Significance
The jawless vertebrates (hagfish and lampreys) possess an alternative adaptive immune system in which variable lymphocyte receptors (VLRs) constructed of leucine-rich repeats are used to recognize foreign antigens. Three VLR genes have been identified in lampreys (VLRA, VLRB, and VLRC), but only two (VLRA and VLRB) have been found in hagfish. Here, we identified and characterized a third hagfish VLR gene. Our analysis indicates that the third hagfish VLR is the ortholog of lamprey VLRA, while the previously identified hagfish “VLRA” is the counterpart of lamprey VLRC. The demonstration of three orthologous VLR genes in hagfish and lampreys suggests that this anticipatory receptor system evolved in a common ancestor of the two jawless vertebrate lineages ∼480 Mya.
Keywords: adaptive immunity, molecular evolution, antigen receptor, agnatha
Abstract
Jawless vertebrates (cyclostomes) have an alternative adaptive immune system in which lymphocytes somatically diversify their variable lymphocyte receptors (VLR) through recombinatorial use of leucine-rich repeat cassettes during VLR gene assembly. Three types of these anticipatory receptors in lampreys (VLRA, VLRB, and VLRC) are expressed by separate lymphocyte lineages. However, only two VLR genes (VLRA and VLRB) have been found in hagfish. Here we have identified a third hagfish VLR, which undergoes somatic assembly to generate sufficient diversity to encode a large repertoire of anticipatory receptors. Sequence analysis, structural comparison, and phylogenetic analysis indicate that the unique hagfish VLR is the counterpart of lamprey VLRA and the previously identified hagfish “VLRA” is the lamprey VLRC counterpart. The demonstration of three orthologous VLR genes in both lampreys and hagfish suggests that this anticipatory receptor system evolved in a common ancestor of the two cyclostome lineages around 480 Mya.
Phylogenetic studies of immunity indicate the emergence of two types of recombinatorial adaptive immune systems (AISs) in vertebrates (1, 2). All of the extant jawed vertebrates generate a vast repertoire of Ig-domain–based T- and B-cell antigen receptors primarily by the recombinatorial assembly of Ig V-(D)-J gene segments and somatic hypermutation (3). The extant jawless vertebrates, lampreys and hagfish, instead have an alternative AIS that is based on variable lymphocyte receptors (VLRs), the diversity of which is generated through recombinatorial use of leucine-rich repeat (LRR) cassettes (4–6). The germ-line VLR genes are incomplete in that they only contain coding sequences for the leader sequence, incomplete amino- and carboxyl-terminal LRR subunits (LRRNT and LRRCT) and the stalk region (4, 7, 8). However, each germ-line VLR gene is flanked by hundreds of different LRR-encoding sequences, which can be used as templates to add LRR sequences during the assembly of a mature VLR gene (4, 8–11). This gene conversion-like process is postulated to involve the activation-induced cytidine deaminase (AID) orthologs, cytidine deaminases 1 and 2 (CDA1 and CDA2) (8, 12). The combinatorial VLR assembly can generate a vast repertoire of anticipatory receptors comparable in diversity to the repertoire of Ig-domain–based antigen receptors in jawed vertebrates (8, 9).
Three VLR genes have been identified in lampreys (VLRA, VLRB, and VLRC), but only two (VLRA and VLRB) have been identified so far in hagfish (4, 7, 8, 13). Lamprey VLRB-expressing cells can respond to immunization by undergoing lymphoblastoid transformation, clonal expansion, and secretion of their antigen-specific VLRB antibodies (9, 14). VLRA- and VLRC-bearing cells also proliferate in response to antigen stimulation, but do not differentiate into antibody secreting cells; instead they maintain cell surface expression of their receptors, while increasing the expression of proinflammatory cytokines, macrophage migration inhibitory factor (MIF), and interleukin-17 (IL-17) (12). CDA1-expressing progenitors assemble their VLRA and VLRC genes to become VLRA+ and VLRC+ lymphocytes in a thymus-equivalent region of the gills termed the thymoid (15, 16). Conversely, VLRB assembly coincides with CDA2 expression during VLRB+ lymphocyte development in hematopoietic tissues (15). The T- and B-like characteristics of the lamprey lymphocytes imply that jawless vertebrates have humoral and cellular arms of adaptive immunity comparable to those of jawed vertebrates.
