Significance
Successful courtship and reproduction, which are at the center of evolutionary processes, involve complex interactions between neural and endocrine systems. In this study, we describe a group of neuropeptides that we have named “natalisin” (from the Latin natalis for “birth”) because of their function in promoting reproduction in arthropods. Three holometabolous insects, Drosophila melanogaster, Bombyx mori, and Tribolium castaneum were examined to understand the patterns of natalisin expression and to assess the phenotype of natalisin RNAi, and revealed the functions in courtship behavior and egg production. The natalisin receptor identified here warrants expanded study to elucidate the mechanisms of natalisin in arthropod reproduction.
Keywords: GPCR, NTL, NTLR, CG34388, CG6515
Abstract
An arthropod-specific peptidergic system, the neuropeptide designated here as natalisin and its receptor, was identified and investigated in three holometabolous insect species: Drosophila melanogaster, Tribolium castaneum, and Bombyx mori. In all three species, natalisin expression was observed in 3–4 pairs of the brain neurons: the anterior dorso-lateral interneurons, inferior contralateral interneurons, and small pars intercerebralis neurons. In B. mori, natalisin also was expressed in two additional pairs of contralateral interneurons in the subesophageal ganglion. Natalisin-RNAi and the activation or silencing of the neural activities in the natalisin-specific cells in D. melanogaster induced significant defects in the mating behaviors of both males and females. Knockdown of natalisin expression in T. castaneum resulted in significant reduction in the fecundity. The similarity of the natalisin C-terminal motifs to those of vertebrate tachykinins and of tachykinin-related peptides in arthropods led us to identify the natalisin receptor. A G protein-coupled receptor, previously known as tachykinin receptor 86C (also known as the neurokinin K receptor of D. melanogaster), now has been recognized as a bona fide natalisin receptor. Taken together, the taxonomic distribution pattern of the natalisin gene and the phylogeny of the receptor suggest that natalisin is an ancestral sibling of tachykinin that evolved only in the arthropod lineage.
Neuropeptides are ancestral signaling molecules that function as cell–cell communication mediators in multicellular organisms. Large numbers of diverse neuropeptides are involved in the control of animal behavior, development, and physiology. Recent genomic approaches have revealed diverse groups of neuropeptides in different taxa, based on similarities in the amino acid sequences to neuropeptides discovered in earlier physiological and anatomical studies (1–4). Sequenced genomes of many insect species (5) provide an opportunity to explore the evolutionary processes of neuropeptides and their receptors. Furthermore, the tools available in biotechnology that are readily applicable in suitable insect model species have advanced our understanding of the functions of neuropeptides. Drosophila melanogaster has been the best model system, allowing functional studies of neuropeptides and their receptors by the use of highly advanced molecular genetic tools and various publicly available resources (6). A number of other insect species, especially those with sequenced genomes, such as Bombyx mori and Tribolium castaneum, also have been used for investigations into the functions of neuropeptide signals, using piggyBac transformation (7) and RNAi (8, 9).
Previous studies on insect neuropeptides and their G protein-coupled receptors (GPCRs) have described tachykinin-related peptides (TRPs) and two GPCRs as the receptors for the TRPs in D. melanogaster and other insect species (10–14). In vertebrates tachykinin and the TRPs form a group of ancestral neuropeptides that are found in a wide range of animals, from octopus to human (14–18). In insects, multiple paracopies of the TRP gene contain the C-terminal FxGxRamide motif, whereas vertebrate tachykinins typically contain the FxGLMamide motif (Fig. 1A). Two closely related TRP receptors (TRPRs) in D. melanogaster were described previously: Drosophila tachykinin receptor (DTKR, also known as Takr99D or CG7887) and neurokinin K receptor of Drosophila (NKD, also known as Takr86C or CG6515). These receptors were identified in the pregenomics era, even before the TRPs were identified in D. melanogaster, using a hybridization-based homology search followed by functional assays (19, 20). In a subsequent study, however, NKD activity was not recapitulated with the typical TRPs, whereas DTKR was activated by the TRPs of D. melanogaster (21, 22).
Fig. 1.
The C-terminal consensus sequences of NTL and the species tree. (A) The hypothetical evolutionary tree and the C-terminal motifs of NTL, tachykinin, and TRP. The tree is based on the species tree. The C-terminal region in NTL is shown by the sequence logo for the frequencies of specific amino acids in the predicted mature peptides within the gene. The numbers of the paracopies carrying the motif are shown by the repeat numbers. The dotted branch is a hypothetical gene-duplication event where NTL diverges from tachykinin. (B) The D. melanogaster NTL precursor sequence, marked with the putative mature peptides (blue fonts) containing the typical NTL motif. Canonical amidation with di-basic signals (23) is marked by bold and underlined fonts. The putative signal peptide at the N terminus is in italics and underlined. (C) An alignment showing the consensus of the five paracopies of D. melanogaster NTL. The letters with black backgrounds are for identical amino acids within the aligned sequences. The numbers below the consensus logo indicate the position of the residues including the motif.
In the present study, we identified an arthropod-specific neuropeptide gene encoding multiple copies of mature neuropeptides carrying the C-terminal motif FxxxRamide. We named this neuropeptide “natalisin” (NTL) (from the Latin natalis for “birth”) for its function in promoting reproduction, based on the RNAi phenotypes. NTL is conserved only in arthropods and contains a C-terminal motif that is closely related to that of the TRPs. Likewise, we found that NTL activates the GPCR formerly known as NKD, and therefore we recommend revising the name “NKD” to “NTL receptor” (NTLR). D. melanogaster, B. mori, and T. castaneum, representing three different orders of holometabolous insects, were investigated to explicate the biology of NTL and its receptor.
Results and Discussion
Evolution and Diversity of Natalisin in Arthropods.
The NTL precursors in different arthropod species contain multiple repeats of 6- to 25-aa-long sequences (paracopies) that are similar to each other, particularly at the C termini. The putative mature peptides were separated by canonical amidation sites (G) followed by di-basic cleavage signals (combinations of R and K), which also are observed in a number of other neuropeptides (23), including tachykinin precursors. For example, the D. melanogaster NTL precursor encodes five paracopies with the consensus sequence DDPFxPxRa (in which “x” represents any amino acid and lowercase “a” indicates the amidated C terminus) (Fig. 1). A large number of other arthropod species, including crustaceans (Daphnia pulex and Caligus clemensi), likewise have a putative NTL peptide motif containing the sequence FxxxRa at the C termini, with several paracopies (ranging from 2 to 15) (Fig. 1A).
The C-terminal motifs among multiple paracopies showed relatively high degrees of conservation within the species, in addition to that of the general FxxxRa motif (Fig. 1 B and C). In the F-x1-x2-x3-R-a motif, x1 is often W in crustaceans and in hemipteran and most holometabolous insects except dipterans. x2 is P in dipterans and some hemipterans; in lepidopterans it often is G, which is identical to the tachykinin motif at the same position. The consensus of x3 is N in dipteran, lepidopteran, and coleopteran species. The N terminus upstream from the FxxxRa motif also shows mild degrees of conservation. The −1 position often is occupied by P, and positions −2 and −3 often are occupied by acidic amino acids D or E. The NTL motif is closely related to the arthropod TRP, which is characterized by an FxGxRa C-terminal motif but also is clearly distinguished based on species (or Order), with specific amino acids being conserved in the degenerate sites. Dataset S1 presents sequences of the NTL precursors captured in BLAST searches of various arthropod genome databases and predictions of mature NTL peptides.
