Significance
Evolution by natural selection is driven by fitness differences, which define a “fitness landscape” in the space of all possible genetic sequences. Understanding the landscape is critical for understanding and predicting natural selection, yet very little is known about the structure of a real fitness landscape. Here we experimentally determine the complete fitness landscape of small RNA selected to interact with GTP, a building block for early life. We find that the landscape is composed of largely disconnected islands of active sequences. This scenario suggests that natural selection under these conditions would be constrained to local exploration of sequence space but that replaying the initial emergence of a functional RNA could lead to a different outcome.
Abstract
The origin of life is believed to have progressed through an RNA world, in which RNA acted as both genetic material and functional molecules. The structure of the evolutionary fitness landscape of RNA would determine natural selection for the first functional sequences. Fitness landscapes are the subject of much speculation, but their structure is essentially unknown. Here we describe a comprehensive map of a fitness landscape, exploring nearly all of sequence space, for short RNAs surviving selection in vitro. With the exception of a small evolutionary network, we find that fitness peaks are largely isolated from one another, highlighting the importance of historical contingency and indicating that natural selection would be constrained to local exploration in the RNA world.
The nucleotide sequence of an organism’s genome determines its fitness in a given selective environment. All possible sequences of length L constitute a discrete sequence space containing 4L points. Including fitness as another variable creates a “landscape” in sequence space, in which highly fit sequences occupy peaks (1, 2). Evolution can be thought of as a random walk on this landscape with a bias toward climbing peaks (3). Knowledge of the fitness landscape is a fundamental prerequisite for a quantitative understanding of evolution. Although several models of theoretical landscapes have been proposed (reviewed in refs. 4 and 5), there is a lack of empirical data. Landscapes based on RNA secondary structure have been explored computationally, but the relationship to function is unknown (6–10). Experimental efforts at determining a comprehensive fitness landscape are generally stymied by the astronomical size of sequence space, but synthesis of nearly every variant is feasible for relatively short sequences (i.e., RNAs with length L <30 nucleotides). Therefore, complete landscapes for short RNAs could be mapped in principle. Such landscapes are of particular interest for understanding evolution in the RNA world of early life (11–14).
Limited fitness landscapes localized around known functional sequences have been explored for proteins (15–17), viruses (18, 19), and functional nucleic acids, including ribozymes and ribosomal RNA (20–24). The local fitness landscape around a known RNA ligase ribozyme (L = 54) was mapped using high-throughput sequencing (25). However, random sampling of RNA and DNA sequence space can be done by in vitro selection, or SELEX, for de novo discovery of functional molecules (26–29). Such studies generally take very sparse samples of sequence space, owing to their focus on obtaining functional, and therefore longer, sequences. Thorough sampling techniques have been used to explore all possible DNA targets for a known DNA-binding protein (30). Such studies illuminate the biology of extant organisms but do not address the initial evolution of macromolecular activity. The entirety of a macromolecular fitness landscape has not yet been explored. Therefore, fundamental questions about the shape of fitness landscapes remain, such as the absolute frequency and distribution of fitness peaks in sequence space [in one review, estimates range over 45 orders of magnitude (31)]. An important unknown is whether sequence space contains “neutral networks” that connect distant sequences by evolution without loss of fitness (32–34); such networks are thought to be necessary for optimization by natural selection (1).
In this work, we determine the fitness landscape for nearly all possible short RNA sequences (starting pool with L = 24) during in vitro selection for binding to GTP agarose resin as a model selection for the RNA world (SI Appendix, Fig. S1). GTP was presumably an important “nutrient” molecule for the RNA world, so RNAs that could sequester GTP would be associated with greater fitness. Isolated examples of longer GTP aptamers are known (35). The short length studied here does not preclude functional activity, because even shorter RNAs have been found during ribozyme selections (36), although active sequences are more common in longer pools up to a point (37, 38), and activity correlates positively with length [or more precisely, with functional information (39)]. Our results give an experimental determination of a comprehensive fitness landscape. A limited neutral network is present, but most fitness peaks are evolutionarily isolated from one another.
Results
Starting Library.
High-throughput sequencing (HTS) of the synthetic RNA library showed good coverage of sequence space, with a monomer ratio of 0.225:0.282:0.236:0.257 (A:C:G:U) in the sequence reads (Materials and Methods). These ratios suggest that >99.99% of the possible 424 sequences are represented in the library (SI Appendix, Fig. S2) with roughly ∼1,000 copies of each sequence present on average. The experimental duplicates were done using two separate syntheses of the starting pool.
Selection Progress.
Bulk GTP binding of the pool increased as the selection progressed (SI Appendix, Fig. S3A). At the same time, a small cohort of sequences came to represent a disproportionate fraction of the pool by round 3 (SI Appendix, Fig. S3B), suggesting the enrichment of fit sequences and a diminishment of unfit sequences. Relatively few peaks survived by the end of round 4, prompting an end to the selection.
Fitness Landscapes.