In the present study, we sought to determine whether or not hagfish have a third VLR gene. In comparing the sequences of the two known hagfish VLRs with those of the three lamprey VLRs, we noticed that the highly variable inserts in the LRRCT module of the currently designated hagfish “VLRA” are more similar to those of lamprey VLRC than to those of lamprey VLRA (13, 17, 18). This led us to hypothesize that a third hagfish VLR, if present, would be the true counterpart of lamprey VLRA. Through a similarity search against the hagfish database, we identified a fragment of a potential third VLR gene, which was in turn used to clone and sequence a previously uncharacterized VLR gene. Here, we report the characterization of this third VLR type in pacific hagfish (Eptatretus stoutii) and its phylogenetic and structural relationship with previously identified VLRs. Our findings suggest a modified nomenclature for the hagfish VLRs.
Results
Identification and Characterization of a third VLR Gene in Hagfish.
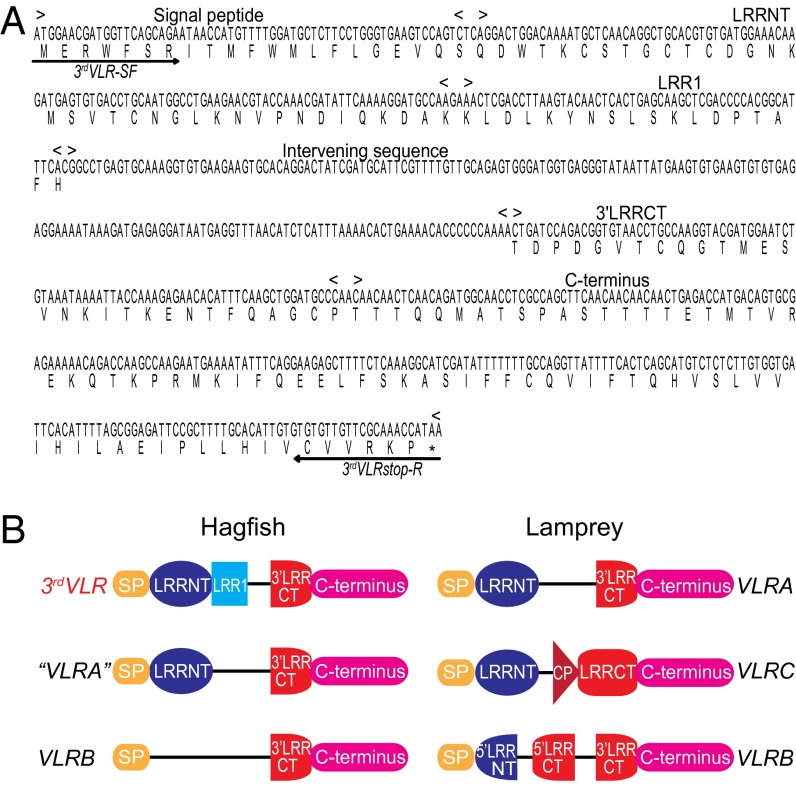
Based on the hypothesis that, if hagfish have a third VLR, it would be orthologous to lamprey VLRA, we used the signal peptides, LRRNT, LRRCT, and the C-terminal regions from 30 mature lamprey VLRAs as queries to perform a tBLASTn-based similarity search of the hagfish EST database. A peptide fragment encoded in the third open reading frame (ORF) of an EST clone (GenBank accession no. BAI66885) was found to share 75% identity with the 5′LRRCT region of one of the query sequences (GenBank accession no. ABO21305). The hagfish EST clone containing this peptide fragment was retrieved and hypothetically translated into three ORFs. Whereas a nonproductive VLR-like sequence in ORF1 lacked a connecting peptide (CP) and intact LRRCT motif, the corresponding region in ORF3 could encode a novel CP-5′LRRCT module (Fig. S1). When this CP-5′LRRCT peptide sequence was blasted in the National Center for Biotechnology Information (NCBI) database, the top 50 BLAST hits were all agnathan VLRs; the highest similarity (68% amino acid identity) was with the sea lamprey (Petromyzon marinus) VLRA clone PmVLRA.D4. Accordingly, a reverse primer was designed for 5′-RACE extension to clone the upstream region of this gene using a hagfish leukocyte cDNA library. After obtaining the 5′-region, a primer against the hypothetical signal peptide region was used to perform 3′-RACE to clone its downstream region. We thus obtained a cDNA clone encoding a protein with the identifying characteristics of currently defined VLRs in jawless vertebrates (Fig. 1). The predicted protein consists of a signal peptide (SP) of 21 residues, a 36-residue LRRNT domain, an 18-residue LRR1 followed by four diverse 24-residue LRRVs, a 13-residue CP, a 62-residue LRRCT module, and a 77-residue C-terminal domain including a threonine/proline-rich stalk region and a hydrophobic tail. Notably, the signal peptide and the C-terminus of this protein have only weak sequence similarity to previously identified hagfish VLRs (Fig. 1). This unique hagfish VLR was provisionally named the 3rdVLR.