The distinctive genes encoding each NTL and TRP were clearly present in insect and crustacean genomes but not in arachnids. In the spider mite Tetranychus urticae, there was no separate gene for NTL in the genome. Instead, two genes were annotated for TRP: TRP1 and TRP2 (4). Among the predicted processed peptides of the TRPs, three of the five putative mature peptides encoded by the two genes were similar to NTL in that they carry a P in the X2 position (RFIPLRa) and a P in the −1 position (SRPFAAMRa, and ARPFAAMLa), whereas two peptides in each gene (AAFTGMRa and SAFNGMRa) contained the typical insect TRP motif FxGxRa (Dataset S1). Therefore, in the spider mite both NTL and TRP appear to be encoded by each gene in a mixed array of the paracopies. The two TRP/NTL genes in the spider mite suggest that a divergence between the TRP and NTL paracopies of the ancestral gene was the first step in the emergence of the NTL signaling system.
The mosquito and octopus species carry genes encoding peptides highly similar to vertebrate tachykinin, with the FxGLMa motif (Fig. 1A), and both species express this mRNA in the salivary glands (24, 25). These two salivary gland peptides likely are used for attacking vertebrate hosts or prey and likely are products of convergent evolutions.
Conserved Expression Patterns of Natalisin in Holometabolous Insects.
We examined three holometabolous insects representing three insect orders—D. melanogaster (Diptera), B. mori (Lepidoptera), and T. castaneum (Coleoptera)—to investigate the NTL expression patterns by using quantitative RT-PCR, immunohistochemistry, and in situ hybridization. The highest transcript levels of NTL were found in the larval CNS and in the early larval stages in the tissue- and stage-specific quantitative PCR, respectively, in T. castaneum (Fig. S1). Similarly, the expression pattern in D. melanogaster, based on the gene expression data in Flybase, showed gene expression in the larval CNS and in the adult brain (26).
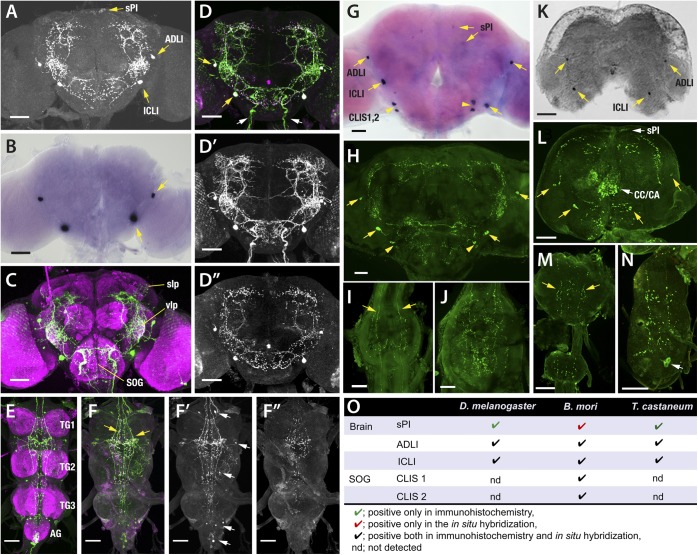
Further investigations used antibodies raised against two Drosophila NTL peptides, DmNTL4 and DmNTL5, raised in rabbit and mouse, respectively. Both antibodies labeled virtually identical cells in the CNS of D. melanogaster. The anti-DmNTL5 antibody was used in D. melanogaster studies because it had a better signal-to-noise ratio; anti-DmNTL4 was used in B. mori and T. castaneum because of its more robust labeling. The brain showed four pairs of NTL-positive somas and their projections, and the thoracic ganglia contained many NTL-positive varicosities, which are the descending processes of the brain NTL neurons. The brain NTL neurons were named according to the locations of their somas: anterior dorso-lateral interneurons (ADLI), inferior contralateral interneurons (ICLI), and small pars intercerebralis (sPI) neurons (Fig. 2A). The most prominent pairs of neurons were the ICLI and ADLI; moderate levels of immunoreactivity were observed in the two pairs of smaller dorso-medial sPI interneurons (Fig. 2A). To confirm the results from antibody staining, we used a Digoxygenin (Dig)-labeled DNA probe for in situ hybridization and detected strong expression of NTL mRNA in the ICLI and ADLI neurons but not in the sPI neurons (Fig. 2B).
Fig. 2.
Immunohistochemistry and in situ hybridization of adult CNS of D. melanogaster (A–F), B. mori (G–J), and T. castaneum (K–N), and a summary table (O). (A) The brains of UAS-NTL-IR/+ females stained with anti-DmNTL5 antiserum. The yellow arrows indicate three major classes of NTL-positive neurons, which we named small pars-intercerebralis neurons (sPI), the anterior dorso-lateral interneuron (ADLI) and the inferior contralateral interneurons (ICLI). (B) The female brain stained with an in situ DNA probe against NTL mRNA. Yellow arrows indicate ADLI and ICLI neurons. (C) The brain of an NTL-Gal4/UAS-mCD8-EGFP female stained with anti-GFP (green) and mAb nc82 (magenta). (D) The brain of an NTL-Gal4/UAS-mCD8-EGFP male stained with anti-GFP (green) and anti-NTL (magenta). The yellow arrows indicate neurons positive for both anti-GFP and anti-NTL. The white arrows indicate neural processes positive for anti-GFP alone, but not for anti-NTL, which enter into the subesophageal ganglion through pharyngeal nerves. Anti-GFP (D′) and anti-NTL (D′′) channels are shown separately. (E) The ventral ganglia of an NTL-Gal4/UAS-mCD8-EGFP male, stained with anti-GFP (green) and anti-nc82 (magenta). (F) The ventral ganglia of an NTL-Gal4/UAS-mCD8-EGFP male, stained with anti-GFP (green) and anti-NTL (magenta). The yellow arrows indicate neural processes and varicosities positive for both anti-GFP and anti-NTL. The white arrows in F′ indicate neurons positive for anti-GFP alone. Anti-GFP (F′) and anti-NTL (F′′) channels are shown separately. (G) In B. mori, in situ hybridization with an NTL probe revealed strong expression in two pairs of cells in the brain: ADLI and ICLI. Weak expression was observed in two pairs of sPI neurons. Two pairs of contralateral interneurons also were detected in the SOG (CLIS1 and 2). (H) Immunohistochemical staining with anti-DmNTL4 showed strong immunoreactivity in the same ADLI, ICLI, and CLIS1,2 neurons. (I and J) ICLI and CLIS1,2 neurons project descending contralateral axons arborizing in each ventral ganglion. (K) In situ hybridization of the brain of adult T. castaneum showing positive reactions in the pairs of ADLI and ICLI cells. (L) The brain of adult T. castaneum stained with anti-DmNTL4 (green). The yellow arrows indicate NTL-positive somas. (M) Descending processes (yellow arrows) running in the third thoracic and the first abdominal ganglia. (N) Terminal abdominal ganglion stained with anti-DmNTL4. White arrow indicates NTL-immunolabeling resistant to systemic NTL-RNAi. (O) A table summarizing NTL expression in the brain of three examined species. AG, abdominal ganglion; slp, superior lateral protocerebrum; SOG, subesophageal ganglion; TG thoracic ganglion; vlp, ventro-lateral protocerebrum. (Scale bars, 50 µmin A–F′′ and K–N; 100 µm in G–J.)