To reconstruct the fitness landscape after each round, the relative fitness of each sequence identified by HTS was estimated from its relative frequency in the sequence reads for that sample, corrected for biases in RNA synthesis and ligation. Sequences related to the HTS adapter sequences were removed from analysis. In addition, because an abundant sequence would produce several spurious point mutants through sequencing errors, frequencies were also corrected for the estimated contribution from this process. Corrected relative frequencies are referred to as “fitness” (Materials and Methods). The correction procedure did not substantially alter the pattern of frequencies observed per round (SI Appendix, Fig. S4). Sequences were grouped into families, corresponding to fitness peaks, if a small number of mutations could convert the sequence into the most fit sequence of that peak (≤3 single-base substitutions, insertions, or deletions, i.e., an edit distance of ≤3) (40). The selection experiment was performed twice independently, to ascertain the reproducibility of the results. The structure of the fitness landscape is shown in Fig. 1A.
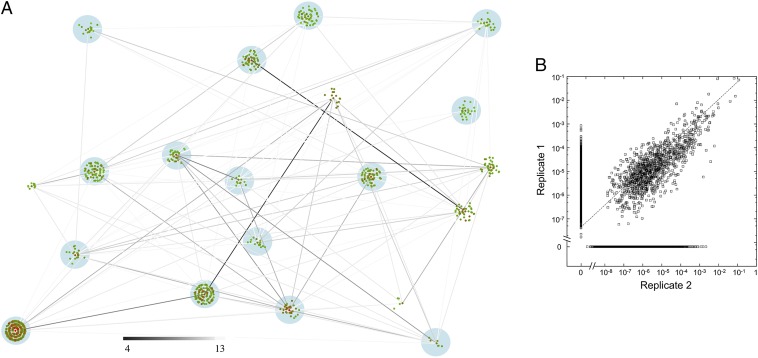
Fig. 1.
Fitness landscape. (A) Network representation. Each sequence is represented by a point whose color indicates relative fitness (red is high; green is low). Related sequences are clustered around the fittest sequence, with successive rings of single, double, and triple mutants. The darkness of the lines indicates the number of mutations needed to convert one peak sequence into another (see legend). The data shown are all detected sequences from round 3 for one replicate experiment; peaks highlighted in blue were also found in the second replicate (SI Appendix, Figs. S5–S8 give peak identification and other rounds). (B) Reproducibility of fitness in two replicate experiments. All peaks are shown; sequences detected in only one replicate are shown at fitness 0 in the other replicate. The dotted line is the line of best fit for log-transformed data (ln y = 0.33 +0.74*ln x; r = 0.79 for sequences detected in both experiments).
Nonfunctional, unfit sequences are likely to comprise the vast majority of possible sequences. These represent a substantial source of random noise in the HTS data, which presumably decreases over the course of the selection. We therefore required that each peak contain at least two unique sequences. Very few sequences appeared in isolation, that is, without closely related sequences being simultaneously identified (SI Appendix, Table S1). To verify the significance of the detected peaks, we performed a replicate selection experiment. Because the number of possible spurious sequences is very large, the chance that the same spurious peak would be detected in more than one experiment is extremely small. The significant peaks found after each selection round are given in Fig. 2 (also SI Appendix, Table S2). Fifteen peaks were found in common between the two experiments. The fraction of peaks found in common was highest in round 3, and we therefore focused our attention on the data from this round. Overall, the fitness for a given sequence in both experiments was correlated (Pearson’s r = 0.79 for sequences detected in both experiments; SI Appendix, Table S3 shows individual peaks) (Fig. 1B). Reproducibility between experiments also indicates that sequence space was largely covered, because sparse sampling in the initial library would yield different results in a repeated experiment.
Fig. 2.
Fitness peaks. The sequence and rank (1 = highest) of the highest fitness sequence in each peak is given for each replicate experiment, rounds 2–4. Color indicates fitness from high (red) to low (yellow-green); SI Appendix, Table S2 gives relative fitness values. Asterisk indicates that the highest fitness sequence of the peak was slightly different in the other replicate [i.e., lacking the three terminal nucleotides (UGG) on the 3′ end].
Fitness Peaks and Functional Information.
