Significance
The ability of the immune system to distinguish self from foreign (“self-tolerance”) is largely established in the thymus, a primary lymphoid organ where T cells develop. Intriguingly, T cells encounter most tissue-specific constituents already in the thymus, thus imposing a broad scope of tolerance before T cells circulate through the body. This preemption of the “immunological self” is afforded by the “promiscuous” expression of numerous tissue-specific antigens in medullary thymic epithelial cells. Here, we identified principles by which promiscuous gene expression at the single-cell level adds up to the full diversity of self-antigens displayed at the population level.
Keywords: human thymic epithelial cells, central tolerance, promiscuous gene expression
Abstract
Promiscuous expression of numerous tissue-restricted self-antigens (TRAs) in medullary thymic epithelial cells (mTECs) is essential to safeguard self-tolerance. A distinct feature of promiscuous gene expression is its mosaic pattern (i.e., at a given time, each self-antigen is expressed only in 1–3% of mTECs). How this mosaic pattern is generated at the single-cell level is currently not understood. Here, we show that subsets of human mTECs expressing a particular TRA coexpress distinct sets of genes. We identified three coexpression groups comprising overlapping and complementary gene sets, which preferentially mapped to certain chromosomes and intrachromosomal gene clusters. Coexpressed gene loci tended to colocalize to the same nuclear subdomain. The TRA subsets aligned along progressive differentiation stages within the mature mTEC subset and, in vitro, interconverted along this sequence. Our data suggest that single mTECs shift through distinct gene pools, thus scanning a sizeable fraction of the overall repertoire of promiscuously expressed self-antigens. These findings have implications for the temporal and spatial (re)presentation of self-antigens in the medulla in the context of tolerance induction.
Central T-cell tolerance in the thymus is an essential step in the complex process of induction, maintenance, and regulation of immunological self-tolerance. The nascent T-cell repertoire is probed against self-antigens presented by MHC class I and II on thymic antigen-presenting cells (APCs). As a result of these TCR peptide/MHC interactions, T cells whose self-reactivity exceeds a certain threshold will be either deleted or, as an alternative fate, deviate into the Treg lineage (1, 2). The cellular and molecular regulation of the various fate decisions within the T-cell lineage is only partially understood. The repertoire of self-antigens and the diversity of thymic APCs are important determinants in these selection events. Thymic APCs include various subsets of thymic dendritic cells (DCs), macrophages, thymic epithelial cells (TECs) and B cells, which present partly overlapping and partly complementary self-antigen repertoires (3).
The pool of self-antigens presented in the thymus is highly diverse in its composition and tissue derivation. Major contributors to this intrathymic antigen diversity are medullary thymic epithelial cells (mTECs) by virtue of expressing a host of tissue-restricted antigens (TRAs), which represent essentially all tissues of the body. This phenomenon has been termed promiscuous gene expression (pGE), allowing self-antigens, which otherwise are expressed in a spatially or temporally restricted manner, to become continuously accessible to developing T cells (4). The scope of central tolerance is to a large extent dictated by this pool of promiscuously expressed genes and even lack of a single TRA in mTECs can result in spontaneous organ-specific autoimmunity (5–8). How a differentiated epithelial cell type can override the temporal and spatial constraints of tissue-specific gene expression is currently poorly understood. The transcriptional regulator autoimmune regulator (Aire) has been shown to play a central role in pGE. Aire acts at the epigenetic level via binding to hypomethylated H3K4 residues and enhances transcription and mRNA splicing (9, 10). More recently, it has been shown that Aire binds more widely in the genome than would have been predicted by selective targeting me0H3K4. Aire-binding sites in the genome essentially overlapped with polymerase II (POL II) binding to transcriptional start sites (11). Aire, however, only controls a fraction of the genes expressed in mTECs, implying the existence of additional molecular mechanisms to ensure comprehensive tolerance against peripheral tissues.
Apart from the unusual molecular action of Aire in the regulation of pGE, another intriguing feature of pGE is its mosaic pattern. Although the pool of promiscuously expressed genes encompasses more than 1,000 TRAs, at a given time, each self-antigen is expressed by only 1–3% of mTECs (12, 13). However, the sets of genes expressed in single mTECs ultimately add up to a “complete” and stable representation of the promiscuous gene pool at the population level. We proposed this mosaic pattern to be the evolutionary result of balancing different parameters that determine the outcome of central tolerance: (i) expression of a maximal number of TRAs; (ii) sufficient TRA epitope density per cell to trigger a tolerogenic fate in developing T cells; and (iii) a critical number of mTECs expressing a given TRA to ensure efficient scanning for autoreactivity in newly generated T cells (4). It is still unclear which mechanisms dictate pGE at the single-cell level, that is, (i) whether it is stochastic, as previously proposed by us and others (13, 14), or subject to rules of coregulation; (ii) whether it is a cell-autonomous process or controlled by external signals; or (iii) whether it is stable during clonal expansion and terminal differentiation of mTECs (15).
To understand how gene expression at the single-cell level faithfully adds up to the full complement of pGE, we developed an experimental approach that allowed us to assess whether expression of a given TRA imposes restriction on the overall pGE pattern at the single-cell level. We performed population and single-cell gene expression analysis of ex vivo-isolated subsets of human mTECs displaying a particular surface TRA. Our studies revealed a considerable degree of gene coregulation in single cells, which encompassed intra- and interchromosomal coexpression groups. We present evidence that the different TRA subsets align along a colinear differentiation sequence implying that single mature MHCIIhi mTECs continue to cycle through distinct sets of promiscuously expressed genes. These findings have implications for the temporal and spatial representation of a diverse self-antigen repertoire within subdomains of the medulla in the context of tolerance induction.
Results
Isolation of TRA-Expressing Human mTEC Subsets.
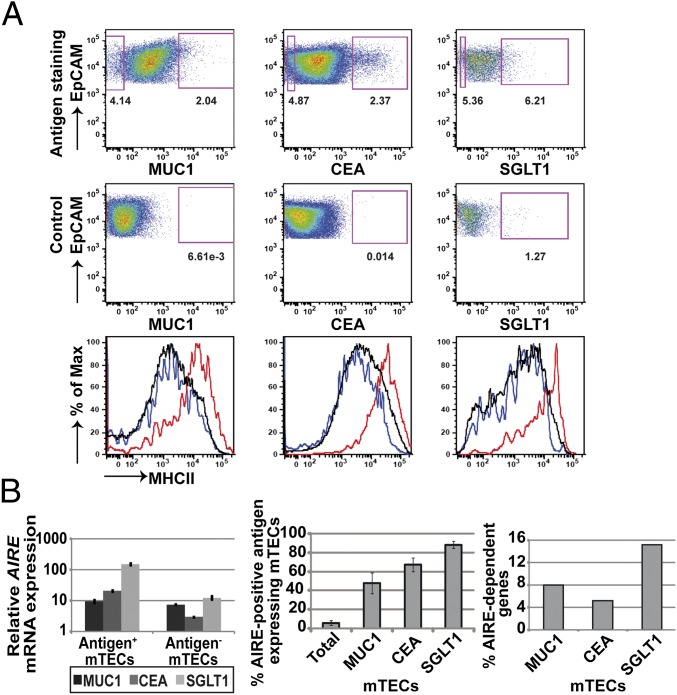
To assess to which extent pGE in single mTECs is random or restricted by gene coregulation, we isolated human mTEC subsets expressing a particular TRA. These TRAs had to fulfill several requirements: (i) cell-surface expression of a monomeric molecule (multiple chains of multimeric receptors are unlikely to be promiscuously coexpressed in the same mTEC); (ii) expression at low frequency, as typical for pGE; and (iii) availability of a suitable monoclonal antibody (mAb). We chose three TRAs namely Mucin (MUC)1, carcinoembryonic antigen-related cell adhesion molecule (CEACAM)5 (in short, CEA), and sodium/glucose cotransporter (SGLT)1 to which these criteria applied (12). The frequency of these TRA subsets ranged between 1% and 6%. Whereas MUC1+ and CEA+ subsets could be isolated from every processed sample, expression of SGLT1 was more variable precluding recovery from several samples. The sort strategy has been verified by the enrichment of mRNA specific for the respective antigen in the corresponding subset (Fig. S1A). All subsets expressed comparatively high levels of MHCII, a hallmark of mature mTECs (Fig. 1A). AIRE expression both at the mRNA and protein level was highest in the SGLT1 subset, followed by CEA and then MUC1 (Fig. 1B and Fig. S1B). Note that SGLT1 is an Aire-dependent gene in mice (16). Moreover, the content of “AIRE-regulated” human genes [i.e., orthologs of murine Aire-dependent genes (16, 17)] was significantly increased in the SGLT1 gene pool (see below), in line with up-regulated AIRE expression in this subset (Fig. 1B). Because the up-regulation of AIRE typifies mTEC differentiation from the immature to the mature stage, in mouse and human, we consider the three TRA subsets to represent sequential stages of this differentiation process.
