Significance
Protein–protein interactions are essential for cellular regulation, but how changes in individual interactions influence cellular physiology or cause disease remains poorly characterized. Although selective and potent inhibitors of protein–protein interactions are powerful tools, developing such reagents is challenging. This is because signaling networks are composed of members of highly conserved protein domain families, with the Src-homology 2 (SH2) domain as an archetype. To address this challenge, we used protein design to successfully generate a set of reagents, termed monobodies, directed to the SH2 domains of SH2 domain-containing phosphatase 2 (SHP2), for which no specific inhibitors had been identified. These monobodies selectively and potently inhibit SHP2 function and demonstrate utility in dissecting the signaling networks of cancer cells.
Keywords: engineered proteins, protein interaction inhibitor, tyrosine phosphatase, interaction proteomics
Abstract
The dysregulated tyrosine kinase BCR-ABL causes chronic myelogenous leukemia in humans and forms a large multiprotein complex that includes the Src-homology 2 (SH2) domain-containing phosphatase 2 (SHP2). The expression of SHP2 is necessary for BCR-ABL-dependent oncogenic transformation, but the precise signaling mechanisms of SHP2 are not well understood. We have developed binding proteins, termed monobodies, for the N- and C-terminal SH2 domains of SHP2. Intracellular expression followed by interactome analysis showed that the monobodies are essentially monospecific to SHP2. Two crystal structures revealed that the monobodies occupy the phosphopeptide-binding sites of the SH2 domains and thus can serve as competitors of SH2–phosphotyrosine interactions. Surprisingly, the segments of both monobodies that bind to the peptide-binding grooves run in the opposite direction to that of canonical phosphotyrosine peptides, which may contribute to their exquisite specificity. When expressed in cells, monobodies targeting the N-SH2 domain disrupted the interaction of SHP2 with its upstream activator, the Grb2-associated binder 2 adaptor protein, suggesting decoupling of SHP2 from the BCR-ABL protein complex. Inhibition of either N-SH2 or C-SH2 was sufficient to inhibit two tyrosine phosphorylation events that are critical for SHP2 catalytic activity and to block ERK activation. In contrast, targeting the N-SH2 or C-SH2 revealed distinct roles of the two SH2 domains in downstream signaling, such as the phosphorylation of paxillin and signal transducer and activator of transcription 5. Our results delineate a hierarchy of function for the SH2 domains of SHP2 and validate monobodies as potent and specific antagonists of protein–protein interactions in cancer cells.
The tyrosine kinase BCR-ABL is causal for chronic myelogenous leukemia (CML) (1). BCR-ABL is established by a balanced chromosomal translocation event [t9;22 (q34;q11)] that fuses the breakpoint cluster region (BCR) gene with the Abelson tyrosine kinase gene (ABL1), resulting in kinase dysregulation and constitutive activation (1, 2). BCR-ABL interacts with dozens of proteins to form a large multiprotein complex, orchestrating a multitude of signaling events that block myeloid differentiation, inhibit proapoptotic, and activate proliferative pathways in CML cells (3, 4).
Small-molecule tyrosine kinase inhibitors (TKIs) of BCR-ABL, such as imatinib, have served as a paradigm for the development of molecularly targeted therapeutics for cancer (5). In the clinical setting, these TKIs have succeeded in turning CML from a fatal disease to a chronic but manageable condition. Unfortunately, acquired drug resistance, along with the persistence of quiescent CML stem cells that tolerate TKI therapy, have prevented TKIs from being curative (6). Thus, the focus needs to shift toward identifying and validating additional targets within the BCR-ABL complex that are critical for BCR-ABL action (7).
The Src-homology 2 (SH2) domain-containing phosphatase 2 (SHP2) has been identified as a potential therapeutic target in CML (8, 9). SHP2 comprises two N-terminal SH2 domains (termed N-SH2 and C-SH2), a central protein tyrosine phosphatase (PTP) domain, and a C-terminal tail containing tyrosine phosphorylation sites (Fig. 1A). The crystal structure of SHP2 and biochemical data demonstrate that in its basal state, SHP2 phosphatase is autoinhibited by intramolecular interactions between the N-SH2 and PTP domains (10). Engagement of the SH2s by phosphotyrosine (pY)-containing motifs relieves this autoinhibition and leads to increased catalytic activity of SHP2 (11). Counterintuitively, SHP2 tyrosine phosphatase activity contributes to activation of Ras-ERK signaling and is critical for normal hematopoietic cell development and function (12). In the context of BCR-ABL, SHP2 is bound and activated via its SH2 domains by the adaptor protein Grb2-associated binder 2 (GAB2) (13). GAB2 is phosphorylated on multiple tyrosine residues in CML cells (14).
