Significance
In this paper we describe a unique method for selection of agonist antibodies that regulate stem cell fate. Unbiased combinatorial libraries (not preselected by phage panning) were used and, thus, no constraints were placed on the discovery process. Remarkably, we found that a single agonist antibody against the alpha chains of integrins can induce human stem cells to become dendritic cells. These studies are important because they help deconvolute an otherwise complex developmental process. This approach is analogous to forward genetics and should be widely used to identify previously undescribed proteins involved in important cellular pathways.
Keywords: intracellular combinatorial antibodies, phenotypic selections
Abstract
When combinatorial antibody libraries are rendered infectious for eukaryotic cells, the integrated antibody genotype and cellular phenotype become permanently linked and each cell becomes a selection system unto itself. These systems should be ideal for the identification of proteins and pathways that regulate differentiation so long as selection systems can be devised. Here we use a selection system based on the ability of secreted antibodies to alter the morphology of colonies expressing them when grown in soft agar. Importantly, this approach is different from all previous studies in that it used a pure discovery format where unbiased libraries that were not preselected against any known protein were used as probes. As such, the strategy is analogous to classical forward genetic approaches except that it operates directly at the protein level. This approach led to the identification of integrin-binding agonist antibodies that efficiently converted human stem cells to dendritic cells.
Recently we developed combinatorial antibody libraries that are infectious for eukaryotic cells by incorporating them into lentiviruses (1). After infection, genotype and phenotype become linked because the antibody genes are incorporated into the genome and expressed and the cell, itself, becomes the reporter. If the target is unknown, the discovery potential is similar to forward genetic approaches with the advantage that target identification is greatly simplified.
When functional antibodies are desired, proteins present in cells have an advantage as targets over isolated molecules because they have physiologically relevant topologies and, most importantly, their proper integration into cellular pathways allows for phenotypic selections. We suggested that phenotypic screening using infectious antibody libraries is a general method and could, in principle, be used to select agonists for known or unknown proteins that are present in any cellular pathway for which a selection system is available. As such, the method should be limited only by the creativity one can bring to the screening process. To date, we have isolated antibodies that are agonists for cytokine receptors as well as those that induce transdifferentiation of stem cells (2, 3).
One of the general observations in biology is that cells change morphology during development and differentiation. If an unbiased selection format that harnesses this general feature of cell development could be devised, it could be a novel screening method for selection of antibodies that induce differentiation by using heretofore-unknown pathways and/or activating known pathways using novel protein–protein interactions. Because the target space is likely to be large and the pathways mostly unknown, this is much more difficult than our previous selections of antibody agonists that are cytokine phenocopies from preselected libraries that were biased for binding to known receptors. Also, when unbiased libraries are used in the search for unknown protein targets, the frequency of recovered agonists will necessarily be lower than that seen with preselected libraries. On the other hand, because the targets are unknown, the potential for new discoveries is high.
The growth and morphology of colonies of cells growing in soft agar have been used widely as measures of the malignant state of cancer cells that have lost anchorage dependence or to assay for the effects of growth factors on cellular proliferation (4–7). We reasoned that this approach could be extended to the study of hematopoietic cells, where the morphology of their colonies would vary as a function of their differentiated state. Thus, agonist antibodies that induce morphological changes in cell colonies could be selected from cells infected with antibody libraries. This approach could inform as to whether the scope of a method that has already been used successfully for selection of cytokine phenocopies can be extended to the more general biological problem of correlating protein components of pathways with cellular phenotypes (1–3).
Here we show that infection of cells with unbiased combinatorial antibody libraries causes growth of colonies in soft agar that exhibit a variety of morphologies. Using mass spectroscopy, the targets for the recovered antibodies were shown to be channels or integrins. In each case, the antibodies function as agonists, as revealed by their activation of downstream pathways. The integrin agonist antibody that was isolated from the morphogenic screen induces human stem cells to differentiate into cells of the macrophage lineage to form dendritic cells. The power of the overall selection process was shown by the fact that it was accompanied by convergent evolution in which the selected antibodies had the integrin-binding sequence Arg-Gly-Asp (RGD) in their complementarity determining regions-heavy chain 3 (CDR-H3). When the RGD sequence was mutated to Arg-Gly-Glu (RGE), the agonist activity was lost.
