Significance
Ether lipid levels are higher in tumors, but their specific function in cancer has remained unclear. We show here that the metabolic enzyme alkylglyceronephosphate synthase (AGPS), a critical step in the synthesis of ether lipids, is up-regulated across multiple types of aggressive human cancer cells and primary tumors. Inactivation of AGPS leads to significant impairments in cancer pathogenicity through not only lowering the levels of cellular ether lipids, but also by altering fatty acid, eicosanoid, and glycerophospholipid metabolism, resulting in an overall reduction in the levels of several oncogenic signaling lipids.
Keywords: cancer metabolism, metabolomics, lipid signaling lysophosphatidic acid, eicosanoids
Abstract
Aberrant lipid metabolism is an established hallmark of cancer cells. In particular, ether lipid levels have been shown to be elevated in tumors, but their specific function in cancer remains elusive. We show here that the metabolic enzyme alkylglyceronephosphate synthase (AGPS), a critical step in the synthesis of ether lipids, is up-regulated across multiple types of aggressive human cancer cells and primary tumors. We demonstrate that ablation of AGPS in cancer cells results in reduced cell survival, cancer aggressiveness, and tumor growth through altering the balance of ether lipid, fatty acid, eicosanoid, and fatty acid–derived glycerophospholipid metabolism, resulting in an overall reduction in the levels of several oncogenic signaling lipids. Taken together, our results reveal that AGPS, in addition to maintaining ether lipids, also controls cellular utilization of fatty acids, favoring the generation of signaling lipids necessary for promoting the aggressive features of cancer.
Cancer cells have fundamentally altered metabolism that drives their pathogenic features (1, 2). One hallmark of cancer cells is a heightened de novo lipogenic signature that serves as a critical foundation for generating lipids required for cell proliferation (3, 4). For nearly half a century, it has also been known that cancer cells possess dramatically higher levels of ether lipids compared with normal cells (5–8). Ether lipids have an alkyl or alkenyl chain on one or more carbons of the glycerol backbone bonded through an ether or vinyl linkage, rather than the usual ester linkage. The physiological roles of ether lipids are not well understood, but they have been implicated in maintaining physicochemical properties of cell membranes, such as membrane fluidity, membrane fusion events, and lipid raft microdomains (9, 10). Certain ether lipids, such as lysophosphatidic acid-ether (LPAe) or platelet-activating factor-ether (PAFe), are signaling molecules that have been shown to possess bioactive and even oncogenic properties through binding specific receptors (11–15). In the late 1960s, Snyder and Wood first reported that rodent and human tumors possess significantly higher levels of ether lipids relative to normal tissue. Over the ensuing decades, dramatic elevations in ether lipid content have been confirmed for a wide range of cancer cells and primary tumors from several tissues of origin and have been correlated with the proliferative capacity and tumorigenic potential of cancer cells (5–8). Nonetheless, whether elevated ether lipids are causally linked, or merely associated with, cancer pathogenicity has remained unclear.
In this study, we show that inactivation of the critical enzyme for ether lipid synthesis, alkylglycerone phosphate synthase (AGPS), lowers ether lipid levels and impairs cancer pathogenicity, whereas AGPS overexpression elevates ether lipid levels and increases cancer cell motility, survival, and tumor growth. We also show that AGPS has a larger role beyond generating ether lipids to include controlling fatty acid, eicosanoid, and acylglycerophospholipid metabolism to favor generation of oncogenic signaling lipids, such as LPAe, lyosphosphatidic acid (LPA), and eicosanoids, which fuel aggressive and tumorigenic features of cancer (12, 13, 16). Our studies thus reveal a heretofore unrecognized role of AGPS and ether lipids in shifting the balance of fatty acid utilization from structural membrane lipids toward generation of oncogenic signaling lipids.
Results and Discussion
Cancer Cells Exhibit Heightened AGPS Expression and Ether Lipid Metabolism.