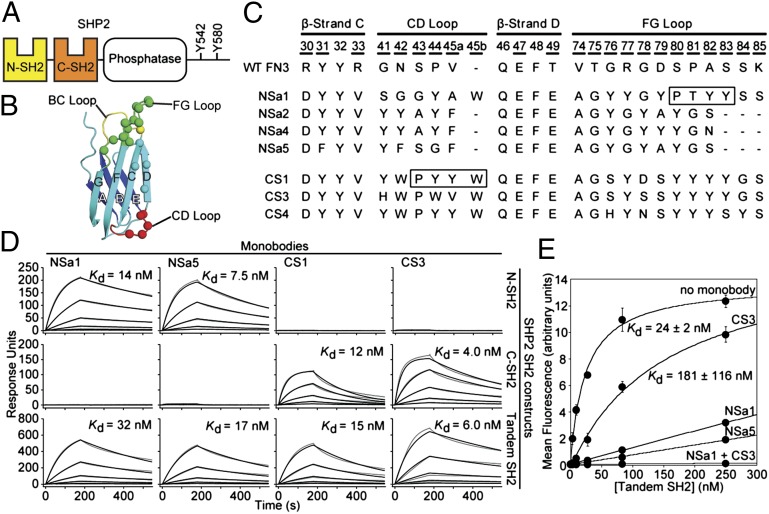
Fig. 1.
Generation of SHP2 SH2-binding monobodies. (A) Domain organization of SHP2. (B) Schematic of the FN3 scaffold. The β-strands are labeled A–G, and diversified residues are shown as colored spheres. (C) Amino acid sequences of selected monobody clones and WT FN3. Residue numbers for diversified positions are underscored. The N series and C series monobodies are directed to the N-SH2 and C-SH2 domains, respectively. The boxes denote segments that bind to the peptide-binding site of the SH2 domains in the crystal structures. (D) SPR sensorgrams and dissociation constants for monobody binding to SHP2 SH2s. Parameters from the best fit of a 1:1 binding model (black traces) to the raw data (gray traces) are given in SI Appendix, Table S1. (E) Binding of the tandem SHP2 SH2 domains to a GAB2 fragment containing tandem pY sites (residues Y614 and Y643) in the absence and presence of saturating concentrations of the indicated monobodies.
Importantly, both SHP2 and GAB2 have been shown to be required for BCR-ABL–induced myeloid transformation and leukemia cell proliferation, thus implicating the GAB2–SHP2 axis as an important signaling event in CML (8, 9). However, many molecular details of GAB2–SHP2 interactions have remained elusive, including which of the two SH2 domains is required for activation of SHP2 activity and whether activated SHP2 can be inhibited by decoupling it from GAB2. Furthermore, gain-of-function mutations of SHP2 have been identified in juvenile myelomonocytic leukemia and in several solid tumors (15), suggesting much greater merit in studying SHP2 signaling beyond CML. To better define how SHP2 contributes to BCR-ABL–induced signaling and dissect the regulatory functions of its SH2 domains, we sought to develop potent and highly specific reagents to inhibit the SHP2 SH2 domains.
No small-molecule inhibitors have yet been reported for the SH2 domains of SHP2. Screening of a pY peptide library yielded optimal binding peptides with Kd values in the submicromolar range (16); however, these peptides were not specific, unable to significantly discriminate between the N-SH2 and C-SH2 domains of SHP2 or from the SH2 domains of the closely related homolog SHP1. Furthermore, the SH2 family is commonly considered an “undruggable” target, with specificity remaining elusive because of the highly homologous nature of its 121 family members in humans (17).
In attempts to meet this challenge of specificity, protein-based inhibitors have emerged as an attractive alternative to small molecules and peptides. One of the best-established platforms is the monobody, a synthetic β-sandwich protein based on the tenth human fibronectin type III (FN3) domain (18). Importantly, unlike conventional antibodies, the folding of the FN3 scaffold does not depend on disulfide formation, making monobodies ideally suited as genetically encoded intracellular inhibitors. We have used monobodies to generate potent and highly specific binders to a variety of targets, including the SH2 domain of BCR-ABL (19–21). Here we describe the development of monobodies that specifically target and potently inhibit each SH2 domain of SHP2, and assess the functional consequences of the monobodies in vitro and in leukemia cells.
Results
Selection of Monobodies to the N-SH2 and C-SH2 Domains of SHP2.
To generate monobodies that bind the N-SH2 or C-SH2 domain of SHP2, we performed selections against the N-SH2 and C-SH2 domains using an FN3 monobody library (dubbed the “side” library) in which surface residues along the β-sheet of the monobody were diversified in addition to residues in the loop regions (Fig. 1B), as in the original (“loop only”) library (21). After three rounds of library sorting against the N-SH2 and C-SH2 domains by phage display, the enriched library for each target was subjected to gene shuffling and then transferred into the yeast surface display format for further sorting (21). Twelve clones exhibiting strong binding for each SH2 domain were selected for sequencing, yielding four unique clones for N-SH2 and three unique clones for C-SH2 (Fig. 1C). Measurements in the yeast surface display format revealed that all clones had Kd values <100 nM to their cognate targets (SI Appendix, Fig. S1). These monobodies did not show detectable binding to six other SH2 domains, indicating that they are highly specific (SI Appendix, Fig. S2).