Fig. 1.
Defining human antigen-expressing mTEC subsets. (A) MUC1+, CEA+, and SGLT1+ mTECs represent minor subsets of total mTECs. All three different mTEC subsets expressed high levels of MHCII (red) compared with antigen-negative mTECs (blue) and complete mTECs (black line). Individual samples used are shown as running sample numbers: MUC1 (#133), CEA (#138), SGLT1 (#174). Mean frequencies: MUC1+ mTECs, 2.7 ± 1.4 (n = 16); CEA+ mTECs, 1.4 ± 0.7 (n = 15); SGLT1+ mTECs, 3.3 ± 1.7 (n = 9). (B) Relative mRNA expression levels of AIRE in the three mTEC subsets and percentage of AIRE protein-positive mTECs. AIRE steadily increased from MUC1- to SGLT1-positive cells, with the latter being nearly completely AIRE-positive. The AIRE mRNA levels within each mTEC subset are shown in MUC1 (#97), CEA (#116), and SGLT1 (#174). For AIRE protein staining, all three antigens and AIRE were costained. The error bars indicate the SD of three samples (#164, #165, #166). The percentage of “AIRE-dependent” genes in the up-regulated gene pool increased from MUC1+ to SGLT1+ mTECs. Definition of “AIRE dependency” in humans was based on the Aire dependency of their mouse orthologs. The pool of AIRE-dependent genes was calculated based on array expression data for each antigen; MUC1, n = 3 (#94, #96, #97); CEA, n = 2 (#116, #118); SGLT1, n = 3 (#169, #174, #177).
Defining Gene Coexpression Groups in TRA Subsets.
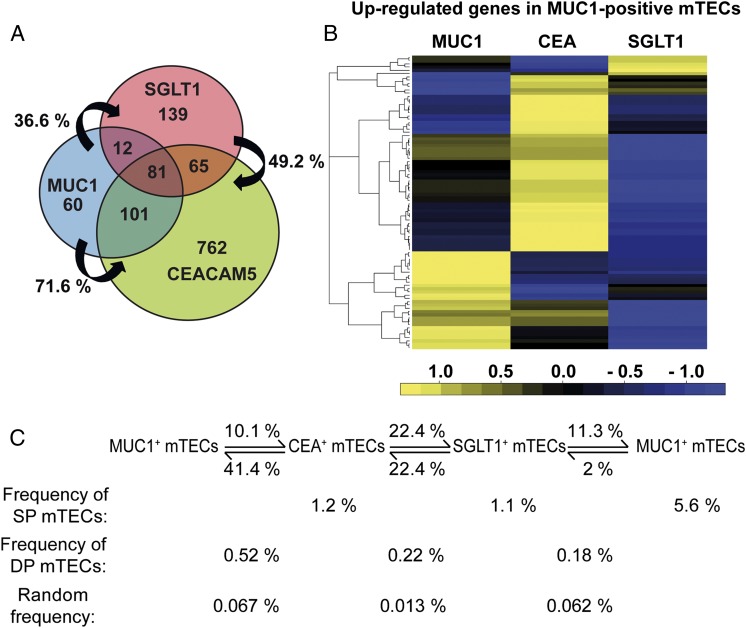
Does expression of a given TRA impose restriction on pGE at the population and single-cell level? To address this issue, we separated each TRA subset into an antigen-positive and -negative fraction according to the sort gates indicated in Fig. 1A. The typical yield of antigen-positive cells ranged between 18,000 and 43,000 cells starting with a total of approximately 3 × 107 CD45-depleted thymic cells. Both fractions were subjected to transcriptome analysis using whole-genome microarrays. Genes which differed between the positive vs. the negative fraction by a log fold change (fc) of ≥2 were considered to be up-regulated and defined as a TRA-related coexpression group. Coexpression groups from different individuals showed a high concordance of overlap (Fig. S1C). The three TRA coexpression groups were both complementary and overlapping (Fig. 2 A and B). A surprisingly high degree of overlap of 70% was noted between the MUC1 and the CEA groups. Interestingly, the overlap was asymmetric (i.e., the MUC1 group was to a much higher degree contained within the CEA group than vice versa). The least overlap was observed between the MUC1 and SGLT1 groups. This particular pattern of mutual overlap was also mirrored at the protein level (i.e., whereas 41% of CEA+ mTECs coexpressed MUC1, only 10% of MUC1+ mTECs also coexpressed CEA; Fig. 2C and Fig. S1D).
Fig. 2.
Overlapping coexpression groups in antigen-expressing mTEC subsets. (A) Mutual overlap of up-regulated gene pools within MUC1+, CEA+, and SGLT1+ mTECs, respectively. For each mTEC subset, amplified cDNA of two to three individuals was separately hybridized to whole-genome BeadArrays. Transcripts were considered up-regulated between antigen-positive and antigen-negative mTECs if the log ratio of the average signal intensities of the corresponding probe over all patients was greater or equal than 2. The logarithm was calculated to base 2. The array results showed varying degrees of overlap between the three subsets. The expression data for each antigen was averaged among individuals; MUC1, n = 3 (#94, #96 #97); CEA, n = 2 (#116, #118); SGLT1, n = 3 (#169, #174, #177). The percentage of overlap between individuals was calculated as the sum of gene overlap between two individuals times 100 divided by the sum of genes of the individual having the smaller gene pool of the two. (B) The top 100 most differentially expressed genes between the MUC1+ and MUC1− mTECs were selected. The heat map shows the top genes in the MUC1 group to be further up-regulated in one of the other two groups, whereas others were down-regulated. Colors show expression relative to the average (black), higher (yellow), or lower (blue) relative expression, respectively. The expression data for each antigen were averaged among individuals (#94, #96, #97). (C) Protein coexpression patterns of the three antigens defining the mTEC subsets in one individual (#141). The calculation of random coexpression frequencies was based on the percentage of each antigen-positive subset within complete mTECs.