To investigate the anatomy of NTL-expressing neurons in D. melanogaster, we generated the transgenic line NTL-Gal4, carrying ∼2.2 kb of the 5′ upstream region of the ntl gene, and examined the Gal4 expression patterns in NTL-Gal4/UAS-mCD8-GFP flies. In this fly line, prominent GFP expression was found in ICLI and ADLI neurons but not in sPI neurons (Fig. 2 C and D). In addition, NTL-Gal4 expression was detected in a pair of neural processes innervating the subesophageal ganglion (SOG), presumably from peripheral neurons, and in small neurons in ventral ganglia, all of which lacked anti-NTL staining (white arrows in Fig 2D). ICLI neurons arborize the anterior ventro-lateral protocerebrum (AVLP) and the anterior SOG, and their processes also wrap around the mushroom body pedunculus and calyx (Fig. 2C and Fig. S2). In addition, ICLI neurons project descending contralateral axons along the ventral nerve cord (Fig. 2 E and F). The neural processes of ADLI neurons also enter into the AVLP neuropile and appear to intermingle with those of the ICLI neurons.
Anti-DmNTL immunoreactive sPI cells were negative in the in situ hybridization and NTL-Gal4 expression (Fig. 2 A–C). This incongruent result led us to examine the immunoreactivity in pan-neuronal NTL-RNAi (UAS-Dicer2/+;nSynb-Gal4/UAS-NTL-IR) and control (UAS-NTL-IR/+) flies. The immunoreactivity was abolished with the pan-neuronal NTL-RNAi, not only in ICLI and ADLI neurons but also in sPI neurons (Fig. S3), supporting the existence of NTL expression in sPI neurons.
We detected NTL expression in the CNS of adult B. mori. As was the case in Drosophila, high transcript levels were observed in paired neurons in the anterior and inferior regions of the brain (Fig. 2G). Weak expression also was found in two pairs of small neurons in the sPI and in two pairs of neurons in the anterior dorso-lateral region of the SOG (Fig. 2G). Immunohistochemistry with DmNTL4 antibody revealed that the ADLI of the brain projects a single arborizing axon into the vicinity of the antennal lobe (Fig. 2H). The most complex innervation shows a single pair of ICLI in the brain. Each neuron projects one contralateral axon into the opposite hemisphere of the brain, where it forms a branching loop around the mushroom body (Fig. 2H), and its descending axon innervates the entire ventral nerve cord and terminates in the terminal abdominal ganglion (TAG) (Fig. 2 I and J). Paired anterior SOG cells also project descending contralateral axons along the entire CNS and therefore were termed contralateral interneurons of the SOG (CLIS1 and -2) (Fig. 2 H–J).
In adult T. castaneum (Fig. 2 K–N), patterns of immunoreactivity similar to those observed in D. melanogaster and B. mori were found in the brain using the anti-DmNTL4 antibody. NTL expression was detected in ADLI and ICLI neurons and in variable numbers of small sPI neurons (three to six pairs) depending on the individual. Varicosities in the brain expand to projections into the complex containing the corpora cardiaca and corpora allata (CC/CA) and to the antennal glomeruli. We observed a pair of descending processes to every segmental ganglion. In situ hybridization revealed strong positive reactions only in ADLI and ICLI neurons, with no reactions in the sPI neurons or in the cells of the TAG. Immunohistochemistry after RNAi in the adult brain confirmed that the RNAi abolished the immunoreactivity in ADLI and ICLI and their arborizations. However, the immunoreactivity in TAG neurons and in the projections to the CC/CA complex remained after RNAi treatment (Fig. S4), suggesting a false-positive reaction in those cells (white arrows in Fig. 2 K and L). Staining in descending axons to segmental ganglia was weakened by the RNAi and presumably was caused by peptides remaining after suppression of the mRNA. Specific expression of NTL in the sPI is inconclusive because of large individual variations of NTL immunoreactivity in the sPI.
To summarize the results from the experiments conducted in the three orders of insects (Fig. 2O), two pairs of brain cells, ADLI and ICLI, were commonly detected in all three species by both in situ hybridization and immunohistochemistry experiments, although the projection patterns appear to be slightly different. The presence of NTL expression in the paired sPI neurons was supported by the suppression of immunoreactivity following the pan-neuronal NTL-RNAi in D. melanogaster and conserved expression in all three species; however, with some techniques the sPI neurons were negative for NTL expression. Two pairs of CLIS1 in the SOG were immunoreactive only in B. mori. No NTL cells were found in the segmental ganglia in any three of the tested species; only the descending projections were positive.
Functions of Natalisin Assessed by RNAi.
NTL is expressed primarily in the central interneurons that extensively innervate the ventrolateral protocerebrum (VLP), where olfactory, gustatory, auditory, and visual pathways converge (27–30). Thus, we examined the role of NTL and NTL-expressing neurons in mating behavior, which requires flies to integrate and coordinate virtually all sensory modalities to execute multiple motor programs. The neuropeptides SIFamide and corazonin recently have been shown to be involved in the mating behaviors of D. melanogaster (31, 32).