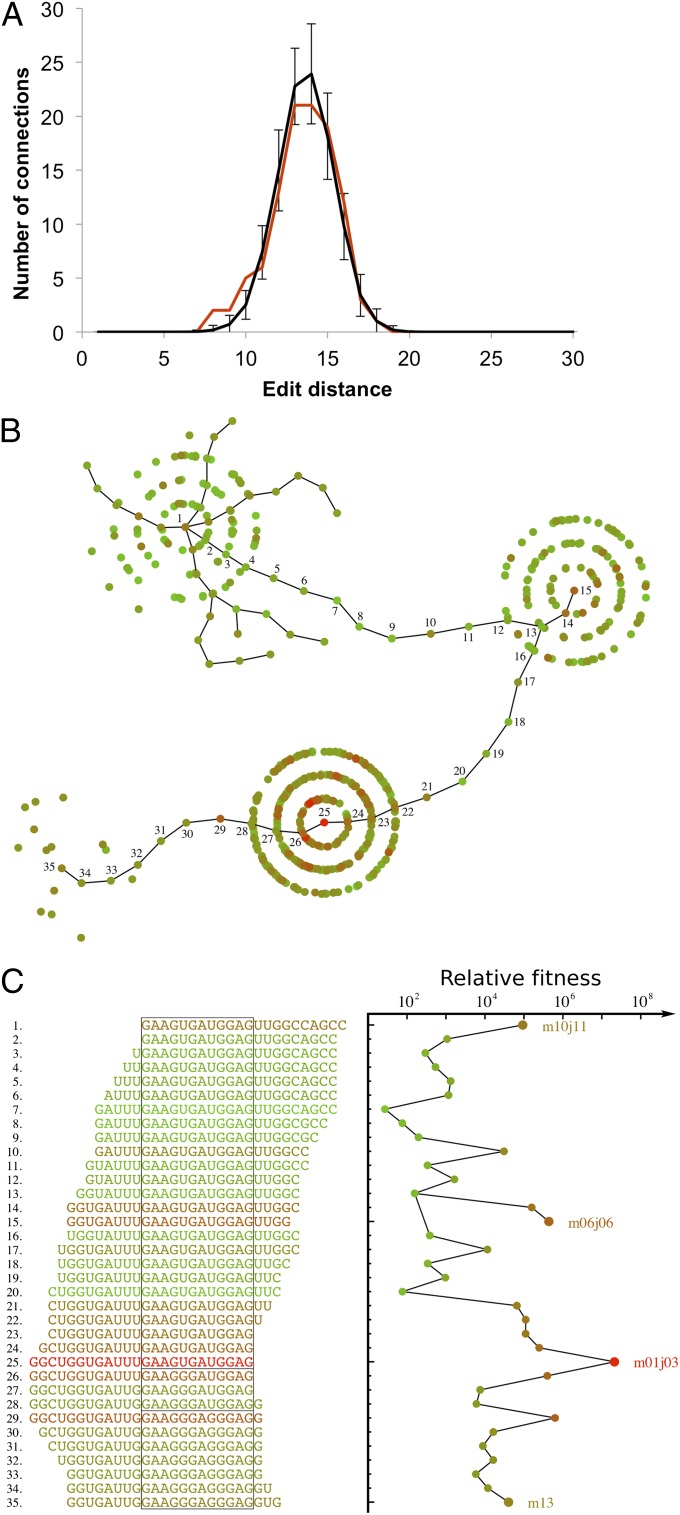
To determine whether certain areas of sequence space were enriched for functional peaks, we compared the distribution of interpeak distances with a control distribution generated from random sampling of the same number of sequences. The observed distribution was similar to the control distribution (Fig. 3A), with the exception that peaks in a small subset were more closely related to one another than expected by random chance (Evolutionary Pathways, discussed below).
Fig. 3.
Connectivity of fitness peaks. (A) The distribution of interpeak edit distances for the 15 peaks found in common between both experiments is shown in red. The control distribution for randomly chosen sequences is shown in black, with SDs given by the error bars. Note the difference between distributions at low edit distance. SI Appendix, Fig. S9 shows similar plots for individual experimental replicates. (B) Potential evolutionary pathways form limited connections among fitness peaks. Each point corresponds to an observed sequence (round 3), with color corresponding to relative fitness, as given in C. Sequences along the connecting pathways are numbered; other sequences from the involved peaks are also shown. Dead-end pathways are also present; a few such “tendrils” are shown surrounding peak m10j11 for illustration. (C) Plot of relative fitness along the pathways with sequences given along the vertical axis. Color corresponds to fitness (red is high; green is low). Peak sequences are labeled in the plot. Pathways among the three main peaks were identified in both replicates; the smallest peak (m13) was only identified in one replicate.
Within individual peaks, average and median fitness dropped as the edit distance from the peak center increased (SI Appendix, Fig. S10). In general, the drop in fitness was most pronounced for the single mutants; additional mutations tended to affect fitness to a lesser degree.
We found a statistically significant correlation between peak rank according to fitness and number of unique sequences in the peak, suggesting that higher fitness peaks are more robust to mutation (Spearman’s ρ ∼ −0.5 to −0.6; SI Appendix, Fig. S11). However, previous work analyzing RNA aptamers and ribozymes indicated that greater activity is correlated with greater specification of the sequence, that is, “functional information,” suggesting that more active sequences are correspondingly more difficult to find in random sequence space (39). We calculated the functional information for the 15 peaks found in common between the two experiments. Functional information measured from one experiment correlated well with that measured in the second experiment but did not seem to be well correlated with peak fitness or number of sequences within a peak in these experiments (SI Appendix, Fig. S12). One possible reason for the difference between the studies is the length, and therefore the potential activity, of the selected sequences; whereas the present selection began with 24-mers, the prior study used a longer region (64 nt) (39), which would presumably allow tighter binding and therefore have greater power to resolve a correlation with activity. Also, functional information in ref. 39 was determined by selecting from a separate mutagenized library for each aptamer, rather than simultaneously in one pot as in the present study, so differences in threshold fitness might also influence the calculations.
Evolutionary Pathways.
Although most peaks were not very closely related, we studied whether potential evolutionary pathways could be found within the experimental dataset that connected peak sequences. We did find pathways connecting a subset of peak sequences (m10j11, m06j06, and m01j03) in both replicates (Fig. 3B). Upon inspection, these peaks were found to share a common 12-base motif (Fig. 3C), with most variants having insertions or deletions at the 5′ or 3′ ends. This motif was also identified independently using the Gibbs Motif Sampler (no other motifs were found among the 15 peaks found in common between the two experiments). For the majority of peak pairs, it was not possible to find step-by-step mutational pathways through the observed sequence data.
