Significance
In this study, we have identified a type of protein that participates in the formation of the tubular endoplasmic reticulum. This protein, protrudin, harbors hydrophobic hairpins that generate high curvature in endoplasmic reticulum tubules, as well as several domains that may link the tubular endoplasmic reticulum to the plasma membrane or other organelles.
Abstract
Hereditary spastic paraplegias are inherited neurological disorders characterized by progressive lower-limb spasticity and weakness. Although more than 50 genetic loci are known [spastic gait (SPG)1 to -57], over half of hereditary spastic paraplegia cases are caused by pathogenic mutations in four genes encoding proteins that function in tubular endoplasmic reticulum (ER) network formation: atlastin-1 (SPG3A), spastin (SPG4), reticulon 2 (SPG12), and receptor expression-enhancing protein 1 (SPG31). Here, we show that the SPG33 protein protrudin contains hydrophobic, intramembrane hairpin domains, interacts with tubular ER proteins, and functions in ER morphogenesis by regulating the sheet-to-tubule balance and possibly the density of tubule interconnections. Protrudin also interacts with KIF5 and harbors a Rab-binding domain, a noncanonical FYVE (Fab-1, YGL023, Vps27, and EEA1) domain, and a two phenylalanines in an acidic tract (FFAT) domain and, thus, may also function in the distribution of ER tubules via ER contacts with the plasma membrane or other organelles.
Hereditary spastic paraplegias (HSPs) are a group of inherited neurological disorders with the cardinal feature of a length-dependent axonopathy of corticospinal motor neurons, resulting in lower-extremity spasticity and weakness. Historically, these disorders were classified as pure or complicated based upon the presence (complicated) or absence (pure) of associated features such as cognitive dysfunction, distal amyotrophy, retinopathy, thin corpus callosum, and neuropathy. More recently, a genetic classification scheme has taken hold, with HSPs commonly identified by their spastic gait (SPG) loci, SPG1 to -57, in order of identification (1, 2). Despite this extensive genetic heterogeneity, a small number of common cellular themes have emerged, including perturbations in endoplasmic reticulum (ER) shaping/distribution, mitochondrial function, myelination, lipid/cholesterol metabolism, and endocytic sorting (3, 4).
Disruption in the tubular ER network in cells has emerged as a key HSP pathogenic theme, because over half of patients have autosomal dominant SPG3A, -4, -12, or -31, each caused by mutations in genes encoding proteins that bind one another and function in tubular ER-network formation (3, 4). The SPG3A protein atlastin-1 and its yeast (synthetic enhancer of Yop1p; Sey1p) and plant (RHD3) orthologs are large, multimeric GTPases that harbor two closely spaced hydrophobic domains that likely form a membrane-spanning hairpin; this hairpin is required for tubular ER localization. Atlastin GTPases mediate homotypic fusion of ER tubules to form the polygonal ER network (5–8). SPG31 and SPG12 proteins receptor expression-enhancing protein 1 (REEP1) and reticulon 2, respectively, are members of the reticulon/REEP/Yop1p superfamily of proteins that contain elongated, partially membrane-spanning, hydrophobic hairpin domains and shape high-curvature ER tubules (9, 10). Lastly, the SPG4 protein spastin is an oligomeric ATPase, and the larger M1 spastin isoform harbors a hydrophobic hairpin and localizes to ER tubules. These domains are important for self-interactions of these proteins and also with one another (11).
The protrudin protein binds spastin and is involved in neurite outgrowth. In addition to three hydrophobic segments, protrudin harbors Rab-binding, two phenylalanines in an acidic tract (FFAT), and FYVE domains. It was originally reported to be mutated in SPG33 (12, 13). However, this HSP locus is controversial, because the only mutation identified, in a single family (p.G191V), is present in the SNP database and not considered pathogenic (14). Studies evaluating G191V mutant proteins have shown apparent discrepancies with regards to interactions with spastin and induction of neurite outgrowth (12, 14). Also, altered subcellular localization and impaired yolk sac extension of zebrafish expressing G191V mutant protrudin have been reported but without affecting neurite outgrowth (15).
Functionally, protrudin has been linked to changes in recycling of endosomes, and its roles in cellular process elongation have been ascribed to effects on bidirectional membrane trafficking in a Rab11-dependent manner (13). However, protrudin also localizes to the ER and binds vesicle-associated membrane-protein associated protein (VAP)-A, an ER-localized VAP. Depletion of VAP-A causes mislocalization of protrudin and inhibition of neurite outgrowth induced by nerve growth factor in PC12 cells (16). Protrudin proteins with FYVE domain mutations causing reduced phosphoinositide-binding affinity fail to promote neurite outgrowth in cultured neurons (17).
Here, we show that the hydrophobic domains of protrudin likely adopt hairpin topologies, similar to those in the atlastins, as well as the ER-shaping reticulons and REEPs. Protrudin interacts with these protein families through the hydrophobic segments. Depletion of protrudin in cells alters ER-network morphology and the sheet-to-tubule balance. Thus, protrudin may exemplify a protein class involved in shaping the ER network.
Results
Protrudin Is a Membrane-Bound, Oligomeric Protein That Localizes to Tubular ER.