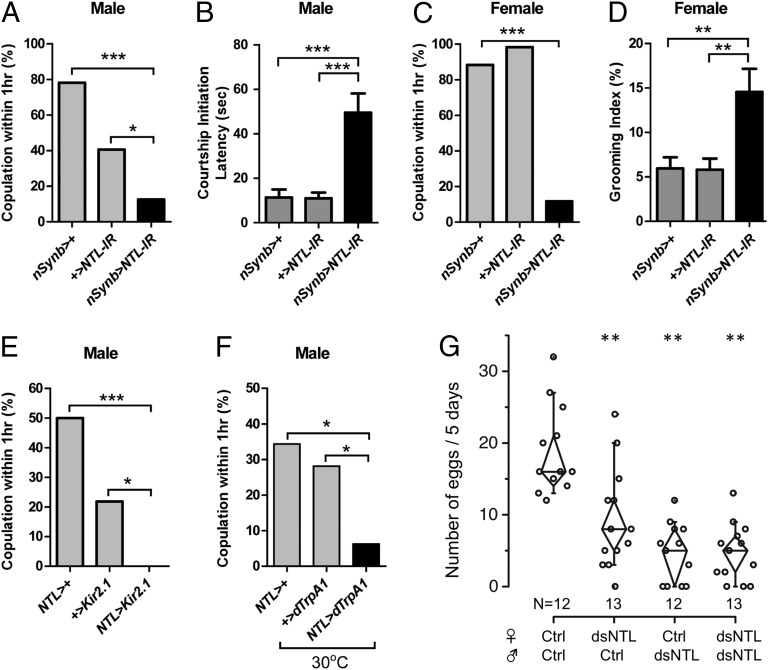
First, we examined the mating rate in NTL-RNAi flies (UAS-Dicer2; nSynb-Gal4/UAS-NTL-IR). Knocking down NTL expression in the nervous system strongly suppressed mating frequency. During an hour-long mating assay, in which individual test males were paired with wild-type Canton S (CS) females, only ∼10% of NTL-RNAi males succeeded in copulation, whereas controls showed a copulation success rate of 40–80% (Fig. 3A). We noted that the control UAS-NTL-IR alone had a considerably lower copulation success rate (∼40%) than the Gal4 control (∼80%). A leaky NTL-RNAi expression is unlikely to be responsible for the relatively low mating rate because there was no discernible decrease in anti-NT staining in the UAS control brain (Fig. S3). The positions of the insertions and numbers of miniwhite (w) transgene, an engineered w construct used as a transgene marker, may be the cause of this variation, because both Gal4 and UAS lines are derived from the same w1118 genetic background. The w gene is known to be important in male courtship, because w− males lacking the light-screening eye pigments cannot track females visually and show lower light-on copulation success rates than w+ males (33, 34). The lower gene dosage for w in the UAS control carrying one copy of miniw (UAS-NT-IR/+) than in Gal4 control with two copies (UAS-Dicer2; nSynb-Gal4/+) may explain the different copulation success rates in the two controls.
Fig. 3.
NTL RNAi phenotypes in D. melanogaster (A–F) and in T. castaneum (G). (A) Mating frequencies and (B) courtship initiation latency (mean ± SEM) of males of the indicated genotypes in single-pair assays with CS virgin females. Mating frequencies: *P < 0.05, ***P < 0.001; Fisher’s exact test. n = 60 each bar. Courtship initiation latency: ***P < 0.001; ANOVA. n = 20–23. The nSynb-Gal4 driver carries UAS-Dicer2. (C) Mating frequencies and (D) grooming indices of females of indicated genotypes in single-pair assays with naive CS males. Mating frequencies: ***P < 0.001; Fisher’s exact test. n = 60 each bar. Grooming index: **P < 0.01; ANOVA. n = 20–22. (E and F) Mating frequencies of males of the indicated genotypes (gray bars) in single-pair assays with CS virgin females. ***P < 0.001, *P < 0.05 versus respective controls; Fisher’s exact test; n = 32 each bar. For the dTrpA1 experiment (F), flies were subjected to the temperature shift 30 min before and during the assay. (G) Reduced egg numbers after injection of 100 ng of dsRNA in the pupal stage of T. castaneum. The control was injected with saline alone. **P < 0.01 versus control; Student t test.
To investigate the nature of the mating defects in NTL-RNAi flies, we examined the following male courtship parameters in high resolution: latency of courtship initiation; frequency of wing extension as a measure of courtship song; courtship index as a measure of courtship enthusiasm; and frequency of copulation attempts (Fig. 3B and Fig. S5). Compared with the controls, NTL-RNAi males exhibited moderate but significant levels of delay in initiating courtship (Fig. 3B). In our assay, control males started to court females with 11 ± 4 s latency, whereas RNAi males initiated courtship with 59 ± 8 s latency. Aside from the small delay in the latency of courtship initiation, NTL-RNAi males showed no discernible defect in courtship parameters compared with control males. Thus, it still is unclear whether this rather weak phenotypic change is a major cause of the robust mating defect.
When paired with CS males for 1 h, NTL-RNAi virgin females did not succeed in mating (Fig. 3C). To investigate the mating phenotype further, we examined whether NTL-RNAi virgins reject males actively by measuring the frequency of ovipositor extrusion. Although NTL-RNAi virgin females showed extremely low mating frequency, they did not actively reject courting males. Instead, they displayed significantly elevated levels of grooming, which likely prevented the males from being able to access the females for attempted copulation (Fig. 3D). Control virgin females also showed grooming behavior, but the duration of this behavior was significantly shorter than that of the NTL-RNAi virgins.
To rule out the possibility that the observed mating phenotypes stem from an overall reduction in vigor and/or the loss of motor coordination, we performed the bang recovery test (35) and confirmed that the recovery after rapping of the vial was not different in any of the tested genotypes. In climbing speed, NTL-RNAi flies were slower than one control (UAS-NTL-IR/+) but were equivalent to the other control (UAS-Dicer2; nSynb-Gal4/+) (Fig. S6).
Next, we studied the role of NTL-producing neurons in mating behavior. Consistent with the NTL-RNAi results, silencing the neural activities of NTL neurons by expressing the mammalian inward-rectifying potassium channel (Kir2.1) with NTL-Gal4 almost completely suppressed the mating rate in males (Fig. 3E). Furthermore, we observed that the acute activation of NTL neurons with a temperature-gated cation channel, dTrpA1, significantly suppressed the male mating rate (Fig. 3F). With these results, we concluded that successful male mating requires the precise coordination of NTL-expressing neuron activities during courtship. In contrast, the analogous manipulations of NTL-Gal4 neurons did not affect female mating. This result suggests that the NTL neurons that are important in the female mating process do not express NTL-Gal4. Because NTL-Gal4 is expressed in ICLI and ADLI neurons in both sexes but not in sPI neurons (Fig. 2 A and C), sPI neurons are likely to be important in female mating behavior. We examined the role of NTL-Gal4 neurons in additional aspects of female reproductive behaviors, such as egg laying and remating, by either silencing (NTL-Gal4/UAS-Kir2.1 or NTL-Gal4/UAS-Shits, 30 °C) or overactivating them (NTL-Gal4/UAS-dTrpA1, 30 °C), but we observed no discernible anomalies compared with the controls (Fig. S7).
Based on the phenotypes assessed using RNAi in D. melanogaster, the NTL-expressing neurons ICLI and/or ADLI are likely to be involved in male mating behaviors. Interestingly, the phenotype of reduced receptivity observed in females was observed only using the pan-neuronal NTL-RNAi but not when silencing NTL-Gal4 neurons. Therefore, we interpreted these results as showing that the female phenotype is caused by the NTL-RNAi expressed in sPI cells, which lacked NTL-GAL4 expression but showed NTL immunoreactivity that was abolished by the pan-neuronal NTL-RNAi. Alternatively, it also is possible that the female phenotype is caused by knockdown of other unknown gene(s) together with ntl. Further studies, such as the generation and analysis of ntl-null mutants, are required to address this possibility.