Biochemical Characterization and Fitness.
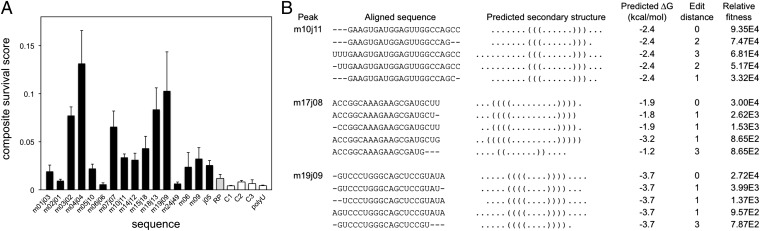
The apparent fitness of a sequence during the selection is likely to be influenced by several experimental factors in addition to the interaction with GTP agarose resin, such as the efficiency of preparation for Illumina sequencing (ligation to adapter sequences and amplification during PCR). We assayed binding to agarose resin, binding to GTP agarose resin, and PCR efficiency for 14 peak sequences, the initial pool, and four control sequences (SI Appendix, Text S1 for methods; Table S4 gives a list of sequences). Peak sequences tended to be recovered in greater amounts as flow-through from agarose resin and also as elution from the GTP agarose resin, compared with the initial pool and control sequences (SI Appendix, Fig. S13 and Table S5). Ligation efficiency for each sequence was estimated as described in Materials and Methods, and no clear difference was seen between the peak sequences and other sequences (SI Appendix, Fig. S13 and Table S5). PCR efficiency was measured for each sequence and tended to be higher for peak sequences than for others (SI Appendix, Fig. S13 and Table S5). The relative efficiencies in each process were multiplied together to obtain a crude survival score for each sequence (Fig. 4A). Although this decomposition of fitness is oversimplified, most peak sequences showed significantly higher survival scores compared with the controls. Interestingly, peak sequences from round 3 with lower survival scores did not survive into round 4 (Fig. 2), suggesting that further selection removed these lower-activity sequences.
Fig. 4.
Fitness and secondary structures for individual RNA sequences. (A) Several peak sequences (black bars), the initial pool (gray bar), and negative control sequences (white bars) were assayed individually for survival through the negative selection (agarose resin) and positive selection (GTP agarose resin) steps as well as relative enrichment during PCR. Relative fitnesses through each step and the expected ligation bias were combined into a composite survival score (Materials and Methods and SI Appendix, Table S5); error bars represent the SD. (B) Predicted secondary structures for the three peaks with lowest minimum free energy. The five sequences with highest fitness from each peak are shown with aligned secondary structures in bracket notation. Edit distance from the peak center and the relative fitness of each sequence are also shown.
Predicted Secondary Structures.
The minimum free-energy secondary structures of the sequences identified in the peaks were predicted using a nearest-neighbor thermodynamic model. Of the 15 peak sequences shared in common between the replicate selection experiments, 11 were predicted to have folds with free energies of −0.4 to −3.7 kcal/mol (SI Appendix, Fig. S14). Four did not have predicted folds, which would be consistent with an induced-fit mechanism of folding in the presence of ligand (41, 42). For peaks with substantial folding energies, the predicted secondary structure tended to be conserved within the peak (Fig. 4B). The structural conservation index (SCI) has been proposed as a measure of such conservation; noncoding, structured RNAs tend to have an SCI >0.5 (43, 44), as did several of our peaks (SI Appendix, Fig. S14). However, the utility of the SCI may be limited here, because it does not consider fitness differences within a peak and cannot account for folding by induced fit.
Discussion
In this work, we determine properties of a fitness landscape of short RNA molecules. We define fitness in terms of the relative survival of sequences during an in vitro selection on immobilized GTP, selecting for sequences that might sequester this nutrient molecule. We began with a pool that nearly saturated sequence space with ∼1,000-fold coverage. The selection process whittled away the vast majority of sequences and a small fraction survived. We avoided PCR amplification between rounds to minimize amplification bias. However, in this approach, sequences that are lost during the selection cannot be recovered (e.g., by mutation during amplification of related sequences). In principle, this limits the resolution for defining the fitness landscape in sequence space. Nevertheless, we observed reasonable concordance between the experimental replicates, indicating that loss of sequences did not obscure the landscape. Interestingly, the yield of recovery of a radiolabeled RNA sequence increases as the total amount of RNA decreases (SI Appendix, Fig. S15), possibly owing to increased availability of binding sites on the GTP agarose resin, suggesting that sequences that survived the initial rounds would have been better able to survive subsequent rounds.
Although we avoided PCR between selection rounds, the fitness of a sequence is likely to be influenced by multiple factors, such as survival during sample preparation and PCR efficiency before sequencing. Indeed, the relationship between representation during in vitro selection and molecular activity is not consistent (45). Fitness in the RNA world would have been even more complex and determined by replication efficiency, survival probability, and environmental variables. However, in vitro laboratory selections can model molecular activities that influence “true” fitness and thus can be informative about prebiotic fitness landscapes.