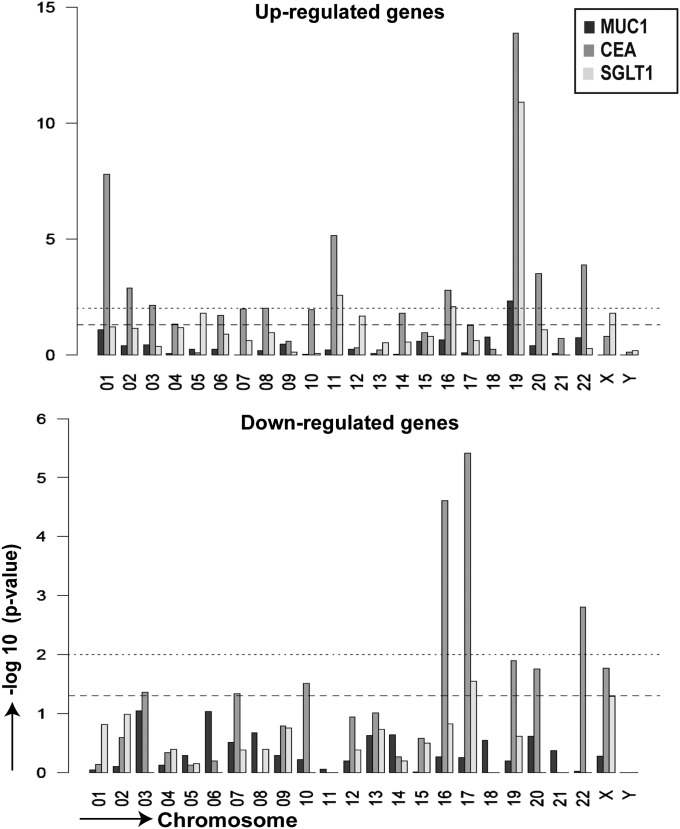
The genes comprising the three coexpression groups showed no structural or functional commonalities or preferential tissue affiliations. They were enriched in TRAs and also clustered in the genome, two features previously described for promiscuously expressed genes irrespective of coexpression (16–19) (Fig. S2). In contrast to the pGE pool in unselected MHCIIhi mTECs, there was, however, a clear preference for genes located on chromosome (chr) 19 in all three groups and a relative depletion of genes on chr 17 for the CEA and SGLT1 gene pools (Fig. 3).
Fig. 3.
Chromosomal preference among up- and down-regulated genes. The preferential chromosomal localization of genes up- and down-regulated in MUC1+, CEA+, and SGLT1+ sorted mTECs was analyzed by Fisher’s exact test. The plot represents the negative log10 of the P value vs. chromosome number. The lower and upper dotted lines correspond to P = 0.05 and P = 0.01, respectively. In all three groups, there was a significant preference for up-regulated genes to localize to chr 19 and in 2 groups for down-regulated genes to localize to chr 17. The data for each antigen was averaged among individuals; MUC1, n = 3 (#94, #96, #97); CEA, n = 2 (#116, #118); SGLT1, n = 3 (#169, #174, #177).
Comparing Expression Patterns Among Coexpression Groups.
The three coexpression groups could either arise independently of each other or reflect a transition between the respective groups (Fig. 2A). To probe this issue, we asked how the 100 top-ranking genes of a coexpression group (with respect to differential expression between antigen-positive and -negative mTECs) changed their relative expression hierarchy between the different groups. The analysis should reveal the relative relatedness between the three groups. Thus, among the top-ranking genes of the MUC1 group, about one-third were down-regulated in both the CEA and SGLT1 groups, whereas about half were transiently further up-regulated in the CEA group and, again, down-regulated in the SGLT1 group. Only a small subset among the top 100 MUC1 genes showed up-regulation in the SGLT1 group (Fig. 2B). These shifts of gene-expression patterns document the close relatedness among the three groups. Next, we applied this approach to a single gene cluster.
Gene Coexpression at the Single-Cell Level.
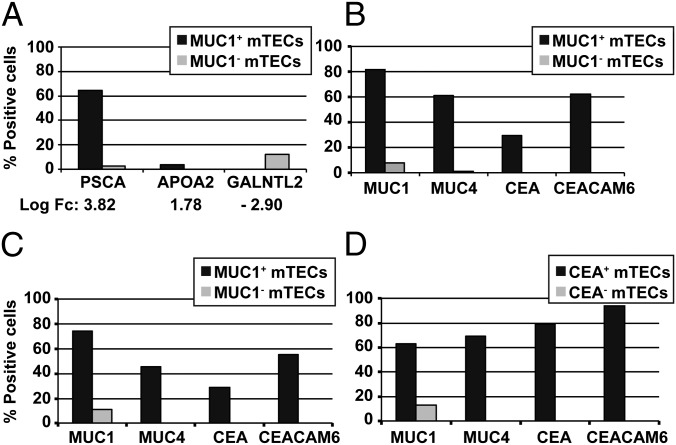
How does coexpression as detected by gene arrays at the population level translate into coexpression frequencies at the single-cell level? To address this question we first arbitrarily selected three genes, which ranked at different positions on the microarray in terms of fc on the list of up-regulated genes in the MUC1+ fraction and determined their expression frequency at the single-cell level. Of single MUC1+ mTECs, 65% expressed prostate stem cell antigen (PSCA), corresponding to a log fc of 3.82; 3% expressed apolipoprotein (APO)A2, corresponding to a log fc of 1.78; and none expressed UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase-like protein 2 (GALNTL2), corresponding to a −log fc of 2.90 (Fig. 4A). This documents a clear concordance between average expression level, as detected on microarrays, and the proportion of single cells expressing the respective TRA.
Fig. 4.
Intra- and interchromosomal gene coexpression at the single-cell level. (A) Correlation between the hierarchy in coexpressed genes at the population level based on fc in microarrays and the frequency of single mTECs coexpressing the corresponding genes. Basal frequency in single mTECs: PSCA, 12%; APOA2, 0%; and GALNTL2, 12%. (B–D) Coexpression patterns of single MUC1+/MUC1− and CEA+/CEA− mTECs. MUC1+/MUC1− mTECs from two different individuals (#103, #105) showed a similar pattern of coexpression of MUC4 (chr 3) and members of CEA gene family locus (chr 19). CEA+/CEA− sorted mTECs (#143) showed a similar pattern of coexpression of the same genes, though at different frequencies. Note the high concordance between protein and mRNA expression for both antigens. Basal frequency in single mTECs: MUC1, 17%; MUC4, 2%; CEA, 8%; and CEACAM6, 8%. On average, 80–82 antigen-positive or -negative single cells and 88 single unseparated mTECs were analyzed for each individual.
Given the overlap of the MUC1 and CEA gene-coexpression groups and costaining of both proteins at the cell surface (Fig. 2 A and C) and in situ (12), we determined the correlation between protein and mRNA expression at the single-cell level. Of MUC1 protein-positive mTECs, 82% expressed the MUC1 gene, and 30% expressed the CEA gene at the mRNA level (Fig. 4 B and C). Inversely, of sorted CEA protein-positive mTECs, 80% expressed the CEA gene, and 64% expressed the MUC1 gene (Fig. 4D). To probe whether members of the same gene family might be coexpressed, we included the MUC4 gene (chr 3) and the CEACAM6 gene (chr 19), both of which were also highly coexpressed (Fig. 4 B–D). Each of these four genes was only expressed in 1–17% of all mTECs [as determined by single-cell (SC)-PCR]; hence, the results obtained in selected mTEC subsets showed a highly significant enrichment (Tables S1–S3). However, coexpression of the four genes at the single cell entailed a degree of variability, presumably reflecting a stochastic component of pGE (13, 14) (Fig. S3 A and B). The SC-PCR data also revealed the aforementioned asymmetry between both groups (i.e., MUC1+ mTECs were less enriched for CEA+ mTECs than CEA+ mTECs for MUC1+ mTECs). Our data document that single mTECs coexpress to a high degree functionally unrelated TRAs.