We then selected clones NSa1 and NSa5 for N-SH2 and clones CS1 and CS3 for C-SH2 for further characterization (Fig. 1C), owing to their high solubility as purified proteins. Surface plasmon resonance (SPR) measurements showed that the four monobodies bound to their cognate targets with low nanomolar Kd values (Fig. 1D and SI Appendix, Table S1). The monobodies also bound to a tandem SH2 construct encompassing both N-SH2 and C-SH2 with similar affinity, demonstrating that their binding was not inhibited by the presence of the neighboring SH2 domain. Consistent with analysis using a yeast surface display, NSa1 and NSa5 exhibited no detectable binding to C-SH2, and likewise CS1 and CS3 exhibited no detectable binding to N-SH2 (Fig. 1D). Furthermore, these monobodies did not appreciably bind the tandem SH2 domains of SHP1, the closest homolog of SHP2, as tested in a competition assay (SI Appendix, Fig. S2).
Taken together, these data show that the four monobodies have high affinity and high specificity to their cognate SH2 domains in vitro. The monobodies have 100- to 300-fold lower Kd values to the SHP2 SH2 domains compared with single pY-containing peptides derived from their bona fide cellular ligands (22), suggesting that they can outcompete interactions of the SHP2 SH2 domain with cognate pY ligands. Indeed, NSa1 and NSa5, and to a lesser extent CS3, inhibited the interaction of a GAB2 tandem phosphopeptide to the tandem SH2 domains of SHP2 (Fig. 1E). These results demonstrate the greater role of N-SH2 compared with C-SH2 in the interaction of SHP2 and the GAB2 tandem phosphopeptide, and also suggest that the monobodies can potently inhibit cellular interactions involving the SHP2 SH2 domains.
Interactome Analysis Shows High Specificity of Monobodies in Cells.
To evaluate the specificity of the four monobodies in complex cellular proteomes using liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS), we stably expressed the monobodies as tandem affinity purification (TAP)-tagged proteins in HEK 293 and BCR-ABL–expressing K562 cells (19) (Fig. 2A). We confirmed that monobodies interacted with endogenous full-length SHP2 (SI Appendix, Fig. S3). Cell lines expressing the monobody HA4 targeting the ABL SH2 domain served as positive controls, giving rise to a very similar dataset with a degree of sensitivity comparable to that of our previous analysis (19) (SI Appendix, Table S2 and Dataset S1).
Fig. 2.
The SHP2-targeting monobody interactome. (A) Schematic representation of a TAP-tagged monobody, including the B1 domain of staphylococcal protein G, tobacco etch virus (TEV) protease recognition site, streptavidin-binding peptide (SBP), and Myc-tag N-terminal to the monobody. (B) Immunoblot analysis of total cell lysates of K562 and HEK293 stably expressing the TAP-tagged monobody clones. HA4, a previously characterized monobody binding the SH2 domain of ABL, served as a control. (C) TAP of NSa5 monobody complexes from K562 cells. TE, total extract; SN1, supernatant IgG beads; TEV, eluate after TEV cleavage; SN2, supernatant streptavidin beads; E1, eluate from streptavidin beads; BB, boiled streptavidin beads to control the efficiency of elution. The bait protein was identified by immunoblotting using an anti-Myc antibody. TAP blots for all other monobodies are shown in SI Appendix, Fig. S3. (D) (Upper) Monobody complexes after TAP (10% of the E1 fractions) from K562 and HEK293 cells, separated by SDS/PAGE and visualized with silver staining. (Lower) Anti-SHP2 immunoblot analysis of the same samples.
All cell lines expressed the monobodies at comparable levels, and the monobodies were efficiently retrieved after the second affinity purification step (Fig. 2 B and C and SI Appendix, Fig. S4). Captured proteins were visualized by silver staining and identified by LC-MS/MS (Fig. 2D and SI Appendix, Table S2 and Dataset S1). In two biological replicates, SHP2 was by far the most abundant of the identified proteins (SI Appendix, Table S2), consistent with the dominant ∼70-kDa band detected in the NSa1, NSa5, and CS3 samples from both cell lines (Fig. 2D). Even for CS1, peptides mapping to SHP2 were identified with a spectral count of >100 in both replicates from both cell lines (SI Appendix, Table S2). Immunoblot analysis confirmed the abundance of SHP2 in the NSa1, NSa5, and CS3 samples from both cell lines (Fig. 2D, Lower) and ABL/BCR-ABL in the HA4 samples (SI Appendix, Fig. S5), but significantly less SHP2 in the CS1 samples (Fig. 2D). Of note, no other SH2 domain-containing proteins were identified reproducibly in the NSa1, NSa5, CS1, and CS3 samples from both cell lines, despite the expression of 90 SH2 domains in these cells (SI Appendix, Table S2). These results demonstrate that the NSa1, NSa5, and CS3 monobodies are essentially monospecific to SHP2 in cells. To the best of our knowledge, these are the most selective reagents targeting SH2 domains, approaching the highest specificity achievable.
Crystal Structures of Monobody/SH2 Complexes Reveal Unusual Modes of Interactions.