Our results show a peak density of ∼10−13 in the total space of possible sequences (∼15 peaks in 424 sequences) for the particular selection undertaken here. This density depends on the definition of fitness, including the environmental conditions, and the threshold for detection. Our selection detected relatively low affinity sequences (Kd in the low millimolar range; SI Appendix, Fig. S16), presumably owing to the high concentration of GTP used for elution, suggesting that this is an upper limit for peak density in this type of selection under relatively constant conditions. The peak density measured here should be considered a single snapshot of the fitness landscape, because the density would increase if multiple conditions were tested and the peaks were aggregated together. The distribution of peaks in sequence space was largely indistinguishable from random expectation. Such a sparse distribution is expected to fall below the percolation threshold, signifying a structure of isolated fitness “islands” (5, 46). This distribution contrasts with the predicted landscape for RNA secondary structure, which is characterized by large neutral networks (9). Indeed, only a few evolutionary pathways connecting a small subset of peaks were found in our data. At a mechanistic level, the generally disconnected nature of the fitness landscape may correspond to multiple possible modes of binding (47), although nucleotide-binding aptamers also sometimes share a similar mode of binding (48). Our study determined the connectivity of the landscape directly, without reference to the mode of binding. The finding of low connectivity implies that the role of historical contingency (e.g., the RNA sequences that happened to be present during the origin of life) might have been relatively large in a constant landscape, because substantial movement through sequence space would lead to low fitness sequences that represent evolutionary dead ends. Mechanisms causing large leaps through sequence space, such as ligation, recombination, or multiple simultaneous mutations (49–52), would be important for crossing fitness valleys. Perturbations to the landscape, such as environmental change, might also uncover evolutionary pathways. Indeed, the RNA landscape itself is likely to be highly dependent on conditions such as ionic composition (e.g., the concentration of Mg2+ and other divalent cations), presence of cofactors, temperature, and water activity, and therefore may be quite dynamic in response to changing conditions (53, 54). Fitness peaks for one functional activity can be near in sequence space to peaks for another activity, potentially leading to adoption of new function (55–58), suggesting that changing selection conditions could have an important role. Further studies are required to evaluate the generality of these findings, their robustness to environmental conditions, and possible mechanisms for optimization within the fitness landscape. Nevertheless, our study suggests that replaying the “tape of life” at the very origin of life might lead to quite different results.
Materials and Methods
Synthesis of the Library.
The library of 24-mer random RNA oligonucleotides was synthesized by University Core DNA Services at the University of Calgary (UCDNA) using the following composition; A:C:G:U = 0.95:1.43:0.66:0.96. This ratio was found to maximize the evenness of the distribution of the four bases, as measured by Illumina sequencing of the initial library (the procedure is discussed below). The library was synthesized at 1-μmol scale (yielding 2.8–6.6 mg, on the order of 1017 molecules). The desalted RNA was resuspended in 100 μL of water before use. Separate syntheses were used for replicate experiments. The nucleotide frequencies for the initial library were 24.4% A, 29.5% C, 23.3% G, and 22.8% U for one replicate and 22.5% A, 28.2% C, 23.6% G, and 25.7% U for the other.
Selection.
Two successive purification steps composed each round of selection (SI Appendix, Fig. S1). The RNA samples were first subjected to a negative selection on an agarose resin. The unbound RNA from that step was then loaded into a GTP agarose resin (γ-phosphate–linked; >6 μmol/mL). Both resins were obtained from Innova Biosciences. The appropriate volume of fresh resin (50–200 μL) was packed into Pierce Spin (initial round) or Micro-Spin columns (subsequent rounds) (Thermo Scientific) and washed and equilibrated with water and at least three volumes of binding buffer [20 mM Tris⋅HCl (pH 7.5), 300 mM NaCl, and 5 mM MgCl2].
Before the selection, 100 pmol of the RNA pool were radioactively labeled at the 5′ end using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Perkin-Elmer) following the manufacturer’s instructions. The resulting radiolabeled RNA was precipitated with ethanol, resuspended, and mixed with the remaining cold RNA pool. Gel analysis of the radiolabeled RNA showed that the amount of free radiolabel in the resuspended pellet was negligible.
The RNA pool was mixed with an appropriate amount of 5× binding buffer and water to obtain a solution in 1× binding buffer at a volume equal to the resin volume at that step. The resin volume was 200 μL in round 1 and 50 μL in subsequent rounds. The solution was incubated at 65 °C for 5 min, cooled down at room temperature for 5 min, and loaded onto the agarose column. After 15 min of incubation, the unbound RNA was removed from the column by brief centrifugation at 1,000 × g in a microfuge. The recovered solution was then loaded into the GTP agarose resin and incubated for 15 min at room temperature without agitation. Unbound material was removed in five washing steps using one volume of binding buffer (1×) in each step, by centrifugation at 1,000 × g as described. The RNA bound to the column was eluted during a 30-min incubation of the resin with one volume of binding buffer (1×) containing 25 mM GTP (Sigma-Aldrich) followed by centrifugation at 1,000 × g.
After elution, samples were brought to a volume of 500 μL by addition of water and desalted on a Sephadex PD midi Trap G-25 column (GE Healthcare). Fractions were collected (∼1 mL). The first fractions eluted containing the radiolabeled RNA were collected and dried in a speed-vac. Pellets were resuspended in 50 μL of water and two aliquots of 5 μL each were taken either for deep sequencing or for relabeling with [γ-32P]ATP, following the procedure described above, to be used in the next round of selection. No PCR amplification was performed on the RNA carried to the next round.