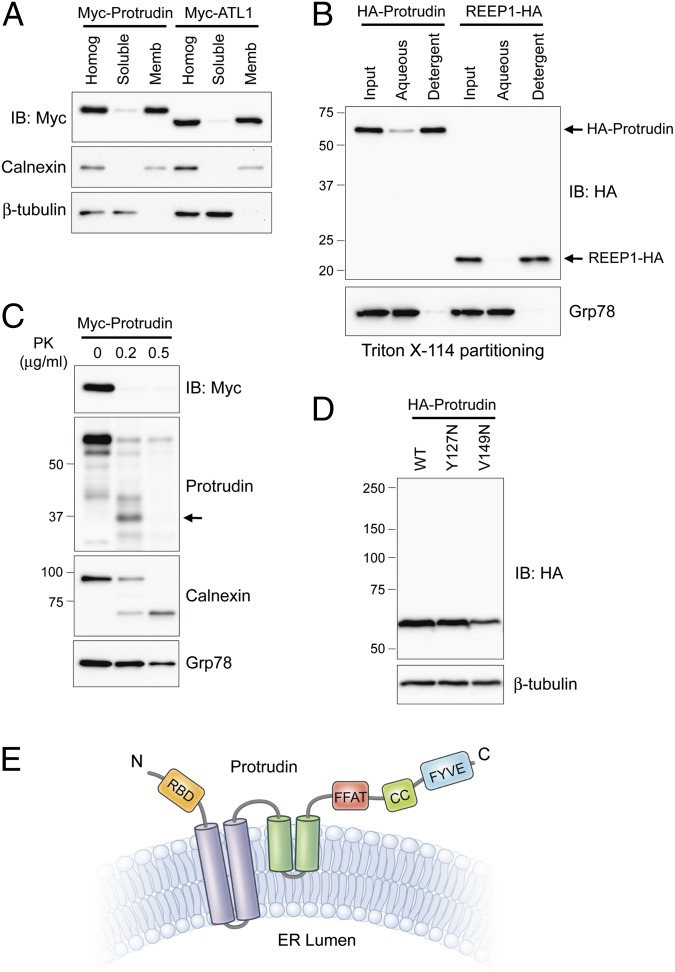
To examine the cellular distribution of protrudin, we fractionated lysates from Myc-protrudin–expressing cells. Protrudin was abundant in the membrane fraction that contains calnexin (Fig. 1A). Upon Triton X-114 phase separation, protrudin partitioned to the detergent phase enriched in integral membrane proteins. REEP1 was also in the detergent phase, as reported previously (Fig. 1B) (11). To investigate membrane topology, protrudin tagged at the N terminus with Myc-epitope was expressed in cells, and intact microsomes were prepared and treated with proteinase K (PK). We were able to selectively identify N- and C-termini of protrudin with Myc-epitope and protrudin (residues 217–411) antibodies, respectively. Whereas PK barely digested the luminal ER protein Grp78 (a control for intact microsomes), it extensively digested both termini of protrudin and the cytoplasmic domain of calnexin (Fig. 1C).
Fig. 1.
Membrane topology of protrudin. (A) Myc-tagged protrudin or atlastin-1 were transfected into HEK293T cells as indicated. Homogenates (Homog) were resolved into soluble and membrane (Memb) fractions, and aliquots were immunoblotted (IB). Calnexin and β-tubulin are markers for membrane and cytosolic fractions, respectively. (B) Membrane fractions from cells expressing HA-protrudin or REEP1-HA were phase partitioned with Triton X-114 and then immunoblotted for HA-epitope or Grp78. (C) Protease protection assays. Microsomes from Myc-protrudin–expressing cells were treated with PK. Aliquots were immunoblotted with Myc-epitope (N terminus of protrudin) or anti-protrudin (C-terminal region of protrudin) antibodies. Calnexin (partly luminal) and Grp78 (completely luminal) were used to monitor proteolysis. A major proteolytic fragment of protrudin is denoted with an arrow. (D) Cells were transfected with wild-type (WT) Myc-protrudin or else Y127N and V149N mutants, and lysates were immunoblotted as shown. (E) Membrane topology model for protrudin. CC, coiled-coil; RBD, Rab-binding domain. Migrations of molecular-mass standards (in kilodaltons) are to the left in B–D.
Protrudin is a >400-aa residue protein existing in several RNA splice forms, all harboring three hydrophobic segments. Two are spaced very closely at residues 67–87 and 89–109, and the third is elongated, spanning residues 180–214 (numbering for human isoform b). A Pro residue (Pro200) within the third segment prefigures a hairpin topology. In fact, partial proteolysis with PK reveals a protrudin fragment of ∼36 kDa lacking the Myc-tagged N terminus (Fig. 1C), most compatible with cytoplasmic cleavage between the first two hydrophobic segments (i.e., within residues 110–179). Even so, we cannot completely exclude the possibility of cleavage at the C terminus that spared some anti-protrudin antibody epitopes. To probe further whether the hydrophilic region between the second and third hydrophobic domains is exposed to the cytoplasm, we engineered protrudin proteins with V127N and V149N mutations, creating consensus N-linked glycosylation sites. These did not alter the migration of protrudin on SDS/PAGE gels of the proteins, consistent with our prediction that this hydrophilic region is not exposed to the ER lumen (Fig. 1D). We propose the membrane topology of protrudin presented in Fig. 1E.
Previously, protrudin was shown to oligomerize (18). Similarly, we observed probable dimer and tetramer forms of recombinant protrudin by immunoblotting (Fig. S1A). We further examined protrudin oligomerization using two different epitope-tagged recombinant proteins. Lysates from cells coexpressing HA- and Myc-tagged protrudin were immunoprecipitated, and HA- and Myc-protrudin proteins coprecipitated with both Myc- and HA-epitope antibodies, confirming that protrudin can homooligomerize (Fig. S1B). To map the domain responsible for oligomerization, we examined truncated protrudin proteins. We found that residues 1–92 are sufficient for self-interaction, and the C terminus (residues 206–411) could not bind even full-length protrudin (Fig. S1C). It has been suggested that protrudin self-interacts using TM3 (18). In our examination of many combinations of interactions among full-length and membrane domain-lacking mutant protrudin, there were no apparent differences (Fig. S1D).
