Significance
Healthy skin is colonized by a diversity of microbiota. Little is known regarding how the host immune response influences the skin microbiota. We demonstrate a role for complement, a key component of innate immunity, in host–microbe interactions of the skin. Inhibiting a key component of the complement cascade reduced diversity and altered composition of the skin microbiota, parallel to a reduction in skin inflammatory cell infiltration and downregulation of skin defense and immune gene expression. Further, we find that the commensal skin microbiota regulates the expression of complement genes in the skin. These results suggest an interactive role between complement and the microbial ecosystem of the skin and could have important implications for inflammatory and/or infectious skin disorders.
Abstract
The skin is colonized by a plethora of microbes that include commensals and potential pathogens, but it is currently unknown how cutaneous host immune mechanisms influence the composition, diversity, and quantity of the skin microbiota. Here we reveal an interactive role for complement in cutaneous host–microbiome interactions. Inhibiting signaling of the complement component C5a receptor (C5aR) altered the composition and diversity of the skin microbiota as revealed by deep sequencing of the bacterial 16S rRNA gene. In parallel, we demonstrate that C5aR inhibition results in down-regulation of genes encoding cutaneous antimicrobial peptides, pattern recognition receptors, and proinflammatory mediators. Immunohistochemistry of inflammatory cell infiltrates in the skin showed reduced numbers of macrophages and lymphocytes with C5aR inhibition. Further, comparing cutaneous gene expression in germ-free mice vs. conventionally raised mice suggests that the commensal microbiota regulates expression of complement genes in the skin. These findings demonstrate a component of host immunity that impacts colonization of the skin by the commensal microbiota and vice versa, a critical step toward understanding host–microbe immune mutualism of the skin and its implications for health and disease. Additionally, we reveal a role for complement in homeostatic host–microbiome interactions of the skin.
The skin is our interface to the outside world and encounters continuous assault by foreign and potentially pathogenic organisms. The skin also harbors populations of nonpathogenic, commensal microorganisms, which have important functions in skin health and disease, including colonization resistance to block invasion of opportunistic or pathogenic microbiota, and regulation of immunity and inflammation (1–3). Culture-independent analyses of the healthy skin microbiota, based on sequencing of the bacterial small-subunit 16S rRNA gene, allow greater resolution in characterizing microbial community structure and have revealed the great topographical and temporal complexity at this barrier surface (4–8). Some environmental and host factors have been identified to influence commensal skin microbial communities (9–12). However, the role of cutaneous immune defense in shaping and maintaining the skin microbial ecosystem is currently unknown. Here, we hypothesize that complement, a central component of innate immunity, influences host–microbe interactions at the skin surface.
To avert microbial invasion and infection, yet simultaneously avoid damaging inflammation or autoimmunity, the host must rely upon carefully calibrated defense mechanisms at the cutaneous barrier. The complement system, a network of more than 50 plasma and membrane-associated proteins, not only acts as a first line of defense against microbes but is a key mediator of inflammation and immune responses (13). Tight regulation of complement activation is required for the proper functioning of the system, and, although excessive complement activation contributes to a wide variety of inflammatory and autoimmune diseases (13), complement-deficiency states result in impaired host defense and increased risk of infection (14). Notably, in the skin, complement dysregulation, deficiency, and genetic polymorphisms have been associated with a number of diseases, including psoriasis, atopic dermatitis, pemphigus vulgaris, bullous pemphigoid, systemic lupus erythematosus, lichen planus, xeroderma pigmentosum, and recurrent cutaneous infection (15–18).
Complement is triggered by one of three pathways (classical, alternative, or lectin), which all converge in the activation of the third complement component (C3). Following activation, the release of biologically active proteins promote diverse defense mechanisms such as microbial opsonization and phagocytosis, direct lysis of target microbial cells through the membrane attack complex (MAC), and the generation of effector molecules that mediate recruitment and activation of inflammatory cells (13). This latter function, mediated by the complement C3a and C5a fragments, has been implicated in the modulation of innate and acquired immune responses via cross-talk between the C5a receptor (C5aR) and pattern recognition receptor signaling (19). In this study, we focused on complement component C5a, the most potent anaphylatoxin produced during complement activation, and its role in cutaneous host–microbiome interactions of mice maintained under specific pathogen-free conditions. Signaling of C5a through its receptor triggers proinflammatory and immunoregulatory actions, including enhanced leukocyte chemotaxis, neutrophil–endothelial cell adhesion, vascular permeability, granule secretion, and proinflammatory cytokine and chemokine release (13).
By using culture-independent high-throughput sequencing of bacterial 16S rRNA genes, we show that systemic inhibition of C5aR signaling leads to significant changes in the skin microbiota over time, including reduced diversity and altered taxonomic composition. We also find that disrupting C5aR signaling leads to decreased infiltration of the skin by inflammatory cells, and that this is accompanied by down-regulation of immune and defense genes in the skin, including antimicrobial peptides, cytokines and chemokines, and pattern recognition receptors. Conversely, we demonstrate that the commensal skin microbiota regulates the expression of complement genes in the skin. Taken together, these results suggest an interactive role between complement and the microbial ecosystem of the skin. These findings demonstrate that cutaneous host–microbe interactions are dynamic, and have important implications for skin disorders that incorporate microbial dysbiosis and immune dysregulation as part of their pathogenesis.
Results
Complement C5aR Signaling Influences the Composition and Diversity of the Skin Microbiota.