Fig. 1.
Sequence comparison between a productive hagfish 3rdVLR and representatives of other types of VLRs. Deduced amino acid sequences of VLRs from Pacific hagfish (Es, Eptatretus stoutii), inshore hagfish (Eb, Eptatretus burgeri), sea lamprey (Pm, Petromyzon marinus), Arctic lampery (Lc, Lethenteron camtschaticum), and European brook lamprey (Lp, Lampetra planeri) are shown in the alignment. Conserved cysteines are highlighted in light green and other conserved residues in yellow, whereas “−” indicates a gap and “#” represents the highly variable inserts in LRRCT. The fifth LRRV module of LpVLRC is not shown to simplify the alignment for illustrative purposes. GenBank accession numbers of representative VLR sequences: Es3rdVLR, KF314050; PmVLRA, ABO27114; LcVLRA, BAJ14924; Eb“VLRA”, ABB59067; Es“VLRA”, ABB59097; PmVLRC, KC244079; LcVLRC, BAJ14926; LpVLRC, AGD98752; PmVLRB, AAT70348; LcVLRB, BAJ14925; EsVLRB, ABB59059; and EbVLRB, BAI66964.
Structure of the Germ-Line 3rdVLR.
The germ-line structure of the hagfish 3rdVLR gene was determined by RT-PCR in combination with genomic PCR. Erythrocyte DNA was used as template for the genomic PCR because these cells maintain their VLR genes in germ-line configuration. As is the case for hagfish “VLRA” and VLRB (7), our genomic PCR analysis suggested that the hagfish 3rdVLR is a single-copy gene. The hagfish germ-line 3rdVLR is also incomplete, but distinctive from previously identified VLRs (Fig. 2). It consists of two coding regions separated by a 173-bp intervening sequence (Fig. 2A). The first coding region encodes the signal peptide, LRRNT, and LRR1 modules, wheras the second coding region encodes the 3′-end of LRRCT and the C-terminus domain (Fig. 2B).
Fig. 2.
Germ-line gene of Pacific hagfish 3rdVLR and its comparison with previously described agnathan VLR genes. (A) Germ-line gene sequence of the hagfish 3rdVLR (GenBank accession no. KF314110). The gene (719 nucleotides in length) possesses two coding regions separated by an intervening sequence. Arrowed lines indicate the forward and reverse primer regions used for 3′-RACE and cDNA cloning. (B) Schematic depiction of the hagfish and lamprey VLR genes (not drawn to scale). Note the distinctive structure of the hagfish 3rdVLR germ-line gene.
Hagfish 3rdVLR Diversity.
To evaluate the diversity of hagfish 3rdVLRs, 64 mature cDNA clones from two Pacific hagfish were sequenced. Sequence analysis indicated that none of these clones coded for an identical protein sequence (Fig. S2). The 3rdVLR sequences contain three to six 24-residue LRRV modules (average, four), which is very similar to the hagfish “VLRA” module composition (average, four; range from two to six) but larger than the average module composition of hagfish VLRB (average, 2.8; range from one to six). Interestingly, this relationship is also true for the three lamprey VLRs (Fig. S3). Of the 256 LRRV modules encoded by the 64 hagfish 3rdVLR cDNA clones, 94% (241/256) had unique sequences. Similarly, 42% (27/64) of the LRR1 modules and 45% (29/64) of the CP modules had unique sequences. Of note, even though the germ-line 3rdVLR encodes a complete LRR1 module, this sequence can be replaced by sequences of divergent LRR1-coding genomic donor cassettes. Like lamprey VLRA and VLRB and hagfish VLRB (4, 7–9), the LRRCT region of the hagfish 3rdVLRs displays a high level of sequence diversity (72% unique LRRCTs, 46/64), due to use of unique 5′-LRRCT donor cassettes during the assembly process. The sequence diversity of the 3rdVLR of hagfish thus appears comparable to that of previously known VLRs.
Hagfish 3rdVLR Is a Lamprey VLRA Ortholog.