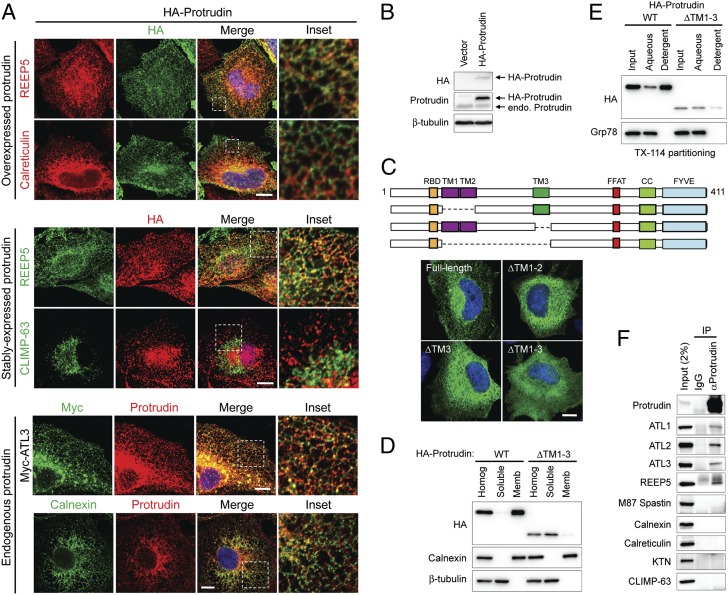
Subcellular localizations of transiently expressed, stably expressed, and endogenous protrudin were examined by costaining with organelle markers. HA-protrudin localized to mesh-like structures throughout the cytoplasm, distributing along tubules that also labeled for endogenous REEP5 and calnexin, as well as recombinant Myc–atlastin-3 (Fig. 2A). The ER can be divided into continuous but morphologically distinct tubular and sheet-like structures (19), and, interestingly, stably expressed HA-protrudin (Fig. 2 A and B) did not colocalize with the ER sheet protein CLIMP-63. Protrudin has three hydrophobic domains, a FFAT motif responsible for binding the ER protein VAP-A, a coiled-coil domain, and a FYVE domain with Zn2+-binding motifs at its C-terminal end (Fig. 1E) (16). To assess whether hydrophobic hairpins of protrudin were required for ER localization, deletion constructs lacking various hydrophobic domains (ΔTM1-2, ΔTM3, and ΔTM1-3) were expressed in cells. Proteins lacking either TM1-2 or TM3 localized to the ER, although the labeling appeared less reticular overall. A mutation of Pro200 within TM3 resulted in a less-tubular ER localization, consistent with our prediction that this Pro residue may cause a “kink” to form a hairpin structure (Fig. 1E and Fig. S2). Most prominently, ΔTM1-3 protrudin was dispersed throughout the cytoplasm (Fig. 2C). Membrane localizations were also examined by subcellular fractionation. Whereas wild-type protrudin showed complete membrane association, the ΔTM1-3 mutant was detected in the soluble fraction (Fig. 2D). Similarly, upon Triton X-114 phase separation, the ΔTM1-3 mutant partitioned to the aqueous phase (Fig. 2E). These results reveal that membrane-bound motifs within the N-terminal half of protrudin are responsible for its ER localization, as for the atlastins and Yop1p/REEP/reticulon proteins (11).
Fig. 2.
Protrudin localizes to the ER via its intramembrane domains and interacts with ER-shaping proteins. (A) From top to bottom, HeLa cells transiently expressing HA-protrudin were immunostained for endogenous REEP5 or calreticulin (red), along with HA-epitope (green). HeLa cells stably expressing HA-protrudin were immunostained for endogenous REEP5 or CLIMP-63 (green) and HA-epitope (red). Cells expressing Myc–atlastin-3 were coimmunostained for Myc-epitope (green) and endogenous protrudin (red). HeLa cells were costained for endogenous calnexin (green) and endogenous protrudin (red). Insets in the merged images (with DAPI staining) are enlarged to the right. (B) Lysates from cells stably expressing HA-protrudin were immunoblotted as shown. Endo., endogenous. (C, Upper) Schematic diagram of human protrudin and ΔTM deletion constructs. CC, coiled-coil; RBD, Rab-binding domain. (C, Lower) Cells expressing HA-tagged wild-type (WT) protrudin and the indicated deletion mutants were stained for HA-epitope (green) and DAPI (blue). (D) Wild-type and mutant protrudin proteins as in C were expressed in HEK293T cells. Homogenates (Homog) were separated into soluble and membrane (Memb) fractions and immunoblotted. (E) Membrane fractions from cells expressing wild-type and mutant protrudin proteins were phase partitioned with Triton X-114 and then immunoblotted for HA-epitope or Grp78. (F) Endogenous protrudin in HEK293T cells was immunoprecipitated (IP) with anti-protrudin antibodies or control IgG, and precipitates were immunoblotted as shown. ATL, atlastin; KTN, kinectin. (Scale bars: 10 μm.)