To investigate the impact of complement signaling on the skin microbiome while controlling for experimental variables such as maternal transmission and environmental influences, we used a longitudinal study design with a peptide inhibitor that antagonizes C5aR (20). Before treatment with the C5aR antagonist (C5aRA) or an inactive control (iC5aRA), C57BL/6J mice were individually housed for 2 wk and skin microbiota were collected to determine the baseline (BL) microbiome composition. Mice were then treated with C5aRA or control iC5aRA for 2 wk (n = 15 mice per treatment over two independent experiments). Following the 2-wk treatment course, skin microbiota were collected and compared with the BL microbiota. To assess microbial communities colonizing the skin before and after treatments, we sequenced the V1–V3 region of the 16S rRNA gene. A total of 193,422 quality sequences (Materials and Methods) generated through 454 pyrosequencing were used for this analysis, with an average of 3,335 sequences per sample. Sequences were assigned taxonomies and clustered in operational taxonomic units (OTUs) at a pairwise distance of 97% sequence identity.
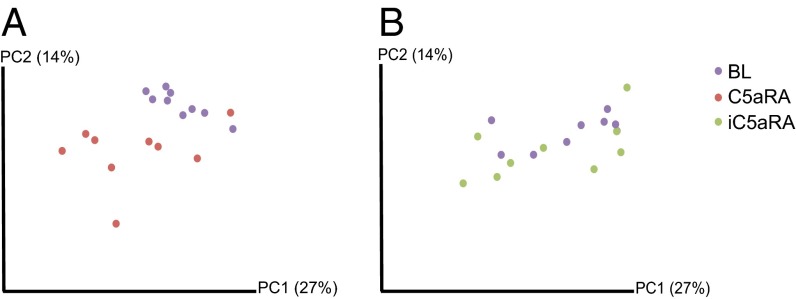
We first quantified differences between BL skin microbiota and posttreatment microbiota by using the UniFrac metric of β-diversity (21), which measures evolutionary distance between microbiotas. Calculation of the weighted UniFrac metric, which takes into account the presence/absence of bacterial taxa along with the abundances of those taxa, followed by principal coordinate analysis (PCoA) was used to infer the distances between the samples. When combining data from both independent experiments, a separation is observed primarily dictated by the experiment, suggesting experiment-dependent effects on the skin microbiome (Fig. S1). To better visualize changes that arise from treatment, we calculated the PCoA separately for each experiment. A clearer distinction is visible following treatment with C5aRA (Fig. 1A) compared with treatment with iC5aRA (Fig. 1B) when comparing posttreatment microbiomes vs. pretreatment BL microbiomes. By using the nonparametric Adonis test, we compared samples that were “positive” for C5aR signaling (BL and iC5aRA-treated samples) vs. those that were “negative” for C5aR signaling (C5aRA-treated samples) by using unweighted UniFrac, weighted UniFrac, Bray–Curtis, and Jaccard distances. For each independent experiment, we observed significant differences (P < 0.05) when comparing microbiomes by C5aR status (Table S1 provides P and R2 values).
Fig. 1.
Antagonism of complement C5aR results in an overall change in the skin microbiota. Skin microbiota samples were collected from C57BL/6J mice before treatment to determine BL microbiota (purple dots), and then treated with C5aRA (red dots) or iC5aRA (green dots). Following a 2-wk treatment course, skin microbiota were collected again to compare with BL microbiota. Depicted are PCoA plots of the weighted UniFrac metric comparing skin microbiota of (A) C5aRA-treated mice and their BL, and (B) iC5aRA-treated mice and their BL. Percentage of variation explained by the principal coordinates is indicated in parentheses on the axes. Depicted is one of two independent experiments (n = 9 mice per treatment group). Table S1 shows R2 and P values of the Adonis test, which assigns strength and statistical significance to the sample groupings visually observed. Fig. S1 illustrates experiment-dependent microbiome effects.
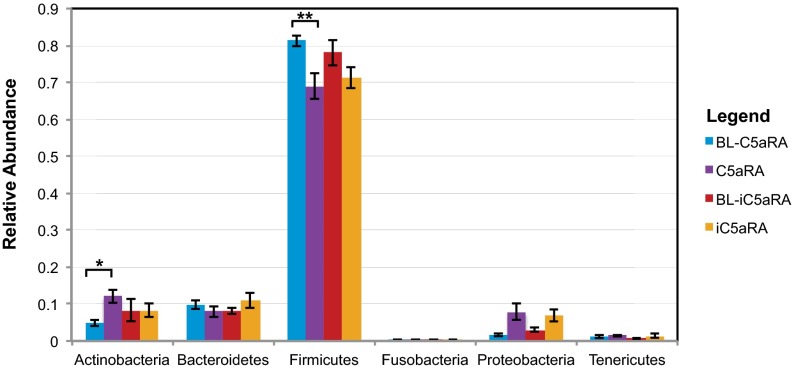
To determine if these observed differences in skin microbiome were caused by specific bacterial taxa that may be affected by complement C5aR signaling, we calculated relative abundance of taxa. By using a paired Student t test to compare each treatment sample to its own BL sample, only those taxa that were significantly altered in mice treated with C5aRA, but not in those mice treated with iC5aRA, were selected as significant. At the phylum level, Actinobacteria is significantly increased (P < 0.007) whereas Firmicutes is significantly decreased (P < 0.008) upon C5aRA treatment but not iC5aRA treatment (Fig. 2A). Phylum Proteobacteria also significantly increased in relative abundance following C5aRA treatment (P < 0.011); nevertheless, the same change is found following iC5aRA treatment (P < 0.034), indicating that these changes in abundance are related to temporal variation of the skin microbiota.
Fig. 2.
Impact of C5aR signaling on the skin microbiota taxonomical composition. Mean relative abundance (y axis) of the six most abundant phyla (x axis) is depicted. Paired t tests were calculated for each mouse comparing the phylum-level taxa of its BL skin microbiota and its posttreatment (C5aRA or iC5aRA) skin microbiota. Only those phyla that significantly changed following C5aRA treatment (P < 0.05), but not iC5aRA treatment, were selected as significant. Error bars represent SEM (n = 14 mice for each group over two independent experiments; *P = 0.008 and **P = 0.007).