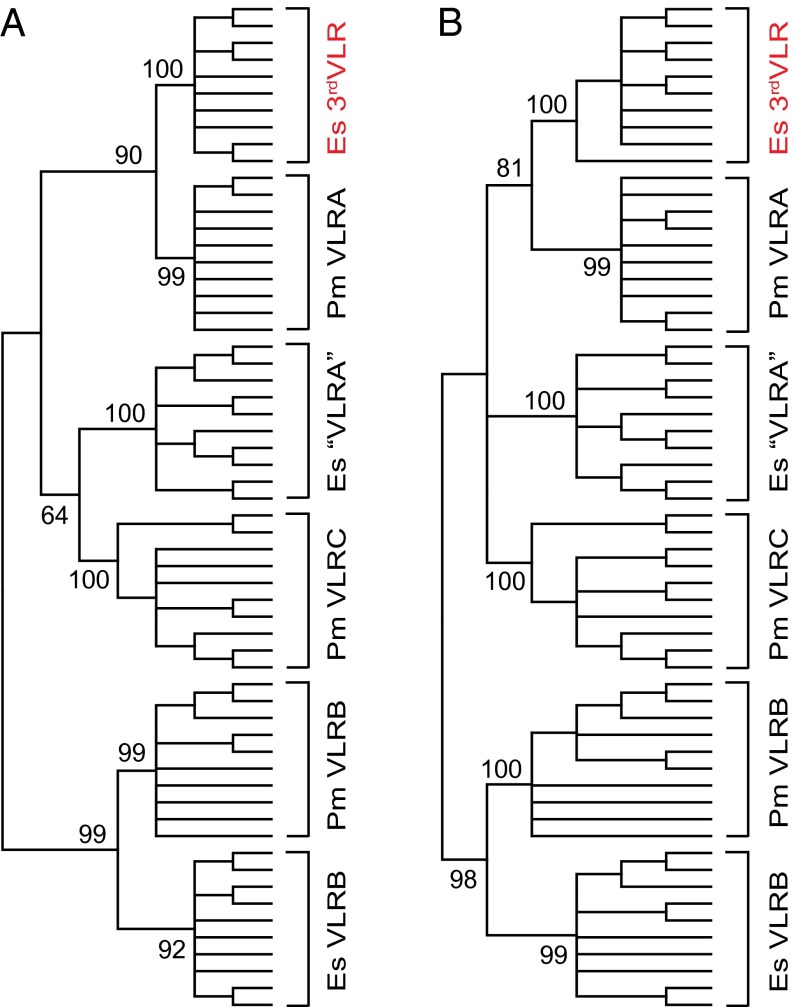
To examine the evolutionary relationship of hagfish 3rdVLR with the known VLRs in jawless vertebrates, we conducted phylogenetic analyses using the maximum likelihood (ML) and neighbor-joining (NJ) methods (19, 20). The bootstrap support for the hagfish “VLRA” and lamprey VLRC-containing cluster was low (64%) in the ML tree (Fig. 3A) and below 60% in the NJ phylogenetic analysis (Fig. 3B). However, the hagfish 3rdVLR clustered with lamprey VLRA with 90% bootstrap support in the ML tree, indicating an orthologous relationship (Fig. 3A), which is also supported by their clustering together with high bootstrap support in the NJ phylogenetic tree (Fig. 3B). Similarly, hagfish VLRB clustered with lamprey VLRB with 99% bootstrap support in the ML tree (Fig. 3A) and 98% bootstrap support for the NJ tree (Fig. 3B).
Fig. 3.
Phylogenetic relationship of Pacific hagfish 3rdVLR and other agnathan VLR genes. The phylogenetic tree is constructed by the (A) maximum likelihood and (B) neighbor-joining methods using amino acid sequences of the diversity region that could be reliably aligned (LRRNT, LRR1, terminal LRRV, CP, and LRRCT). The tree is condensed at the 60% bootstrap value level. The bootstrap supports for interior branches are shown. GenBank accession numbers. of VLR sequences are: hagfish 3rdVLR, KF314046–KF314055; hagfish “VLRA”, ABB59089–ABB59098; hagfish VLRB, ABB59051–ABB59060; lamprey VLRA, ABO21293–ABO21302; lamprey VLRC, KC244100–KC244109; and lamprey VLRB, ABO15209–ABO15218.
Hagfish 3rdVLR and Lamprey VLRA Share Common LRRCT Features.
All of the VLRs contain four cysteines that form two sets of disulfide bridges in both the LRRNT and LRRCT. These serve to cap the hydrophobic core of the solenoid structure of VLRs (21). With the exception of lamprey VLRA, however, the LRRCT of previously defined VLRs belongs to the class 1 according to the C-terminal cysteine-containing flanking domain classification (22) in that they share a characteristic four-cysteine motif C1-X-C2-Xm-C3-Xn-C4 (where X stands for any amino acid other than cysteine and m and n stand for variable numbers) (Fig. S4). The exception is LRRCT of lamprey VLRA, which contains a distinctive four cysteine motif in which an extra serine or glycine is inserted between the first and second cysteines, C1-(S/G)-X-C2-Xm-C3-Xn-C4. Interestingly, the four-cysteine motif in the LRRCT of the hagfish 3rdVLR (C1-X-S-C2-Xm-C3-Xn-C4) matches with lamprey VLRA (Fig. S4) in that the first and second cysteines are separated by two amino acids instead of the single amino acid spacing found in other VLRs.