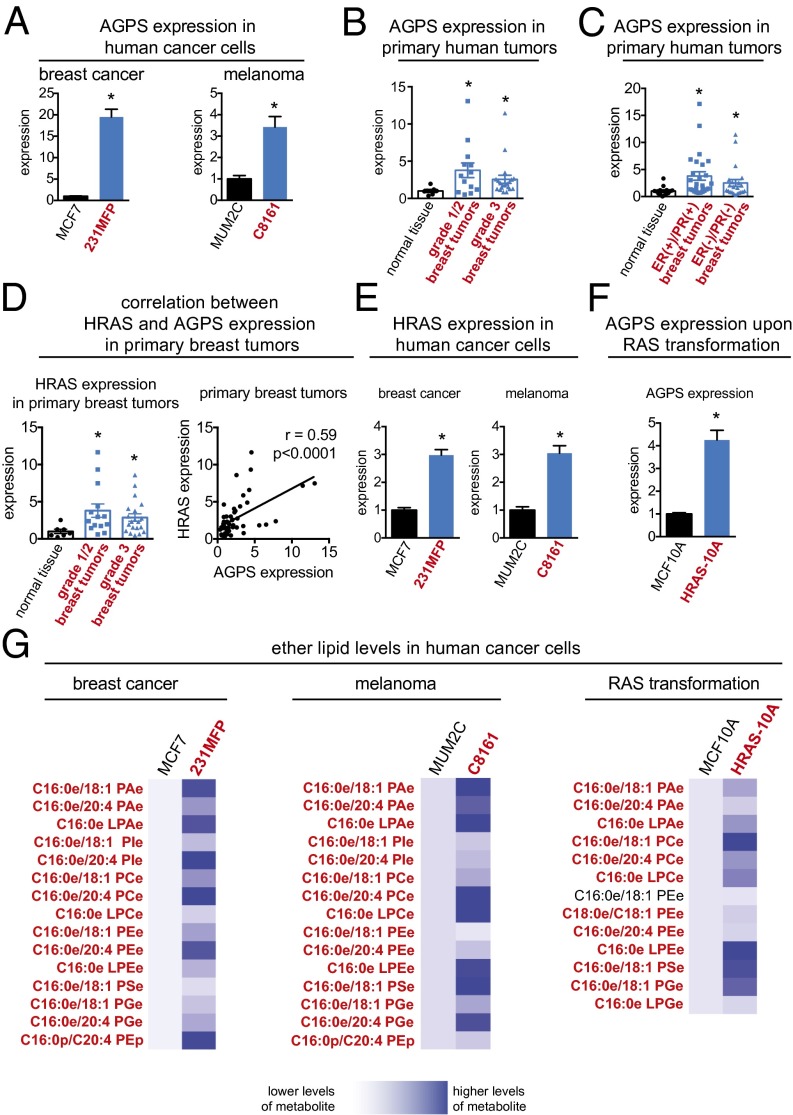
AGPS converts acyl-glycerone-3-phosphate into alkyl-glycerone-3-phosphate, which is a requisite step in the generation of all ether lipids. We hypothesized that cancer cells and tumors, which possess elevated levels of ether lipids, would have heightened AGPS expression. Consistent with this premise, we find that AGPS is highly expressed across aggressive breast (231MFP), melanoma (C8161), and prostate cancer (PC3) cells compared with less aggressive cancer cells (MCF7, MUM2C, and LNCaP, respectively) (Fig. 1A; Fig. S1A). We have reported that these aggressive cancer cells have heightened migratory, invasive, and tumorigenic properties compared with less aggressive cells (17–19). We also show that AGPS expression is elevated two- to fourfold in Nottingham grade I (low-grade), II (intermediate-grade), and III (high-grade) primary human breast tumors (Fig. 1B), as well as in estrogen receptor-positive/progesterone receptor-positive [ER(+)/PR(+)] and ER-negative/PR-negative [ER(−)/PR(−)] breast tumors (Fig. 1C) compared with normal breast tissue, indicating that AGPS expression may be heightened early in breast cancer development. Accordingly, we find that the expression of the commonly dysregulated transforming Harvey-Rat sarcoma oncogene (HRAS) (20) is heightened in low-, moderate-, and high-grade primary human breast tumors and correlates significantly with AGPS expression (Fig. 1D). We also find that HRAS expression is higher in the aggressive 231MFP breast and C8161 melanoma cancer cells compared with the less-aggressive MCF7 breast and MUM2C melanoma cancer cells (Fig. 1E). Consistent with this association between HRAS and AGPS, HRAS transformation of MCF10A mammary epithelial cells (21) induces up-regulation of AGPS expression (Fig. 1F). We thus show that transformation of cells by HRAS is one regulatory route through which cells may up-regulate AGPS expression. We also show that the aggressive human breast 231MFP, melanoma C8161, and prostate PC3 cancer cells, as well as HRAS-transformed MCF10A cells, possess significantly higher ether lipid levels compared with their less aggressive MCF7, MUM2C, and LNCaP counterparts, or MCF10A control cells (Fig. 1G; Fig. S1 B–F; Dataset S1). Heightened lipid species include phosphatidic acid-ether (PAe), LPAe, phosphatidyl inositol-ether (PIe), phosphatidylcholine-ether (PCe), lysophosphatidylcholine-ether (LPCe), phosphatidylserine-ether (PSe), phosphatidylglycerol-ether (PGe), lysophopshatidylglycerol-ether (LPGe) lipids, and plasmalogen ether lipids, such as phosphatidylethanolamine-plasmalogen (PEp) (Fig. 1G; Fig. S1 B–F). We annotate ether lipids in this study by the specific ether (e) or plasmalogen (p) species and alkyl chain length and type (carbon number:degree of unsaturation—e.g., C16:0e is a palmityl-ether linkage with no unsaturation) in Fig. 1G and Fig. S1 B–F. Our results show that AGPS expression and ether lipid levels are heightened in multiple types of aggressive human cancer cells and upon RAS transformation and that AGPS expression is increased in primary breast tumors.
Fig. 1.