To understand the structural basis for how the monobodies recognize their targets, we determined the crystal structures of the NSa1/N-SH2 and CS1/C-SH2 complexes at 2.3-Å resolution (Fig. 3 A and B and SI Appendix, Table S3). The overall folds of the monobodies and SHP2 SH2 domains were very similar to those of other monobodies and SHP2 SH2 structures (Cα rmsd <0.5 Å, excluding mutated loops, and <1 Å, respectively). Both monobodies occupy the pY-binding pockets of the SH2 domains, consistent with the ability of NSa1 and CS3 (which is closely related to CS1) to inhibit SH2–pY interactions (Fig. 1E). NSa1/N-SH2 and CS1/C-SH2 bury ∼900 Å2 and 1,000 Å2, respectively (Fig. 3 A and B, Right), much larger interfaces than those for pY–SH2 interfaces, which are typically ∼500 Å2 (Fig. 3C). The monobodies make contacts with residues on the surfaces of the SH2 domains that differ between SHP1 and SHP2 (SI Appendix, Fig. S6), explaining their ability to discriminate these closely related proteins.
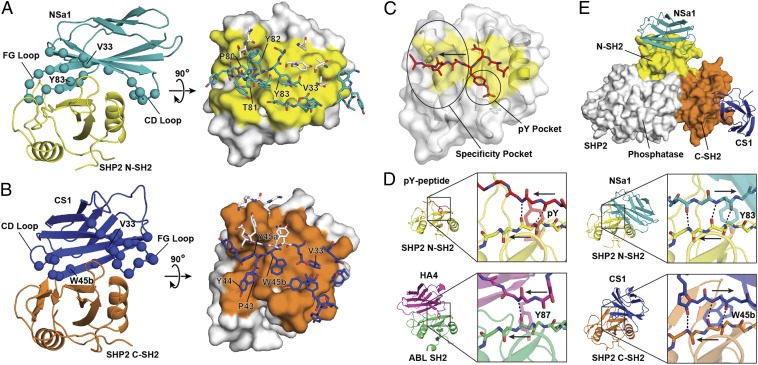
Fig. 3.
Crystal structures of SHP2 SH2/monobody complexes [Protein Data Bank (PDB) ID codes 4JE4 and 4JEG]. (A) The monobody NSa1/N-SH2 complex. (Left) Cartoon representation with NSa1 in teal, N-SH2 in yellow, and residues diversified in the library shown as spheres. (Right) Orthogonal view of the interface, in which N-SH2 is represented as a surface model with the epitope in yellow and residues within the paratope of monobody NSa1 are represented as sticks. Residues that were not diversified in the library are shown as white sticks. (B) The CS1/SHP2 C-SH2 complex. The labeling scheme is similar to that in A, except that monobody CS1 is in blue and C-SH2 is in orange. (C) Interface of a phosphopeptide with SHP2 N-SH2 (31) (PDB ID code 3TL0) showing the typical positions of the pY-binding and specificity pockets of SH2. The arrow shows the direction of the bound peptide. (D) The binding modes of a pY peptide, HA4, NSa1, and CS1 to their respective cognate SH2 domains. Intermolecular hydrogen bonds are shown as dashed lines, and the directions of the strands are labeled with the arrows in the expanded boxes. (E) Overlay of NSa1/N-SH2 and CS1/C-SH2 complexes on autoinhibited SHP2 (10) (PDB ID code 2SHP). SHP2 is represented as a surface model with the N-SH2 and C-SH2 domains colored yellow and orange, respectively, and the phosphatase domain in white. Monobodies NSa1 and CS1 are represented as cartoon models colored teal and blue, respectively.
All of the isolated monobodies contain the R33V mutation in β-strand C (Figs. 1C and 3 A and B). Reverting this position back to Arg severely impaired binding of the NSa1 and CS3 monobodies (SI Appendix, Fig. S7), demonstrating the importance of this mutation in recognizing the SH2 domains and also explaining the superior performance of the “side” library over the “loop only” library. Out of the 23 positions diversified in the library, 20 were located within the N-SH2/NSa1 complex interface and 13 were located within the C-SH2/CS1 complex interface, closely matching our interface design (Fig. 3 A and B, Right). Taken together, these results support the authenticity of the binding interfaces observed in the crystal structures.
Modeling the two monobodies onto the crystal structure of autoinhibited, full-length SHP2 by superimposing the SH2 domains revealed no obvious steric clashes (Fig. 3D), suggesting that these monobodies would not substantially perturb the autoinhibited form of SHP2. Although some synthetic phosphopeptides have been reported to stimulate SHP2 phosphatase activity (16, 23) we found that the monobodies, or tandem phosphopeptides derived from SHP2 ligands, did not significantly activate the phosphatase activity of SHP2 in vitro (SI Appendix, Fig. S8). The absence of strong activation by the monobodies is consistent with our modeling described above.