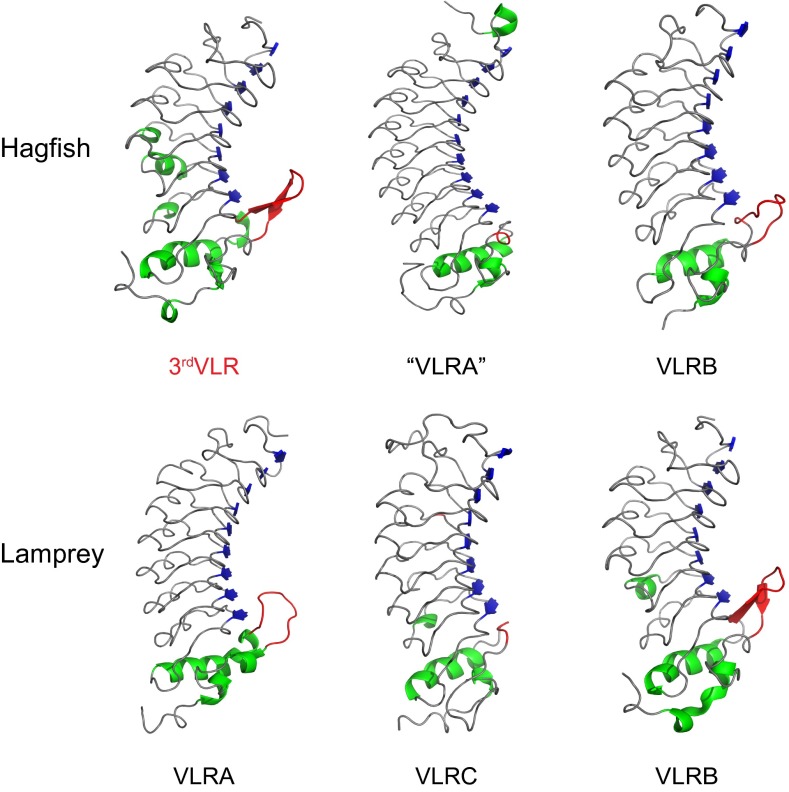
In the lamprey VLRAs and VLRBs and in hagfish VLRBs, the LRRCT regions contain a highly variable insert that can form a protruding loop. The importance of this loop in antigen recognition has been elucidated by several solved VLR-antigen structures (18, 23–25). However, the variable inserts in lamprey VLRCs (median length, 2 residues) and hagfish “VLRA”s (median length, 3 residues) are too short to form this protrusion (Fig. S5) (13, 17). Our 3D structure modeling predicts that the hagfish 3rdVLR adopts a solenoid structure like the other VLRs and has the potential to form a protruding loop at its LRRCT portion (Fig. 4) due to its relatively large LRRCT insert (median length, 10 residues). Analyses of the inserts indicate that the 3rdVLR insert lengths range from 9 to 13 residues (Fig. S5), which is comparable to the insert lengths in lamprey VLRAs (from 10 to 13 residues) (8, 24).
Fig. 4.
Comparison of the predicted 3D structure of hagfish 3rdVLR (GenBank accession no. KF314090) and other agnathan VLRs. The crystal structures of hagfish “VLRA” (PDB ID: 2O6Q), hagfish VLRB (PDB ID: 2O6S), lamprey VLRA (PDB ID: 3M18), and lamprey VLRB (PDB ID: 3E6J) were retrieved from Protein Data Bank (PDB). The 3D structure of lamprey VLRC (GenBank accession no. KC244064) is also predicted for comparison. β-Sheet and α-helix structures are shown in blue and green colors, respectively. Loops located in the LRRCT portion are indicated in red. Note that the hagfish 3rdVLR, lamprey VLRA, hagfish VLRB, and lamprey VLRB all possess a protruding loop in the LRRCT region, whereas hagfish “VLRA” and lamprey VLRC do not.
Expression Patterns of the Three Hagfish VLRs.
We used qPCR to examine the transcript expression profiles for hagfish 3rdVLR, “VLRA”, and VLRB in different tissues (Fig. S6). Hagfish 3rdVLR and the previously identified “VLRA” have similar expression patterns, wherein their transcripts were most abundant in blood leukocytes, followed by liver, intestine, and skin; only trace levels were found in kidneys and gills. The expression levels of hagfish VLRB were generally higher, especially in liver and intestine; expression was also detected in blood and skin samples.