By using the same strategy, complement C5aR-related changes in relative abundance at the genus level were determined. We first selected those 154 genera that were present in >10% of samples and comprised >1% of total relative abundance. When comparing BL vs. C5aRA and iC5aRA, those six genera that significantly differed after C5aRA treatment but not iC5aRA treatment included Propionibacterium, unclassified Coriobacteriaceae, unclassified Clostridiales, Turicibacter, Allobaculum, and Fusobacterium when a false discovery rate (FDR) <0.05 was applied to correct for multiple comparisons. We therefore concluded that taxonomical changes that are not caused by normal longitudinal variation occur in the skin microbiota when C5aR signaling is inhibited, thus suggesting a role for complement in shaping the skin microbiota.
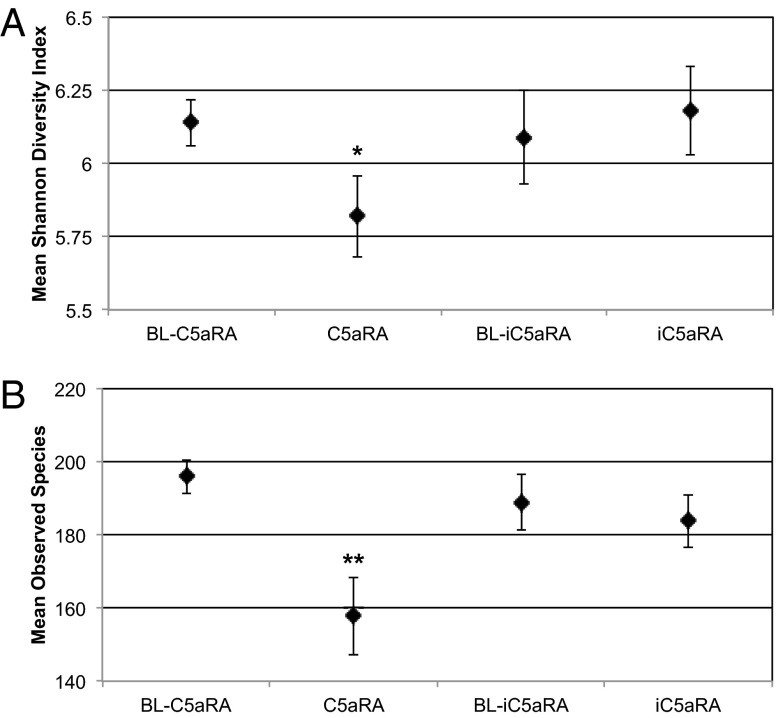
To determine if complement C5aR signaling has an effect on the overall diversity of the skin microbiota, we calculated the Shannon diversity index, a measure of α-diversity for each sample (Fig. 3A). The paired Student t test was applied to determine if significant changes in Shannon diversity occurred upon treatment with C5aRA compared with treatment with iC5aRA. The Shannon diversity index decreased from a mean of 6.14 to 5.81 (P = 0.037) upon treatment with C5aRA, but no significant change was observed in the iC5aRA treatment samples (P = 0.66). A similar trend was observed when comparing the number of observed OTUs before and after treatment (Fig. 3B), in that treatment with C5aRA decreased the number of observed OTUs from a mean of 195.9 to 157.86 (P = 0.005), whereas no significant change was observed in iC5aRA-treated mice (P = 0.61). This is in line with a previous report that C5aR−/− mice are colonized with lower diversity of gut microbiota compared with their WT littermates (22). These findings suggest that complement may in part be responsible for maintaining microbial diversity at the skin surface.
Fig. 3.
Diversity and richness of skin microbiota decreases when C5aR signaling is inhibited. (A) The mean Shannon diversity index, a measure of α-diversity that takes into account OTU richness and evenness, was used to compare skin microbiota before treatment (BL) to C5aRA- and iC5aRA-treated skin microbiota. Higher Shannon diversity index indicates higher diversity. (B) Mean number of species-level OTUs observed before treatment (BL) and following treatment with C5aRA or iC5aRA. Error bars represent SEM (n = 14 mice for each group over two independent experiments; *P = 0.037 and **P = 0.005).
In addition to specific bacterial taxa and bacterial diversity, we examined bacterial load to determine if complement C5aR signaling may influence the quantity of bacteria on the skin. We used quantitative PCR of 16S rRNA genes to estimate relative differences in bacterial load following treatment with C5aRA and iC5aRA. We did not detect any significant differences following either treatment, and therefore concluded that complement C5aR signaling does not influence the absolute numbers of bacteria on the skin, but rather influences the composition and diversity of the bacteria that colonize the skin.
Blockade of C5aR Alters Expression of Cutaneous Immune and Defense Genes While Influencing Inflammatory Cell Infiltration.