Closer inspection of the binding interfaces revealed unusual modes of interaction between the monobodies and SH2 domains. A previously developed monobody, HA4, directed to the ABL SH2 closely mimics the canonical mode of the pY–SH2 interaction in which a Tyr residue occupies the pY pocket and residues immediately C-terminal to the Tyr run along the peptide-binding groove, or the so-called “specificity pocket” (Fig. 3 C and D) (24). pY peptides generally run perpendicular to the central β-sheet of the SH2 domain (Fig. 3 C and D). Surprisingly, segments of NSa1 and CS1 bound within the peptide-binding site of SH2 run in the opposite direction to that of pY peptides and the HA4 monobody and thereby extend this antiparallel β-sheet (Fig. 3D). In NSa1, residues 80–82 in the FG loop bind the specificity pocket, and Y83 mimics pY (Fig. 3 A and D). Residues 81–83 of NSa1 (boxed in Fig. 1C) dock onto the central β-sheet through backbone hydrogen bonds, and P80 located three positions N-terminal to the Y83 has the cis peptide bond, allowing the FG loop to make a sharp turn. In CS1, residues 43–45b (boxed in Fig. 1C) in the CD loop occupy the binding pockets, and this segment also docks onto and extends the antiparallel β-sheet (Fig. 3 B and D). Interestingly, a Trp (W45b), rather than Tyr, makes contact with the pY pocket, although it occupies only a small portion of the pocket. Furthermore, P43 is located three residues N-terminal to the pY equivalent W45b, with a cis peptide bond involving a sharp kink in the backbone, just like P80 of NSa1 discussed above. Although NSa1 and CS1 use distinct segments for interacting their cognate SH2 domains, their modes of interaction to the peptide-binding site are strikingly similar.
To the best of our knowledge, the interfaces of the NSa1/N-SH2 and CS1/C-SH2 complexes represent a unique pY-independent mode of interaction with the SH2 domain. This rare binding mode may contribute to the ability of these monobodies to discriminate their cognate targets from the other SH2 domains. This new mode of peptide–SH2 interaction also helps explain why the canonical mode of pY peptide–SH2 interaction is favored. Approximately half of the binding energy of pY peptides comes from pY (22). In the canonical orientation, the pY side chain adopts the most energetically favorable conformer and forms extensive close interactions with the SH2 domain. In contrast, the side chains of Y83 in NSa1 and of W45b in CS1 adopt much less favorable side chain conformers located slightly out of the pY-binding pocket. Thus, although the reverse orientation improves hydrogen bonding and general packing across the peptide fragment, it is more detrimental for pY interactions. Because the monobodies do not contain a pY residue, their binding modes are not restricted by the dominant anchoring that pY provides. Furthermore, the numerous contacts to regions outside the peptide-binding sites might diminish the importance of contacts of the monobodies to the peptide-binding site, which in turn may have stabilized the unusual binding mode.
Monobodies Inhibit Activating Phosphorylation Events on SHP2.
We next studied the biological effects of our monobodies on SHP2 in cells. Expression of the NSa1, NSa5, or CS3 monobody along with BCR-ABL in cells produced a significant decrease in the intensity of a prominent tyrosine phosphorylated band of ∼90 kDa, in contrast to no such decrease with a nonbinding control monobody and only a small decrease with the CS1 monobody (Fig. 4A). Because the size of this band is in line with SHP2 expressed from the construct used, and because residues Y542 and Y580 of SHP2 are phosphorylated on activation (Fig. 4 B and C) (25, 26), we interpreted these results as an indication that the monobodies might diminish SHP2 phosphorylation or activation by disrupting its interactions with GAB2 (Fig. 4C; see also Figs. 1E and 5A). Immunoprecipitation of SHP2 showed that its phosphorylation was indeed reduced in the presence of monobodies (Fig. 4B).
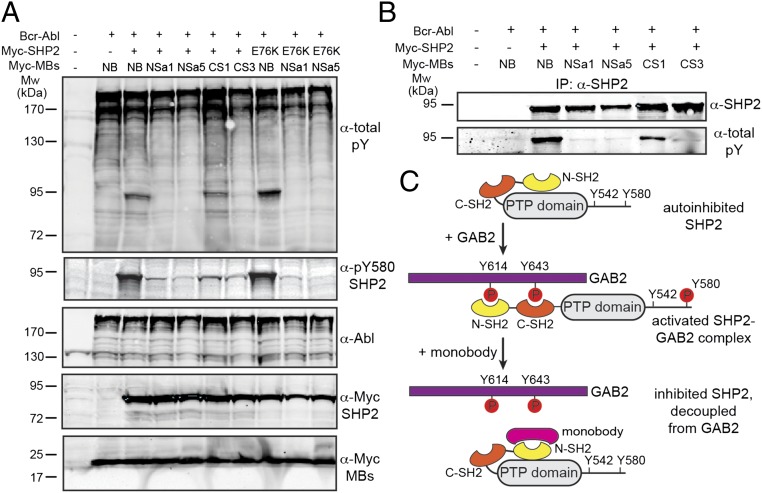
Fig. 4.
Monobodies inhibit SHP2 phosphorylation in cells. (A) HEK293 cells were transiently transfected with the indicated expression constructs, and total cell lysates were immunoblotted with the indicated antibodies. E76K is an activating point mutation in SHP2, and NB is a nonbinding control monobody (the Y87A mutant of HA4; ref. 19). (B) SHP2 was immunoprecipitated from HEK293 cell lysates that had been transfected with the indicated expression constructs and immunoblotted with anti-SHP2 and anti-pY (total pY) antibodies. (C) Schematic representation of the autoinhibited SHP2 conformation and activated SHP2 bound to GAB2, and the perturbation of this interaction by monobodies. The model is based on conformational changes on SHP2 activation reported previously (10, 12) and the data presented in Figs. 1E and 5A.
