarily internal use. Public access via mutant mouse reports on NMF Web site. | Database open to public; only project members have write-access to appropriate tables. |
5. Neuromice.org—a community resource for neurological mutant mice
New mutant mouse lines are important tools for functional gene identification and only have their full scientific potential realized through investigation by the greater scientific community. In order to create an efficient and effective means to distribute mutant mice, a distribution consortium, known as Neuromice.org, was created.
The Neuromice.org consortium was established as part of the Trans-NIH Initiative to create new research resources, such as mutant mouse models of neurological and behavioral disorders for the neuroscience research community. As with the mutagenesis programs, seven NIH Institutes support Neuromice.org—National Institute of Mental Health (NIMH), National Institute on Deafness and Other Communication Disorders (NIDCD), National Institute of Neurological Disorders and Stroke (NINDS), National Institute on Aging (NIA), National Eye Institute (NEI), National Institute on Drug Abuse (NIDA), and National Institute on Alcohol Abuse and Alcoholism (NIAAA). The consortium, directed by Dr. Joseph S. Takahashi, consists of three mutagenesis and phenotypic screening facilities, the Neuromutagenesis Project of the Tennessee Mouse Genome Consortium, the Neuroscience Mutagenesis Facility at The Jackson Laboratory, and the Neurogenomics Project at Northwestern University. These three mutagenesis and phenotypic screening facilities combine their knowledge, resources, and efforts to maintain, characterize, and distribute recently identified mouse lines with alterations in nervous system function and/or behavior. All three sites are using ENU mutagenesis and high-throughput phenotyping protocols to screen mice, thus identifying mutations affecting a broad array of neural and behavioral domains. Although the consortium involves three sites, information on mice from each of the three sites is available through a central database and Web site at http://www.neuromice.org.
The Web site, http://www.neuromice.org, is a diversified resource. It offers an extensive, searchable, and customizable phenotype and genotype database. Currently, there are more than 100 mouse lines available for distribution. They reflect a diversity of phenotypic abnormalities, such as alcohol response, epilepsy, and circadian rhythm. Sixty-five mouse lines have been mapped to a chromosome and 22 mouse lines have the gene identified. High-throughput phenotyping protocols are available for researchers interested in adopting or developing these techniques. Scientists can search the database by keyword, phenotypic domain, availability status, mouse line, gene/locus, and chromosome. By registering with Neuromice.org and creating a login identity, scientists can create customized automatic searches. The database is a portal to other mouse resource databases. The Web site is a virtual storefront offering customer service, online ordering, and distribution to scientists worldwide. Scientists can order live or cryopreserved mice from each of the three research centers on one site: http://www.neuromice.org.
6. Conclusions
The extension of ENU mutagenesis to specifically identify recessive mutant mice with neurological phenotypes has been a challenge in terms of building up the personnel and phenotyping expertise and the informatics infrastructure to help run the various programs. Each of the three facilities has developed unique ways to meet this challenge and have identified mutant mice with a wide range of neurobehavioral phenotypes. Furthermore, these efforts speak to an industrial-sized approach to identifying single gene mutations which underlie mouse nervous system structure and function that is a highly relevant model to human disease.
Acknowledgments
We would like to thank all the members of each Mutagenesis Center for their contributions to this community effort. This work is generously supported by cooperative agreements U01-MH61971 (TMGC), U01-MH61915 (NU), U01-NS41215 (TJL).
References
- 1.Battey J, Jordan E, Cox D, Dove W. An action plan for mouse genomics. Nat Genet. 1999;21:73–75. doi: 10.1038/5012. [DOI] [PubMed] [Google Scholar]
- 2.Bult C, Kibbe WA, Snoddy J, Vitaterna M, Swanson D, Pretel S, Li Y, Hohman MM, Rinchik E, Takahashi JS, Frankel WN, Goldowitz D. A genome end-game: understanding gene function in the nervous system. Nat Neurosci. 2004;7:484–485. doi: 10.1038/nn0504-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hentges K, Thompson KPA. The flat-top gene is required for the expansion and regionalization of the telencephalic primordium. Development. 1999;126:1601–1609. doi: 10.1242/dev.126.8.1601. [DOI] [PubMed] [Google Scholar]