Because protrudin associates with the tubular ER membrane, we examined interactions of protrudin with known ER proteins. Members of the Yop1p/REEP and reticulon families are major factors shaping the high curvature of the tubular ER (9, 10). On the other hand, CLIMP-63, p180, kinectin, and translocons are enriched in flat ER sheets (20). Intriguingly, all three mammalian atlastins and REEP5, a DP1/REEP/Yop1p family member, coprecipitated with endogenous protrudin (Fig. 2F). However, we did not detect interactions with the sheet ER protein CLIMP-63, the ER membrane protein calnexin, the ER luminal protein calreticulin, or the M87 form of spastin (Fig. 2F). There are two major forms of spastin in mammals; the longer M1 spastin contains a hydrophobic domain and is abundant only in brain and spinal cord (21), whereas the soluble M87 form is ubiquitous. M1 spastin, but not M87, interacts with REEP1, atlastins, and reticulons at the ER (11, 22, 23). Because only M87 spastin is detectable in cultured cells, we were unable to examine the endogenous interaction of protrudin with M1 spastin. We conclude that protrudin is an ER membrane protein specifically associated with ER-shaping proteins in the tubular ER.
Protrudin Associates with REEPs, M1 Spastin, and Atlastins Through Its N-Terminal Membrane-Bound Domains.
The clear and specific endogenous interactions among protrudin and the atlastins and ER-shaping proteins prompted us to investigate the interactions in detail. First, we examined the interactions with the DP1/Yop1p family. There are six REEPs (REEP1 to -6) in mammals, categorized into REEP1-4 and REEP5-6 based on phylogenetic analyses and structural features (11). REEP5/DP1 is an abundant protein shaping the curvature of the tubular ER, and REEP1 is involved in microtubule interactions as well (9, 11). Both types of REEP proteins interacted with protrudin when coexpressed (Fig. S3 A and B). To determine the domain within protrudin responsible for REEP interactions, we coexpressed protrudin deletion mutants and REEPs (Fig. S3 C–F). Both REEPs interacted with the N terminus of protrudin containing the hydrophobic membrane-binding domains (residues 1–205; Fig. S3 C–E) but not with protrudin mutants lacking various hydrophobic domains (Fig. S3 C and F).
Spastin interacts with protrudin through its N terminus (12). Therefore, the function of spastin at the ER is thought to be restricted to the larger M1 isoform (Fig. S4A). In fact, recombinant M1 spastin was codistributed with endogenous REEP5 and recombinant HA-protrudin along ER tubules, whereas M87 spastin was dispersed throughout the cytoplasm (Fig. S4B). HA-tagged protrudin interacted only with Myc-tagged M1 spastin, not the M87 form, indicating that the membrane domain in M1 spastin is crucial not only for its ER localization but also for interaction with protrudin (Fig. S4C). Moreover, the N terminus of protrudin harboring the membrane domains (residues 1–205) was necessary and sufficient for M1 spastin interaction (Fig. S4D).
We examined the interaction of protrudin with atlastin family proteins. All three human atlastins implicated in the fusion of ER tubules to form three-way junctions (5, 6) (atlastin-1, atlastin-2, and atlastin-3) interacted with endogenous and recombinant protrudin (Fig. 2F and Fig. S5A). The interaction with atlastins required protrudin residues 1–205, as for the REEPs and M1 spastin (Fig. S5B). Tandem membrane domains located near the C terminus of atlastins play a pivotal role in its association with the ER membrane (5, 6, 24) and are necessary and sufficient for the interaction of atlastins with protrudin (Fig. S5C).
A natural question is whether SPG3A missense mutations in ATL1 affect this interaction. However, we could detect no differences in protrudin interactions among wild-type, GTP-binding, or SPG3A-causing atlastin-1 mutants (Fig. S5D). Although it is doubtful that p.G191V is a causative mutation for the HSP subtype SPG33 (12, 14), we searched for any defects induced by this mutation because it sits within the third hydrophobic domain. The mutant form interacted with atlastin-1 to a similar degree similar as wild-type protrudin (Fig. S5E), and it localized to ER when expressed alone or with atlastin-1 (Fig. S5 F and G). Taken together, these data indicate that protrudin interacts with ER membrane proteins that play a role in tubular ER shaping and network formation through its membrane domains. Because many of these proteins are encoded by major causal genes for HSP (4), protrudin could be implicated in HSP-relevant pathways, although we have no evidence that the G191V mutation causes HSP.
Protrudin Functions in ER Network Formation.
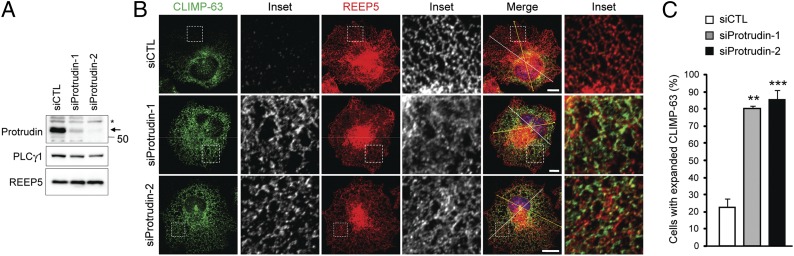
The localization of protrudin to the tubular ER network and its association with ER-shaping proteins suggested to us that protrudin might be involved in ER morphogenesis. We suppressed the expression of protrudin by RNA interference, testing eight different target sequences for siRNA-mediated depletion. The two most effective siRNAs were used subsequently (Fig. 3A).
Fig. 3.
Protrudin plays a role in forming the tubular ER network. (A) Two different siRNAs against human protrudin were transfected into HeLa cells for 72 h, and cell lysates were immunoblotted. An asterisk (*) indicates a cross-reacting band, and an arrow identifies protrudin. Migrations of molecular-mass standards are to the right. (B) siRNA-transfected HeLa cells were immunostained for CLIMP-63 (green) and REEP5 (red). Insets are enlarged to the right of each image. (Scale bars: 10 μm.) Line-scan plots are shown in Fig. S6A. (C) Numbers of cells with expanded CLIMP-63 signal area were quantified in control and protrudin-depleted conditions (n = 3; >200 cells per experiment). Means ± SD are shown. Paired Student t test: **P < 0.01; ***P < 0.001.