AGPS is highly expressed in aggressive cancer cells, primary human tumors, and RAS-transformed cells. (A) AGPS gene expression is heightened across aggressive breast (231MFP) and melanoma (C8161) cancer cells (in red) compared with less aggressive breast (MCF7) and melanoma (MUM2C) cancer cells (in black), as measured by qPCR. (B) AGPS gene expression is significantly higher in Nottingham grade 1 (low-grade) and grade 2 (intermediate-grade), as well as grade 3 (high-grade) primary human breast tumors compared with normal breast tissue as measured by qPCR. (C) AGPS gene expression is also significantly higher in ER(+)/PR(+) and ER(−)/PR(−) human breast tumors compared with normal breast tissue. (D) HRAS expression is higher in grade 1/2 and 3 primary human breast tumors compared with normal breast tissue. HRAS expression from matching normal tissue and breast tumors is significantly correlated with AGPS expression with a Pearson correlation coefficient of r = 0.59. (E) HRAS expression is higher in aggressive breast and melanoma cancer cells compared with less aggressive cells. (F) HRAS overexpression in MCF10A nontransformed mammary epithelial cells increases AGPS expression. (G) Aggressive breast and melanoma cancer cells and HRAS-transformed MCF10A cells possess significantly higher levels of multiple species of ether lipids compared with less aggressive or empty vector-infected MCF10A control cells, respectively. Data in A–F are presented as mean ± SEM; n = 4 per group for A, E, and F, n = 7–26 per group for B–D, and n = 4–5 per group for G. Significance in A–F is presented as *P < 0.05 compared with less-aggressive cancer cells (A and E), normal breast tissue (B–D), or empty vector-infected MCF10A cells (F). Significance in G is presented as P < 0.05 for lipid designations that are bolded in red comparing 231MFP to MCF7, C8161 to MUM2C, or HRAS-10A to MCF10A groups.
AGPS Is a Critical Enzyme in Cancer Pathogenicity.
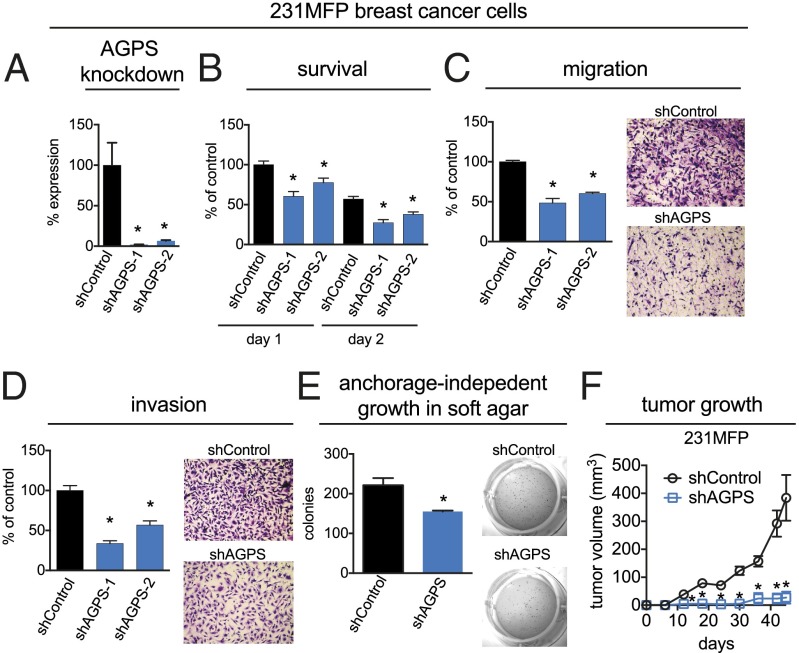
We next sought to determine the extent to which AGPS was necessary for maintaining the pathogenic features of cancer cells. We generated two independent stable short hairpin knockdown lines of AGPS (shAGPS-1 and shAGPS-2) with greater than 90% knockdown in breast 231MFP and melanoma C8161 cancer cells (Fig. 2A; Fig. S2A). AGPS inactivation results in a significant decrease in cellular motility, invasiveness, and anchorage-independent growth in soft agar in both breast and melanoma cancer cells and reduces cell survival in breast cancer cells (Fig. 2 B–E; Fig. S2 B–E). AGPS inactivation also reduces breast 231MFP and melanoma C8161 tumor xenograft growth in immune-deficient mice (Fig. 2F; Fig. S2F).
Fig. 2.
AGPS ablation leads to impairments in breast cancer pathogenicity. (A) AGPS expression was stably knocked down in 231MFP breast cancer cells using two independent shRNA oligonucleotides (shAGPS-1 and shAGPS-2), resulting in >90% reduction in AGPS expression compared with shControl cells determined by qPCR. (B–E) AGPS inactivation in 231MFP cells decreases serum-free cell survival (B), cell migration (C), invasion (D), and anchorage-independent growth in soft-agar (E). Serum-free cell survival was determined by measuring cell viability in serum-free media 24 h after seeding 10,000 cells using a WST cell viability assay. Migration and invasion assays were performed by transferring cancer cells to serum-free media for 4 h before seeding 50,000 cells into inserts with 8-μm pore size containing membranes coated with collagen (10 μg/mL) or BioCoat Matrigel, respectively. Migrated or invaded cells were fixed and stained after 8 h, and these cells were counted over four independent fields of view at 400× magnification and averaged for each biological replicate. For soft-agar assays, 4,000 cells were seeded in agar and colonies were counted by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) staining after 4 wk. (F) AGPS knockdown impairs tumor growth in immune-deficient SCID mice compared with shControl cells; 2 × 106 231MFP cells/100 μL were injected s.c. into the flank of female mice, and tumor growth was measured using calipers. Data are presented as mean ± SEM; n = 3–7 per group. Significance is presented as *P < 0.05 compared with shControl.
We also stably overexpressed AGPS in the less aggressive breast (MCF7) and melanoma (MUM2C) cancer cells to determine whether AGPS was sufficient to confer pathogenic features (Fig. S2G). AGPS overexpression increased cell migration and serum-free cell survival in situ and tumor xenograft growth in vivo (Fig. S2 H and I). Our results indicate that AGPS not only is important to maintain aggressive and tumorigenic features, but also is sufficient to confer these effects in less aggressive human cancer cells.