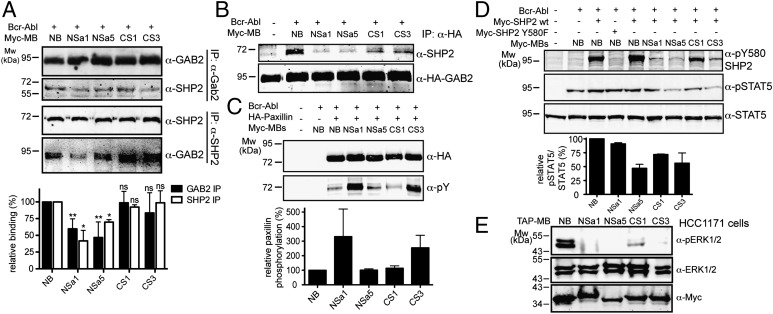
Fig. 5.
Expression of monobodies decouples GAB2 and SHP2 and modulates downstream signaling. (A) The interaction of endogenous SHP2 and GAB2 was monitored by reciprocal immunoprecipitation experiments on overexpression of monobodies and BCR-ABL. The bar graph quantifies the SHP2–GAB2 interaction and its perturbation by monobody expression from six (for GAB2) and two (for SHP2) independent immunoprecipitation experiments. ns, not significant. *P < 0.05; **P < 0.005. (B) The interaction of endogenous SHP2 with GAB2 in the presence of monobodies was studied in HEK293 cells stably overexpressing HA-tagged GAB2. (C and D) HEK293 cells were transiently transfected with the indicated expression constructs, and total cell lysates were immunoblotted with the indicated antibodies. The bar graphs quantify the phosphorylation of paxillin (C) and STAT5 (D) and its perturbation by monobody expression (n = 2). (E) HCC1171 cells stably expressing the TAP-tagged monobody clones and total cell lysates immunoblotted with the indicated antibodies.
Mutational analysis identified Y580 as the predominant site affected by monobody expression and Y542 and Y580 as the two principal SHP2 phosphorylation sites under our experimental conditions (Fig. 4A and SI Appendix, Fig. S9). Importantly, tyrosine phosphorylation of the constitutively active E76K mutant of SHP2, a common mutation in the N-SH2 domain in Noonan syndrome and juvenile myelomonocytic leukemia, was also inhibited by the NSa1 and NSa5 monobodies (Fig. 4A, rightmost three lanes). Taken together, these data show that targeting the peptide-binding interface of either SH2 domain of SHP2 with the monobodies inhibits the phosphorylation of SHP2 on critical tyrosine residues.
SHP2 Monobodies Inhibit SHP2–GAB2 Interactions and Modulate Downstream Signaling.
We next set out to examine the consequences of SHP2 monobody expression in cells. Binding of its SH2 domains to GAB2 is an important mechanism of SHP2 activation (13) and we found that NSa1 and NSa5 strongly reduced the effective affinity of the interaction between a GAB2 tandem phosphopeptide and the tandem SH2 domains of SHP2 (Fig. 1E). Expression of NSa1 or NSa5 monobodies in cell lines and immunoprecipitation of endogenous SHP2 or GAB2 yielded a strongly reduced GAB2–SHP2 interaction, whereas expression of CS1 or CS3 produced no reduction in this interaction (Fig. 5A). In line with this observation, the N-SH2–targeting monobodies strongly inhibited the interaction of endogenous SHP2 with overexpressed GAB2 (Fig. 5B). These results suggest that monobodies targeting the N-SH2 domain but not the C-SH2 domain may potently decouple SHP2 from GAB2 and thus from the BCR-ABL protein complex, thereby inhibiting SHP2 activity (Fig. 4C).
To investigate the effects of monobodies on downstream signaling of SHP2, we evaluated the phosphorylation of paxillin, signal transducer and activator of transcription 5 (STAT5), and extracellular regulated kinase 1/2 (ERK1/2), which are known to be affected by SHP2 activity (27–29). Paxillin phosphorylation increased significantly with expression of NSa1 and CS3, whereas expression of NSa5, CS1, or a nonbinding control monobody had little effect (Fig. 5C and SI Appendix, Fig. S10). In contrast, expression of NSa5 and CS3 decreased the phosphorylation of STAT5 in the presence of BCR-ABL (Fig. 5D and SI Appendix, Fig. S10). Finally, expression of NSa1, NSa5, and CS3 almost completely abolished ERK1/2 phosphorylation in HCC1171 lung cancer cells carrying the activating V45L mutation in the SHP2 N-SH2 domain (15). Taken together, our findings indicate that targeting of the N-SH2 domain of SHP2 with monobodies strongly reduces its interaction with GAB2 and has profound effects on downstream signaling.