Using systemic RNAi to knock down NTL expression in T. castaneum resulted in significant reductions in egg numbers. Combinations of single-pair mating in which either sex is treated with dsRNA also showed similar degrees of reduction in egg laying (25–50% of the control) (Fig. 3G) regardless of which sex was treated. We were unable to detect other defects caused by the RNAi in the early larval or pupal stages, such as developmental defects, alterations in the egg-hatching rate, alterations in mating frequencies or duration, or detectable morphological malformations. In contrast to the case of D. melanogaster, therefore the reduced fecundity in T. castaneum is unlikely to be caused by aberrant mating behavior. Nevertheless, we conclude NTL-RNAi severed reproductive capability in both D. melanogaster and T. castaneum, although the precise function of NTL in each species needs to be investigated further.
Identification of the NTL Receptor.
We investigated a GPCR, CG6515, as the best candidate receptor for the neuropeptide NTL. The CG6515, formerly named NKD or TakR86C, was described as the receptor for tachykinin (20). However, a previous study reported that the CG6515 is not activated by typical DmTRPs carrying the C-terminal motifs FxGMRa or FxGLRa but shows low levels of activity in response to DmTK6, which has the atypical TRP C-terminal sequence FVAVRa (21).
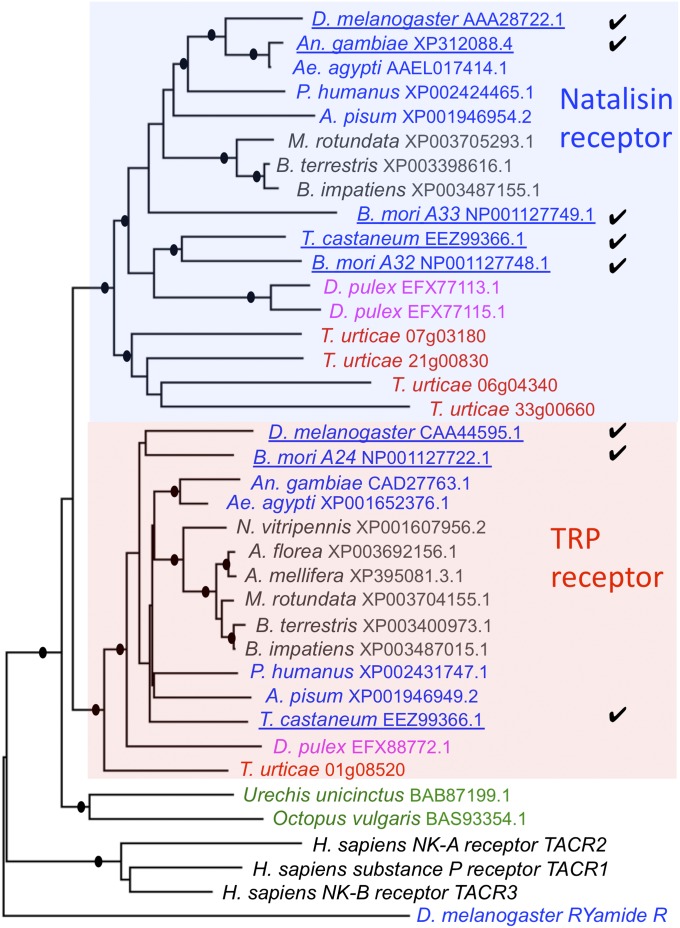
The phylogenetic analyses of TRPRs and NTLRs strongly support the divergence of the NTLR cluster from the arthropod TRPR (Fig. 4). The TRPRs in other invertebrates and the tachykinin receptors in vertebrates form the root of the NTLR and TRPR families in arthropods, further supporting the relative timing of the emergence of the NTL system. B. mori has two copies of genes in the NTLR group. The Hymenoptera Apis mellifera and Nasonia vitripennis lack the NTLR and do not contain a sequence for the NTL ligand. Other hymenopterans—Megachile rotundata and two Bombus species—contain a GPCR in the NTLR cluster, but the ligand NTLs are not found in these species. In further basal lineages, the crustacean D. pulex has two genes grouped in the NTLR cluster that appear to be products of a recent gene duplication. In comparison, the genome sequence of the spider mite, T. urticae, possesses four genes in the NTLR group (Fig. 4).
Fig. 4.
Phylogeny showing possible evolutionary relationships among the NTL and TRP receptors. Insects are in blue fonts, with the exception of hymenopterans, which are in black. Crustaceans are in magenta, arachnids are in red, and protostomians are in green fonts. The filled circles at the nodes represent bootstrapping supports greater than 75% in 500 replications. The D. melanogaster RYa receptor was used as the outgroup.
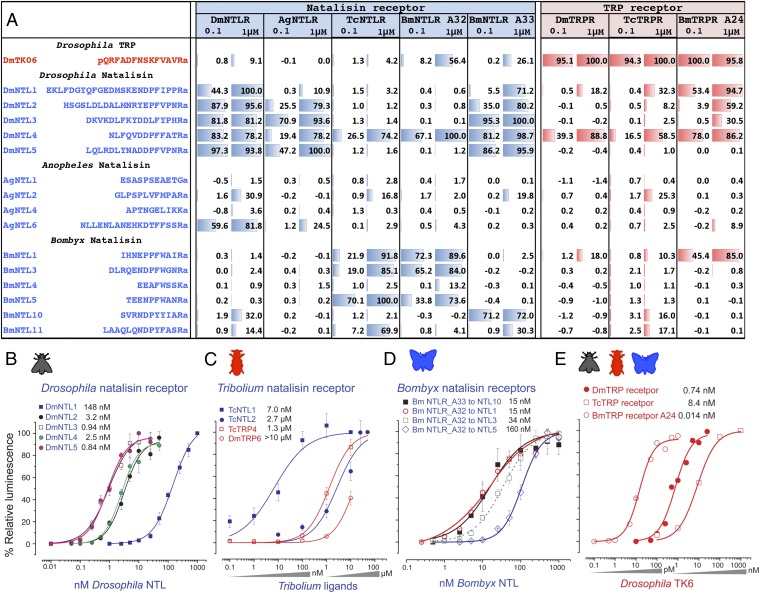
To investigate the ligand–receptor interactions in the NTL and TRP complexes, we performed functional assays of the receptors. We tested the TRPRs of D. melanogaster, T. castaneum, and B. mori, the NTLRs of D. melanogaster, Anopheles gambiae, and T. castaneum, and two receptors of B. mori, BNGR A32 and A33 (Fig. 5). The following NTL ligands were used: all five NTLs in D. melanogaster; four variants of six total C-terminally amidated putative peptides encoded by the ntl gene of An. gambiae; both of the predicted NTLs of T. castaneum; and six NTLs out of 11 total putatively amidated peptides encoded by the ntl gene of B. mori (Fig. 5 and Dataset S1). We specifically tested the TRP ligand DmTK6 (Fig. 5A), which previously was found to be the only Drosophila TRP ligand that activates the NTLR at a high concentration (21), and the TRP ligand TcTRP4 (APSGFFGMRa), which contains the typical TRP motif.
Fig. 5.