Functional Metabolomics Reveals Widespread Alterations in Cellular Lipid Levels on AGPS Knockdown or Overexpression.
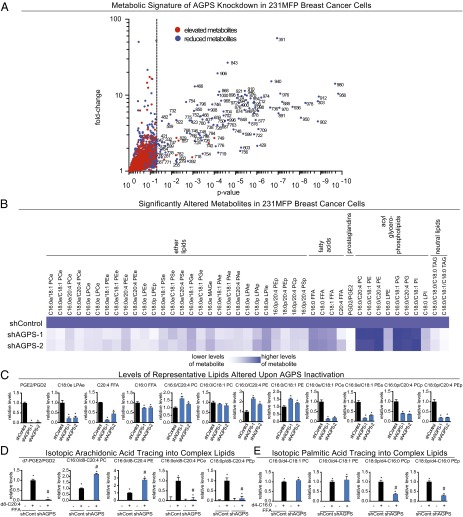
We next wanted to understand the mechanism through which AGPS drives cancer pathogenicity. We performed both targeted and untargeted metabolomic analyses to comprehensively identify alterations in cancer cell metabolites upon AGPS knockdown in 231MFP breast and C8161 melanoma cells (Fig. 3 A–C; Figs. S3 A–C and S4 A–C; Dataset S1). We used single reaction monitoring (SRM)-based targeted metabolomic analysis to quantify the levels of ∼100 common lipids, and used untargeted metabolomic profiling to comparatively profile levels of an additional >3,000 ions. We used the METLIN database to generate putative identifications for many of the altered metabolites and quantified these metabolites by SRM analysis (22). We then filtered for metabolites that were changed significantly in both shAGPS lines (shAGPS-1 and shAGPS-2). Consistent with the function of AGPS as an early critical step in ether lipid synthesis, AGPS inactivation in 231MFP and C8161 cancer cells showed global reduction in the levels of multiple structural ether lipid and plasmalogen species, including PCe, LPCe, PEe, LPEe, PSe, PGe, monoalkylglycerol ether (MAGe), PAe, PIe, lysophosphatidyl inositol-ether (LPIe), PEp, lysophosphatidyl ethanolamine-plasmalogen (LPEp), phosphatidylcholine-plasmalogen (PCp), and phosphatidyl serine-plasmalogen (PSp) (Fig. 3 A–C; Figs. S3 A–C and S4 A–C; Dataset S1). We also find lower levels of the signaling ether lipids LPAe and LPA-plasmalogen (LPAp) that, similar to their acyl-LPA counterparts, stimulate LPA receptors (12, 13) (Fig. 3 B and C; Fig. S3; Dataset S1). Intriguingly, AGPS knockdown also lowers levels of saturated and unsaturated free fatty acids (FFAs) in both breast and melanoma cancer cells, with a striking reduction in the levels of arachidonic acid–derived signaling eicosanoids, prostaglandin E2/D2 (PGE2/PGD2) (16), in 231MFP breast cancer cells (Fig. 3C; Fig. S4C). We also observe increases in the levels of several diacylated glycerophospholipids, with a particular enrichment in arachidonoyl-containing acyl-glycerophospholipids (e.g., C16:0/C20:4 PC, C16:0/C20:4 PE, and C16:0/C20:4 PG), coupled with lower levels of several acyl-lysophospholipids (e.g., LPA, LPE, and LPC) (Fig. 3 B and C; Fig. S4 B and C; Dataset S1). Overall, this lipidomic signature suggests that AGPS knockdown has broader effects beyond ether lipid pathways that include fatty acid, eicosanoid, and acyl-glycerophospholipid metabolism.
Fig. 3.
Metabolomic profiling of AGPS knockdown in breast cancer cells reveals widespread alterations in the levels of ether lipids, acylglycerophospholipids, fatty acids, and eicosanoids. (A–C) Metabolomic analyses of AGPS knockdown 231MFP cells compared with shControl cells. (A) AGPS knockdown shows significant (P < 0.05) alterations in the levels of many metabolites. Each point on the volcano plot corresponds to a distinct ion detected in shAGPS-1 and shControl 231MFP cells. Metabolites that are decreasing or increasing in levels on AGPS knockdown are represented as blue and red dots, respectively. The x axis denotes P value of each ion between shAGPS and shControl groups, in which metabolites levels that are significantly altered (P < 0.05) are displayed to the right of the dotted line. The y axis indicates the relative fold-change in the levels of the metabolite between shControl and shAGPS groups. (B) The heat map shows all identified metabolites that are significantly altered in levels (P < 0.05) upon AGPS knockdown in 231MFP cells. These metabolites were quantified by SRM-based targeted metabolomics. Darker blue shading on the heat map corresponds to higher relative levels of metabolite, whereas white or lighter blue shading indicates lower levels. (C) Representative lipids from B are shown as bar graphs. AGPS knockdown not only lowers the levels of ether lipids, but also fatty acids, eicosanoids, and neutral lipids, and raises the levels of several diacylated glycerophospholipids. (D) Targeted metabolomic analysis of isotopic d8-C20:4 FFA (arachidonic acid) incorporation into certain complex cellular lipids. (E) Targeted metabolomic analysis of isotopic d4-C16:0 FFA (palmitic acid) incorporation into certain complex cellular lipids. Data in A–E are presented as mean ± SEM; n = 4–5 per group. Significance is expressed in C as *P < 0.05 compared with shControl groups. Significance in D and E is presented as *P < 0.05 between d4-C16:0 FFA and d8-C20:4 FFA groups compared with nonisotopic fatty acid treatment groups, and #P < 0.05 between d4-C16:0 and d8-C20:4 FFA-treated shAGPS vs. matching shControl groups. All raw data for relative levels and absolute levels are shown in Dataset S1. Further quantitative details for LPAe and LPAp lipids are shown in Fig. S3. The metabolomic profile of AGPS inactivation in C8161 melanoma cells is provided in Fig. S4, and the metabolomic profile of AGPS overexpression in MCF7 and MUM2C cells is provided in Fig. S5.