Discussion
We have developed monobodies that bind the SH2 domains of SHP2 with high affinity and extreme specificity, thereby enabling the precisely targeted perturbation of protein–protein interactions at a resolution of protein domains in cells. We believe that our methodology is among the most rigorous described to date for testing the specificity of protein–protein interactions. An important observation derived from our results is the low specificity of the CS1 monobody in cells despite its comparable in vitro binding and specificity characteristics with the other monobodies. This finding emphasizes the importance of unbiased characterization of cellular specificity of engineered binders beyond the testing for cross-reactivity using close homologs in in vitro or cell-based assays. We propose that affinity purification-MS approaches such as that described here should become standard tools for assessing the cellular specificity of binding molecules.
Unlike RNA interference approaches, our monobody-based approach does not depend on the depletion of an entire protein. Thus, results obtained with monobody-based perturbation are particularly informative for advancing our understanding of the cellular functions of target molecules and their druggability (20). Furthermore, monobodies also may serve as tools for targeting a particular state of a signaling protein and thereby provide insight into its regulatory mechanisms. The monobodies described herein are likely to substantially affect the function of only the active state of SHP2, in which the SH2 domains are engaged in protein interactions, whereas the latent autoinhibited state is only slightly perturbed (SI Appendix, Fig. S9).
All of the monobodies we selected turned out to be competitors for pY–SH2 interactions, although we did not include any positive selection for this interaction surface, consistent with our previous results with monobodies for structurally diverse targets (21, 30). These results further confirm the strong tendency of monobodies to target functional surfaces involved in protein–protein or protein–ligand interactions. The three high-affinity monobodies (HA4, NSa1, and CS1) directed to the pY- binding site of SH2 domains have dramatically distinct modes of interaction, clearly eliminating the possibility that the monobody scaffold is predisposed to binding to SH2 domains (Fig. 3D). Thus, these results support the idea that monobodies are particularly suitable as genetically encoded intracellular inhibitors of protein functions.
We have defined the roles of the SHP2 SH2 domains in the context of the CML signaling network. It is important to note that aberrant GAB2-SHP2 signaling also contributes to oncogenicity in different solid tumors; gain-of-function mutations in SHP2 have been identified in non-small cell lung carcinoma, colon cancers, neuroblastomas, and melanoma (15). Most of these mutations are located in the SH2 domains of SHP2, and both experimental evidence (Fig. 4A) and structural modeling (SI Appendix, Fig. S11) indicate that the monobodies developed in this study could target these SHP2 mutants. In line with this, we observed strong inhibition of ERK phosphorylation in a cell line expressing the activating SHP2 V45L mutation. Furthermore, amplifications and/or overexpression of GAB2 or GAB1 (leading to hyperactivated SHP2) have been reported in ovarian, gastric, breast, and lung cancers (14). All of the foregoing findings suggest that our approach is broadly applicable. Given the unavailability of selective small-molecules targeting the SHP2 phosphatase domain, investigating whether targeting the SH2 domains of SHP2 with monobodies can be of therapeutic benefit will be of interest.
In conclusion, our present results provide ample evidence that monobodies are excellent tools for effectively and specifically interfering with protein–protein interactions and signaling networks in cancer cells, and that knowledge of monobody-based interference can advance mechanistic understanding and guide the design of anticancer strategies.
Materials and Methods
Generation and characterization of monobodies were performed following published procedures (21). For interactome analysis, HEK293 and K562 cells stably expressing TAP-tagged monobodies were generated by retroviral infection and FACS sorting, as described previously (19). Further details on the materials and methods used in this study are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank L. Bailey, S. Crosson, R. Gilbreth, and S. Mukherjee for advice on crystallographic data analysis; K. Cook, J. Wojcik, and J. Barkinge for support with peptide binding; B. J. Hoey for support with surface plasmon resonance measurements; M. Bentries-Alj for the HCC1171 cells; S. Dhe-Paganon for a kinase construct; and the École Polytechnique Fédérale de Lausanne’s Proteomics, Protein Expression, and Flow Cytometry Core Facilities for expert support with MS analysis, large-scale expansion of TAP cell lines, and cell sorting, respectively. This work was supported by the Swiss Institute for Experimental Cancer Research Foundation (E.B.G., S.G., and O.H.), National Center of Competence in Research Molecular Oncology (E.B.G. and O.H.), European Molecular Biology Organization (Short Term Fellowship ASTF 497-2010, to O.H.), and National Institutes of Health (NIH; Grants R01-GM072688 and R01-GM090324, to S.K.). F.S. was supported by NIH Training Grant T32GM008720-S. This work includes research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, supported by NIH Grants 5P41RR015301-10 and 8 P41 GM103403-10. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy’s Office of Science by the Argonne National Laboratory, was supported by the Department of Energy under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of the NSa1/N-SH2 and CS1/C-SH2 complexes have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4JE4 and 4JEG, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303640110/-/DCSupplemental.