Compared with control cells, protrudin-depleted cells showed a significant increase in the density of apparent polygonal structures, especially noticeable in the cell periphery, with ER tubules identified by REEP5 staining (Fig. 3B). However, it was often difficult to discern whether some of these structures were sheets as opposed to a dense tubular network. Thus, we costained cells for REEP5 and the ER-sheet protein CLIMP-63 (20). CLIMP-63 immunoreactivity was not within the tubular ER in control cells but appeared prominently interspersed with REEP5 labeling in protrudin-depleted cells (Fig. 3B). Protrudin-depleted cells showed multiple CLIMP-63 peaks in line-scan plots, whereas control cells showed more restricted peaks in perinuclear regions (Fig. S6A). Thus, in protrudin-depleted cells CLIMP-63 was distributed more extensively throughout the peripheral ER, and ER sheets were expanded (Fig. 3C).
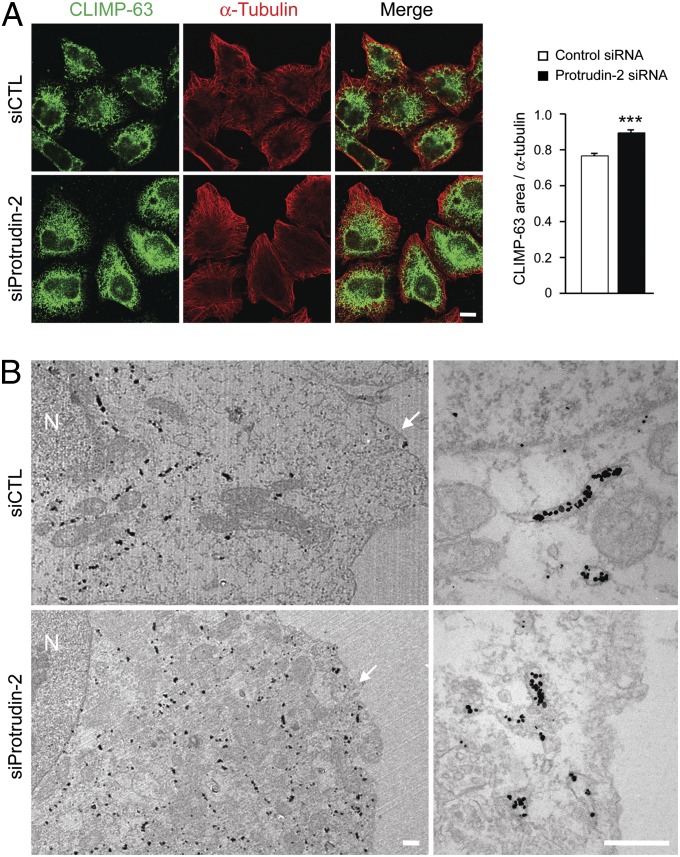
Overexpression of CLIMP-63, which serves as a luminal spacer for ER sheets, induces more and larger sheets and inhibits tubule formation, whereas overexpression of reticulons conversely increases ER tubules (20). To examine further any alterations in ER sheets, we costained cells for endogenous CLIMP-63 and the cytoskeletal protein α-tubulin. The ratio of CLIMP-63/α-tubulin staining area in protrudin-depleted cells was increased (Fig. 4A), indicating that ER sheets were expanded. Similar findings were obtained examining CLIMP-63 distributions using immunogold electron microscopy (Fig. 4B). Importantly, levels of ER proteins including CLIMP-63 appeared unaffected by depletion (Fig. S6B) or overexpression (Fig. S6C) of protrudin, indicating that alterations in ER sheets are not an indirect effect induced by modulating levels of ER sheet or tubular proteins.
Fig. 4.
Protrudin depletion modulates the ER sheet-to-tubule balance. (A) HeLa cells were transfected with control (siCTL) (n = 33) or protrudin (n = 22) siRNAs for 72 h and then immunostained with CLIMP-63 (green) and α-tubulin (red). All studies were done at the same time, with the same procedures and microscope settings. Pixel areas of CLIMP-63 were normalized to those of α-tubulin using ImageJ tools. Data are presented in arbitrary units. Means ± SEM are shown. Student t test: ***P < 0.001. (Scale bar: 10 μm.) (B) siRNA-transfected HeLa cells were immunogold-labeled for a luminal epitope of CLIMP-63, with visualization by electron microscopy. N, nucleus. Arrowheads denote the plasma membrane. (Scale bars: 500 nm.)
In rescue experiments, we depleted protrudin then expressed siRNA-resistant, wild-type protrudin (with four silent mutations) or siRNA-resistant deletion mutants. Expression of wild-type protrudin efficiently rescued the altered ER morphology induced by depletion of protrudin (Fig. S7). Not surprisingly, the isolated C terminus (residues 206–411) and membrane domain-lacking protrudin mutants (ΔTM1-3), both cytosolic, failed to rescue the phenotype (Fig. S7). However, expression of the isolated N terminus sufficient for ER localization (residues 1–205) also did not suppress the protrudin siRNA-mediated ER phenotype (Fig. S7), indicating that the C-terminal region containing FFAT, coiled-coil, and FYVE domains is essential for the function of protrudin in ER-network formation.