Ligand-receptor specificities of the NTLRs and TRPRs. (A) Table showing the ligand activities that were calculated based on the relative activity compared with the highest response of the receptor for each set of ligands. (B) Dose–response curves for the DmNTL1 to 5 activation of the DmNTLR. (C) Dose–response curves for the TcNTLs and TRPs. (D) Dose–response curves for BmNTL activation of the BmNTLRs A32 and A33. (E) Dose–response curves for DmTK6 activation of the TRPRs from Drosophila, Tribolium, and Bombyx. The amino acid sequence for TcNTL1 is ASGQEEFGPFWANRa, for TcNTL2 is DDNDINDNEPFYVTRa, and for TcTRP4 is APSGFFGMRa.
The NTLRs were all strongly activated by the NTLs from the same species or from heterospecies (Fig. 5A). The DmNTLR responded to DmNTL3 and DmNTL5, containing the C-terminal motifs DDLFYPHRa and DDPFVPNRa, respectively, at subnanomolar levels (Fig. 5 A and B). The receptor from An. gambiae was activated by the DmNTLs and AgNTL6 (Fig. 5A), although AgNTL5, with the C-terminal sequence DEYFFPNRa containing a consensus motif, was not tested in the present study (Fig. 1). The TcNTL1 (ASGQEEFGPFWANRa) activated TcNTLR at nanomolar levels, but TcNTL2 activated it only weakly (Fig. 5C).
DmTK6, which previously was described as being the only D. melanogaster tachykinin that activated the NTLR (21), was tested again in the present study. DmTK6 showed little to no activation of the NTLRs from the different species tested in the present study (Fig. 5A). TcTK4, which contains the typical TRP motif APSGFFGMRa, also showed very low levels of activity (EC50, 1.3 μM) on the TcNTLR (Fig. 5C and Fig. S8).
Conversely, the TRPRs from different species generally were specific to the TRP ligands but were activated by a number of NTLs at high concentrations. The DmTRPR (CG7887) was activated by DmNTL4 at high concentrations of 0.1 and 1 μM. The BmTRPR (BNGR A24) was activated by BmNTL1 at the same high concentrations and heterospecifically by a number of DmNTLs. However, the strong activations of the TRPRs by DmTK6 were observed with EC50s in the low nanomolar or picomolar ranges (Fig. 5 A and E). The TcTRPR also showed a clear discrimination between TRP and NTL, with more than 1,000-fold differences in the EC50s (Fig. 5C and Fig. S8).
In the lepidopteran lineage, a further divergence of the NTL peptides was distinguished by the FxxxRa and YxxxRa motifs (Dataset S1), which may reflect an evolutionary process based on the ligand–receptor activities. Unlike other insect species that have only one copy of the NTLR, B. mori has two receptors in the NTLR group, BNGR A32 and A33 (36). BmNTLR A32 was specific to BmNTL1, 3, and 5, which have the C-terminal FxxxRa consensus sequence, whereas BmNTLR A33 was specific to BmNTL10 and BmNTL11, which have the YxxxRa consensus sequence (Fig. 5 A and D). In the predicted NTL precursor of B. mori, eight peptides carry the FxxxRa motif, and two peptides, located on the C-terminal end of the precursor, contain the YxxxRa motif. This NTL precursor structure and the associated motifs also are conserved in other lepidopteran species, such as Danaus plexippus and Manduca sexta; both contain two putative C-terminally located peptides with the YxxxRa motif in the precursor, and the remaining peptides contain the FxxxRa motif (Dataset S1). The two differently conserved motifs in the precursor of NTL within the lepidopteran species and the presence of two receptors that distinguish between the two ligand motifs suggest that the two signaling systems diverged at an early evolutionary stage in the Lepidoptera.
In general, the ligand–receptor interactions investigated by functional assays demonstrated the specificity of NTL and TRP to their own receptors, with moderate to low degrees of cross-activation. These include DmNTL4 activating the DmTRPR and BmNTL1 activating the BmTRPR, but in both cases only at the high concentrations (Fig. 5A). The specificities of the partnerships between the ligand and receptor appear to be strongly established, although the possible pleiotropy of ligand–receptor interactions, which was proposed to be an important process in function and evolution (37, 38), could not be excluded completely. Nevertheless, we observed that the phenotype of receptor NTLR-RNAi is highly similar to that of the ligand NTL-RNAi in T. castaneum with a significantly reduced fecundity (Fig. S9). With the mutually supported data, we conclude that NTL and the authentic NTLR are involved in the reproductive functions in insects. The phylogeny of the NTLR, combined with the taxonomic distribution pattern of NTL ligands, strongly supports the idea that the evolutionary origin of the peptidergic NTL system is likely to be the TRP/TRPR system, which has a deeper evolutionary history.
Materials and Methods
Sequence Analyses.
RT-PCR confirmed the expression of ntl genes in T. castaneum, B. mori, and D. melanogaster. The primers used in the present study can be found in S1 Materials and Methods. The sequence logos for the NTL C-terminal motifs of each species were generated by Weblogo (39). Sequence alignments of the putative translations were made using the Clustal W module in MEGA5 (40). Phylogenetic analysis was performed using MEGA5 for sequences including the putative first to the seventh transmembrane domains, using the neighbor-joining method, pairwise deletions for gaps, uniform rates of mutation, and 500 bootstrap tests.
Fly Stocks and Tribolium RNAi.
NTL-Gal4 was generated as described (41). Briefly, a 2,249-bp-long 5′ upstream region of the ntl gene was amplified with a genomic DNA PCR (forward primer, 5′-gtgggtctgctgcctcttac; reverse primer, 5′-gctgcgtttttggctcttag), cloned into pENTR (Invitrogen), and recombined into pBPGal4.2::VP16Uw. NTL-Gal4 was inserted into a specific site of the second chromosome (VIE-72A, a gift from B. J. Dickson) using the phiC31 system. nSynb-Gal4, carrying UAS-Dicer2, was kindly provided by B. J. Dickson (Institute of Molecular Pathology, Vienna). UAS-NTL-IR (Vienna Drosophila RNAi stock center transformation ID 19547) was obtained from the Vienna Drosophila RNAi stock center. Canton S was used as a wild-type partner in the mating pairs of behavioral tests. Systemic RNAi in the T. castaneum GA1 strain was performed as previously described (8, 9, 42, 43). Two different stages, approximately the fourth to fifth larval stage and early pupal stage, were used for injections of dsRNA.
GPCR Assays.
Plasmids for the mammalian cell expression of D. melanogaster GPCRs CG7887 and CG6515 (44) and B. mori GPCRs BNGR A24, A32, and A33 (36) were kindly provided by P. Taghert (Washington University, St. Louis) and H. Kataoka (University of Tokyo, Tokyo), respectively. The T. castaneum NTLR was cloned from TC004977 but with modifications made for the 3′ end exons (GenBank accession number KF192693). The coding sequence of the An. gambiae NTLR (AgNTLR, AGAP002824-PA) was obtained from VectorBase (http://vectorbase.org), and its coding sequence, with a mammalian 5′ Kozak sequence, was synthesized by Bioneer and cloned into pcDNA3.1(+) (Invitrogen). CHO-K1 cells cultured in the DMEM/F-12 medium (Welgene) were transfected with plasmids carrying the corresponding receptor, a codon-optimized aequorin and the wild-type Gq protein. For transient transfection, Fugene6 was used, according to the manufacturer’s instructions. The procedures used for the receptor assays were described previously (45). HPLC-purified synthetic peptides predicted from Drosophila, Aedes, and Bombyx NTL genes were obtained from Anygene, and Tribolium peptides were obtained from GeneScript.