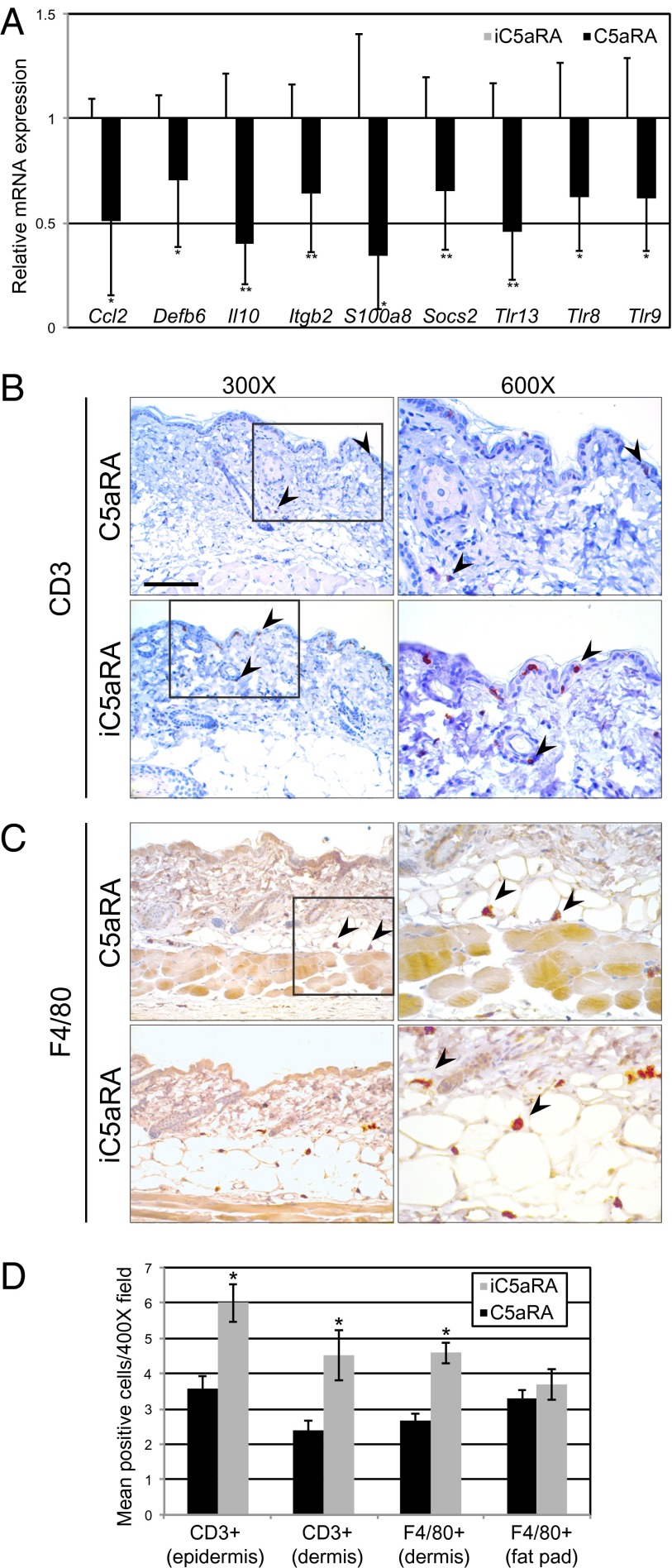
To gain additional insight into the effect that complement C5aR inhibition may have on host–microbe interactions at the skin surface, we examined expression of cutaneous genes that encode mediators of inflammation and innate immunity. Interestingly, all genes identified as differentially expressed in skin of C5aRA-treated mice relative to skin of iC5aRA-treated mice were down-regulated (Fig. 4A). Genes that were significantly (P < 0.05) differentially expressed after treatment with C5aRA included those encoding antimicrobial peptides (Defb6, S100a8), cytokines (Il10), chemokines (Ccl2), cell adhesion molecules (Itgb2), and pattern recognition receptors (Tlr8, Tlr9, Tlr13). These results are consistent with known effector functions of C5aR signaling, including trafficking and activation of leukocytes, enhancement of adhesion, and crosstalk with Toll-like receptors.
Fig. 4.
Inhibition of complement C5aR decreases cutaneous expression of innate immune and inflammatory mediators and skin inflammatory cell infiltration. (A) After 2-wk treatment with C5aRA or iC5aRA, skin was collected and RNA extracted for gene expression analysis. Skin mRNA levels were determined by quantitative real-time PCR (normalized to B2m) and expressed as fold change in C5aRA-treated transcript levels relative to iC5aRA-treated transcript levels, which were assigned an average value of 1. Data are means ± SD (n = 9 C5aRA-treated mice and n = 8 iC5aRA-treated mice from two independent experiments; *P < 0.05 and **P < 0.01). Table S2 shows all genes assayed and primer/probe sets. (B and C) Skin sections (6 µM thick) of mice treated with C5aRA and iC5aRA for 2 wk were stained with antibodies specific for (B) CD3 and (C) F4/80 to identify infiltrating lymphocytes and macrophages, respectively. Depicted are representative images at magnifications of 300× (Left) and the same image at a magnification of 600× (Right). Fig. S2 shows control and H&E staining. (D) For all staining, three to five fields per section were analyzed at magnification of 400× and positive cells were counted. Data are expressed as average number of immunoreactive cells in the field and are representative of one experiment consisting of three mice for each treatment and two skin biopsies per mouse. Error bars represent SEM (*P < 0.05). (Scale bars: Left, 100 µm; Right, 200 µm.)
To further explore the effect that C5aR signaling has on inflammatory cell populations of the skin, we assessed infiltrating cell populations in C5aRA- and iC5aRA-treated skin by using immunohistochemistry. We assessed lymphocytes by using immunohistochemistry against CD3 (Fig. 4B), and we assessed macrophages by using immunohistochemistry against F4/80 (Fig. 4C). We observed that treatment with C5aRA but not iC5aRA resulted in significantly decreased (P < 0.05) numbers of CD3+ cells in the epidermis and dermis and decreased numbers of F4/80+ cells in the dermis (Fig. 4D). The amount of F4/80+ cells infiltrating the fat pad was not significantly different between the two treatments (Fig. 4D). Taken together, these results demonstrate that inhibition of C5aR signaling decreases cutaneous expression of chemokines and cytokines, antimicrobial peptides, and pattern recognition receptors, which parallels decreased lymphocytes and macrophages infiltrating the skin.
Induction of Complement Gene Expression by the Commensal Skin Microbiota.