The FYVE domain is of particular interest because it mediates interactions with the plasma membrane (17), which might play a role in ER-network formation. We generated a mutant lacking the FYVE domain, and whereas protrusions were induced by overexpression of wild-type HA-protrudin, as described previously (13), expression of HA-tagged ΔFYVE protrudin did not induce these (Fig. S8). Because expression levels of the ΔFYVE mutant were lower (Fig. S8A), we generated a missense mutation, C405S, targeting a critical Cys residue in the Zn2+-binding motif. This mutant similarly failed to induce cellular protrusions, even when expressed at levels equivalent to the wild-type protein (Fig. S8 B and C). By contrast, the C405S mutant was able to suppress ER changes triggered by protrudin depletion to the same degree as wild-type protrudin (Fig. S9A).
Lastly, protrudin is distinctive for the presence of a Rab-binding domain at its N terminus that binds GDP-Rab11 (13). Previous colocalization studies examining protrudin and Rab11 (particularly the GDP-locked form) reported significant colocalization in PC12 cells following nerve growth factor stimulation (13). However, the mutant lacking the Rab-binding domain was able to suppress the ER morphology changes induced by protrudin depletion (Fig. S9B).
Discussion
We describe a function of protrudin in ER morphogenesis. First, we have shown that endogenous and recombinant protrudin localize to the ER, with intramembrane hydrophobic hairpins mediating its tubular ER localization and interactions with ER-shaping proteins, a number of which are mutated in common forms of HSP (4). Protrudin-depleted cells showed an aberration in ER-network morphology, with an apparent increased density of interconnections and expansion of sheets, indicating that protrudin may act antagonistically to the atlastin GTPases.
This ER-related function was unexpected, because protrudin was originally implicated in a Rab11-dependent recycling pathway, promoting the growth of neurites (13). However, Saita et al. (16) identified an interaction of protrudin with the ER-localized VAP-A, and Gil et al. (17) localized some protrudin at ER, which increased dramatically upon deletion of the FYVE domain. Matsuzaki et al. (25) used mass spectrometry to identify proteins interacting with protrudin, and a number were ER proteins, as well as the KIF5 molecular motor. Protrudin facilitated the interaction of KIF5 not only with Rab11 but also with ER proteins VAP-A/B and reticulon 3 (25). This is of particular interest because KIF5A is mutated in another HSP, SPG10 (26), and reticulon 2 is mutated in SPG12 (27). Thus, protrudin potentially links a large number of autosomal dominant HSPs: SPG3A, SPG4, SPG10, SPG12, and SPG31. Furthermore, a p.P56S mutant VAP-B in patients with familial amyotrophic lateral sclerosis (ALS8) alters ER morphology, generating unusual ER sheets (28) and a nuclear envelope defect characterized by separation of the outer and inner nuclear membrane (29). Multiple cytoplasmic domains in protrudin may mediate its effects as an adaptor to facilitate the proper interactions of ER tubules with molecular motors such as KIF5, distributing ER properly in large, highly polarized cells such as the neurons affected in HSPs and amyotrophic lateral sclerosis.
Mechanistically, the increases in sheet formation, and possibly increased formation and density of ER polygons upon protrudin depletion, are reminiscent of changes seen upon human and Drosophila atlastin overexpression (6, 7), as well as upon depletion of the lunapark protein Lnp1p in yeast (30). Interestingly, protrudin shares some structural features with lunapark proteins, including two closely spaced membrane domains, as well as Zn2+-binding motifs, and both interact with REEP/reticulon proteins (30). This suggests that related protein classes may act antagonistically to atlastins in ER-network formation. It has been proposed that lunapark may function to close polygons by competing with Sey1p for interaction with reticulons (30). Because endogenous protrudin coprecipitates with atlastins, as well as ER-shaping proteins, a similar mechanism might be in play.
Even so, protrudin and lunapark proteins are distinct. Protrudin harbors two additional motifs: a FFAT domain and a FYVE domain. FYVE domains target proteins to membranes via interactions with phosphoinositides, and the protrudin FYVE domain has a binding preference for phosphatidylinositol-3,4-bisphosphate, phosphatidylinositol-4,5-bisphosphate, and phosphatidlyinositol-3,4,5-trisphosphate, in contrast to canonical FYVE domains that bind phosphatidyl-3-phosphate selectively. Since ER interactions with other organelles are important for functions such as lipid transfer (31), protrudin may function within the tubular ER as a lipid sensor.
Because the isolated protrudin FYVE domain localizes to the plasma membrane, an intriguing possibility could be that ER-resident protrudin is recruited to the plasma membrane to participate in ER-plasma membrane interactions. In this regard, Osh proteins and VAPs, the latter of which interact with protrudin, are involved in ER–plasma membrane tethering, as well as regulation of phosphoinositide metabolism and cell signaling (32, 33). Interestingly, this interaction is not required for the major ER morphology changes described here.
Another intriguing role for protrudin, which harbors a Rab-binding domain, is provided by studies linking Rab GTPases to changes in ER morphology. In addition to the work of Audhya et al. (34) reporting effects of Rab5 on ER morphology, a recent study has implicated Rab10 in dynamic shaping of the ER network (35). It will be interesting to see how these and other Rabs contribute to shaping the ER network (36). Importantly, although Shirane and Nakayama (13) focused on the role of GDP-Rab11 binding to protrudin, they noted that this Rab-binding domain is similar to that of GDI-α and GDI-β, both of which bind the GDP-bound form of many Rabs. Even so, the Rab-binding domain of protrudin, like the FYVE domain, appeared dispensable for suppressing the ER morphological changes induced by protrudin siRNA. Thus, it seems likely that the FFAT, coiled-coil, and/or other interaction domains of protrudin play crucial roles in determining ER morphology.
Materials and Methods
DNA Constructs.
