Abstract
B-cell failure at the onset of type 2 diabetes is caused by a decline in β-cell function in the postprandial state and loss of pancreatic β-cell mass. Recently, we showed an association between increased insulin secretion and a single nucleotide polymorphism (SNP), SNP rs12686676, in the NR4A3 gene locus encoding the nuclear receptor Nor-1. Nor-1 is expressed in β-cells, however, not much is known about its function with regard to insulin gene expression and insulin secretion. Nor-1 is induced in a glucose-/incretin-dependent manner via the PKA pathway and directly induces insulin gene expression. Additionally, it stimulates insulin secretion possibly via regulation of potentially important genes in insulin exocytosis. Moreover, we show that the minor allele of NR4A3 SNP rs12686676 fully rescues incretin resistance provoked by a well-described polymorphism in TCF7L2. Thus, Nor-1 represents a promising new target for pharmacological intervention to fight diabetes.
Keywords: Nor-1, Insulin gene expression, Insulin secretion, Incretin resistance, TCF7L2
1. Introduction
B-cell failure at the onset of type 2 diabetes (T2D) is caused by different mechanisms. On the one hand, β-cell function in the postprandial state declines and, on the other hand, pancreatic β-cell mass decreases over time [1,2]. Together with other factors, this promotes hyperglycemia. The main cause leading to T2D is overweight caused by western lifestyle (high-caloric diet and physical inactivity), however, a certain susceptibility to T2D is thought to be due to single nucleotide polymorphisms (SNPs). Interestingly, the majority of known T2D risk genes have an impact on the β-cell [3].
In 2009, we were able to show an association between insulin secretion and a common SNP in the NR4A3 gene locus encoding the metabolically important nuclear receptor Nor-1 [4], i.e., SNP rs12686676. Our studies revealed increased insulin secretion in subjects carrying this SNP's minor allele.
The nuclear receptor 4A (NR4A) subgroup consists of Nur77 (encoded by NR4A1) [5], Nurr1 (encoded by NR4A2) [6], and Nor-1 (encoded by NR4A3) [7]. These nuclear receptors are thought to be ligand-independent and to function as early-response genes regulating important cellular processes, e.g., cell proliferation, differentiation, and survival. Due to their expression in tissues with high energy demand, such as liver [8], brain, skeletal muscle [7], and adipose tissue [9], the function of the NR4A members has been analyzed mainly in these tissues. Nur77 regulates hepatic gluconeogenesis [10] and induces genes associated with glucose metabolism in skeletal muscle [11]. Nor-1 is involved in the regulation of lipid and energy metabolism in skeletal muscle [12]. Nur77 and Nor-1 have been shown to be induced by insulin in skeletal muscle and are implicated in insulin resistance [13]. In the liver, Nr4a3 is a CREB target and regulates hepatic glucose metabolism [10]. In most of these tissues, the members of the NR4A subgroup are induced via β-adrenoceptor activation which is tightly coupled to intracellular cAMP levels [14].
Upon expression, NR4A receptors can bind as monomers [15] and homodimers [16] to the octanucleotide 5′-AAAAGGTCA-3′ (NGFI-B response element, NBRE). Even though there are no endogenous ligands known up to now, several pharmacological compounds have been reported to activate the NR4A subgroup, like 6-mercaptopurine (6-MP) via the AF-1 domain [17] or 1,1-di(3′-indolyl)-1-(p-substituted phenyl)methanes via the aberrant C-terminal ligand-binding domain [18].
However, not much is known about the function of these nuclear receptors with regard to insulin gene expression and insulin secretion. We and others were able to show the expression of Nor-1 and Nur77 in β-cells [4,19,20]. Furthermore, our recent human study showed increased insulin secretion in individuals carrying the minor allele of SNP rs12686676 in NR4A3 [4]. Therefore, the aim of our study was to determine the molecular role of Nor-1 in glucose-stimulated insulin secretion (GSIS) and insulin gene expression.