Results
Strategy of Antibody Selection from Unbiased Combinatorial Antibody Libraries.
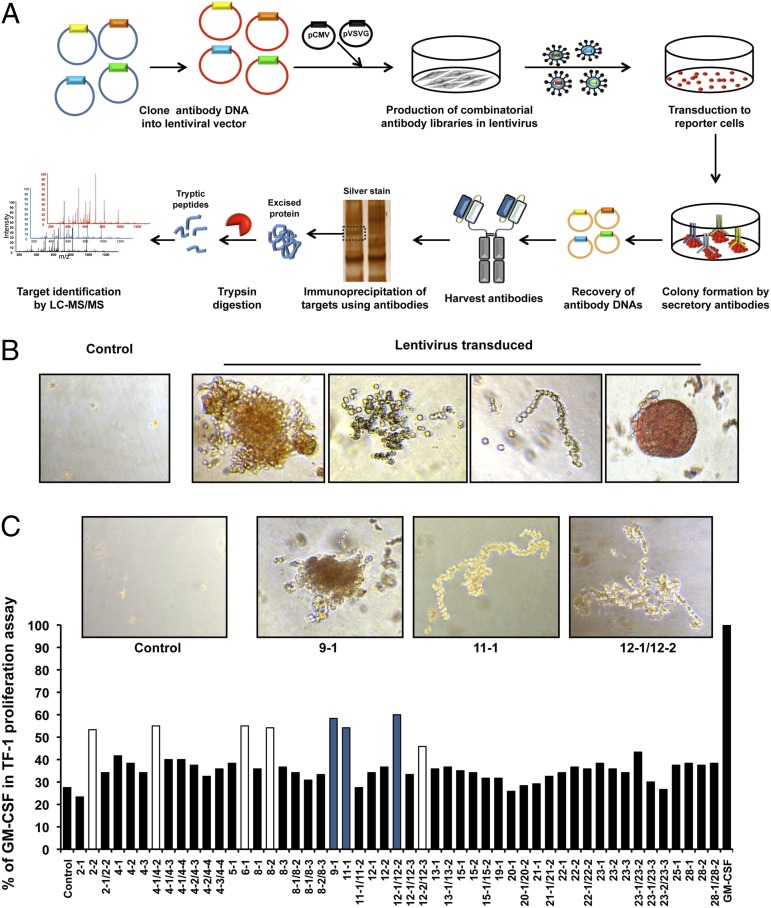
To select antibody agonists for molecules that are not known to influence cellular phenotypes, we devised a method based on infectious combinatorial antibody libraries that does not use conventional antibody panning as an initial step (Fig. 1A). A naïve human combinatorial scFv-Fc antibody library that contained 108 members was incorporated into lentiviral vectors. TF-1 erythroblast cells infected with the library were incorporated into methylcellulose agar for morphogenic screening. The antibody format that was used caused the antibodies to be secreted into the agar surrounding the cells (1). Compared with the control, the cells infected with members of the antibody library formed colonies with unique morphologies that included amorphous, scattered, linear, or round phenotypes (Fig. 1B).
Fig. 1.
Scheme for the unbiased selection of active antibodies against cellular targets from combinatorial libraries. (A) Lentiviral antibody libraries were constructed from naïve human combinatorial antibody libraries. Plasmids of lentiviral antibody libraries and virus packaging were used to transiently transfect HEK293T cells. TF-1 erythroblast cells were infected with lentiviral antibody libraries. The resulting cells were grown on methylcellulose agar for morphogenic screening. Thirty colonies that showed unusual and different morphologies were picked after 2 wk and antibody genes were recovered by PCR. After verification of the activity of the antibodies by the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) cell proliferation assay, each antibody was incubated with cell lysates followed by collection of the complexes on protein A/G beads. The beads were collected and washed, and the bound proteins were eluted by acid (pH 2.8) and separated on SDS/PAGE gels that were silver-stained. The bands of gel were sliced and analyzed by nano-LC-MS/MS after trypsinization. (B) Representative morphologies of colonies. (C) All possible combinations of antibody genes isolated from the same colonies were transfected into HEK293T cells. TF-1 cell proliferation was tested using the conditioned medium from HEK293T cells 48 h after transfection of antibodies.