Consistent with the conjecture of Welsh et al. that lack of AGPS activity is responsible for the general deficiency of ether lipids in MCF7 breast cancer cells (23), we find that AGPS overexpression in MCF7 cells is sufficient to increase the levels of several ether lipids. We also show that oleic and arachidonic acid levels are also increased (Fig. S5A; Dataset S1). In AGPS overexpressing MUM2C melanoma cancer cells, we see a similar increase in the levels of ether lipids, as well as reduced levels of arachidonoyl-containing glycerophospholipids (Fig. S5B; Dataset S1). PGE2/PGD2 levels were not detectable in either mock or AGPS-overexpressing cells, suggesting that additional enzymatic pathways may be required, such as cyclooxygenase enzymes (16), to generate these lipids.
Isotopic Fatty Acid Tracing Reveals Alterations in Arachidonate Utilization on Inactivation of AGPS.
Our results showing that AGPS knockdown leads to lower arachidonic acid and eicosanoid levels and an increase in certain arachidonoyl-containing acyl-glycerophospholipids implies a shift in fatty acid use toward specific diacylated structural lipids and away from lysophospholipids and eicosanoids. Consistent with this premise, isotopic fatty acid tracing with deuterated d8-arachidonic acid in 231MFP breast cancer cells reveals heightened d8-incorporation into acylglycerophospholipids and reduced incorporation into ether lipids and eicosanoids on AGPS knockdown (Fig. 3D). In contrast, isotopic tracing with d4-palmitic acid did not show increased d4-incorporation into acylglycerophospholipids, despite reduced incorporation into ether lipids upon AGPS knockdown (Fig. 3E). These results indicate preferential incorporation of arachidonic acid into acylglycerophospholipids, compared with saturated fatty acids.
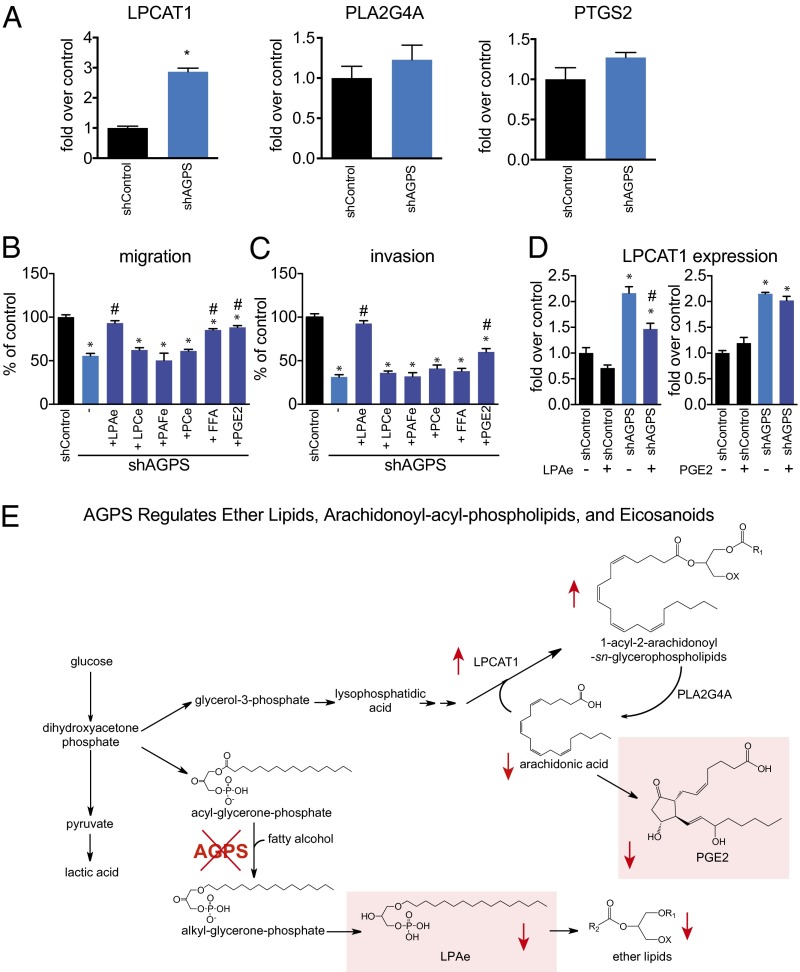
This shift in fatty acid utilization may be attributed to either increased lysophospholipid acyltransferase activity, reduced phospholipid hydrolysis, or lower cyclooxygenase activity. Upon testing for these possibilities by quantitative PCR (qPCR), we found that AGPS knockdown led to increased levels of LPC acyl transferase (LPCAT1), but no change in cytosolic phospholipase A2 (PLA2G4A) or cyclooxygenase 2 (PTGS2) expression (Fig. 4A). These results indicate that AGPS knockdown directs fatty acid use to generating arachidonoyl acylglycerophospholipids through increased acylation of fatty acids onto glycerol-phosphate and lysophospholipids, depleting the arachidonic acid pool that generates eicosanoids.