Comparing Coexpression Patterns Within a Single Gene Cluster.
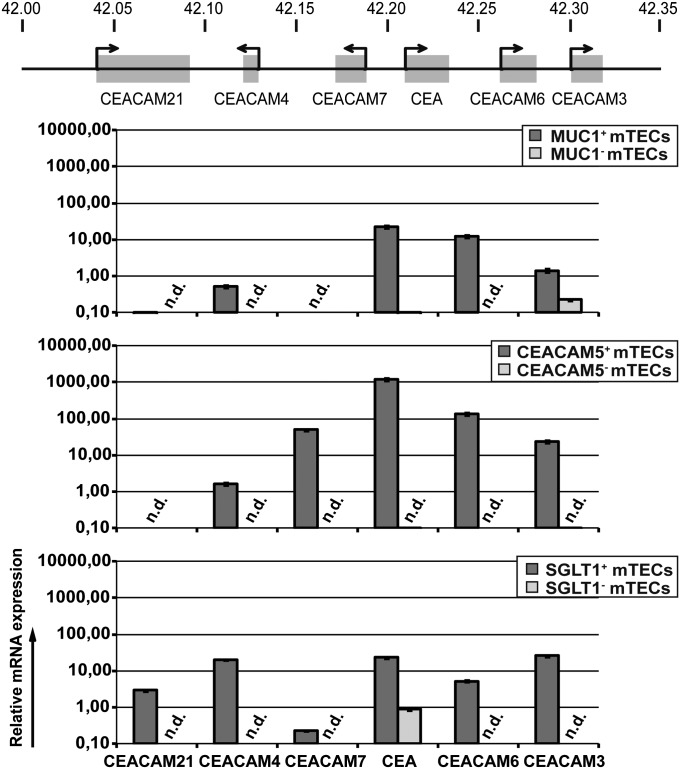
The CEACAM family consists of 12 members spread along chr 19, 6 of which form a contiguous cluster spanning 250 kb (Fig. 5). Notably CEA+ mTECs coexpressed the adjacent CEACAM6 gene to a higher degree than MUC1+ mTECs (Fig. 4 B–D). We therefore asked whether appropriate enrichment of minor mTEC subsets (i.e., selecting for a gene within a given cluster) might reveal extended or even contiguous expression of this cluster, which would not be evident in the bulk population (13, 14). Accordingly, we compared the expression of the six clustered CEACAM genes in all three coexpression groups by quantitative RT-PCR. Because of the high homology among these genes, we were not able to design gene-specific primer pairs to perform multiplex SC-PCR. The MUC1+ subset expressed three of six members of this cluster; the CEA+ mTECs expressed five contiguous members. The SGLT1+ mTECs, in addition, expressed CEACAM21, while downregulating CEACAM7 (Fig. 5). Note that the expression levels were 10- to 100-fold higher in the CEA+ subset. This differential representation of the CEACAM locus in the three subsets also reflected the respective overlap of the three gene coexpression pools (Fig. 2). Hence, in this case, promiscuous expression of a given TRA goes along with coexpression of the immediate gene neighborhood. Such coexpressed neighborhoods could only be fully revealed by enrichment of the appropriate mTEC subset.
Fig. 5.
Differential representation of the CEACAM locus in the coexpression groups. Different expression patterns of the CEACAM gene locus were observed in MUC1+ (#97), CEA+ (#118), and SGLT1+ (#174) sorted mTECs. Note that CEA+ sorted mTECs showed contiguous expression of five genes, and 10- to 100-fold higher gene-expression levels than the other groups. Expression levels were normalized to GAPDH, relative to complete human thymus (set to relative expression of 1, which was considered as the expression cutoff). n.d., not detectable.
Coexpressed Gene Loci Are Colocalized.
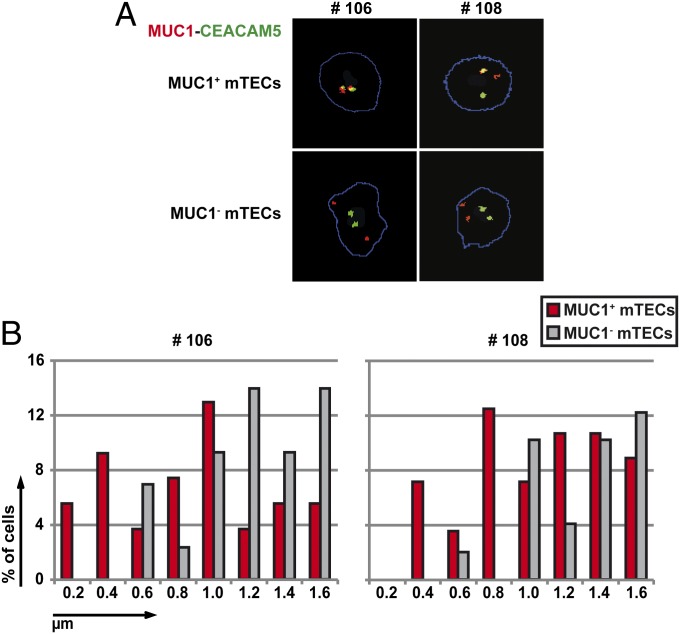
Coordinated expression of functionally related genes has been associated with their colocalization in nuclear subdomains [e.g., transcription factories or nuclear speckles (20)]. The distinct and reproducible coexpression patterns in single mTECs prompted us to analyze whether such colocalization in 3D space may also apply to functionally unrelated, promiscuously expressed genes. We performed two-color DNA-fluorescence in situ hybridization (FISH) analysis of the MUC1 and CEA loci in sorted MUC1+/MUC1− and CEA+/CEA− mTECs. Strikingly, the MUC1+ mTECs of three different individuals contained a significantly higher fraction of cells in which the MUC1 and CEA loci were colocalized in 3D space compared with MUC1− mTECs (Fig. 6B and Fig. S4). Similar results were obtained in CEA+ vs. CEA− mTECs (Fig. S4). We observed both mono- and biallelic colocalization (Fig. 6A). Thus, coexpression of the MUC1 and CEA loci might operate via colocalization within the same nuclear subcompartment, a mechanism that has been described to operate in case of tissue-specific gene regulation (21).
Fig. 6.
Coexpressed genes colocalized to nuclear subdomains. (A) MUC1-specific (red) and CEA-specific (green) probes were used for two-color DNA-FISH analysis of ex vivo-isolated human MUC1+ and MUC1− mTECs. Examples of monoallelic and biallelic colocalizations are shown, both of which were highly enriched in the MUC1+ compared with the MUC1− mTEC fraction. (B) Distribution of distances in MUC1+ vs. MUC1− mTECs between the MUC1 and CEA gene loci using DNA-FISH. In both samples studied (#106, #108), the MUC1+ fraction showed significant colocalization of the two loci (≤0.8 µm) compared with MUC1− mTECs. Each plot represents cells from one individual; a minimum of 50 cells was analyzed using the semiautomatic image analysis for each sample. (x axis: distances in 0.2-µm increments.)