Discussion
This study identifies a third hagfish VLR gene whose germ-line configuration and sequence are unique. Our analysis of cDNA clones indicates that the sequence diversity of the newly identified hagfish VLRs is comparable to that observed for other VLRs (8, 9). Together with the previously described “VLRA” and VLRB genes, the third hagfish VLR is transcribed by cells in the blood, liver, intestine, and skin, suggesting a potential role in immune defense.
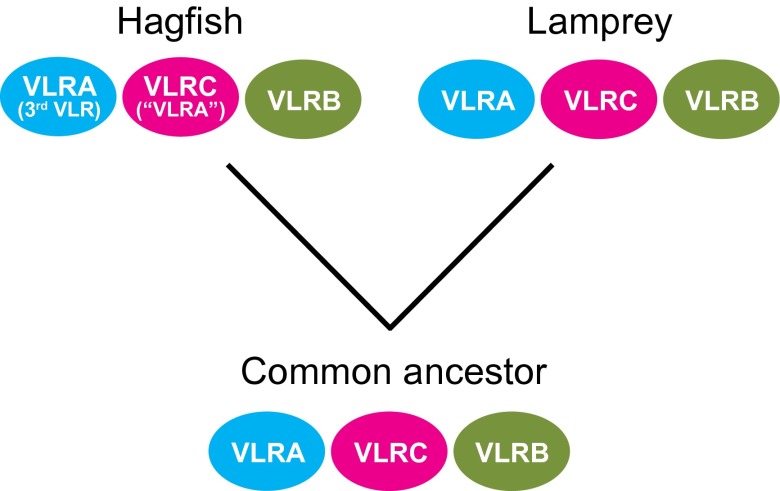
Previous studies have shown that the hagfish VLRB gene is orthologous to the lamprey VLRB gene (7). Our analysis of the cysteine configuration in the LRRCT motif, length variations of the LRRCT inserts, and phylogenetic relationships of the third hagfish VLR indicate that it is orthologous to lamprey VLRA, whereas the previously designated hagfish “VLRA” is more closely related to lamprey VLRC. Based on our comparative analysis of the three types of cyclostome VLR genes, we propose a modified nomenclature for the hagfish VLRs in which the unique third VLR in hagfish is the true lamprey VLRA counterpart, and the previously identified “VLRA” in hagfish is orthologous to the lamprey VLRC.
Structural comparison of the three VLRs in lamprey and hagfish also support the revised nomenclature proposal. VLRs use amino acid variations on their concave surfaces, which are composed of the LRRNT, LRR1, LRRVs, and CP motifs, along with the highly variable LRRCT loops to bind antigens (18). Together with the results of previous studies, our analysis reveals structural differences among the three VLR isotypes, which imply specialization in terms of ligand recognition (18, 23–25). The VLRAs and VLRCs in lampreys and hagfish have a higher average number of LRRV modules than do the VLRBs, thus indicating they have a larger concave surface for antigen binding. Another structural difference in the three VLR types relates to the highly variable LRRCT inserts. The VLRAs and VLRBs typically have a protruding loop because of their relatively large LRRCT inserts, whereas the LRRCT inserts in VLRCs are too short to form this loop (refs. 13 and 17 and this study). Furthermore, the length variation of LRRCT inserts in hagfish and lamprey VLRAs is less pronounced than in the VLRBs. These structural differences may suggest different antigen-binding modes for the three VLR isotypes. The single example in which a structural comparison was conducted of hen egg white lysozyme (HEL) binding by a VLRA.R2.1 and by a VLRB.2D favors this interpretation (24), although further analysis of the antigen-binding modes of the different VLRs is clearly needed.
Our recent studies indicate that lampreys have two primordial T-like lineages of lymphocytes that express VLRA and VLRC, respectively, and a prototypic B-like lineage of lymphocytes, much like the αβ T, γδ T, and B cells in jawed vertebrates. In some ways the lamprey VLRA- and VLRC-expressing cells resemble the αβ and γδ T cells of jawed vertebrates, whereas the VLRB-bearing cells are more morphologically and functionally similar to B cells (12, 14, 16). Hagfish VLRC (“VLRA” in the old nomenclature) and VLRB are also expressed by two distinct lymphocyte populations (26), and the hagfish VLRB protein is secreted (27). The future characterization of the cells that express the three types of hagfish VLRs described here promises to provide deeper insight into the evolution of this tripartite lymphocytic differentiation pattern. Our findings indicate that the three VLR isotypes existed in the common ancestor of lampreys and hagfish (Fig. 5).