Fig. 4.
AGPS fuels cancer pathogenicity in breast cancer cells through altering fatty acid utilization to favor the generation of oncogenic signaling lipids. (A) AGPS knockdown results in altered levels of not only ether lipids, but also reduced levels of fatty acids and eicosanoids and increased levels of acylglycerophospholipids. This may be due to heightened acyltransferase or reduced phospholipase or cyclooxygenase activity. AGPS ablation results in a significant increase in the expression of lysophosphatidyl choline acyltransferase 1 (LPCAT1) without significantly altering the expression of cytosolic phospholipase A2 (PLA2G4) or cyclooxygenase 2 (PTGS2), as determined by qPCR. (B and C) The migratory (B) and invasive (C) impairments in shAGPS 231MFP cells are significantly rescued upon treatment of cells with low concentrations (100 nM) of C18:0e LPA, PGE2, and C16:0 FFA, but not other ether lipids such as C16:0e LPCe, C16:0e/C20:4 PCe, and C16:0e PAFe. Treatment with the aforementioned lipids was initiated concurrently with the seeding of cells for assessment of cancer cell migration (8 h) and invasion (24 h). (D) Treatment with C18:0e LPAe (100 nM), but not PGE2 (100 nM), significantly rescued the heightened expression of LPCAT1 conferred by AGPS knockdown, as assessed by qPCR. (E) Model depicting the metabolic role of AGPS in exerting control over ether lipid metabolism, fatty acid metabolism, and glycerophospholipid metabolism. We show that AGPS knockdown in 231MFP breast cancer cells leads to reduced levels of ether lipids, including the oncogenic LPAe signaling lipid (boxed in red), as well as an alteration in arachidonic acid utilization toward membrane phospholipids through LPAe-mediated LPCAT1 up-regulation, at the expense of generating oncogenic prostaglandins (boxed in red). R1 denote acyl or alkyl chains and X refers to lipid head-group such as phosphocholine, phosphoethanolamine, or phosphate. Data are presented as mean ± SEM; n = 3 per group. Lipid rescue of migratory and invasive impairments on AGPS knockdown in C8161 melanoma cells and C18:0 LPA rescue experiments are provided in Fig. S6. Significance is represented as *P < 0.05 compared with shControl and #P < 0.05 compared with DMSO-treated shAGPS groups.
AGPS Affects Cancer Pathogenicity Through Multiple Lipid Signaling Pathways.
Among the many lipidomic changes conferred by AGPS knockdown, we were particularly intrigued by the lower levels of LPAe and PGE2 in 231MFP breast cancer cells and LPAe and LPA in C8161 melanoma cancer cells. Both LPAe and its acyl-LPA counterpart act as oncogenic signaling lipids that bind LPA receptors to drive multiple aspects of cancer (13). PGE2 is also a signaling lipid that acts through EP2 receptors to fuel proliferative, malignant, and tumorigenic features of cancer (16). Consistent with their function, C18:0e LPAe fully rescues and PGE2 partially rescues the migratory and invasive deficits conferred by AGPS knockdown in 231MFP breast cancer cells (Fig. 4 B and C). LPAe, but not PGE2, fully rescues the migratory and invasive impairments in shAGPS C8161 cells, consistent with the lipidomic signature showing that eicosanoids are not detected in these cells. We find that C18:0 LPA also partially or fully rescues the migratory impairments in shAGPS 231MFP and C8161 cells, respectively (Fig. S6 C and D), likely because either acyl-LPA or LPAe can rescue overall reduced LPA receptor signaling caused by lower LPAe or LPA levels. We also show that palmitate (C16:0 FFA) partially rescues the pathogenic deficits of AGPS knockdown, likely through the generation of acyl-LPA (Fig. 4 B and C; Fig. S6 C and D). Indeed, we show that d4-C16:0 FFA labeling of 231MFP cancer cells results in the generation of d4-C16:0 LPA (Fig. S6E). We note that no other ether lipids such as PAF, C16:0e/C20:4 PCe, or C16:0e LPCe rescue the oncogenic impairments, confirming the specificity of our proposed mechanism.
Interestingly, C18:0e LPAe, but not PGE2, significantly rescues the up-regulation of LPCAT1 expression in AGPS knockdown breast cancer cells, suggesting that the broader alterations in fatty acid, acylglycerophospholipid, and PGE2 levels may be downstream of reduced LPAe signaling (Fig. 4D). In AGPS-overexpressing cancer cells that show higher levels of LPAe but undetectable levels of PGE2, we show that the increased migration observed in AGPS-overexpressing MCF7 and MUM2C cancer cells is significantly reversed by an LPA receptor antagonist Ki16425, but not by the cyclooxygenase inhibitor ibuprofen (Fig. S7).
We profiled expression of LPCAT1 in primary human breast tumors to determine whether LPCAT1 expression was lower compared with normal breast tissue. Instead, we find that LPCAT1 expression is significantly higher in primary breast tumors compared with normal breast tissue (Fig. S8). Our finding that reduced LPAe signaling leads to an up-regulation in LPCAT1 expression may occur only upon knockdown of AGPS or depletion of cellular ether lipids, under conditions in which cancer cells have adapted to a state of higher ether lipid levels.