Interconversion of TRA-Specific Subsets in Vitro.
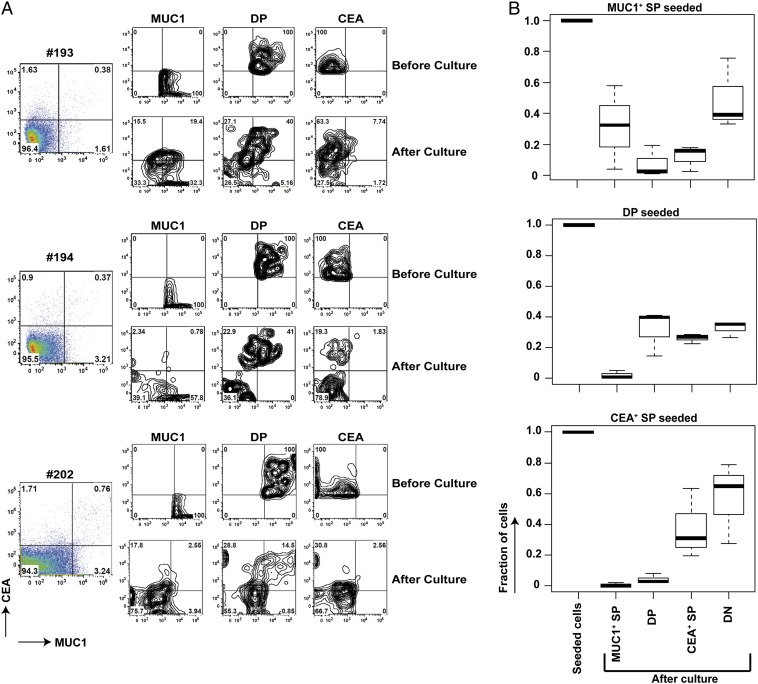
The different levels of AIRE expression, and the different content of “AIRE-regulated” genes in the three subsets, insinuated a developmental sequence with MUC1+ mTECs being the least and SGLT1+ mTECs the most differentiated subset. To assess a potential precursor–product relationship among these subsets, we purified MUC1 SP (single-positive), CEA SP, double-negative (DN), and double-positive (DP) mTECs and placed them into a simplified 3D-culture system in the presence of receptor activator of NF-κB ligand (RANKL). After 50–55 h of culture, the recovered cells were rephenotyped. Intriguingly, the phenotype of the antigen-expressing subsets followed the predicted developmental sequence; MUC1 SP turned into DP, CEA SP, and DN mTECs; DP turned into CEA SP and DN mTECs; CEA SP turned into DN mTECs; whereas DN cells retained their phenotype (Fig. 7). Notably, we did not observe the “reverse” conversion (i.e., CEA SP or DP mTECs did not convert into MUC1 SP mTECs). The data suggest that different TRA-specific subsets progress within the mature mTEC subset in the absence of an intact thymic microenvironment. Given the relatively low recovery rate of mTECs after the 3 d of culture, we cannot strictly exclude selective survival to at least partially account for the observed phenotype shift. This, however, is rather unlikely, because preferential survival of any subset should have resulted in the same conversion pattern irrespective of the seeding population.
Fig. 7.
Interconversion of TRA specific subsets in vitro. (A) Sorted DN, MUC1 SP, CEA SP, and DP mTECs from an individual sample were cultured in a short-term simplified 3D culture model. Within 50–55 h, the purified subsets converted along the sequence: MUC1 SP > DP > CEACAM5 SP > DN. Typically, 5,000–300,000 cells were seeded. The three datasets (#193, #194, #202) are representative of four independent experiments. The sort purity (reanalysis) was as follows (n = 4): DN, 99.05% (± 0.64%); MUC1 SP, 95.35% (± 1.34%); DP, 88.45% (± 8.70%); CEA SP, 80.85% (± 0.07%). The percentage of cells retrieved after culture was as follows: DN, 2.95% (± 1.93%); MUC1 SP = 3.06% (± 0.19%); DP, 32.53% (± 8.36%); CEA SP, 9.19% (± 5.75%). The percentage of cells retrieved after culture was based on the input of sorted cells. These numbers underestimate the conversion efficiency of those cells that eventually seed the culture, given substantial cell death during the first 12 h of culture as a result of the extended isolation procedure. (B) Boxplots showing the distribution of percentages of the three purified subsets pre- and postculture. Whereas there are variations between the three individuals, as reflected by the height of the boxes (i.e., the interquartile range), each seeded subset yields its distinct conversion pattern within the four quadrants of the 2D FACS analysis.
Discussion
In this study, we show that pGE at the single-cell level is not purely stochastic but reveals distinct patterns of gene coexpression. Coexpression groups were identified via isolation of minor human mTEC subsets expressing a particular surface TRA. Genes coexpressed with the selected TRA localized preferentially to certain chromosomes and may also encompass contiguous gene clusters if located on the same chromosome. Coexpressed genes had no apparent functional or structural commonalities. Moreover, in the case of two highly coexpressed loci on chr 1 and chr 19, we show preferential colocalization in 3D nuclear space. Intriguingly, the three identified coexpression groups may represent successive differentiation stages within the mature mTEC subset.
Gene coexpression was only revealed by focusing our analysis on restricted mTEC subsets, as defined by expression of a particular TRA. A previous study analyzing complete mTECs in mice failed to detect coexpression patterns within the casein gene locus at the single-cell level, leading to the conclusion that pGE might be a stochastic process (13). This conclusion was reported independently by applying a similar approach (14). Because of the mosaic expression of TRAs in mTECs, any coexpression pattern confined to a minor subset of mTECs, however, would be difficult to distinguish from the heterogeneous background noise. The necessity to enrich for a specific subset to reveal a particular gene coexpression pattern was exemplified by the differential representation of the CEACAM locus in the three mTEC subsets. Those mTECs expressing a member of the locus itself also showed the highest expression levels of five contiguous CEACAM genes. Coexpression of genes located in clusters rather than individual genes could also be seen in two functionally unrelated genes down-stream of SGLT1. These observations are compatible with the notion that promiscuously expressed genes per se and, in particular, Aire-dependent genes tend to cluster in the genome (16, 19).
Which mechanism enacts the coexpression of a limited gene set in TRA-positive mTECs? Coexpressed genes displayed neither obvious functional or structural commonalities nor common transcription factor (TF)-binding motifs in their promoters but showed enrichment for TRAs, which were clustered in the genome, features shared with pGE in general. The only distinctive feature of coexpression groups identified so far was their chromosomal predilection. All three groups were enriched for genes on chr 19. Two out of three groups were enriched for genes on the chromosome on which the selecting TRA was localized; conversely, the CEA and SGLT1 groups were highly depleted for genes on chr 17. The frequent colocalization of the MUC1 and CEA loci in MUC1+ and CEA+ cells was reminiscent of similar findings showing a correlation between gene coregulation and nuclear colocalization when analyzing tissue-specific gene programs. These examples include the interaction between the IFN-γ promoter and the Th2 locus control region during Th1 vs. Th2 cell differentiation (22), the interaction between X-inactive specific transcript (Xist) and XIST antisense RNA (Tsix) elements [critical control sequences in the X inactivation center (Xic) during X chromosome inactivation (23)], selection of olfactory receptor genes in different sensory neurons (24), and the colocalization of Kruppel-like factor (Klf)1-dependent genes in transcription factories in the erythroid cell lineage (21). Gene coregulation in the context of pGE might rely on the same strategy, although tissue-specific TFs are unlikely to be involved. In all instances tested to date, tissue-specific TFs were dispensable for pGE (25–28). Irrespective of the molecular mechanisms promoting colocalization of individual gene loci, the overall preference for certain chromosomal localizations (both in cis and trans with respect to the selecting TRA) reveals yet another layer of regulation of pGE at the level of chromosomal topology.