Fig. 5.
Evolutionary scenario of VLR genes in the two cyclostome lineages. The revised nomenclature for hagfish “VLRA” and VLRC is indicated.
The relatively primitive anatomy of hagfish compared with the lampreys initially led to their classification as more basal jawless vertebrates (28). However, molecular sequence data suggests a monophyletic relationship between hagfish and lampreys (29) and recent studies of hagfish embryos suggest that their morphological divergence may reflect a loss of features as cyclostomes divergently evolved under different selective pressures (30). Whereas the presence of three related VLR types in these cyclostome representatives suggests that a common ancestor of the hagfish and lamprey lineages possessed three VLR-based anticipatory receptors, many differences can be anticipated between the immune systems of hagfish and lampreys, given the >400 My of independent evolution of the two lineages (31).
Materials and Methods
Animals.
Pacific hagfish E. stoutii (30–60 cm long) were purchased from Marinus and maintained at 14–17 °C in artificial sea water (Oceanic Systems). All experiments were approved by the Institutional Animal Care and Use Committee at Emory University.
Data Mining and Similarity Search.
The signal peptides, LRRNT, LRRCT, and 3′-terminus portions from 30 mature lamprey VLRAs (GenBank accession nos. ABO21280–ABO21309 and ABO27114) were used as queries for tBLASTn search against the hagfish EST database (http://transcriptome.cdb.riken.go.jp/vtcap/blast/blast.html) (32). The correct ORF was identified by alignment of the query sequence and the hypothetical translation of the hit sequence. The signal peptide region was determined using SignalP (33). The sequence conservation was assessed by WebLogo (http://weblogo.berkeley.edu/logo.cgi) (34).
cDNA Library Construction and Cloning.
Hagfish were anesthetized in MS-222 (Sigma) before collecting blood from the tail sinus (7). Blood samples were loaded onto a 10% (wt/vol) OptiPrep gradient (Axis-Shield) and centrifuged at 800 × g for 15 min to obtain white blood cells in the buffy coat. Total RNA extracted from buffy coat cells using the RNeasy kit (Qiagen) was used to construct a 5′- and 3′-RACE cDNA library (Clontech). Full-length hagfish 3rdVLR was cloned from the 5′- and 3′-RACE cDNA library by a two-step PCR. The first step was to clone the 5′-region of the hagfish 3rdVLR by using a 3rdVLR-R1 primer (Fig. S1 and Table S1) against the partial sequence identified from the hagfish EST database in combination with the 5′-RACE adaptor primer included in the kit. In step two, the 3′-RACE PCR was performed to clone the 3′-region of hagfish 3rdVLR. The 3′-RACE adaptor primer together with a 3rdVLR-SF primer (forward primer against the signal peptide region of hagfish 3rdVLR as indicated in Fig. 2A and Table S1) was used in this step. The germline and mature hagfish 3rdVLRs were cloned using the primers 3rdVLR-SF and 3rdVLRstop-R (reverse primer according to the position of stop codon as indicated in Fig. 2A and Table S1).
Phylogenetic Analysis.
Sequences were aligned by using the CLUSTALW program (35) with adjustment by manual correction where necessary. NJ (20) and ML (19) trees were constructed using MEGA software 5.0 with the pairwise deletion option (36). The evolutionary distances were computed using the Jones–Taylor–Thornton (JTT) matrix-based method (37). Bootstrap values were generated from 1,000 replicates.
Three-Dimensional Structure Modeling.
The crystal structure files of lamprey and hagfish “VLRA”, VLRB were extracted from Protein Data Bank (PDB). Three-dimensional models of lamprey VLRC and hagfish 3rdVLR were predicted through PHYRE2 Protein Fold Recognition Server (21) and visualized by Pymol software (http://www.pymol.org/).
VLR Transcription Analysis.
Expression of 3rdVLR, “VLRA”, and VLRB was examined by quantitative real-time PCR (qPCR) analysis of mRNA from cells in various tissues using RNeasy kit with on-column DNA digestion by DNase I (Qiagen). First-strand cDNA was synthesized with random hexamer primers and SuperScript III (Invitrogen). qPCR was conducted with SYBR Green on a 7900HT ABI Prism detection system (Applied Biosystems). Cycling conditions were 1 cycle at 50 °C for 2 min, 1 cycle at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. The values for the VLR genes were normalized to the expression of GAPDH. Primers used in this experiment are listed in Table S1.
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism software 5.0. The Kruskal–Wallis with post hoc Dunn’s tests was used to evaluate the LRRV module difference between different VLRs. Statistical significance was set up to P < 0.05.