As a positive control, the class-III GTP aptamer (39) with the sequence 5′-gagccagaagagcacguauacgcaaggcuc was synthesized by UCDNA; 50 pmol were kinased, ethanol-precipitated, and resuspended in 50 μL of 1× binding buffer. The RNA was subjected to the same binding procedure on agarose and GTP agarose resins as described above.
High-Throughput Sequencing.
Samples were ligated to adapters, amplified by PCR, and sequenced with single end reads on the Illumina HiSeq platform. Details are given in SI Appendix, Text S1.
Calculation of Composite Survival Score.
The average composite survival score (ac) was calculated as the product of the fractions obtained in negative and positive selections, the relative score for ligation bias, and the fraction of PCR amplification. SD of the composite score (sc) was calculated according to the following formula:
where s and a are, respectively, the SD and average of the negative selection (sn, an), GTP selection (sg, ag), and PCR yield (sp, ap) values and l is the ligation bias (assumed to be a constant).
Estimation of Fitness from Observed Sequence Frequency.
The fitness of a sequence was estimated from its observed frequency in the sequence reads, corrected for expected sequencing errors, biases during adapter ligation, and the expected abundance of the sequence in the initial pool given biases during synthesis. A complete description of the fitness estimation is given in SI Appendix, Text S1.
Computational Analysis of Evolutionary Networks.
Evolutionary distance was defined as the minimum number of operations required to convert one sequence into another, in which operations could be single-base insertions, deletions, or substitutions. This distance is also known as the edit distance (40). Sequences were organized into families, or fitness peaks, containing at least two sequences, according to the following algorithm. First, a sequence was randomly chosen from the pool of sequence reads. The fitness of this sequence was then compared with the fitness of all its neighboring sequences, that is, all sequences that could be converted into the chosen sequence with three or fewer single-base operations. Neighbors were found by performing pairwise sequence comparisons base-by-base until a difference was found, at which point single-base edits were introduced until either the second sequence was converted into the first or more than three edits were required. This method was exhaustive in testing all possible combinations of edits (3L possible substitutions, 4(L + 1) possible insertions, L possible deletions) to relate two sequences up to a distance of 3, and the lowest number of edits was taken to be the edit distance. Sequences without any neighbors were designated as isolated sequences. If a nonisolated sequence had a higher relative fitness than all of its neighbors, it was defined as a peak center. A peak consists of the peak center and all of its neighbors. Edit distance thresholds other than 3 were also tried, but it was found that distances below 3 created artificial separations within sequence families, whereas distances greater than ∼8 created spurious connections between peaks (causing peaks to collapse together). Intermediate distances of 4 to 8 were also tried, but these did not change the number of peaks or their features, and seemed to add only sequences of relatively low fitness. Because the probability of detecting the same peak in two different experiments by chance is exceedingly small (∼10−10 assuming peaks contain 1,000 sequences), we set a fitness cutoff so as to maximize the overlap of peaks between the two replicates. Peaks below this fitness level were not analyzed further. All analyzed peaks contained at least one sequence above this cutoff. This threshold allowed us to focus our analysis on the most important peaks, 20 from replicate 1 and 22 from replicate 2, with 15 peaks common to both replicates. Isolated sequences above this threshold were also recorded.
A metric based on the Hamming distance was also considered as the measure of evolutionary distance. For this metric, the usual Hamming distance was used if the two sequences were of identical length. For sequences of different length, the minimum Hamming distance among the possible alignments (without gaps) was used. Using this metric, peaks tended to contain fewer sequences, but no essential difference with the analysis using the edit distance was seen.
Motif analysis and secondary structure prediction were done using the Gibbs Motif Sampler (59), rnafold from the Matlab 2011b Bioinformatics toolbox, and RNAz (43). Details are given in SI Appendix, Text S1.
Calculation of Functional Information.
To calculate functional information for a peak, sequences were aligned, including gaps, and the information content at each site was calculated based on a previously described measure (39). SI Appendix, Text S1 gives details.
Supplementary Material
Acknowledgments
We thank Sudha Rajamani, Jack Szostak, Martin Nowak, David Liu, Eugene Shakhnovich, and James Carothers for discussions. I.A.C. was a Bauer Fellow at Harvard University and is a Simons Investigator (Grant 290356 from the Simons Foundation). J.I.J. was a Foundational Questions in Evolutionary Biology (FQEB) fellow at Harvard University sponsored by the John Templeton Foundation. R.X.-B. received a fellowship from the Human Frontiers Science Program. This work was supported by National Institutes of Health Grant GM068763 to the National Centers of Systems Biology and Grant RFP-12-05 from FQEB.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307604110/-/DCSupplemental.