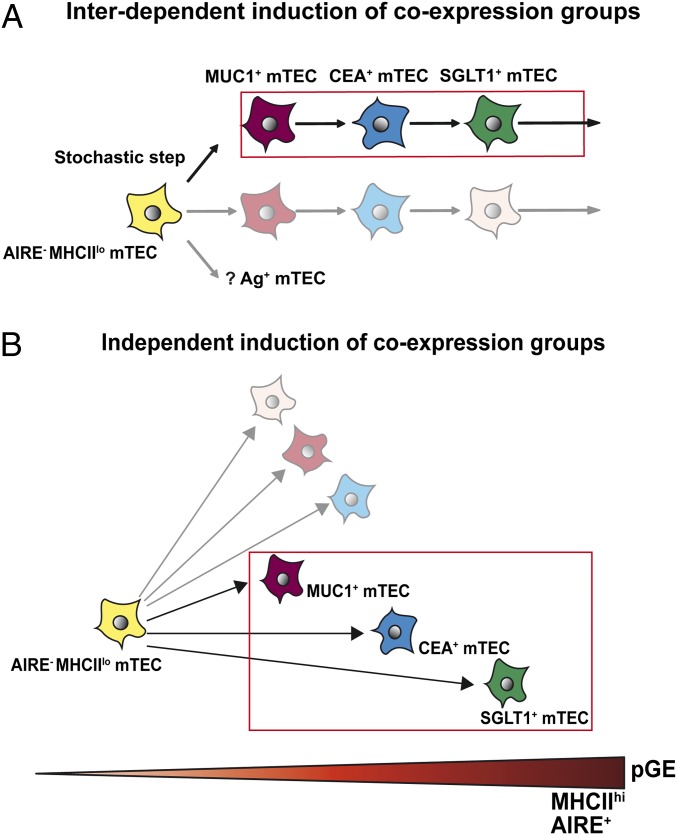
Several models can be considered to explain the genealogy of coexpression groups. The different coexpression groups could arise independently of each other, represent a single colinear differentiation sequence or a combination of both (Fig. 8). We favor the notion that the three groups represent a colinear differentiation sequence, based on (i) the increase of AIRE expression/AIRE-dependent gene regulation in ascending order from MUC1 to CEA to SGLT1 [note that all three coexpression groups display high levels of MHCII and, thus, are unlikely to map to the MHCIIlo/int post-Aire stage of mTEC differentiation as recently described in mice (29, 30)], (ii) the substantial overlap—both at the protein and mRNA level—among the coexpression groups, and (iii) the results of the short-term in vitro conversion assay. This model implies that promiscuous expression of a given set of genes is transient and not locked in, as typically observed for terminally differentiated cell lineages (31, 32). Evidence for such transiency of pGE has been previously reported for two TRAs, glutamate decarboxylase (GAD)67 and connexin 57 in mouse (26). Given the close correspondence between the PCR results at the population and the single-cell level, our results infer that single mTECs “shift” through different coexpression groups and, thus, may cover a sizeable portion of the overall pGE pool during their lifetime. How such fleeting coexpression patterns would be regulated at the molecular level remains unclear. Given the fact that the three TRAs analyzed encompass only 24% of MHCIIhi or 8% of total mTECs and considering the observed chromosomal bias, there have to be additional coexpression groups. However, the considerable overlap among the three coexpression groups suggests that only a limited number of delineable groups exist.
Fig. 8.
Genealogy of coexpression groups. (A) The model of shifting coexpression groups posits that the three coexpression groups represent a colinear differentiation sequence originating from a common precursor. Note that all three groups add up to 24% of all MHCIIhi mTECs, implying the existence of additional coexpression groups. It is proposed that a stochastic step initiates different coexpression patterns in individual cells, followed by sequential induction of coexpression patterns in a deterministic and hierarchical fashion in different sublineages. (B) Alternatively, coexpression groups arise independently of each other from a common precursor. Delineable coexpression groups are color-coded.
Fluctuating pGE should result in a graded density of T-cell epitope display on mTECs, which might offer an explanation for the correlation between TCR affinity and niche size for intrathymic selection of Treg cells with mTECs displaying low levels of self-antigen epitopes extending the niche to high affinity TCRs (33, 34). A prerequisite of this proposition is a tight quantitative and temporal correlation between the expression level of mRNA and corresponding protein (or peptide intermediates thereof) and subsequent T-cell epitope display by surface MHC. This requires the turnover of MHC receptors in mature mTECs to be somewhat synchronized with the fluctuation of pGE. Indeed, the half-life (t1/2) of MHC II (IAb) on mTECs in C57BL/6 mice, which has been estimated to be around 24 h, is considerably shorter than the lifespan of mature mouse mTECs in the range of 14–21 d (35, 36).
Our findings have implications for the generation of antigen diversity in time and space within the thymic microenvironment. The sequential expression and presentation of different sets of genes at the single-cell level would substantially reduce the number of mTECs required to represent the “full” antigen repertoire. Thus, a diverse antigen repertoire might be already displayed in subdomains of the medulla. In turn, it would be sufficient for nascent T cells to scan such subdomains provided they reencounter the same mTEC over time. Recent in vivo imaging data showing that autoreactive thymocytes in the presence of the cognate antigen roamed in confined areas are compatible with such a scenario (37).
Materials and Methods
Human Thymic Tissue.
Human thymus samples were obtained in the course of corrective cardiac surgery at the Department of Cardiac Surgery, Medical School of the University of Heidelberg. This study has been approved by the Institutional Review Board of the University of Heidelberg.
Isolation of Thymic Epithelial Cells.
Human thymic epithelial cells were purified as described previously (18), with some modifications. In brief, the thymi were digested sequentially with three rounds of collagenase/dispase for 20 min, each at 37 °C, followed by trypsin for 10 min, each at 37 °C, in a water bath with magnetic stirring. The trypsin fractions were pooled and filtered through 60-μm gauze. mTECs were preenriched by magnetic cell sorting, followed by FACS.
Magnetic cell sorting was performed using anti-CD45 Microbeads (Miltenyi Biotech). The labeled CD45+ cells were depleted using the autoMACS Pro Separator (Miltenyi Biotech). This enriched stromal cell fraction was stained with a biotinylated anti-epithelial cell adhesion molecule Ab [EpCAM-bio, clone HEA125; kindly provided by G. Moldenhauer, German Cancer Research Center (DKFZ); sav-PE (BD Biosciences)], CDR2-Alexa488 [cortical dendritic reticulum antigen 2 (DKFZ); Alexa Fluor 488 Protein Labeling kit (Molecular Probes)], Alexa 647- or Alexa 680-conjugated mAb HLA-DR [clone L243 (kindly provided by G. Moldenhauer); Alexa Fluor 647 or 680 Protein Labeling kit (Molecular Probes)], and CD45-PerCP (clone 21D; BD Biosciences). Depending on which antigen-expressing mTECs were isolated, either Alexa 647- or Alexa 680-conjugated mAb specific for MUC1 [clone 214D4 (kindly provided by W. Germeraad, University of Maastricht, Maastricht, The Netherlands); Alexa Fluor 647 or 680 Protein Labeling kit (Molecular Probes)], Alexa 647- or Alexa 680-conjugated mAb specific for CEACAM5 [clone PARLAM-4 (kindly provided by W. Germeraad); Alexa Fluor 647 or 680], or polyclonal anti-SGLT1 (ab14686; Abcam)/anti-rabbit Alexa 647 or Alexa 680 were used. mTECs were sorted as CD45− CDR2− EpCAM+ cells. The antigen-expressing mTEC gate ranged between 1% and 6%. Dead cells were excluded by propidium iodide (0.2 μg/mL). Cell sorting was performed on a FACS Aria (BD Biosciences).