Antibodies and Immunohistochemistry.
Antibodies against NTLs (rabbit anti-DmNTL4 and mouse anti-DmNTL5) were generated using synthetic peptides conjugated with keyhole limpet hemocyanin through additional cysteine residues at their N termini (Abfrontier). Additional antibodies used include rabbit anti-GFP (1:1,000) (A11122; Invitrogen), mouse anti-nc82 (1:20–1:50) (Developmental Studies Hybridoma Bank), DyLight 488 (Jackson ImmunoResearch), Alexa 488-conjugated goat anti-rabbit (A11008; Invitrogen) and Alexa 568-conjugated goat anti-mouse (A11004; Invitrogen).
For immunohistochemistry, the CNS from 1- to 5-d-old virgin male and female flies, moths, or beetles was dissected in PBS (pH 7.4) or saline (140 mM NaCl, 5 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 4 mM NaHCO3, Hepes 5 mM, pH 7,2). The tissues were fixed for 1–2 h at room temperature in 4% (wt/vol) paraformaldehyde in PBS. After extensive washing, the tissues were incubated in a primary antibody (1:1,000 for anti-DmNTL4 and anti-DmNTL5) for 48 h at 4 °C and in a secondary antibody for 24 h at 4 °C. The CNS was mounted in glycerol or VECTORSHIELD (H-1000; Vector Laboratory). Images were acquired with a Zeiss LSM 700/Axiovert 200 M or Leica TCS SPE-II confocal microscope and were processed using Image J (46).
In Situ Hybridization.
Dissected tissues were fixed in 4% paraformaldehyde, washed with 70% (vol/vol) ethanol and PBS with Tween 20, treated with Proteinase K, and hybridized in a hybridization solution containing a Dig-labeled probe overnight at 48 °C. After several washes, the tissues were incubated overnight with alkaline phosphatase-labeled anti-Dig antibody and were stained with nitro-blue tetrazolium/5-bromo-4-chloro-3'-indolyphosphate (Roche).
Behavioral Assays.
All flies were raised on standard medium at 23 °C on a 12-h:12-h dark:light cycle and 60% relative humidity. All assays were performed at zeitgeber time 6:00–12:00 on at least two independent days. GraphPad Prism (GraphPad Software, Inc.) was used for the statistical analyses. For mating and courtship assays, we followed procedures described previously (47). Males and females were collected at eclosion. Males were aged individually for 5 d, and females were aged for 4 d in groups of 10–15. For the mating assay, a single virgin female and a naive male were paired in a 10-mm diameter chamber and were videotaped for either 1 h (for mating frequency) or 10 min (for male courtship parameters and female grooming index). The mating chamber has a sliding divider which allows the flies to habituate to the chamber before the assay start, which occurs when the divider is removed. For the female grooming index, the total duration of grooming is calculated as a fraction of the observation period (10 min or until copulation is achieved). To calculate male courtship parameters, we followed the protocol previously described (48).
Supplementary Material
Acknowledgments
We thank John Ruberson for comments on the manuscript; Ladislav Roller for assistance with confocal microscopy; H. Kataoka and P. Taghert for GPCR clones; H-S. Yoon, J. Mun, and J. Song for technical assistance; and the Gwangju Institute of Science and Technology (GIST) Systems Biology Research Center funded by the 2013 GIST Systems Biology Infrastructure Establishment Grant for use of the confocal imaging facility. Y-J.K. was supported by Basic Science Research Programs 2011-0019291 and 2011-0018559 through the National Research Foundation of Korea and Research & Development innovation cluster development program funded by the Ministry of Science, ICT and Future Planning of the Republic of Korea. D.Ž. and I.D. were supported by Slovak Grant Agencies Agentúra na Podporu Výskumu a Vývoja (APVV-0827-11) and Research & Development Operational Program funded by the European Regional Development Fund (ITMS: 26240220044). L.S. and Y.P. were supported by National Institutes of Health Grant Number R01AI090062. This paper is contribution no. 13-380-J from the Kansas Agricultural Experiment Station.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession number KF192693).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310676110/-/DCSupplemental.
References
- 1.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11(6):1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. 2008;18(1):113–122. doi: 10.1101/gr.6714008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roller L, et al. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38(12):1147–1157. doi: 10.1016/j.ibmb.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Veenstra JA, Rombauts S, Grbić M. In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochem Mol Biol. 2012;42(4):277–295. doi: 10.1016/j.ibmb.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Robinson GE, et al. Creating a buzz about insect genomes. Science. 2011;331(6023):1386. doi: 10.1126/science.331.6023.1386. [DOI] [PubMed] [Google Scholar]
- 6. doi: 10.3389/fendo.2012.00151. Caers J, et al. (2012) More than two decades of research on insect neuropeptide GPCRs: An overview. Front Endocrinol 3:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daubnerová I, Roller L, Zitnan D. Transgenesis approaches for functional analysis of peptidergic cells in the silkworm Bombyx mori. Gen Comp Endocrinol. 2009;162(1):36–42. doi: 10.1016/j.ygcen.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arakane Y, et al. Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech Dev. 2008;125(11-12):984–995. doi: 10.1016/j.mod.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Begum K, Li B, Beeman RW, Park Y. Functions of ion transport peptide and ion transport peptide-like in the red flour beetle Tribolium castaneum. Insect Biochem Mol Biol. 2009;39(10):717–725. doi: 10.1016/j.ibmb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Nässel DR. Neuropeptides in the nervous system of Drosophila and other insects: Multiple roles as neuromodulators and neurohormones. Prog Neurobiol. 2002;68(1):1–84. doi: 10.1016/s0301-0082(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 11.Nässel DR, Winther AME. Drosophila neuropeptides in regulation of physiology and behavior. Prog Neurobiol. 2010;92(1):42–104. doi: 10.1016/j.pneurobio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Schoofs L, et al. Locustatachykinin III and IV: Two additional insect neuropeptides with homology to peptides of the vertebrate tachykinin family. Regul Pept. 1990;31(3):199–212. doi: 10.1016/0167-0115(90)90006-i. [DOI] [PubMed] [Google Scholar]
- 13.Schoofs L, Holman GM, Hayes TK, Nachman RJ, De Loof A. Locustatachykinin I and II, two novel insect neuropeptides with homology to peptides of the vertebrate tachykinin family. FEBS Lett. 1990;261(2):397–401. doi: 10.1016/0014-5793(90)80601-e. [DOI] [PubMed] [Google Scholar]