After 2 wk in culture, we harvested 30 colonies that showed unique morphologies that included round, amorphous, scattered, and linear phenotypes. By contrast, control cells exhibited only a few background small clumps of cells. The antibody genes present in the cells from each colony with a unique morphology were recovered by PCR (Fig. 1A). A total of 20 antibody genes were recovered. The genes encoding the antibody heavy-chain CDR3 (CDR-H3) regions were analyzed by DNA sequencing to determine the number of different antibodies present in each clone (Table S1). Each colony had between one and four different antibodies (Table S1). Soluble antibodies encoded by the lentiviruses were generated by transient transfection of HEK293T cells to generate either single or a combination of two antibodies that originated from the different clones. In total, 51 antibodies or antibody combinations were tested for their ability to influence the proliferation of TF-1 cells. Eight antibodies had ∼50% of the activity of GM-CSF (Fig. 1C). Interestingly, three of the most active antibodies were bispecific. We selected arbitrarily three antibodies (9-1, 11-1, and 12-1/12-2) for further study because they originated from colonies with the most unusual morphologies (Fig. 1C).
Identification of Target Proteins.
Before target identification, we wished to ensure that the selected antibodies were agonists. The purified antibodies produced by transfection of the antibody genes encoding three active antibodies (9-1, 11-1, and the 12-1/12-2 combination) into 293F cells were tested to determine whether they activated cellular functions. They all increased the proliferation of TF-1 cells and induced phosphorylation of key signaling molecules such as AKT and ERK in these cells (Fig. S1 A and B). The 9-1 antibody uniquely activated the phosphorylation of STAT5, as did GM-CSF. Importantly, neither 12-1 nor 12-2 alone had any effect above control, but a combination of the two significantly increased cell proliferation.
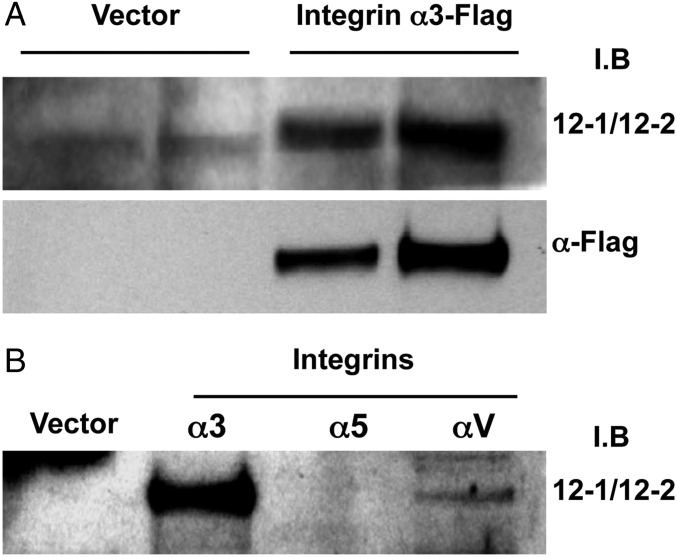
To identify the proteins that the agonist antibodies bound to, the isolated antibodies were used as probes to isolate the targets that were then identified by mass spectrometry analysis (Fig. S1 C and D and Table S2). Two criteria were used to identify the target. A peptide of the correct sequence had to be found, and it had to originate from a protein of the expected molecular weight. The antibodies from the amorphous, scattered, and linear colonies were shown to bind to the proton-gated channel HVCN1, the integrin α3, and the transient receptor potential cation channel TRPM7, respectively. Because integrins are well-characterized and known for their importance in pathophysiology, we selected this target for further study. We used Western blot analysis of lysates from cells overexpressing human integrin α3 to confirm that the 12-1/12-2 bispecific antibody interacts with the human integrin α3-chain subunit (Fig. 2A). An analysis of binding to other integrin α-chains showed that the antibody is not absolutely specific for α3-chains in that it binds to αV as well, albeit less strongly (Fig. 2B).
Fig. 2.
Verification of the interaction between 12-1/12-2 antibodies and integrins. (A) Lysates of HEK293T cells that overexpressed human integrins containing α3-chains were separated on SDS/PAGE and Western-blotted using 12-1/12-2 antibodies or anti-Flag antibody. (B) Lysates from cells overexpressing integrin chains α5 and αv as well as α3.
Antibody-Induced Differentiation of Human Bone Marrow CD34+ Cells into Migratory Cells.