For intracellular AIRE staining, directly conjugated AIRE-PE-Cy7 antibody [clone 6.1; kindly provided by P. Peterson (University of Tartu, Tartu, Estonia)] was used. The staining was performed using the FoxP3 staining buffer set (Miltenyi Biotec) according to the manufacturer’s instructions and measured on a LSRII flow cytometer (BD Biosciences).
RNA Preparation and cDNA Synthesis.
The RNA from single-cells was isolated using the High Pure RNA Isolation Kit (Roche). The isolated RNA was reverse-transcribed into cDNA with Oligo(dT)20 Primer and SuperScript II Reverse Transcriptase (Invitrogen), followed by RNase H digestion (Fermentas).
μMACS SuperAmp Technology for Illumina BeadArrays.
The μMACS SuperAmp protocol optimized for rare cell populations (Miltenyi Biotec) was used for RNA amplification of sorted mTECs. The amplified cDNA was labeled and hybridized to microarrays for gene-expression profiling using Illumina’s whole-genome BeadArrays. For technical details, see the manufacturer’s TechNote (38).
Quantitative PCR.
Real-time PCR reactions were performed in a final volume of 25 µL using either unamplified cDNA or unbiotinylated SuperAmp cDNA with optimal concentrations of forward and reverse primers (50–900 nM) using Power SYBR Green PCR Master Mix (Applied Biosystems). Reactions were run on a sequence-detection system (GeneAmp 7300; Applied Biosystems) in duplicates, and expression values were normalized to GAPDH expression, relative to complete human thymus using the comparative threshold cycle (Ct) method. Primers were purchased from MWG and, whenever possible, were designed to span at least one intron. Sequence information on primer pairs used is available upon request.
Single-Cell Sort and Single-Cell PCR.
Single-cell sorting and PCR were performed as described previously (13). In short, single cells were sorted with a FACSDiVa (Becton Dickinson) at 16 psi in single-cell mode using the automatic cell deposition unit. Cells were collected in 5 μL of PBS-DEPC (diethylpyrocarbonate, 0.1%) using 0.2-mL PCR eight-well strips (Nerbe) arranged in a 96-well format and stored at −80 °C. Single-cell PCR reverse transcription, first PCR amplification, and real-time quantitative PCR were performed using the DNA engine Dyad (MJ Research), and the 7300 Real-Time PCR System (Applied Biosystems). Fourteen cycles were used for the first PCR providing the best correlation between input cDNA and resulting Ct values. The data were analyzed in a qualitative fashion. In the case of an atypical melting curve, the product from the respective well was reamplified by the appropriate primer combination and verified by sequencing. Primer sequences are available upon request.
Bioinformatic Analysis of Microarrays.
Quantile normalization (39) of the microarray data followed by limma differential gene-expression analysis was performed to identify differentially expressed genes between the antigen-positive and -negative mTEC subsets. Genes with a log fc of ≥2 or ≤2 were considered to be significantly differentially expressed. All calculations were performed in R version 13.1 (40).
AIRE-regulated human genes were defined using mouse orthologs from the Ensembl database (release 62), annotated to the corresponding Ensembl gene identifier.
Tissue-restricted antigens were defined using the public database at http://symatlas.gnf.org (41). A gene was defined as a TRA if its expression was above 5× the median expression over all tissues in less than five tissues. TRAs were defined at the gene level; we mapped differentially expressed transcripts from the array analysis to the corresponding gene identifiers to compare them with the defined TRAs. The significance of TRA enrichment (P value) was calculated using Fisher’s exact test.
To determine whether the up-regulated genes in MUC1, CEA, and SGLT1 arrays were clustered, the 10-gene window algorithm was used as described previously (16). Briefly, a running window of 10 consecutive genes in a chromosomal region was tested for the number of TRAs. Window position of peaks was identified and counted as cluster of size n (n > 1). In some cases, immediately neighbored clusters were less than 10 genes apart; an assembly step was appended to the algorithm to combine such clusters. The significance of the clustering was determined by repeating the same procedure 1,000 times in each case with a list of randomly selected genes of the same size as the experimental dataset, and the results were compared with the number of clusters found.
Chromosomal preference calculation for the up- and down-regulated genes was performed using Fisher’s exact test, taking into consideration the distribution of the genes covered by the probes on the chip to different chromosomes.
Log fc expression values of genes that showed more than twofold up-regulation in MUC1-positive vs. MUC1-negative mTECs, or CECAM5-positive vs. CEACAM5-negative mTECs, or SGLT1-positive vs. SGLT1-negative mTECs, were clustered by hierarchical clustering (complete linkage algorithm, Euclidean distance metric) of values that had been transformed to zero mean and unit standard deviation (SD). Visualization was by heat maps, where yellow denoted high expression and blue denoted low expression relative to the mean over the three datasets.
Three-Dimensional DNA-FISH.
Three-dimensional DNA-FISH was performed on ex vivo-isolated human mTECs as described previously (26). Labeled BAC clones (RPCI-11-263K19, RPCI-11-343B1) spanning genes of interest were purchased from Empire Genomics. Z-stack images were acquired with a Leica TCS SP5 and Zeiss LSM 780 confocal microscopes using the 63× oil objective.
Semiautomatic image analysis was performed using a custom-made ImageJ plugin and Matlab software (MathWorks). The 3D Euclidean distance was measured between the spot centers of the confocal slices with the largest area for each FISH probe, which was considered as the probe center. The significance (P value) was calculated using Fisher's exact test.
For fully automatic 3D DNA-FISH image analysis, first, 3D segmentation of the cell nucleus and the FISH signals was performed, and, second, distances between the 3D centroids of the segmented FISH signals were computed. The method for 3D nucleus segmentation is based on multilevel Otsu thresholding and 3D watershed transform after Euclidean distance transform. Segmentation of the FISH signal comprises a tophat transform followed by Renyi entropy thresholding and watershed transform. For each FISH channel the two segmented FISH signals with largest volume were selected, 3D distances between all signals of different channels were computed, and, finally, the minimum distances were calculated.
Three-Dimensional Culture on Alvetex Scaffold.
Alvetex scaffold (Reinnervate) was used for short-term 3D culture of sorted human mTEC subsets. The 3D Alvetex scaffold was rinsed in 70% (vol/vol) ethanol in water, washed in media according to the manufacture’s instruction, and placed in a flat-bottom 96-well plate. The scaffold was held in place with a plastic insert with varying inner diameter (1.2–4 mm) adjusted to the cell input to ensure a similar cell density within the scaffold. Around 5,000–300,000 sorted mTECs were immediately seeded onto the scaffold in a minimal amount of RPMI media (10–15 µL) supplemented with 5% FCS and RANKL (0.1 μg/mL; R&D Systems). Cells were allowed to settle into the scaffold for about 30 min at 37 °C. Thereafter, the cultures were submersed in 150–200 µL media. After incubation for 50–55 h, the cultures were processed for FACS analysis. Cells were retrieved from the Alvetex scaffold using a trypsin-EDTA solution with shaking at 100–200 rpm per min at 37 °C for 15–20 min; after which, the cells were washed and stained for phenotype analysis.