Overall, AGPS expression is heightened in aggressive human cancer cells and primary human breast tumors, and AGPS inactivation significantly impairs aggressive features of cancer cells and attenuates tumor growth, not only through lowering the levels of oncogenic ether lipids, but also through altering arachidonic acid utilization away from protumorigenic eicosanoids and lysophospholipids (Fig. 4E).
Conclusion
We show here that AGPS serves as a metabolic node in cancer cells that maintains levels of structural and signaling ether lipids and also modulates fatty acid utilization pathways to favor generation of key oncogenic lysophospholipids and eicosanoids, which promote survival, migratory, invasive, and tumorigenic features. We used advanced metabolomic platforms to gain broad overview into changes in cancer cell lipid metabolism that are controlled by AGPS, but there are many metabolic alterations that we have not yet been able to identify. These yet uncharacterized AGPS-mediated changes to the lipidome may yield further insights into the roles of AGPS in cancer. Although we have established the importance of AGPS in controlling the levels of oncogenic signaling lipids, such as LPAe, LPA, and eicosanoids, there may be other enzymes or regulatory pathways, even within the ether lipid metabolic pathway, that can be targeted to achieve similar effects. For example, we have reported that monacylglycerol lipase is up-regulated across aggressive cancer cells and primary tumors where it controls release of FFAs from monoacylglycerols to fuel generation of FFA-derived oncogenic signaling lipids, such as LPA and PGE2 to drive malignant features of cancer (18). Chiang et al. demonstrated that blocking of KIAA1363, a deacetylase of 2-acetyl MAGe, lowers levels of its neutral ether lipid product MAGe, which decreases the levels of its downstream metabolite LPAe, leading to impaired cellular migration and tumor growth (15). It will be of future interest to determine the effects of polypharmacologically blocking several of these pathways simultaneously in cancer cells to achieve maximal anticancer activity. Alternatively, controlling the supply of fatty alcohols in cancer cells may also modulate levels of ether lipids similarly to controlling AGPS expression (24).
In conclusion, we provide a potential mechanistic link between the historical associations between aggressive or hyperproliferative tumors and higher levels of ether lipids (7). Our studies indicate a previously unrecognized cross-talk between ether lipid, fatty acid, eicosanoid, and acylglycerophospholipid metabolism in cancer cells that leads to an optimal landscape of lipid signaling pathways that subserve cancer pathogenesis. Manipulating this landscape may yield novel strategies for thwarting the lipid signals that drive cancer pathogenicity and may provide unique approaches to cancer therapy.
Materials and Methods
Cell Lines.
The C8161 and MUM2C lines were provided by Benjamin Cravatt (The Scripps Research Institute, La Jolla, CA). MCF10A and HRAS–transformed cells were obtained from Stefano Piccolo at the University of Padua (25). The 231MFP cells were generated from explanted xenograft tumors of MDA-MB-231 cells, as described previously (26). PC3 and LNCaP cells were obtained from ATCC.
Cell Culture Conditions.
HEK293T cells were cultured in DMEM media containing 10% (vol/vol) FBS and maintained at 37 °C with 5% (wt/vol) CO2. C8161 cells were cultured in RPMI1640 media containing 10% FBS and glutamine and maintained at 37 °C at 5% (wt/vol) CO2. 231MFP cells were cultured in L15 media containing 10% FBS and glutamine and were maintained at 37 °C in 0% CO2. MCF10A cells were cultured as previously described (25).
qPCR.
qPCR was performed using the manufacturer’s protocol for Fischer Maxima SYBRgreen, with 10 μM primer concentrations. Primer sequences were derived from Primer Bank (27).
Breast Tumor Array qPCR.
Breast cancer tumor array 1 was purchased from Origene, and qPCR was performed on the array using the protocol described above.
Constructing AGPS Knockdown Cells.
We used shRNA using two independent silencing oligonucleotides to knockdown the expression of AGPS using previously described procedures (18). For construction of stable shRNA knockdown lines, lentiviral plasmids (pLKO.1) containing shRNA (Open Biosystems) against human AGPS were transfected into HEK293 cells using Fugene (Roche). Lentivirus was collected from filtered culture media and delivered to the target cancer cell line with polybrene. These target cells were subsequently selected over 3 d with 1 μg/mL puromycin. The short-hairpin sequences used for constructing the two independent AGPS knockdowns were as follows: shRNA#1: AATTCGCTCAAACATTCCTTC; shRNA #2: AAGGATTTCTTCTCTAGCAGC. The control shRNA was targeted against GFP with the target sequence GCAAGCTGACCCTGAAGTTCAT. We confirmed knockdown by qPCR.
Overexpression Studies in Human Cancer Cell Lines.
Stable AGPS overexpression was achieved by subcloning the AGPS gene into the pMSCVpuro vector (Clontech), generating retrovirus using the AmphoPack-293 Cell Line, as described above with the RNA interference studies. The human AGPS was subcloned into the pMSCVpuro (Clontech) using Xho1 and AgeI restriction sites using the following primers: 5′GTACGTACCTCGAGAGGCGGCGGCTG-3′ and 5′GTACGTACGAATTCGTTTCTGTTTCC-3′.
Cancer Phenotypic Studies.
Migration, invasion, cell proliferation, and survival studies were performed as described previously (18). Details are providing in SI Materials and Methods.
Tumor Xenograft Studies.