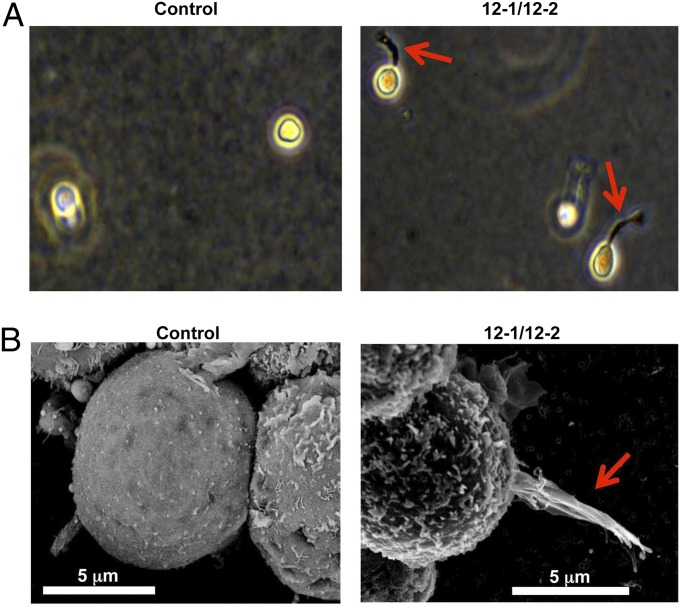
TF-1 cells are widely used for studies related to the functions of various cytokines (8, 9). However, TF-1 cells are of the erythroblast lineage and, although useful for initial studies of agonist activity, are not suitable for study of a broad range of phenotypes. Thus, we studied the potential of our antibodies to regulate cellular fates in human bone marrow CD34+ stem cells. To study this, we incorporated human bone marrow CD34+ cells into methylcellulose agar containing 12-1/12-2 antibodies. After 1 wk in culture, cells exposed to 12-1/12-2 antibodies displayed signs of differentiation in that they showed marked formation of distinct podia (Fig. 3A). These features were not seen in control cells. To further characterize the nature of these podia, the cells were analyzed by scanning electron microscopy (SEM) (Fig. 3B). Again, compared with control cells, those treated with antibodies 12-1/12-2 showed increased formation of podia that exhibited classical morphology (10, 11) (Fig. 3B).
Fig. 3.
Antibody increases podia formation from human bone marrow CD34+ cells. (A) CD34+ cells were cultured in methylcellulose agar with antibodies (50 μg/mL) or PBS for 6 d. Morphologies of cells were analyzed by bright-field microscopy. (B) CD34+ cells grown in methylcellulose agar were analyzed by SEM. The arrows indicate podia of CD34+ cells.
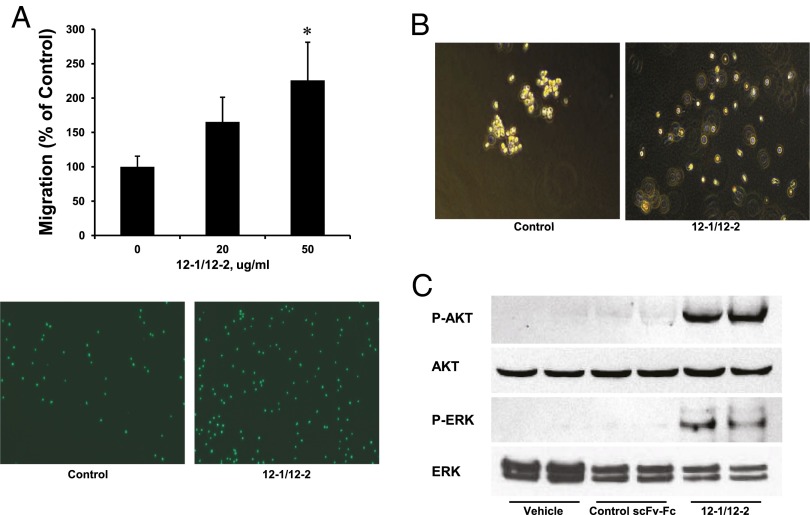
One major phenotype known to accompany integrin signaling is an increase in cell migration. We used a transwell migration assay to show that treatment of CD34+ cells with 12-1/12-2 antibodies increased their migration (Fig. 4A). The antibodies also caused the cells to scatter in soft agar (Fig. 4B). This migratory behavior correlates with the induction of podia and the scattered phenotype that allowed them to be selected in the first place. This migratory phenotype in the CD34+ cells was accompanied by the strong phosphorylation of AKT and ERK that are events known to be keys to downstream integrin signaling (Fig. 4C) (12, 13).