References
- 1.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96(10):3343–3356. [PubMed] [Google Scholar]
- 2.Hantschel O. Structure, regulation, signaling, and targeting of abl kinases in cancer. Genes Cancer. 2012;3(5-6):436–446. doi: 10.1177/1947601912458584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5(3):172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 4.Brehme M, et al. Charting the molecular network of the drug target Bcr-Abl. Proc Natl Acad Sci USA. 2009;106(18):7414–7419. doi: 10.1073/pnas.0900653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Druker BJ. Translation of the Philadelphia chromosome into therapy for CML. Blood. 2008;112(13):4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton A, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119(6):1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Hare T, Zabriskie MS, Eiring AM, Deininger MW. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat Rev Cancer. 2012;12(8):513–526. doi: 10.1038/nrc3317. [DOI] [PubMed] [Google Scholar]
- 8.Scherr M, et al. Enhanced sensitivity to inhibition of SHP2, STAT5, and Gab2 expression in chronic myeloid leukemia (CML) Blood. 2006;107(8):3279–3287. doi: 10.1182/blood-2005-08-3087. [DOI] [PubMed] [Google Scholar]
- 9.Sattler M, et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell. 2002;1(5):479–492. doi: 10.1016/s1535-6108(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 10.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92(4):441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 11.Vogel W, Lammers R, Huang J, Ullrich A. Activation of a phosphotyrosine phosphatase by tyrosine phosphorylation. Science. 1993;259(5101):1611–1614. doi: 10.1126/science.7681217. [DOI] [PubMed] [Google Scholar]
- 12.Mohi MG, Neel BG. The role of Shp2 (PTPN11) in cancer. Curr Opin Genet Dev. 2007;17(1):23–30. doi: 10.1016/j.gde.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Gu H, Pratt JC, Burakoff SJ, Neel BG. Cloning of p97/Gab2, the major SHP2-binding protein in hematopoietic cells, reveals a novel pathway for cytokine-induced gene activation. Mol Cell. 1998;2(6):729–740. doi: 10.1016/s1097-2765(00)80288-9. [DOI] [PubMed] [Google Scholar]
- 14.Wöhrle FU, Daly RJ, Brummer T. Function, regulation and pathological roles of the Gab/DOS docking proteins. Cell Commun Signal. 2009;7:22. doi: 10.1186/1478-811X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentires-Alj M, et al. Activating mutations of the Noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64(24):8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 16.Imhof D, et al. Sequence specificity of SHP-1 and SHP-2 Src homology 2 domains: Critical roles of residues beyond the pY+3 position. J Biol Chem. 2006;281(29):20271–20282. doi: 10.1074/jbc.M601047200. [DOI] [PubMed] [Google Scholar]
- 17.Machida K, et al. High-throughput phosphotyrosine profiling using SH2 domains. Mol Cell. 2007;26(6):899–915. doi: 10.1016/j.molcel.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 18.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284(4):1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 19.Wojcik J, et al. A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat Struct Mol Biol. 2010;17(4):519–527. doi: 10.1038/nsmb.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grebien F, et al. Targeting the SH2-kinase interface in Bcr-Abl inhibits leukemogenesis. Cell. 2011;147(2):306–319. doi: 10.1016/j.cell.2011.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koide A, Wojcik J, Gilbreth RN, Hoey RJ, Koide S. Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J Mol Biol. 2012;415(2):393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladbury JE, et al. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: A reappraisal. Proc Natl Acad Sci USA. 1995;92(8):3199–3203. doi: 10.1073/pnas.92.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lechleider RJ, et al. Activation of the SH2-containing phosphotyrosine phosphatase SH-PTP2 by its binding site, phosphotyrosine 1009, on the human platelet-derived growth factor receptor. J Biol Chem. 1993;268(29):21478–21481. [PubMed] [Google Scholar]
- 24.Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high-affinity phosphotyrosyl peptide to the Src SH2 domain: Crystal structures of the complexed and peptide-free forms. Cell. 1993;72(5):779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- 25.Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG. Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc Natl Acad Sci USA. 1994;91(15):7335–7339. doi: 10.1073/pnas.91.15.7335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keegan K, Cooper JA. Use of the two-hybrid system to detect the association of the protein-tyrosine-phosphatase, SHPTP2, with another SH2-containing protein, Grb7. Oncogene. 1996;12(7):1537–1544. [PubMed] [Google Scholar]
- 27.Li L, et al. A critical role for SHP2 in STAT5 activation and growth factor-mediated proliferation, survival, and differentiation of human CD34+ cells. Blood. 2011;118(6):1504–1515. doi: 10.1182/blood-2010-06-288910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren Y, et al. Roles of Gab1 and SHP2 in paxillin tyrosine dephosphorylation and Src activation in response to epidermal growth factor. J Biol Chem. 2004;279(9):8497–8505. doi: 10.1074/jbc.M312575200. [DOI] [PubMed] [Google Scholar]
- 29.Araki T, Nawa H, Neel BG. Tyrosyl phosphorylation of Shp2 is required for normal ERK activation in response to some, but not all, growth factors. J Biol Chem. 2003;278(43):41677–41684. doi: 10.1074/jbc.M306461200. [DOI] [PubMed] [Google Scholar]
- 30.Gilbreth RN, Koide S. Structural insights for engineering binding proteins based on non-antibody scaffolds. Curr Opin Struct Biol. 2012;22(4):413–420. doi: 10.1016/j.sbi.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, et al. Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain. Biochemistry. 2011;50(35):7637–7646. doi: 10.1021/bi200439v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.