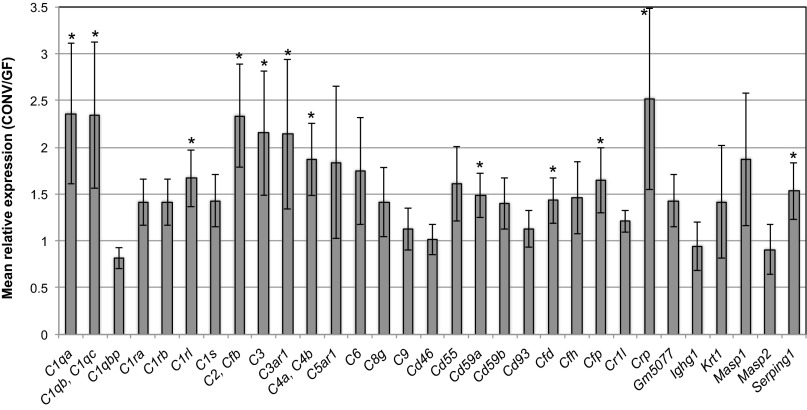
We hypothesized that the relationship between complement and the microbiota was interactive, and that complement not only modulates the skin microbiota, but that the skin microbiota also regulates activation of complement. To test this hypothesis, we reasoned that the colonization of germ-free (GF) mice with commensal microbiota would result in differential gene expression of those cutaneous genes that are regulated by the microbiota. We therefore compared gene expression of GF mouse skin vs. conventionalized (CONV) mouse skin by using an RNA-sequencing (RNA-seq) dataset. This dataset, derived from four C57BL/6J GF and CONV mice each, contained, on average, 68.79 million paired end reads of 100 bp per sample. Following processing, mapping, and assembly by using the Tuxedo protocol (23), we queried all 58 genes falling under the Gene Ontology (GO) terms “complement activation” and “complement binding,” which encompass alternative, classical, and lectin pathways of activation, activation of the MAC, and negative and positive regulation of complement. If complement is regulated by the microbiota, we would observe differential expression of this subset of genes in CONV mouse skin compared with GF mouse skin. At a threshold of >1.5-fold or <0.75-fold change in expression in CONV mice compared with GF, those significantly differentially expressed genes (P < 0.05) in the subset examined included C1qa, C1qb, C1qc, C1rl, C2, Cfb, C3, C4a, C4b, Cd59a, Cfd, Cfp, Crp, and Serping1 (Fig. 5). In all cases, genes that were significantly differentially expressed were up-regulated in CONV compared with GF skin, suggesting that the commensal microbiota has a role in positively regulating expression of genes encoding complement components.
Fig. 5.
Mice colonized with commensal microbiota have higher cutaneous expression of complement genes compared with GF mice. Cutaneous gene expression of GF mice was compared with CONV mice. Depicted on the x axis are all genes categorized under GO terms “complement activation” and “complement binding” that were expressed in skin above the threshold FPKM >1 in at least two of the eight skin samples subjected to RNA sequencing (SI Materials and Methods). Data are expressed as mean expression level (y axis), as measured by FPKM normalized transcripts, of CONV mouse skin relative to GF mouse skin. Error bars represent propagated SE of the ratio CONV/GF (n = 4 GF mice and CONV mice each; *P < 0.05).
Discussion
Here, we provide evidence of a component of the immune system that impacts the commensal skin microbial ecosystem. Until now, most of our understanding of host–microbe immune mutualism was derived from studies of the gastrointestinal tract microbiota (24). Given that rich communities of microorganisms also inhabit the skin (3), similar interactions likely maintain homeostatic relationships with our microbial partners, while preventing pathogen invasion. Many exogenous and endogenous factors have been identified as associated with shifts in the skin microbiota, such as body site (4, 7), sex and handedness (10), age (11), and lifestyle, ethnicity, and/or geography (9). However, the role of major innate immune components on the microbial ecosystem, including complement, is largely unknown.
Likewise, disruption of these highly evolved relationships can have dire consequences for host health. Dysbiosis of the skin microbiota is hypothesized to contribute to the pathogenesis of multiple disorders, including atopic dermatitis, acne, and psoriasis. Decoding the underlying mechanisms that modulate and interact with microbial communities on the skin surface is a critical step toward dissecting the pathogenesis of these diseases, while shedding light on the potential for novel approaches for the treatment and/or prevention of disease.
We demonstrate that complement C5aR signaling influences the composition and diversity, but not the quantity, of the skin microbiota. In particular, we note that complement C5aR signaling appears to maintain microbial diversity and richness of the skin, also observed in the gut of C5aR−/− mice compared with their WT littermates (22). Low microbial diversity has been associated with a number of dysbiotic disease states, including atopic dermatitis of the skin (25). Greater microbial diversity may be advantageous in that it provides colonization resistance to invasion by opportunistic and/or pathogenic organisms. Other innate immune components may also be active in shaping and maintaining skin microbial communities and remain to be investigated, including Toll-like receptors, Nod-like receptors, and antimicrobial peptides. Furthermore, cross-talk between these effector mechanisms may take place and needs to be explored in greater depth. For example, emerging evidence suggests that the cross-talk between Toll-like receptors and complement is extensive and may synergize to enhance host defense, or antagonize to regulate excessive inflammation (19).
Our work also reveals a function in the skin for complement during homeostasis. The direct killing mechanism of complement, through MAC-mediated microbial lysis, has long been appreciated, but our work shows that, under homeostatic conditions, C5aR signaling contributes to modulation of microbial communities of the skin. Interestingly, hereditary deficiency and dysfunction in complement C5 have been associated with recurrent cutaneous infection and abscesses in addition to Leiner disease, presenting in infancy as extensive dermatitis similar to seborrheic dermatitis with increased likelihood of infection (26–28).