Fig. 4.
Antibody increases the motility of human bone marrow CD34+ cells. (A) CD34+ cells were incubated with PBS or antibodies for 16 h in transwell chambers. The values are the mean +/− SE of three independent experiments. *P < 0.05 versus basal values. (Lower) Fluorescent dye-labeled cells from the lower chamber by fluorescent microscopy. (B) CD34+ cells were cultured in methylcellulose agar with antibody or PBS for 6 d. Images are from bright-field microscopy analysis. (C) CD34+ cells were incubated with PBS or antibodies for 10 min at 37 °C and analyzed by Western blots with anti–p-AKT, AKT, p-ERK, and ERK antibodies.
Lineage Specification by Induction of Dendritic Cells.
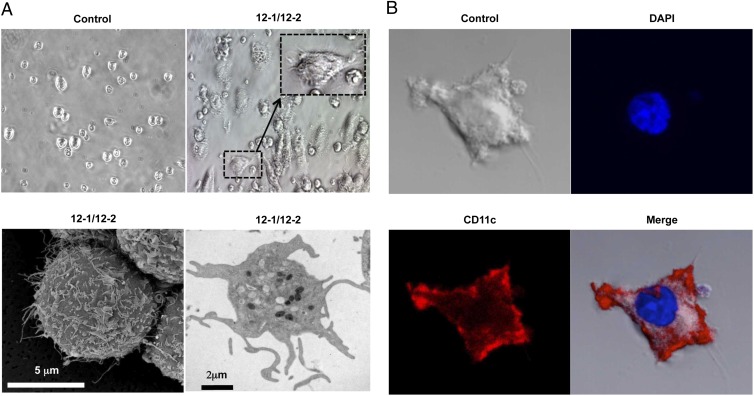
Because bone marrow CD34+ cells contain “pluripotent” stem cells that give rise to all cell types in blood, extracellular stimuli such as immune cytokines cause them to differentiate into various lineages (14–20). Thus, we wondered whether the formation of podia and migratory behavior in stem cells induced by 12-1/12-2 antibodies were associated with any particular lineage specification in these stem cells. A more detailed morphological and immunocytochemical analysis showed that antibodies 12-1/12-2 potently increased the differentiation of CD34+ cells into cells that exhibited a dendritic cell-like morphology that is very different from the parental cells. Analysis by bright-field microscopy showed the presence of long and multiple hairy-like dendritic extensions only on the induced cells (Fig. 5A, dashed box). SEM and transmission electron microscopy (TEM) confirmed that the cells had the typical morphology of dendritic cells that, again, differed markedly from the smooth surfaces of the uninduced CD34+ stem cells (Fig. 5A). To confirm that the induced cells were of dendritic lineage, we carried out an immunocytochemistry analysis using anti-CD11c antibodies as a marker of dendritic cells. The cells strongly expressed the dendritic cell marker CD11c on their surface (Fig. 5B).
Fig. 5.
Antibody induces the differentiation of dendritic cells from human bone marrow CD34+ cells. CD34+ cells were cultured on 24-well tissue-culture plates with 12-1/12-2 antibodies or PBS for 10 d. (A) (Upper) The common type of CD34+ and dendritic cell-like morphology by bright-field microscopy. (Lower) Dendritic cell-like morphology by SEM and TEM. (B) PBS- or antibody-treated CD34+ cells were stained with phycoerythrin (PE)-conjugated anti-human CD11c antibody or DAPI and analyzed by confocal microscopy.
Convergent Evolution.