The human protrudin cDNA (GenBank accession no. NM_144588) was cloned into the EcoRI site of mammalian expression vectors pGW1-HA and pGW1-Myc, with N-terminal epitope tags (24). Truncation and internal deletion mutants of protrudin were generated by PCR, and mutagenesis was performed using QuikChange (Stratagene). Expression constructs for HA– and Myc–atlastin-1, -2, and -3; Myc-M1 and -M87 spastin; REEP1-HA; and REEP5-HA have been described previously (5, 11, 24, 37, 38).
Antibodies.
The following primary antibodies were obtained commercially: anti–α-tubulin (Abcam), anti–β-tubulin (Sigma-Aldrich), anti-calnexin (BD Biosciences), anti-calreticulin (Abcam), anti–CLIMP-63 (clone G1/296; Enzo Life Sciences; ref. 39), anti-Grp78 (BD Biosciences), anti-HA (Santa Cruz Biotechnology), anti-kinectin (Santa Cruz Biotechnology), anti–Myc-epitope (Santa Cruz Biotechnology), anti-PLCγ1 (Cell Signaling Technology), anti-protrudin (Proteintech), anti-REEP5 (Proteintech), and anti-spastin (Sigma-Aldrich). Anti–atlastin-1, -2, and -3 antibodies were described previously (5).
Cell Culture, Transfection, and RNA Interference.
HeLa, COS-7, and HEK293T cells were maintained in DMEM supplemented with 10% (vol/vol) FBS (37 °C; 5% CO2). Transfections were performed using GenJet Plus Reagent (SignaGen) for plasmid DNA and Lipofectamine RNAiMAX (Invitrogen) for siRNAs. For rescue experiments, plasmids were transfected 6 h after siRNA transfection. Sequences of siRNAs (Qiagen): siProtrudin-1, 5′-CUACAAGAGGCUGGAGAUCUA-3′; and siProtrudin-2, 5′-AACGGGUUCCUGAGCAAGAAU-3′. Control siRNAs were obtained from Ambion. To generate siRNA-resistant protrudin constructs, four silent mutations were introduced. Stable HeLa cell lines were generated as described previously (40).
Immunoprecipitation and Immunoblotting.
Cells were washed with PBS and then lysed in PBS with 0.5% CHAPS, 2 mM EGTA, and 1 mM EGTA. Complete protease inhibitor and PhosSTOP phosphatase inhibitor mixture (Roche) on ice for 30 min. Lysates were clarified by centrifugation (15,700 × g; 4 °C; 15 min), and supernatants were incubated with primary antibodies against at 4 °C for 16 h, followed by incubation with Protein A/G PLUS-agarose (Santa Cruz Biotechnology) for 2 h. Beads were washed with lysis buffer, and bound proteins were resolved by SDS/PAGE and transferred onto Hybond ECL membranes (GE Healthcare). Membranes were blocked with 5% nonfat milk in TBST (0.05% Tween-20 in Tris-buffered saline) for 30 min and incubated with primary antibodies in 3% BSA/TBST for 16 h. Blots were washed with TBST, incubated with HRP-conjugated secondary antibodies (Santa Cruz Biotechnology) for 1 h, and then washed with TBST. Finally, immunoreactive proteins were revealed using the Immun-Star WesternC Kit (Bio-Rad). Images were captured using a ChemiDoc XRS+ (Bio-Rad).
Membrane-Association Assays and Protease Treatments.
HEK293T cells expressing epitope-tagged protrudin or atlastin-1 were washed with 10 mM Tris⋅HCl (pH 7.5) and then harvested and homogenized in 10 mM Tris⋅HCl (pH 7.5) with 10 mM NaCl and 1.5 mM MgCl2. Lysates were centrifuged (1,300 × g; 4 °C; 3 min), and the postnuclear supernatant (homogenate) was recentrifuged at 100,000 × g. Equal proportions of homogenate, soluble and membrane fractions were resolved by SDS/PAGE and immunoblotted. Triton X-114 phase partitioning was performed as described previously (11). For protease digestion, microsomes from HEK293T cells were treated with PK at 37 °C for 15 min; the reaction was terminated with SDS/PAGE sample buffer.
Immunofluorescence Staining and Confocal Microscopy.
Cells were cultured and immunostained as described previously (40). Cells were imaged with a Zeiss LSM710 confocal microscope using a 63× 1.4 NA Plan-Apochromat lens and ZEN 2009 software (Carl Zeiss Microimaging). To analyze the ER phenotype, cells showing a distribution of endogenous CLIMP-63 not only in the perinuclear regions but also more evenly throughout the cytoplasm were classified as having expanded CLIMP-63.
Electron Microscopy.
For CLIMP-63 immunogold staining, cells were fixed in 4% paraformaldehyde in PBS for 1 h. After washing with PBS, cells were blocked and permeabilized in 5% goat serum and 0.1% saponin in PBS for 40 min, and then placed in blocking buffer with the primary antibody for 1 h. Subsequent preparations have been described previously (24).
Supplementary Material
Acknowledgments
We thank J. Nagle and D. Kauffman for DNA sequencing, J.-H. Tao-Cheng and V. Crocker for electron microscopy, and A. Hoofring and E. Tyler for artwork. This work was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307391110/-/DCSupplemental.