2. Material and methods
2.1. Cells
INS-1E cells (kindly provided by Dr. C. B. Wollheim, University of Geneva, Switzerland) were cultured in a humidified atmosphere containing 5% CO2 in RPMI 1640 (11 mM glucose) supplemented with 10% fetal calf serum, 2 mM glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 60 µM 2-mercaptoethanol, 100 U/ml penicillin and 100 U/ml streptomycin. The following treatments were performed prior to lysis: forskolin (10 µM, 90 min, 3 h, and 24 h), dibutyryl-cAMP (db-cAMP, 1 mM, 90 min), exendin-4 (10 nM, 90 min), glucose (22.5 mM, 90 min), H89 (10 µM, 30 min pre-incubation), 6-MP (50 µM, 48 h).
2.2. Rat islets
Rat islets were isolated from 6-week-old male Wistar rats (Charles River Laboratories, Wilmington, MA, USA). Islets were isolated by collagenase (3 mg/ml, Serva, Heidelberg, Germany) infusion of the pancreas followed by digestion for 10 min at 37 °C and separated from exocrine tissue first by centrifugation through a Histopaque 1.077 g/ml (Sigma-Aldrich, St. Louis, MO, USA) cushion and then by manually collecting islets under the dissection microscope [21]. Islets were cultured overnight in RPMI 1640 containing 5.6 mM glucose. The following treatments were performed prior to lysis: forskolin (1 µM, 90 min), exendin-4 (10 nM, 90 min), glucose (22.5 mM, 90 min), H89 (10 µM, 30 min pre-incubation), 6-MP (50 µM, 24 h). Following the 6-MP treatment, batches of 10 islets each were pre-incubated for 1 h at 37 °C in buffer containing 2.8 mM glucose. For measurement of basal insulin secretion, the islets were incubated for 1 h at 37 °C in buffer containing 2.8 mM glucose. All animal procedures were approved by local government authorities for animal research according to the guidelines of laboratory animal care.
2.3. Human islets
Frozen human islets from 8 donors were purchased from ProCell Biotech (Newport Beach, Ca, USA). Islets were obtained as frozen pellets, resuspended in RLT (Qiagen, Hilden, Germany) buffer and subjected to RNA isolation.
2.4. RNA interference
Small interfering RNA (siRNA) oligonucleotides targeting Nr4a3, Creb1, and Glp1r were purchased as siGENOME-SMART-pool (Thermo Scientific, Rockford, IL, USA). As control, we used an unrelated siRNA targeting firefly luciferase in all experiments as reported earlier [22]. Transfection was performed with Dharmafect 4 (Dharmacon, Lafayette, CO, USA) according to the instructions of the manufacturer. 24 h after transfection, the cells either were immediately harvested or the medium was changed. Then, cells were either harvested at the indicated time point or were further incubated with 6-MP for 48 h or with forskolin for 24 h.
2.5. Quantitative real-time RT-PCR (reverse transcription PCR)
Cells and islets were washed once with PBS, lysed with RLT buffer and homogenized using QIAshredder (Qiagen). Total RNA (RNeasy Mini Kit, Qiagen) isolation, transcription to cDNA (Transcriptor First Strand cDNA Synthesis Kit, Roche Diagnostics, Indianapolis, IN, USA) and RT-PCR were performed as described before [23]. The following primer sequences (TIB Molbiol, Berlin, Germany) were used to amplify the indicated genes. Human genes: RPS13 5′-CCCCACTTGGTTGAAGTTGA-3′ and 5′-ACACCATGTGAATCTCTCAGGA-3′; NR4A1 5′-GCCCATGTCGACTCCAAC-3′ and 5′-ACTCATTTGATAGTCAGGGTTCG-3′; NR4A2 5′-GCCCATGTCGACTCCAAC-3′ and 5′-ACTCATTTGATAGTCAGGGTTCG-3′; NR4A3 5′-ACACCCAGAGATCTTGATTATTCC-3′ and 5′-GTAGAATTGTTGCACATGCTCAG-3′; and 5′-CACAATGCCACGCTTCTG-3′. Rat genes: Rps13 5′-CTGACGACGTGAAGGAACAA-3′ and 5′-TCACAAAACGGACCTGTGC-3′; Nr4a1 5′-TGCTCTGGTCCTCATCACTG-3′ and 5′-ACAGCTAGCAATGCGGTTC-3′; Nr4a2 5′-CCACGTGCACTCCAATCC-3′ and 5′-TAGTCAGGGTTTGCCTGGAA-3′; Nr4a3 5′-TGCCTGTCAGCACTGAGTATG-3′ and 5′-GCTGCTTGTGATCTTGTTGC-3′; Glp1r 5′-CTGCTTTGTCAACAATGAGGTC-3′ and 5′-GTCCCTCTGGATGTTCAAGC-3′; Creb1 5′-GACGGAGGAGCTTGTACCAC-3′ and 5′-GCATCTCCACTCTGCTGGTT-3′; Ins1 5′-AGACCATCAGCAAGCAGGTC-3′ and 5′-CTTGGGCTCCCAGAGGAC-3′; Ins2 5′-CGAAGTGGAGGACCCACA-3′ and 5′-TGCTGGTGCAGCACTGAT-3′. PCR conditions can be provided upon request. All RNA data are presented relative to the housekeeping gene Rps13 using the ΔΔCt method.