- 4.Hitotsumachi S, Carpenter DA, Russell WL. Dose-repetition increases the mutagenic effectiveness of N-ethyl-N-nitrosourea in mouse spermatogonia. Proc Natl Acad Sci U S A. 1985;82:6619–6621. doi: 10.1073/pnas.82.19.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isnard P, Core N, Naquet P, Djabali M. Altered lymphoid development in mice deficient for the mAF4 proto-oncogene. Blood. Jul 15;96(2):705–710. [PubMed] [Google Scholar]
- 6.Isaacs AM, Oliver PL, Jones EL, Jeans A, Potter A, Hovik BH, Nolan PM, Vizor L, Glenister P, Simon AK, Gray IC, Spurr NK, Brown SD, Hunter AJ, Davies KE. A mutation in Af4 is predicted to cause cerebellar ataxia and cataracts in the robotic mouse. J Neurosci. 2003;23:1631–1637. doi: 10.1523/JNEUROSCI.23-05-01631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Hum Mol Genet. 1999;8:1955–1963. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- 8.Kasarskis A, Manova K, Anderson KV. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc Natl Acad Sci U S A. 1998;95:7485–7490. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyon MF, Phillips RJ, Fisher G. Use of an inversion to test for induced X-linked lethals in mice. Mutat Res. 1982;9:217–228. doi: 10.1016/0027-5107(82)90225-1. [DOI] [PubMed] [Google Scholar]
- 10.Mural RJ, Adams MD, Myers EW, Smith HO, Miklos GL, Wides R, Halpern A, Li PW, Sutton GG, Nadeau J, Salzberg SL, Holt, Kodira CD, Lu F, Chen L, Deng Z, Evangelista CC, Gan T, Heiman Jt, Li J, Li Z, Merkulov GV, Milshina NV, Naik AK, Qi R, Shue BC, Wang A, Wang J, Wang X, Yan X, Yooseph JYS, Zhao Q, Zheng L, Zhu SC, Biddick K, Bolanos R, Delcher AL, Dew IM, Fasulo D, Flanigan MJ, Huson DH, Kravitz SA, Miller JR, Mobarry CM, Reinert K, Remington KA, Zhang Q, Zheng XH, Nusskern DR, Lai Z, Lei Y, Zhong W, Yao A, Guan, Ji RR, Gu Z, Wang ZY, Zhong F, Xiao C, Chiang CC, Yandell, Wortman JR, Amanatides PG, Hladun SL, Pratts EC, Johnson JE, Dodson KL, Woodford KJ, Evans CA, Gropman B, Rusch DB, Venter E, Wang M, Smith TJ, Houck JT, Tompkins DE, Haynes C, Jacob D, Chin SH, Allen DR, Dahlke CE, Sanders R, Li K, Liu X, Levitsky AA, Majoros WH, Chen Q, Xia AC, Lopez JR, Donnelly MT, Newman MH, Glodek A, Kraft CL, Nodell M, Ali F, An HJ, Baldwin-Pitts D, Beeson KY, Cai S, Carnes M, Carver A, Caulk PM, Center A, Chen YH, Cheng ML, Coyne MD, Crowder M, Danaher S, Davenport LB, Desilets R, Dietz SM, Doup L, Dullaghan P, Ferriera S, Fosler CR, Gire HC, Gluecksmann A, Gocayne JD, Gray J, Hart B, Haynes J, Hoover J, Howland T, Ibegwam C, Jalali M, Johns D, Kline L, Ma DS, MacCawley S, Magoon A, Mann F, May D, McIntosh TC, Mehta S, Moy L, Moy MC, Murphy BJ, Murphy SD, Nelson KA, Nuri Z, Parker KA, Prudhomme AC, Puri VN, Qureshi H, Raley JC, Reardon MS, Regier MA, Rogers YH, Romblad DL, Schutz J, Scott JL, Scott R, Sitter CD, Smallwood M, Sprague AC, Stewart E, Strong RV, Suh E, Sylvester K, Thomas R, Tint NN, Tsonis C, Wang G, Wang G, Williams MS, Williams SM, Windsor SM, Wolfe K, Wu MM, Zaveri J, Chaturvedi K, Gabrielian AE, Ke Z, Sun J, Subramanian G, Venter JC, Pfannkoch CM, Barnstead M, Stephenson LD. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science. 2002;296:1661–1671. doi: 10.1126/science.1069193. [DOI] [PubMed] [Google Scholar]
- 11.Noveroske JK, Weber JS, Justice MJ. The mutagenic action of N-ethyl-N-nitrosourea in the mouse. Mamm Genome. 2000;11:478–483. doi: 10.1007/s003350010093. [DOI] [PubMed] [Google Scholar]
- 12.Rinchik EM, Carpenter DA. N-ethyl-N-nitrosourea mutagenesis of a 6-to 11-cM subregion of the Fah–Hbb Interval of mouse chromosome 7: completed testing of 4,557 gametes and deletion mapping and complementation analysis of 31 mutations. Genetics. 1999;152:373–383. doi: 10.1093/genetics/152.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinchik EM, Russell LB. Germ-line deletion mutations in the mouse: tools for intensive functional and physical mapping of regions for the mammalian genome. In: Davies K, Tilghman S, editors. Genome Analysis. I. CSH Laboratory Press; Cold Spring Harbor, NY: 1990. pp. 121–158. [Google Scholar]
- 14.Rinchik EM. Developing genetic reagents to facilitate recovery, analysis, and maintenance of mouse mutations. Mamm Genome. 2000;11:489–499. doi: 10.1007/s003350010095. [DOI] [PubMed] [Google Scholar]
- 15.Roderick T. Using inversions to detect and study recessive lethals and detrimentals in mice. In: deSerres F, Sheridan W, editors. Utilization of Mammalian Specific-Locus Studies in Hazard Evaluation and Estimation of Genetic Risk. Plenum; New York: 1983. pp. 135–167. [Google Scholar]
- 16.Russell WL, Kelly EM, Hunsicker PR, Bangham JW, Maddux SC, Phillips EL. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc Natl Acad Sci U S A. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Human Genome Issue Science. 2003;291:1145–1434. [Google Scholar]
- 18.The Human Genome Issue Nature. 2003;409:749–958. [Google Scholar]