Statistical Methods.
κ coefficients were calculated with SAS Version 9.2 (SAS Institute).
Additional Supporting Information.
Tables S1–S3 show the correlation analysis of gene expression in antigen-positive and -negative mTECs as detected by single-cell PCR. Fig. S1 defines coexpression groups. Fig. S2 shows the TRA enrichment and clustering of genes up-regulated in MUC1+, CEA+, and SGLT1+ sorted human mTECs. Fig. S3 shows the coexpression patterns of single MUC1+/MUC1− and CEA+/CEA− mTECs. Fig. S4 shows that coexpressed genes are colocalized.
Supplementary Material
Acknowledgments
We thank Klaus Hexel for expert single-cell sorting, Annette Koop-Schneider for statistical analysis, Martina Gärtner for single-cell PCR primer design, and Stefanie Egle and Yvonne Wiencek for excellent technical help. We also thank the German Cancer Research Center (DKFZ) Light Microscopy Facility, Zeiss Application Center for help with confocal microscopy and the DKFZ Genomics Core Facility for microarray hybridization. We thank Prof. Dr. Sebening and Dr. Loukanov (Department of Cardiac Surgery, Medical School of the University of Heidelberg) for providing human thymic tissue and Dr. Stefan Schoenfelder (Babraham Institute) for critical comments on the manuscript. This work was supported by the DKFZ, German Research Foundation (DFG) Sonderforschungsbereich 938 (to S.P.), the European Union Consortium Tolerage (C.M.), the Feinberg Graduate School, a Weizmann Institute Fellowship (to H.S.-G.), Federal Ministry of Education and Research (BMBF)-National Genome Research Net (NGFN+) project ENGINE (N.H. and K.R.), Miltenyi Biotech (S.W.), the Intramural DKFZ fund (B.B.), and the European Research Council 2012-Advanced Investigators Grant (B.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE49625).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308311110/-/DCSupplemental.
References
- 1.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: Learning self-control in the thymus. Nat Rev Immunol. 2005;5(10):772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 2.Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12(3):157–167. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- 3.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9(12):833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 4.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 5. DeVoss J, et al. (2006) Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J Exp Med 203(12):2727–2735, and erratum 204(1):203. [DOI] [PMC free article] [PubMed]
- 6.Gavanescu I, Kessler B, Ploegh H, Benoist C, Mathis D. Loss of Aire-dependent thymic expression of a peripheral tissue antigen renders it a target of autoimmunity. Proc Natl Acad Sci USA. 2007;104(11):4583–4587. doi: 10.1073/pnas.0700259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan Y, et al. Thymus-specific deletion of insulin induces autoimmune diabetes. EMBO J. 2009;28(18):2812–2824. doi: 10.1038/emboj.2009.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv H, et al. Impaired thymic tolerance to α-myosin directs autoimmunity to the heart in mice and humans. J Clin Invest. 2011;121(4):1561–1573. doi: 10.1172/JCI44583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abramson J, Giraud M, Benoist C, Mathis D. Aire’s partners in the molecular control of immunological tolerance. Cell. 2010;140(1):123–135. doi: 10.1016/j.cell.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 10.Org T, et al. AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum Mol Genet. 2009;18(24):4699–4710. doi: 10.1093/hmg/ddp433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraud M, et al. Aire unleashes stalled RNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci USA. 2012;109(2):535–540. doi: 10.1073/pnas.1119351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloosen S, et al. Expression of tumor-associated differentiation antigens, MUC1 glycoforms and CEA, in human thymic epithelial cells: Implications for self-tolerance and tumor therapy. Cancer Res. 2007;67(8):3919–3926. doi: 10.1158/0008-5472.CAN-06-2112. [DOI] [PubMed] [Google Scholar]
- 13.Derbinski J, Pinto S, Rösch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci USA. 2008;105(2):657–662. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Villaseñor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: Probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci USA. 2008;105(41):15854–15859. doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodewald HR, Paul S, Haller C, Bluethmann H, Blum C. Thymus medulla consisting of epithelial islets each derived from a single progenitor. Nature. 2001;414(6865):763–768. doi: 10.1038/414763a. [DOI] [PubMed] [Google Scholar]
- 16.Derbinski J, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202(1):33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298(5597):1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 18.Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med. 2004;199(2):155–166. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnnidis JB, et al. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci USA. 2005;102(20):7233–7238. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: Gene expression in 3D. Curr Opin Genet Dev. 2010;20(2):127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42(1):53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 23.Bacher CP, et al. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat Cell Biol. 2006;8(3):293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 24.Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126(2):403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 25.Guerau-de-Arellano M, Mathis D, Benoist C. Transcriptional impact of Aire varies with cell type. Proc Natl Acad Sci USA. 2008;105(37):14011–14016. doi: 10.1073/pnas.0806616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tykocinski LO, et al. Epigenetic regulation of promiscuous gene expression in thymic medullary epithelial cells. Proc Natl Acad Sci USA. 2010;107(45):19426–19431. doi: 10.1073/pnas.1009265107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Danso-Abeam D, et al. (2013) Aire mediates thymic expression and tolerance of pancreatic antigens via an unconventional transcriptional mechanism. Eur J Immunol 43(1):75–84. [DOI] [PubMed]
- 28.Liu Z, et al. Thymus-associated parathyroid hormone has two cellular origins with distinct endocrine and immunological functions. PLoS Genet. 2010;6(12):e1001251. doi: 10.1371/journal.pgen.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White AJ, et al. Lymphotoxin signals from positively selected thymocytes regulate the terminal differentiation of medullary thymic epithelial cells. J Immunol. 2010;185(8):4769–4776. doi: 10.4049/jimmunol.1002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishikawa Y, et al. Biphasic Aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. J Exp Med. 2010;207(5):963–971. doi: 10.1084/jem.20092144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudra D, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol. 2012;13(10):1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu W, et al. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13(10):972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinterberger M, et al. Autonomous role of medullary thymic epithelial cells in central CD4(+) T cell tolerance. Nat Immunol. 2010;11(6):512–519. doi: 10.1038/ni.1874. [DOI] [PubMed] [Google Scholar]
- 34.Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS. A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity. 2012;37(3):475–486. doi: 10.1016/j.immuni.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray D, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med. 2007;204(11):2521–2528. doi: 10.1084/jem.20070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gäbler J, Arnold J, Kyewski B. Promiscuous gene expression and the developmental dynamics of medullary thymic epithelial cells. Eur J Immunol. 2007;37(12):3363–3372. doi: 10.1002/eji.200737131. [DOI] [PubMed] [Google Scholar]
- 37.Le Borgne M, et al. The impact of negative selection on thymocyte migration in the medulla. Nat Immunol. 2009;10(8):823–830. doi: 10.1038/ni.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pinto S, et al. (2009) Increase Sensitivity of Illumina BeadArray with μMACS SuperAmp Technology, TechNote (Miltenyi Biotec, Heidelberg). Available at www.miltenyibiotec.com/~/media/Images/Products/Import/0002400/IM0002467.ashx.
- 39.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 40. R Core Development Team (2011) R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at www.R-project.org.
- 41.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.