References
- 1.Schüle R, Schöls L. Genetics of hereditary spastic paraplegias. Semin Neurol. 2011;31(5):484–493. doi: 10.1055/s-0031-1299787. [DOI] [PubMed] [Google Scholar]
- 2.Finsterer J, et al. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci. 2012;318(1-2):1–18. doi: 10.1016/j.jns.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 3.Blackstone C, O’Kane CJ, Reid E. Hereditary spastic paraplegias: Membrane traffic and the motor pathway. Nat Rev Neurosci. 2011;12(1):31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackstone C. Cellular pathways of hereditary spastic paraplegia. Annu Rev Neurosci. 2012;35:25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rismanchi N, Soderblom C, Stadler J, Zhu P-P, Blackstone C. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum Mol Genet. 2008;17(11):1591–1604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138(3):549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460(7258):978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 8.Anwar K, et al. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol. 2012;197(2):209–217. doi: 10.1083/jcb.201111115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124(3):573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 10.Hu J, et al. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319(5867):1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 11.Park SH, Zhu P-P, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120(4):1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mannan AU, et al. ZFYVE27 (SPG33), a novel spastin-binding protein, is mutated in hereditary spastic paraplegia. Am J Hum Genet. 2006;79(2):351–357. doi: 10.1086/504927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirane M, Nakayama KI. Protrudin induces neurite formation by directional membrane trafficking. Science. 2006;314(5800):818–821. doi: 10.1126/science.1134027. [DOI] [PubMed] [Google Scholar]
- 14.Martignoni M, Riano E, Rugarli EI. The role of ZFYVE27/protrudin in hereditary spastic paraplegia. Am J Hum Genet. 2008;83(1):127–128. doi: 10.1016/j.ajhg.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, et al. Role of spastin and protrudin in neurite outgrowth. J Cell Biochem. 2012;113(7):2296–2307. doi: 10.1002/jcb.24100. [DOI] [PubMed] [Google Scholar]
- 16.Saita S, Shirane M, Natume T, Iemura S, Nakayama KI. Promotion of neurite extension by protrudin requires its interaction with vesicle-associated membrane protein-associated protein. J Biol Chem. 2009;284(20):13766–13777. doi: 10.1074/jbc.M807938200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gil J-E, et al. Phosphoinositides differentially regulate protrudin localization through the FYVE domain. J Biol Chem. 2012;287(49):41268–41276. doi: 10.1074/jbc.M112.419127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pantakani DV, Czyzewska MM, Sikorska A, Bodda C, Mannan AU. Oligomerization of ZFYVE27 (Protrudin) is necessary to promote neurite extension. PLoS ONE. 2011;6(12):e29584. doi: 10.1371/journal.pone.0029584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SH, Blackstone C. Further assembly required: Construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 2010;11(7):515–521. doi: 10.1038/embor.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata Y, et al. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143(5):774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claudiani P, Riano E, Errico A, Andolfi G, Rugarli EI. Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp Cell Res. 2005;309(2):358–369. doi: 10.1016/j.yexcr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Evans K, et al. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc Natl Acad Sci USA. 2006;103(28):10666–10671. doi: 10.1073/pnas.0510863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanderson CM, et al. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet. 2006;15(2):307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu P-P, et al. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J Biol Chem. 2003;278(49):49063–49071. doi: 10.1074/jbc.M306702200. [DOI] [PubMed] [Google Scholar]
- 25.Matsuzaki F, Shirane M, Matsumoto M, Nakayama KI. Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Mol Biol Cell. 2011;22(23):4602–4620. doi: 10.1091/mbc.E11-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reid E, et al. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) Am J Hum Genet. 2002;71(5):1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montenegro G, et al. Mutations in the ER-shaping protein reticulon 2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J Clin Invest. 2012;122(2):538–544. doi: 10.1172/JCI60560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasana E, et al. A VAPB mutant linked to amyotrophic lateral sclerosis generates a novel form of organized smooth endoplasmic reticulum. FASEB J. 2010;24(5):1419–1430. doi: 10.1096/fj.09-147850. [DOI] [PubMed] [Google Scholar]
- 29.Tran D, Chalhoub A, Schooley A, Zhang W, Ngsee JK. A mutation in VAPB that causes amyotrophic lateral sclerosis also causes a nuclear envelope defect. J Cell Sci. 2012;125(Pt 12):2831–2836. doi: 10.1242/jcs.102111. [DOI] [PubMed] [Google Scholar]
- 30.Chen S, Novick P, Ferro-Novick S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the lunapark family member Lnp1p. Nat Cell Biol. 2012;14(7):707–716. doi: 10.1038/ncb2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012;125(Pt 20):4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stefan CJ, et al. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144(3):389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 33.Manford AG, Stefan CJ, Yuan HL, MacGurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012;23(6):1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178(1):43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.English AR, Voeltz GK. Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol. 2013;15(2):169–178. doi: 10.1038/ncb2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang J, Blackstone C. Rab10 joins the ER social network. Nat Cell Biol. 2013;15(2):135–136. doi: 10.1038/ncb2682. [DOI] [PubMed] [Google Scholar]
- 37.Zhu P-P, Soderblom C, Tao-Cheng J-H, Stadler J, Blackstone C. SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum Mol Genet. 2006;15(8):1343–1353. doi: 10.1093/hmg/ddl054. [DOI] [PubMed] [Google Scholar]
- 38.Yang YS, Harel NY, Strittmatter SM. Reticulon-4A (Nogo-A) redistributes protein disulfide isomerase to protect mice from SOD1-dependent amyotrophic lateral sclerosis. J Neurosci. 2009;29(44):13850–13859. doi: 10.1523/JNEUROSCI.2312-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schweizer A, Rohrer J, Hauri H-P, Kornfeld S. Retention of p63 in an ER-Golgi intermediate compartment depends on the presence of all three of its domains and on its ability to form oligomers. J Cell Biol. 1994;126(1):25–39. doi: 10.1083/jcb.126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, et al. MITD1 is recruited to midbodies by ESCRT-III and participates in cytokinesis. Mol Biol Cell. 2012;23(22):4347–4361. doi: 10.1091/mbc.E12-04-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.