References
- 1.Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225(5232):563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- 2.Wright S. Evolution in Mendelian populations. Genetics. 1931;16(2):97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kauffman S, Levin S. Towards a general theory of adaptive walks on rugged landscapes. J Theor Biol. 1987;128(1):11–45. doi: 10.1016/s0022-5193(87)80029-2. [DOI] [PubMed] [Google Scholar]
- 4.Orr HA. The genetic theory of adaptation: A brief history. Nat Rev Genet. 2005;6(2):119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 5.Gavrilets S. Fitness Landscapes and the Origin of Species. Princeton: Princeton Univ Press; 2004. [Google Scholar]
- 6.Stich M, Lázaro E, Manrubia SC. Phenotypic effect of mutations in evolving populations of RNA molecules. BMC Evol Biol. 2010;10:46. doi: 10.1186/1471-2148-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuster P, Stadler PF. Landscapes: Complex optimization problems and biopolymer structures. Comput Chem. 1994;18(3):295–324. doi: 10.1016/0097-8485(94)85025-9. [DOI] [PubMed] [Google Scholar]
- 8.Fontana W, Schuster P. Continuity in evolution: On the nature of transitions. Science. 1998;280(5368):1451–1455. doi: 10.1126/science.280.5368.1451. [DOI] [PubMed] [Google Scholar]
- 9.Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: A case study in RNA secondary structures. Proc Biol Sci. 1994;255(1344):279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- 10.Forst CV. Molecular evolution of catalysis. J Theor Biol. 2000;205(3):409–431. doi: 10.1006/jtbi.2000.2076. [DOI] [PubMed] [Google Scholar]
- 11.Orgel LE. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol. 2004;39(2):99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 12.Orgel LE. Evolution of the genetic apparatus. J Mol Biol. 1968;38(3):381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 13.Crick FH. The origin of the genetic code. J Mol Biol. 1968;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 14.Woese CR, Dugre DH, Dugre SA, Kondo M, Saxinger WC. On the fundamental nature and evolution of the genetic code. Cold Spring Harb Symp Quant Biol. 1966;31:723–736. doi: 10.1101/sqb.1966.031.01.093. [DOI] [PubMed] [Google Scholar]
- 15.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312(5770):111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 16.Lunzer M, Miller SP, Felsheim R, Dean AM. The biochemical architecture of an ancient adaptive landscape. Science. 2005;310(5747):499–501. doi: 10.1126/science.1115649. [DOI] [PubMed] [Google Scholar]
- 17.Hietpas RT, Jensen JD, Bolon DN. Experimental illumination of a fitness landscape. Proc Natl Acad Sci USA. 2011;108(19):7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinkley T, et al. A systems analysis of mutational effects in HIV-1 protease and reverse transcriptase. Nat Genet. 2011;43(5):487–489. doi: 10.1038/ng.795. [DOI] [PubMed] [Google Scholar]
- 19.Burch CL, Chao L. Evolution by small steps and rugged landscapes in the RNA virus phi6. Genetics. 1999;151(3):921–927. doi: 10.1093/genetics/151.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayden EJ, Wagner A. Environmental change exposes beneficial epistatic interactions in a catalytic RNA. Proc Biol Sci. 2012;279(1742):3418–3425. doi: 10.1098/rspb.2012.0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YH, Dsouza L, Fox GE. Experimental investigation of an RNA sequence space. Orig Life Evol Biosph. 1993;23(5-6):365–372. doi: 10.1007/BF01582086. [DOI] [PubMed] [Google Scholar]
- 22.Lee YH, DSouza LM, Fox GE. Equally parsimonious pathways through an RNA sequence space are not equally likely. J Mol Evol. 1997;45(3):278–284. doi: 10.1007/pl00006231. [DOI] [PubMed] [Google Scholar]
- 23.Zhang ZD, Nayar M, Ammons D, Rampersad J, Fox GE. Rapid in vivo exploration of a 5S rRNA neutral network. J Microbiol Methods. 2009;76(2):181–187. doi: 10.1016/j.mimet.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Schlosser K, Li Y. Diverse evolutionary trajectories characterize a community of RNA-cleaving deoxyribozymes: A case study into the population dynamics of in vitro selection. J Mol Evol. 2005;61(2):192–206. doi: 10.1007/s00239-004-0346-7. [DOI] [PubMed] [Google Scholar]
- 25.Pitt JN, Ferré-D’Amaré AR. Rapid construction of empirical RNA fitness landscapes. Science. 2010;330(6002):376–379. doi: 10.1126/science.1192001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joyce GF. Amplification, mutation and selection of catalytic RNA. Gene. 1989;82(1):83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 27.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 28.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 29.Ross CA, Poirier MA, Wanker EE, Amzel M. Polyglutamine fibrillogenesis: The pathway unfolds. Proc Natl Acad Sci USA. 2003;100(1):1–3. doi: 10.1073/pnas.0237018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren CL, et al. Defining the sequence-recognition profile of DNA-binding molecules. Proc Natl Acad Sci USA. 2006;103(4):867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorsch JR, Szostak JW. Chance and necessity in the selection of nucleic acid catalysts. Acc Chem Res. 1996;29(2):103–110. doi: 10.1021/ar9501378. [DOI] [PubMed] [Google Scholar]
- 32.Schuster P. Mathematical modeling of evolution. Solved and open problems. Theory Biosci. 2011;130(1):71–89. doi: 10.1007/s12064-010-0110-z. [DOI] [PubMed] [Google Scholar]