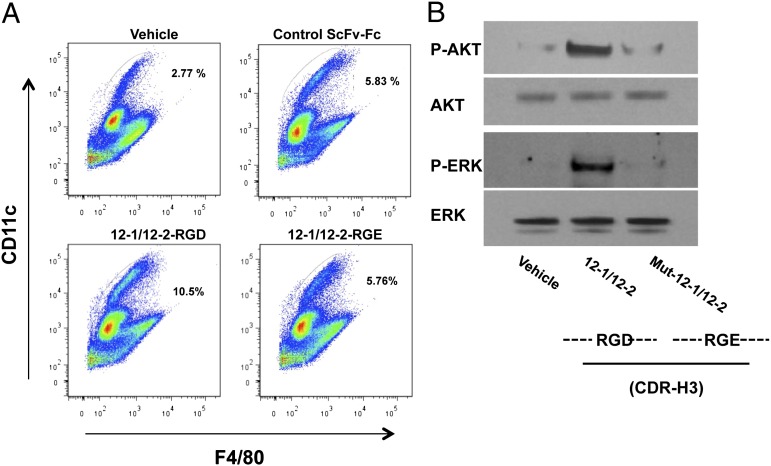
Interestingly, the antibody 12-2 component of the 12-1/12-2 bispecific heterodimer contains the Arg-Gly-Asp (RGD) integrin recognition motif of the natural ligands within the CDR-H3 (Table S1). To examine the contribution of the RGD motif of CDR-H3 to the cellular function of antibodies 12-1/12-2, this sequence was mutated to RGE by site-directed mutagenesis. The D (Asp)-to-E (Glu) exchange within the RGD motif was shown to markedly reduce the induction of dendritic cells by the antibodies. However, the macrophage marker F4/80 was not present (Fig. 6A). Also, the RGD-to-RGE mutation greatly reduced the ability of the antibody to induce phosphorylation of the major AKT and ERK integrin-signaling molecules in CD34+ stem cells (Fig. 6B). These findings strongly suggest that the RGD motif in the 12-1/12-2 complex is critical for full integrin activation by these antibodies.
Fig. 6.
RGD motif of the antibody is critical for the activity of the antibody. (A) CD34+ cells were treated with 12-1/12-2 antibodies (containing RGD) or a mutant form of 12-1/12-2 antibodies (containing RGE), an inactive control antibody, or PBS for 10 d. These cells were stained with a PE-conjugated anti-human CD11c antibody and FITC-conjugated anti-human F4/80 antibody and analyzed by flow cytometry. CD11c and F4/80 are specific markers of dendritic cells and macrophages, respectively. (B) CD34+ cells were incubated with PBS, 12-1/12-2 antibodies, or the mutant form of 12-1/12-2 antibodies for 10 min at 37 °C and analyzed by Western blotting with anti–p-AKT, AKT, p-ERK, and ERK antibodies.
Discussion
In stem cell biology, we have witnessed the rapid generation of powerful approaches that allow regulation of the pluripotency and lineage specification of cells (2, 3, 21–25). These methods are largely based on our ability to alter the genetic program of cells and an ever-increasing knowledge of protein factors that regulate cell differentiation (25). The method described here is an orthogonal approach that may add to this already-robust menu. In this sense, antibodies have certain advantages over other approaches in that they can operate by either gain or loss of function.
There are a variety of formats that allow study of infectious antibody libraries that perturb cellular phenotypes. In the broadest sense, antibody libraries can be selected initially by phage panning for binding to a particular molecule or, as used here, can be unbiased. Each format is interesting, because one can discover new molecules or add to the knowledge of how already-known molecules function (1–3). Arguably, however, the use of unbiased libraries is more important because, as a pure discovery format, it is more likely to lead to heretofore-unrecognized components of cellular pathways. In the limiting case, discovery of any new agonists should yield useful reagents for the study of cellular pathways. However, in terms of medicine, discovery of agonist antibodies that react with proteins associated with the plasma membrane may be more important, because they are more likely to be therapeutically useful.
An interesting aspect of the selection from antibody libraries is that it is an evolutionary process that sorts through large numbers of molecules for fitness. The fitness can be superior binding to an antigen or, as in the case reported here, function. Given the input of large numbers of candidate molecules coupled with the power of selection, it is perhaps not surprising that convergence to solutions that are the result of natural evolution are seen. In general, when convergence is observed, it indicates that the unbiased selection process was powerful because the input diversity was large enough to approach a “best solution” in real time. For instance, in earlier studies, antibodies that bind to magnetite were selected from combinatorial libraries and shown to use essentially the same protein sequence as the natural magnetite-binding protein of bacteria (26). Also, integrin-binding antibodies that use RGD were selected previously by us and others using phage panning (27–29). Importantly, the RGD-containing antibodies previously selected by simple phage binding were antagonists, whereas those selected here using phenotypic screens are agonists. The identification of agonists instead of antagonists illustrates another aspect of our method where identification of agonists is favored because phenotypic selections favor gain of function.