The mechanism by which C5aR signaling modulates the cutaneous microbiota warrants further investigation, but our data suggest that complement is activated even in the absence of active infection, and inhibiting C5aR signaling influences proinflammatory effectors and inflammatory cell recruitment. In particular, the expression of a number of proinflammatory mediators was down-regulated in the skin upon C5aR inhibition. This included the gene encoding the chemokine CCL2, also known as monocyte chemotactic protein-1. CCL2 recruits monocytes, T helper type 17 cells, and basophils to sites of inflammation, and has a known role in the inflammatory skin disease psoriasis (29–31). We also observed down-regulation of Itgb2 (mRNA), encoding the β-integrin β-2 chain, and a component of the LFA-1 integrin, expressed on leukocytes, and Mac-1, expressed on leukocytes and myeloid cells (32). Integrins play important roles in the recruitment of inflammatory cells, but are also implicated in the pathophysiology of inflammatory and autoimmune diseases. For example, efalizumab, an antibody against LFA-1, is used to treat psoriasis and selectively and reversibly inhibits trafficking of T cells (33).
This work suggests that complement is at least in part regulated by the commensal microbiota at the gene expression level. Although the changes in gene expression we observe are small (approximately twofold increase in CONV skin relative to GF skin), complement is a tightly regulated system, and even subtle changes in complement activity can significantly impact risk for inflammatory and infectious disease (34). This observation has important implications, as it suggests a potential strategy for modulating complement activation and regulation. The skin microbiota is a highly accessible and modifiable factor that could potentially be targeted for the treatment of diseases where complement dysregulation or dysfunction is implicated. Additionally, complement therapeutics, especially small peptide inhibitors such as the one used in the present study, are emerging as effective immune modulators. Complement activation has been implicated in psoriasis, especially C5a signaling (35, 36), and, interestingly, the same C5aR antagonist used in the present study underwent clinical trials in psoriasis and was found to improve lesions (20). Furthermore, psoriasis may in part be triggered or maintained by microbial antigens, and the variant guttate psoriasis is known to be triggered by Streptococcus infection (37). In situations of impaired wound healing, in which a self-amplifying cycle of inflammation and microbial bioburden may in part contribute to the pathogenesis (38), we envision that complement inhibition may be an effective treatment strategy. For example, our previous work in a mouse model of impaired wound healing (Leprdb/db) demonstrated that genes encoding complement components and receptors were up-regulated and persistently expressed during impaired wound healing, and these changes in gene expression were highly correlated with relative abundance of specific bacterial taxa that were associated with the impaired healing phenotype (39). Targeting proinflammatory mechanisms may be a useful strategy for altering dysbiotic cutaneous microbial ecosystems and a viable alternative to antibiotic manipulation of the microbiota. Thus, the homeostatic role of complement C5aR signaling in the skin could have significant implications for the treatment and prevention of some skin diseases, including psoriasis and impaired cutaneous wound healing.
Materials and Methods
Mice and C5aR Antagonist Intervention.
All mouse procedures were performed under protocols approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Eight-week-old C57BL/6J mice were obtained from Jackson Laboratories and maintained in individual cages, with ad libitum access to food and water in specific pathogen-free conditions at University of Pennsylvania Perelman School of Medicine. A specific and potent C5aR antagonist (C5aRA), the cyclic hexapeptide Ac-F[OP(D)Cha-WR] (acetylated phenylalanine–[ornithine-proline(D)cyclohexylalanine-tryptophan-arginine]), and an inactive analogue (iC5aRA), Ac-F[OP(D)Cha-A(D)R] (acetylated phenylalanine–[ornithine-proline-(D)cyclohexylalanine-alanine-(D)arginine]), were synthesized as previously described (40, 41). In all experiments, C57BL/6J mice were injected intraperitoneally with C5aRA or iC5aRA control every other day at a dose of 1 mg/kg for a total of 14 d.
Collection of Microbiota, DNA Extraction, Amplification, and Sequencing of 16S rRNA Genes.
Before treatment as described earlier, mice were shaved on the dorsum, and, 24 h later, a BL sample of skin microbiota was collected with a swab (Catch-all Sample Collection Swab; Epicentre) moistened in Yeast Cell Lysis Buffer from the MasterPure Yeast DNA Purification Kit (Epicentre). The swabs were placed directly into 300 μL of the Yeast Cell Lysis buffer and stored at −80 °C until DNA extraction. Following the 14-d treatment with C5aRA or iC5aRA, a skin swab was collected again. A total of 14 mice were treated with C5aRA and 14 were treated with iC5aRA, over two independent experiments. Ready-Lyse Lysozyme solution (Epicentre) was added to swabs and buffer to a final concentration of 20 mg/mL before incubation at 37 °C for 1 h with shaking. Samples were then processed in a TissueLyser (Qiagen) at maximum speed for 2 min, followed by a 30-min incubation at 65 °C for 30 min with shaking. A total of 150 μL of Protein Precipitation Reagent (Epicentre) was added, and samples were spun for 10 min at maximum speed. The supernatant was removed and mixed with 500 μL of isopropanol and applied to a column from the PureLink Genomic DNA Mini Kit (Invitrogen). Subsequently, the protocol from the PureLink kit was followed exactly and DNA was eluted in 50 μL of the Elution Buffer supplied in the kit (Invitrogen).
PCR of 16S rRNA genes was performed on 2 μL of sample DNA using forward primer 27F and a barcoded reverse primer 534R. PCR was performed in duplicate using an AccuPrime Taq DNA Polymerase High Fidelity kit (Invitrogen). The cycling conditions were as follows: 95 °C for 2 min, then 30 cycles of 95 °C for 20 s, 56 °C for 30 s, and 72 °C for 5 min. Negative (no template and mock swab) controls were treated similarly and failed to produce visible PCR product or sequencing reads. Duplicates were combined, and PCR products were purified by using the Agencourt AMPure XP kit according to the manufacturer’s instructions (Beckman Coulter). A total of 50 ng of each sample was pooled, and the pool was then purified using the MinElute PCR Purification Kit (Qiagen) following the manufacturer’s protocol. Sequencing was then conducted on a Roche 454 GS FLX Titanium instrument at the National Institutes of Health Intramural Sequencing Center.