- 33.Rendel MD. Adaptive evolutionary walks require neutral intermediates in RNA fitness landscapes. Theor Popul Biol. 2011;79(1-2):12–18. doi: 10.1016/j.tpb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Ellington AD, Chen X, Robertson M, Syrett A. Evolutionary origins and directed evolution of RNA. Int J Biochem Cell Biol. 2009;41(2):254–265. doi: 10.1016/j.biocel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 35.Davis JH, Szostak JW. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc Natl Acad Sci USA. 2002;99(18):11616–11621. doi: 10.1073/pnas.182095699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turk RM, Chumachenko NV, Yarus M. Multiple translational products from a five-nucleotide ribozyme. Proc Natl Acad Sci USA. 2010;107(10):4585–4589. doi: 10.1073/pnas.0912895107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legiewicz M, Lozupone C, Knight R, Yarus M. Size, constant sequences, and optimal selection. RNA. 2005;11(11):1701–1709. doi: 10.1261/rna.2161305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabeti PC, Unrau PJ, Bartel DP. Accessing rare activities from random RNA sequences: The importance of the length of molecules in the starting pool. Chem Biol. 1997;4(10):767–774. doi: 10.1016/s1074-5521(97)90315-x. [DOI] [PubMed] [Google Scholar]
- 39.Carothers JM, Oestreich SC, Davis JH, Szostak JW. Informational complexity and functional activity of RNA structures. J Am Chem Soc. 2004;126(16):5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levenshtein V. Binary codes capable of correcting deletions, insertions, and reversals. Soviet Physics Doklady. 1966;10(8):707–710. [Google Scholar]
- 41.Lee JH, Jucker F, Pardi A. Imino proton exchange rates imply an induced-fit binding mechanism for the VEGF165-targeting aptamer, Macugen. FEBS Lett. 2008;582(13):1835–1839. doi: 10.1016/j.febslet.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merino EJ, Weeks KM. Fluorogenic resolution of ligand binding by a nucleic acid aptamer. J Am Chem Soc. 2003;125(41):12370–12371. doi: 10.1021/ja035299a. [DOI] [PubMed] [Google Scholar]
- 43.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci USA. 2005;102(7):2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gruber AR, Bernhart SH, Hofacker IL, Washietl S. Strategies for measuring evolutionary conservation of RNA secondary structures. BMC Bioinformatics. 2008;9:122. doi: 10.1186/1471-2105-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlosser K, Lam JC, Li Y. A genotype-to-phenotype map of in vitro selected RNA-cleaving DNAzymes: Implications for accessing the target phenotype. Nucleic Acids Res. 2009;37(11):3545–3557. doi: 10.1093/nar/gkp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gavrilets S, Gravner J. Percolation on the fitness hypercube and the evolution of reproductive isolation. J Theor Biol. 1997;184(1):51–64. doi: 10.1006/jtbi.1996.0242. [DOI] [PubMed] [Google Scholar]
- 47.Carothers JM, Oestreich SC, Szostak JW. Aptamers selected for higher-affinity binding are not more specific for the target ligand. J Am Chem Soc. 2006;128(24):7929–7937. doi: 10.1021/ja060952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carothers JM, Davis JH, Chou JJ, Szostak JW. Solution structure of an informationally complex high-affinity RNA aptamer to GTP. RNA. 2006;12(4):567–579. doi: 10.1261/rna.2251306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leu K, et al. Cascade of reduced speed and accuracy after errors in enzyme-free copying of nucleic acid sequences. J Am Chem Soc. 2013;135(1):354–366. doi: 10.1021/ja3095558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehman N. A recombination-based model for the origin and early evolution of genetic information. Chem Biodivers. 2008;5(9):1707–1717. doi: 10.1002/cbdv.200890159. [DOI] [PubMed] [Google Scholar]
- 51.Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323(5918):1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briones C, Stich M, Manrubia SC. The dawn of the RNA World: Toward functional complexity through ligation of random RNA oligomers. RNA. 2009;15(5):743–749. doi: 10.1261/rna.1488609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsiao C, et al. RNA with iron(II) as a cofactor catalyses electron transfer. Nat Chem. 2013;5(6):525–528. doi: 10.1038/nchem.1649. [DOI] [PubMed] [Google Scholar]
- 54.Athavale SS, et al. RNA folding and catalysis mediated by iron (II) PLoS ONE. 2012;7(5):e38024. doi: 10.1371/journal.pone.0038024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Held DM, Greathouse ST, Agrawal A, Burke DH. Evolutionary landscapes for the acquisition of new ligand recognition by RNA aptamers. J Mol Evol. 2003;57(3):299–308. doi: 10.1007/s00239-003-2481-y. [DOI] [PubMed] [Google Scholar]
- 56.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11(1):29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 57.Huang Z, Szostak JW. Evolution of aptamers with a new specificity and new secondary structures from an ATP aptamer. RNA. 2003;9(12):1456–1463. doi: 10.1261/rna.5990203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schultes EA, Bartel DP. One sequence, two ribozymes: Implications for the emergence of new ribozyme folds. Science. 2000;289(5478):448–452. doi: 10.1126/science.289.5478.448. [DOI] [PubMed] [Google Scholar]
- 59. Thompson WA, Newberg LA, Conlan S, McCue LA, Lawrence CE (2007) The Gibbs Centroid Sampler. Nucleic Acids Res 35(Web Server issue):W232–237. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.