Human tumor xenografts were established by transplanting cancer cells ectopically into the flank of C.B17 SCID mice (Taconic Farms) as described previously (18). Briefly, cells were washed two times with PBS, trypsinized, and harvested in serum-containing medium. Next, the harvested cells were washed two times with serum-free medium and resuspended at a concentration of 2.0 × 104 cells/mL, and 100 μL was injected. Growth of the tumors was measured every 3–6 d with calipers.
Metabolomic Profiling of Cancer Cells.
Metabolite measurements were conducted using modified versions of previous procedures (18, 28). Details are provided in SI Materials and Methods.
Analysis of Isotopic Incorporation of Palmitate and Arachidonate into Lipids.
Isotopically labeled fatty acid incorporation into lipids was measured by labeling cells with d4-palmitic acid or d8-arachidonic acid and measuring isotopic incorporation into lipids. Cells were treated either with palmitate, d4-palmitate, arachidonate, or d8-arachidonate (10 μM). Cells were harvested 4 h after labeling, and the nonpolar metabolome was extracted as previously described and analyzed by SRM-based targeted LC-MS/MS for the presence of isotopically labeled lipids.
Supplementary Material
Acknowledgments
We thank the members of the Nomura Research Group and the Luo laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health Grants R21CA170317, R01CA172667, and R00DA030908 (to D.I.B., A.C., L.S.R., S.M.L., M.M.M., and D.K.N.) and Grant R01CA101891 (to K.L.) and the Searle Scholar Foundation (D.K.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310894110/-/DCSupplemental.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin DI, Cravatt BF, Nomura DK. Global profiling strategies for mapping dysregulated metabolic pathways in cancer. Cell Metab. 2012;16(5):565–577. doi: 10.1016/j.cmet.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 4.Louie SM, Roberts LS, Nomura DK. Mechanisms linking obesity and cancer [published online ahead of print March 5, 2013] Biochim Biophys Acta. 2013 doi: 10.1016/j.bbalip.2013.02.008. doi:10.1016/j.bbalip.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snyder F, Wood R. Alkyl and alk-1-enyl ethers of glycerol in lipids from normal and neoplastic human tissues. Cancer Res. 1969;29(1):251–257. [PubMed] [Google Scholar]
- 6.Roos DS, Choppin PW. Tumorigenicity of cell lines with altered lipid composition. Proc Natl Acad Sci USA. 1984;81(23):7622–7626. doi: 10.1073/pnas.81.23.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howard BV, Morris HP, Bailey JM. Ether-lipids, -glycerol phosphate dehydrogenase, and growth rate in tumors and cultured cells. Cancer Res. 1972;32(7):1533–1538. [PubMed] [Google Scholar]
- 8.Albert DH, Anderson CE. Ether-linked glycerolipids in human brain tumors. Lipids. 1977;12(2):188–192. doi: 10.1007/BF02533292. [DOI] [PubMed] [Google Scholar]
- 9.Wallner S, Schmitz G. Plasmalogens the neglected regulatory and scavenging lipid species. Chem Phys Lipids. 2011;164(6):573–589. doi: 10.1016/j.chemphyslip.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Brites P, Waterham HR, Wanders RJ. Functions and biosynthesis of plasmalogens in health and disease. Biochim Biophys Acta. 2004;1636(2–3):219–231. doi: 10.1016/j.bbalip.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Tsukahara T, Haniu H, Matsuda Y. Effect of alkyl glycerophosphate on the activation of peroxisome proliferator-activated receptor gamma and glucose uptake in C2C12 cells. Biochem Biophys Res Commun. 2013;433(3):281–285. doi: 10.1016/j.bbrc.2013.02.101. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y, Tanaka M, Arai H, Aoki J, Prestwich GD. Alkyl lysophosphatidic acid and fluoromethylene phosphonate analogs as metabolically-stabilized agonists for LPA receptors. Bioorg Med Chem Lett. 2004;14(21):5323–5328. doi: 10.1016/j.bmcl.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3(8):582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- 14.Melnikova V, Bar-Eli M. Inflammation and melanoma growth and metastasis: The role of platelet-activating factor (PAF) and its receptor. Cancer Metastasis Rev. 2007;26(3-4):359–371. doi: 10.1007/s10555-007-9092-9. [DOI] [PubMed] [Google Scholar]
- 15.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13(10):1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10(3):181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci USA. 2002;99(16):10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomura DK, et al. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140(1):49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nomura DK, et al. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol. 2011;18(7):846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: A model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148(1):313–319. [PMC free article] [PubMed] [Google Scholar]
- 22.Tautenhahn R, et al. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30(9):826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welsh CJ, et al. Accumulation of fatty alcohol in MCF-7 breast cancer cells. Arch Biochem Biophys. 1994;315(1):41–47. doi: 10.1006/abbi.1994.1468. [DOI] [PubMed] [Google Scholar]
- 24.Honsho M, Asaoku S, Fujiki Y. Posttranslational regulation of fatty acyl-CoA reductase 1, Far1, controls ether glycerophospholipid synthesis. J Biol Chem. 2010;285(12):8537–8542. doi: 10.1074/jbc.M109.083311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 26.Jessani N, et al. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci USA. 2004;101(38):13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38(Database issue):D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopp F, et al. The glycerophospho metabolome and its influence on amino acid homeostasis revealed by brain metabolomics of GDE1(-/-) mice. Chem Biol. 2010;17(8):831–840. doi: 10.1016/j.chembiol.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.