Analysis of 16S rRNA Sequence Data.
Sequence quality control and analyses were performed using Quantitative Insights Into Microbial Ecology (QIIME) (42). See SI Materials and Methods for sequence processing and analysis details.
Quantitative Real-Time PCR.
Punch biopsy specimens (8 mm) of dorsal skin were obtained from mice treated for 14 d, as described earlier, with C5aRA or iC5aRA. RNA was extracted from skin by using the RNeasy Mini Kit (Qiagen) and quantified with a Qubit (Life Technologies). RNA was reverse-transcribed by using the SuperScript II Reverse Transcriptase Kit (Life Technologies). TaqMan probes and primers (Life Technologies) were used for detection and quantification on a ViiA 7 real-time PCR instrument (Life Technologies) according to the manufacturer’s protocols. Specifically, the TaqMan Mouse Immune Array (Life Technologies) in microfluidic card format was used in addition to the primer and probe sets listed in Table S2. Target mRNA was normalized to the β2 microglobulin (B2M) mRNA. Nine C5aRA-treated mice and eight iC5aRA-treated mice, over two independent experiments, were used for these experiments. Significance was assessed by a two-tailed t test whereby P values <0.05 were considered to be significant.
Quantification of bacterial 16S rRNA genes was performed as previously described (39), and values were normalized to total bacterial 16S rRNA gene copy number. Significance was assessed by using a paired two-tailed t test whereby P values <0.05 were considered to be significant.
RNA Sequencing and Analysis.
We obtained skin from the dorsum of 8- to 10-wk-old male C57BL/6J mice raised in GF conditions (n = 4) in the Penn Gnotobiotic Mouse Facility and raised conventionally in specific pathogen-free conditions (CONV; n = 4). Total RNA was extracted from the skin by using the RNeasy Mini Kit (Qiagen), and poly-A–selected RNA-seq libraries were prepared for 100 bp paired end sequencing on the Illumina HiSEq 2000 using the Tru-Seq (Illumina) mRNA-seq V2 kit, with 500 ng total RNA starting material. See SI Materials and Methods for details of RNA-seq and processing. Transcripts encoding genes that were categorized under GO terms “complement activation” and “complement binding” were searched and pulled from the dataset for analysis of differential expression. Of the 58 total genes represented by those GO terms, we detected 33 genes in our dataset that were expressed at the minimum threshold set. Significance of differential expression was assessed by a two-tailed t test, whereby P < 0.05 was deemed as significant.
Histology and Immunohistochemistry.
Skin biopsies were collected from the dorsal side of the experimental mice and their controls, fixed in 10% (wt/vol) formalin, embedded in paraffin, and sectioned at 6 µm. Prepared tissue sections were subjected to immunostaining for detection of CD3, a pan-lymphocyte marker, and F4/80, a marker for macrophages. Briefly, the sections were deparaffinized with xylene and rehydrated in downgraded alcohol. Heat-inactivated antigen retrieval was performed by incubating the tissue sections in 10 mM sodium citrate buffer, pH 6.0, and subsequently washed in PBS solution. To quench endogenous peroxidase, tissue sections were incubated in 3% (wt/vol) H2O2, washed, and blocked with 10% (vol/vol) normal goat serum for 1 h at room temperature. Thereafter, the sections were incubated with a primary Ab, rabbit anti-mouse CD3 (1:50; Abcam) and rat anti-mouse F4/80 (1:40; Abcam) at 4 °C overnight in parallel with the negative control in which the primary antibody Abs were omitted. Following multiple washes, secondary antibodies, goat anti-rabbit IgG-HRP and goat anti-rat-HRP (both from Abcam) were applied for 1 h at room temperature and then washed. The signal was amplified with DAB, counterstained with hematoxylin, and coverslipped. CD3+ and F4/80+ cells were counted in three to five fields per tissue section at 400× magnification, two tissue sections per mouse, and three mice per treatment (C5aRA or iC5aRA).
Supplementary Material
Acknowledgments
We thank University of Pennsylvania Skin Disease Research Center (SDRC) Core A and Stephen Prouty and Tzvete Dentchev for assistance with and interpretation of histology and immunohistochemistry; the National Institutes of Health (NIH) Intramural Sequencing Center [National Human Genome Research Institute (NHGRI)], the University of Pennsylvania Next Generation Sequencing Core, and Rick Bushman and his laboratory (University of Pennsylvania) for sequencing support; David Artis, Brian Kim, and Jonathan Brestoff (University of Pennsylvania) for germ-free mouse tissues; Brendan Hodkinson (E.A.G. laboratory) for assistance with processing RNA sequencing data; Julia Segre (NIH/NHGRI) for her underlying contributions; and George Hajishengallis and Edimara Reis (University of Pennsylvania) for critical review of the manuscript. This work was supported by NIH Grants AR060873 (to E.A.G.) and AI068730 (to J.D.L.); and the University of Pennsylvania SDRC, which was supported by NIH Grant AR057217. Germ-free mice were provided by the Penn Gnotobiotic Mouse Facility, which is supported by NIH Grants AI095466, AI09560, and AI097333 (to David Artis); Mucosal Immunology Studies Team consortium, which was supported by National Institute of Allergy and Infectious Diseases Grant U01 AI095608; and the University of Pennsylvania Perelman School of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.J.T. is a guest editor invited by the Editorial Board.
Data deposition: The sequence reported in this paper have been deposited in the GenBank database (accession no. KF501886–KF509852).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307855110/-/DCSupplemental.