The data presented here are in accord and add to the experiments and concepts of others. It is widely believed that stem cell fate is regulated in niches by integration of signals from soluble factors and elements of the extracellular matrix (ECM) (13, 30–34). In particular, interactions between the ECM and cell-surface integrins regulate stem cell fates by activating several signaling pathways including ERK and AKT, as was also seen in the present experiments. Thus, a picture emerges of a complex milieu of soluble and insoluble molecules that act in concert. Our experiments may help deconvolute this complex process, because they show that a single event involving a particular protein–protein interaction at the cell surface is sufficient to regulate lineage specification. It seems reasonable to suggest that factors in the niche bring stem cells to the stage where they are competent to receive the ECM signals that regulate their lineage specifications. In our specific case, all of the early processes that are required to form cells of the CD34+ phenotype may be a prelude to the late activation by integrins that determine the ultimate fates, such as the formation of dendritic cells.
Finally, the observation that simple integrin activation at the cell surface is sufficient to induce dendritic cell formation from CD34+ stem cells underscores the utility of our forward proteomics platform for identification. Interestingly, the pathway to dendritic cell formation reported here appears to differ from that initiated by GM-CSF because induction of dendritic cell formation by the antibody is independent of phosphorylation of the key transcription factor, STAT5 (Fig. S1B).
Materials and Methods
The anti-STAT5, anti–p-STAT5 (Tyr694), anti-AKT, anti–p-AKT (Ser473), anti-ERK1/2, and anti–p-ERK1/2 (Thr-202/Tyr-204) antibodies were used to analyze the signal transduction pathways. All of the antibodies were purchased from Cell Signaling. The total lysates of HEK293T cells that overexpressed human integrin α3, α5, or αV genes were obtained from Novus Biologicals. All recombinant cytokines were purchased from R&D Systems. Polybrene was from Millipore. Cell-culture media and FBS were from Gibco-Invitrogen. The methods for cell culture, antibody selection and characterization, and antigen identification are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Malcolm Wood for the EM studies and Linh Hoang for assistance with mass spectrometry analysis.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313671110/-/DCSupplemental.
References
- 1.Zhang H, Wilson IA, Lerner RA. Selection of antibodies that regulate phenotype from intracellular combinatorial antibody libraries. Proc Natl Acad Sci USA. 2012;109(39):15728–15733. doi: 10.1073/pnas.1214275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie J, Zhang H, Yea K, Lerner RA. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc Natl Acad Sci USA. 2013;110(20):8099–8104. doi: 10.1073/pnas.1306263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, et al. Selecting agonists from single cells infected with combinatorial antibody libraries. Chem Biol. 2013;20(5):734–741. doi: 10.1016/j.chembiol.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Maag D, et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc Natl Acad Sci USA. 2011;108(4):1391–1396. doi: 10.1073/pnas.1017831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takezawa K, et al. Sorafenib inhibits non-small cell lung cancer cell growth by targeting B-RAF in KRAS wild-type cells and C-RAF in KRAS mutant cells. Cancer Res. 2009;69(16):6515–6521. doi: 10.1158/0008-5472.CAN-09-1076. [DOI] [PubMed] [Google Scholar]
- 6.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449(7165):1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Indo K, Matsuoka T, Nakasho K, Funahashi I, Miyaji H. Reversible induction of anchorage independent growth from normal mouse epidermal keratinocytes, MSK-C3H-NU, in soft agar medium by 12-O-tetradecanoylphorbol-13-acetate and epidermal growth factor. Cancer Res. 1988;48(6):1566–1570. [PubMed] [Google Scholar]
- 8.Kitamura T, et al. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140(2):323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 9.Chrétien S, et al. Erythropoietin-induced erythroid differentiation of the human erythroleukemia cell line TF-1 correlates with impaired STAT5 activation. EMBO J. 1996;15(16):4174–4181. [PMC free article] [PubMed] [Google Scholar]