Supplementary Material
Acknowledgments
We thank Baohua Wu and Yueju Su for their assistance in statistical analysis. J.L., S.D., B.R.H., M.H. and M.D.C. were supported by National Institutes of Health Grants R01 AI072435 and R01 GM100151 and the Georgia Research Alliance.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information database (GenBank accession nos. KF314046–KF314110).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314540110/-/DCSupplemental.
References
- 1.Litman GW, Anderson MK, Rast JP. Evolution of antigen binding receptors. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 2.Hirano M, Das S, Guo P, Cooper MD. The evolution of adaptive immunity in vertebrates. Adv Immunol. 2011;109:125–157. doi: 10.1016/B978-0-12-387664-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 3.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: Similarities and differences. Adv Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 4.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430(6996):174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- 5.Herrin BR, Cooper MD. Alternative adaptive immunity in jawless vertebrates. J Immunol. 2010;185(3):1367–1374. doi: 10.4049/jimmunol.0903128. [DOI] [PubMed] [Google Scholar]
- 6.Boehm T, et al. VLR-based adaptive immunity. Annu Rev Immunol. 2012;30:203–220. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pancer Z, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102(26):9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogozin IB, et al. Evolution and diversification of lamprey antigen receptors: Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8(6):647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 9.Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310(5756):1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 10.Nagawa F, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007;8(2):206–213. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- 11.Das S, et al. Organization of lamprey variable lymphocyte receptor C locus and repertoire development. Proc Natl Acad Sci USA. 2013;110(15):6043–6048. doi: 10.1073/pnas.1302500110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459(7248):796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasamatsu J, et al. Identification of a third variable lymphocyte receptor in the lamprey. Proc Natl Acad Sci USA. 2010;107(32):14304–14308. doi: 10.1073/pnas.1001910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alder MN, et al. Antibody responses of variable lymphocyte receptors in the lamprey. Nat Immunol. 2008;9(3):319–327. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]
- 15.Bajoghli B, et al. A thymus candidate in lampreys. Nature. 2011;470(7332):90–94. doi: 10.1038/nature09655. [DOI] [PubMed] [Google Scholar]
- 16.Hirano M, et al. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature. 2013 doi: 10.1038/nature12467. 10.1038/nature12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HM, et al. Structural diversity of the hagfish variable lymphocyte receptors. J Biol Chem. 2007;282(9):6726–6732. doi: 10.1074/jbc.M608471200. [DOI] [PubMed] [Google Scholar]
- 18.Velikovsky CA, et al. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat Struct Mol Biol. 2009;16(7):725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 22.Kajava AV. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277(3):519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 23.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321(5897):1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L, et al. A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc Natl Acad Sci USA. 2010;107(30):13408–13413. doi: 10.1073/pnas.1005475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchdoerfer RN, et al. Variable lymphocyte receptor recognition of the immunodominant glycoprotein of Bacillus anthracis spores. Structure. 2012;20(3):479–486. doi: 10.1016/j.str.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kishishita N, et al. Regulation of antigen-receptor gene assembly in hagfish. EMBO Rep. 2010;11(2):126–132. doi: 10.1038/embor.2009.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takaba H, et al. A major allogenic leukocyte antigen in the agnathan hagfish. Sci Rep. 2013;3:1716. doi: 10.1038/srep01716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janvier P. Early Vertebrates. Oxford: Oxford Univ Press; 1996. [Google Scholar]
- 29.Heimberg AM, Cowper-Sal-lari R, Sémon M, Donoghue PC, Peterson KJ. microRNAs reveal the interrelationships of hagfish, lampreys, and gnathostomes and the nature of the ancestral vertebrate. Proc Natl Acad Sci USA. 2010;107(45):19379–19383. doi: 10.1073/pnas.1010350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ota KG, Kuraku S, Kuratani S. Hagfish embryology with reference to the evolution of the neural crest. Nature. 2007;446(7136):672–675. doi: 10.1038/nature05633. [DOI] [PubMed] [Google Scholar]
- 31.Kuraku S, Ota KG, Kuratani S. Jawless fishes (Cyclostomata) In: Hedges SB, Kumar S, editors. The Timetree of Life. New York: Oxford Univ Press; 2009. pp. 317–319. [Google Scholar]
- 32.Takechi M, et al. Overview of the transcriptome profiles identified in hagfish, shark, and bichir: Current issues arising from some nonmodel vertebrate taxa. J Exp Zoolog B Mol Dev Evol. 2011;316(7):526–546. doi: 10.1002/jez.b.21427. [DOI] [PubMed] [Google Scholar]
- 33.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 34.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.