- 14.Van Loy T, et al. Tachykinin-related peptides and their receptors in invertebrates: A current view. Peptides. 2010;31(3):520–524. doi: 10.1016/j.peptides.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Nässel DR. Tachykinin-related peptides in invertebrates: A review. Peptides. 1999;20(1):141–158. doi: 10.1016/s0196-9781(98)00142-9. [DOI] [PubMed] [Google Scholar]
- 16.Satake H, Kawada T, Nomoto K, Minakata H. Insight into tachykinin-related peptides, their receptors, and invertebrate tachykinins: A review. Zoolog Sci. 2003;20(5):533–549. doi: 10.2108/zsj.20.533. [DOI] [PubMed] [Google Scholar]
- 17.Severini C, Improta G, Falconieri-Erspamer G, Salvadori S, Erspamer V. The tachykinin peptide family. Pharmacol Rev. 2002;54(2):285–322. doi: 10.1124/pr.54.2.285. [DOI] [PubMed] [Google Scholar]
- 18.Zhou WY, et al. The evolution of tachykinin/tachykinin receptor (TAC/TACR) in vertebrates and molecular identification of the TAC3/TACR3 system in zebrafish (Danio rerio) Mol Cell Endocrinol. 2012;361(1-2):202–212. doi: 10.1016/j.mce.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Li XJ, Wolfgang W, Wu YN, North RA, Forte M. Cloning, heterologous expression and developmental regulation of a Drosophila receptor for tachykinin-like peptides. EMBO J. 1991;10(11):3221–3229. doi: 10.1002/j.1460-2075.1991.tb04885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monnier D, et al. NKD, a developmentally regulated tachykinin receptor in Drosophila. J Biol Chem. 1992;267(2):1298–1302. [PubMed] [Google Scholar]
- 21.Poels J, et al. Characterization and distribution of NKD, a receptor for Drosophila tachykinin-related peptide 6. Peptides. 2009;30(3):545–556. doi: 10.1016/j.peptides.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Poels J, et al. Functional comparison of two evolutionary conserved insect neurokinin-like receptors. Peptides. 2007;28(1):103–108. doi: 10.1016/j.peptides.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Veenstra JA. Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch Insect Biochem Physiol. 2000;43(2):49–63. doi: 10.1002/(SICI)1520-6327(200002)43:2<49::AID-ARCH1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 24.Anastasi A, Erspamer V. The isolation and amino acid sequence of eledoisin, the active endecapeptide of the posterior salivary glands of Eledone. Arch Biochem Biophys. 1963;101(1):56–65. doi: 10.1016/0003-9861(63)90533-2. [DOI] [PubMed] [Google Scholar]
- 25.Champagne DE, Ribeiro JM. Sialokinin I and II: Vasodilatory tachykinins from the yellow fever mosquito Aedes aegypti. Proc Natl Acad Sci USA. 1994;91(1):138–142. doi: 10.1073/pnas.91.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39(6):715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 27.Lai JSY, Lo SJ, Dickson BJ, Chiang AS. Auditory circuit in the Drosophila brain. Proc Natl Acad Sci USA. 2012;109(7):2607–2612. doi: 10.1073/pnas.1117307109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyamoto T, Amrein H. Suppression of male courtship by a Drosophila pheromone receptor. Nat Neurosci. 2008;11(8):874–876. doi: 10.1038/nn.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otsuna H, Ito K. Systematic analysis of the visual projection neurons of Drosophila melanogaster. I. Lobula-specific pathways. J Comp Neurol. 2006;497(6):928–958. doi: 10.1002/cne.21015. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka NK, Endo K, Ito K. Organization of antennal lobe-associated neurons in adult Drosophila melanogaster brain. J Comp Neurol. 2012;520(18):4067–4130. doi: 10.1002/cne.23142. [DOI] [PubMed] [Google Scholar]
- 31.Tayler TD, Pacheco DA, Hergarden AC, Murthy M, Anderson DJ. A neuropeptide circuit that coordinates sperm transfer and copulation duration in Drosophila. Proc Natl Acad Sci USA. 2012;109(50):20697–20702. doi: 10.1073/pnas.1218246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terhzaz S, Rosay P, Goodwin SF, Veenstra JA. The neuropeptide SIFamide modulates sexual behavior in Drosophila. Biochem Biophys Res Commun. 2007;352(2):305–310. doi: 10.1016/j.bbrc.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 33.Heisenberg M, Wolf R. Vision in Drosophila. Genetics of Microbehaviour. Berlin: Springer; 1984. [Google Scholar]
- 34.Sturtevant AH. Experiments on sex recognition and the problem of sexual selection in Drosoophilia. J Anim Behav. 1915;5(5):351. [Google Scholar]
- 35.Featherstone DE, Yanoga F, Grosjean Y. Accelerated bang recovery in Drosophila genderblind mutants. Commun Integr Biol. 2008;1(1):14–17. doi: 10.4161/cib.1.1.6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamanaka N, et al. Neuropeptide receptor transcriptome reveals unidentified neuroendocrine pathways. PLoS ONE. 2008;3(8):e3048. doi: 10.1371/journal.pone.0003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park Y, Adams ME. Insect G protein-coupled receptors: Recent discoveries and implicaitons. In: Gilbert LI, Gill SS, editors. Insect Pharmacology: Channels, Receptors, Toxins and Enzymes. London: Elsevier, Academic; 2010. [Google Scholar]
- 38.Yamanaka N, et al. Regulation of insect steroid hormone biosynthesis by innervating peptidergic neurons. Proc Natl Acad Sci USA. 2006;103(23):8622–8627. doi: 10.1073/pnas.0511196103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186(2):735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aikins MJ, et al. Vasopressin-like peptide and its receptor function in an indirect diuretic signaling pathway in the red flour beetle. Insect Biochem Mol Biol. 2008;38(7):740–748. doi: 10.1016/j.ibmb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Li B, Beeman RW, Park Y. Functions of duplicated genes encoding CCAP receptors in the red flour beetle, Tribolium castaneum. J Insect Physiol. 2011;57(9):1190–1197. doi: 10.1016/j.jinsphys.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 44.Johnson EC, et al. Identification of Drosophila neuropeptide receptors by G protein-coupled receptors-beta-arrestin2 interactions. J Biol Chem. 2003;278(52):52172–52178. doi: 10.1074/jbc.M306756200. [DOI] [PubMed] [Google Scholar]
- 45.Park Y, Kim YJ, Dupriez V, Adams ME. Two subtypes of ecdysis-triggering hormone receptor in Drosophila melanogaster. J Biol Chem. 2003;278(20):17710–17715. doi: 10.1074/jbc.M301119200. [DOI] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yapici N, Kim YJ, Ribeiro C, Dickson BJ. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature. 2008;451(7174):33–37. doi: 10.1038/nature06483. [DOI] [PubMed] [Google Scholar]
- 48. Ejima A, Griffith LC (2007) Measurement of courtship behavior in Drosophila melanogaster. Cold Spring Harbor Protocols 2007(10):pdb.prot4847. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.