- 10.Fruehauf S, Srbic K, Seggewiss R, Topaly J, Ho AD. Functional characterization of podia formation in normal and malignant hematopoietic cells. J Leukoc Biol. 2002;71(3):425–432. [PubMed] [Google Scholar]
- 11.Lamb RF, et al. Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr Biol. 1997;7(9):682–688. doi: 10.1016/s0960-9822(06)00295-8. [DOI] [PubMed] [Google Scholar]
- 12.King WG, Mattaliano MD, Chan TO, Tsichlis PN, Brugge JS. Phosphatidylinositol 3-kinase is required for integrin-stimulated AKT and Raf-1/mitogen-activated protein kinase pathway activation. Mol Cell Biol. 1997;17(8):4406–4418. doi: 10.1128/mcb.17.8.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285(5430):1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 14.Brugger W, et al. Ex vivo expansion of enriched peripheral blood CD34+ progenitor cells by stem cell factor, interleukin-1 beta (IL-1 beta), IL-6, IL-3, interferon-gamma, and erythropoietin. Blood. 1993;81(10):2579–2584. [PubMed] [Google Scholar]
- 15.Van Epps DE, et al. Harvesting, characterization, and culture of CD34+ cells from human bone marrow, peripheral blood, and cord blood. Blood Cells. 1994;20(2-3):411–423. [PubMed] [Google Scholar]
- 16.Boitano AE, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329(5997):1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satterthwaite AB, Burn TC, Le Beau MM, Tenen DG. Structure of the gene encoding CD34, a human hematopoietic stem cell antigen. Genomics. 1992;12(4):788–794. doi: 10.1016/0888-7543(92)90310-o. [DOI] [PubMed] [Google Scholar]
- 18.Asheuer M, et al. Human CD34+ cells differentiate into microglia and express recombinant therapeutic protein. Proc Natl Acad Sci USA. 2004;101(10):3557–3562. doi: 10.1073/pnas.0306431101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki K, et al. Characterization of monocyte-macrophage-lineage cells induced from CD34+ bone marrow cells in vitro. Int J Hematol. 2007;85(5):384–389. doi: 10.1532/IJH97.06213. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J, Emerson SG. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene. 2002;21(21):3295–3313. doi: 10.1038/sj.onc.1205318. [DOI] [PubMed] [Google Scholar]
- 21.Willerth SM, Rader A, Sakiyama-Elbert SE. The effect of controlled growth factor delivery on embryonic stem cell differentiation inside fibrin scaffolds. Stem Cell Res (Amst) 2008;1(3):205–218. doi: 10.1016/j.scr.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13(5):497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10(4):440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Barbas CF, III, Rosenblum JS, Lerner RA. Direct selection of antibodies that coordinate metals from semisynthetic combinatorial libraries. Proc Natl Acad Sci USA. 1993;90(14):6385–6389. doi: 10.1073/pnas.90.14.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felding-Habermann B, et al. Combinatorial antibody libraries from cancer patients yield ligand-mimetic Arg-Gly-Asp-containing immunoglobulins that inhibit breast cancer metastasis. Proc Natl Acad Sci USA. 2004;101(49):17210–17215. doi: 10.1073/pnas.0407869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 29.Salsmann A, Schaffner-Reckinger E, Kabile F, Plançon S, Kieffer N. A new functional role of the fibrinogen RGD motif as the molecular switch that selectively triggers integrin alphaIIbbeta3-dependent RhoA activation during cell spreading. J Biol Chem. 2005;280(39):33610–33619. doi: 10.1074/jbc.M500146200. [DOI] [PubMed] [Google Scholar]
- 30.Sun Y, Chen CS, Fu J. Forcing stem cells to behave: A biophysical perspective of the cellular microenvironment. Annu Rev Biophys. 2012;41:519–542. doi: 10.1146/annurev-biophys-042910-155306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabodi S, et al. Integrin regulation of epidermal growth factor (EGF) receptor and of EGF-dependent responses. Biochem Soc Trans. 2004;32(Pt 3):438–442. doi: 10.1042/BST0320438. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475(7356):316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armant DR. Blastocysts don’t go it alone. Extrinsic signals fine-tune the intrinsic developmental program of trophoblast cells. Dev Biol. 2005;280(2):260–280. doi: 10.1016/j.ydbio.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SH, Turnbull J, Guimond S. Extracellular matrix and cell signalling: The dynamic cooperation of integrin, proteoglycan and growth factor receptor. J Endocrinol. 2011;209(2):139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.