References
- 1.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337(6098):1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai Y, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15(12):1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104(8):2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grice EA, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18(7):1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaser MJ, et al. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2013;7(1):85–95. doi: 10.1038/ismej.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105(46):17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med. 2012;4(10):77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redel H, et al. Quantitation and composition of cutaneous microbiota in diabetic and nondiabetic men. J Infect Dis. 2013;207(7):1105–1114. doi: 10.1093/infdis/jit005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: A key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettigrew HD, Teuber SS, Gershwin ME. Clinical significance of complement deficiencies. Ann N Y Acad Sci. 2009;1173:108–123. doi: 10.1111/j.1749-6632.2009.04633.x. [DOI] [PubMed] [Google Scholar]
- 15.Kotnik V. Complement in skin diseases. Acta Dermatovenerol Alp Panonica Adriat. 2011;20(1):3–11. [PubMed] [Google Scholar]
- 16.Isolauri E, et al. Altered expression of IgG and complement receptors indicates a significant role of phagocytes in atopic dermatitis. J Allergy Clin Immunol. 1997;99(5):707–713. doi: 10.1016/s0091-6749(97)70034-4. [DOI] [PubMed] [Google Scholar]
- 17.Tagami H. The role of complement-derived mediators in inflammatory skin diseases. Arch Dermatol Res. 1992;284(suppl 1):S2–S9. doi: 10.1007/BF00638232. [DOI] [PubMed] [Google Scholar]
- 18.Scott DG, Cunliffe WJ, Gowland G. Activation of complement-a mechanism for the inflammation in acne. Br J Dermatol. 1979;101(3):315–320. doi: 10.1111/j.1365-2133.1979.tb05625.x. [DOI] [PubMed] [Google Scholar]
- 19.Hajishengallis G, Lambris JD. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol. 2010;31(4):154–163. doi: 10.1016/j.it.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köhl J. Drug evaluation: The C5a receptor antagonist PMX-53. Curr Opin Mol Ther. 2006;8(6):529–538. [PubMed] [Google Scholar]
- 21.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinicke AT, et al. C5aR regulates intestinal microbiota composition and controls the induction of gastrointestinal allergic hypersensitivity. Immunobiology. 2012;217(11):1147. [Google Scholar]
- 23.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guenther LC. Inherited disorders of complement. J Am Acad Dermatol. 1983;9(6):815–839. doi: 10.1016/s0190-9622(83)70195-7. [DOI] [PubMed] [Google Scholar]
- 27.Rosenfeld SI, Kelly ME, Leddy JP. Hereditary deficiency of the fifth component of complement in man. I. Clinical, immunochemical, and family studies. J Clin Invest. 1976;57(6):1626–1634. doi: 10.1172/JCI108433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, et al. Inherited human complement C5 deficiency. Nonsense mutations in exons 1 (Gln1 to Stop) and 36 (Arg1458 to Stop) and compound heterozygosity in three African-American families. J Immunol. 1995;154(10):5464–5471. [PubMed] [Google Scholar]
- 29.Wang H, et al. Activated macrophages are essential in a murine model for T cell-mediated chronic psoriasiform skin inflammation. J Clin Invest. 2006;116(8):2105–2114. doi: 10.1172/JCI27180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomura I, et al. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: A gene microarray analysis. J Allergy Clin Immunol. 2003;112(6):1195–1202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- 31.Pelletier M, et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2010;115(2):335–343. doi: 10.1182/blood-2009-04-216085. [DOI] [PubMed] [Google Scholar]
- 32.Hyun YM, Lefort CT, Kim M. Leukocyte integrins and their ligand interactions. Immunol Res. 2009;45(2-3):195–208. doi: 10.1007/s12026-009-8101-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebwohl M, et al. Efalizumab Study Group A novel targeted T-cell modulator, efalizumab, for plaque psoriasis. N Engl J Med. 2003;349(21):2004–2013. doi: 10.1056/NEJMoa030002. [DOI] [PubMed] [Google Scholar]
- 34.Harris CL, Heurich M, Rodriguez de Cordoba S, Morgan BP. The complotype: Dictating risk for inflammation and infection. Trends Immunol. 2012;33(10):513–521. doi: 10.1016/j.it.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mrowietz U, et al. Psoriasis scales contain C5a as the predominant chemotaxin for monocyte-derived dendritic cells. Exp Dermatol. 2001;10(4):238–245. doi: 10.1034/j.1600-0625.2001.100403.x. [DOI] [PubMed] [Google Scholar]
- 36.Takematsu H, Ohkohchi K, Tagami H. Demonstration of anaphylatoxins C3a, C4a and C5a in the scales of psoriasis and inflammatory pustular dermatoses. Br J Dermatol. 1986;114(1):1–6. doi: 10.1111/j.1365-2133.1986.tb02773.x. [DOI] [PubMed] [Google Scholar]
- 37.McFadden JP, Baker BS, Powles AV, Fry L. Psoriasis and streptococci: The natural selection of psoriasis revisited. Br J Dermatol. 2009;160(5):929–937. doi: 10.1111/j.1365-2133.2009.09102.x. [DOI] [PubMed] [Google Scholar]
- 38.Grice EA, Segre JA. Interaction of the microbiome with the innate immune response in chronic wounds. Adv Exp Med Biol. 2012;946:55–68. doi: 10.1007/978-1-4614-0106-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grice EA, et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defense response. Proc Natl Acad Sci USA. 2010;107(33):14799–14804. doi: 10.1073/pnas.1004204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finch AM, et al. Low-molecular-weight peptidic and cyclic antagonists of the receptor for the complement factor C5a. J Med Chem. 1999;42(11):1965–1974. doi: 10.1021/jm9806594. [DOI] [PubMed] [Google Scholar]
- 41.Markiewski MM, et al. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9(11):1225–1235. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.