2.6. Microarray analysis
INS-1E cells were transfected with control or Nr4a3 siRNA as described earlier and harvested 72 h after transfection. Total RNA was isolated from four independent experiments, replicate samples were pooled according to the test condition and subjected to microarray analysis using GeneChip Rat Gene 1.0 ST Arrays from Affymetrix (Santa Clara, CA, USA). The microarray analysis was performed by MFT Services (Tübingen, Germany), an official Affymetrix service provider. Genes with a more than 2-fold and less than 0.5-fold change in expression were taken into account and the expression level of candidate genes was verified by RT-PCR in individual samples.
2.7. Intracellular insulin content
INS-1E cells were transfected with siRNA and kept in culture medium. The medium was changed to low-glucose medium (RPMI 1640, 2.8 mM glucose) 24 h prior to extraction. Cells were counted, washed once with PBS and subjected to acid ethanol (0.18 mol/l HCl in 70% ethanol) extraction. Extraction was performed overnight at 4 °C. After centrifugation, supernatants were frozen at −20 °C. For activation of Nor-1, INS-1E cells were incubated for 48 h with 6-MP (50 µM) in culture medium, counted, and subjected to acid ethanol extraction. Intracellular insulin content was quantified by ELISA (Mercodia AB, Uppsala, Sweden).
2.8. Insulin secretion
Cells were washed three times with Krebs–Ringer Buffer (KRB) containing 2.8 mM glucose and pre-incubated for 3 h with the same buffer. For basal insulin secretion, cells were incubated for 1 h in the same buffer. GSIS was measured in cells incubated with KRB containing 11 mM glucose. Cells were subjected to acid ethanol extraction, and intracellular as well as secreted insulin were quantified by ELISA (Mercodia). Basal rat islet insulin secretion was measured accordingly, except with 1 h pre-incubation in 2.8 mM Glc. Secreted insulin was normalized for intracellular insulin content.
2.9. Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed using the MAGnify Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. In brief, Nor-1 antibody (H7833) was purchased from R&D Systems (Minneapolis, MN, USA) and coupled to Dynabeads. INS-1E cells were incubated with 6-MP (50 µM) for 48 h to achieve binding of Nor-1 to DNA binding sites. Cells were washed with PBS, trypsinized and counted. Nor-1 binding to chromatin was fixed with 37% formaldehyde. After cell lysis, chromatin shearing and isolation, chromatin from 200,000 cells was used for chromatin immunoprecipitation. Crosslinking was reversed and DNA extracted according to the manufacturer's instructions. The following primers (Invitrogen) were used to amplify Nor-1 binding sites in Ins1 and Ins2 using quantitative real-time RT-PCR: Ins1 NBRE I 5′-CTTCGTTGTGACCTATTTTGGATGA-3′ and 5′-GGTTCAGTAACAAATGCCTGGAG-3′; Ins1 NBRE II 5′-GGAAGGCAACTGATTTCTTTGAGTTA-3′ and 5′-GATACAGATCGGAAAAGAAGAGGTCA-3′; Ins1 NBRE III 5′-ATCCCACACCATCCTGCAAT-3′ and 5′-CTTAGTTGGCCCACAAAAATCTT-3′; Ins2 NBRE I 5′-GGGAAGAAATTGGGCTTGGT-3′ and 5′-TTCAGAGCACTAAAGGTCACTTGGAT-3′; Ins2 NBRE II 5′-GATGTGCACCTTTGGGGTTTA-3′ and 5′-AACTCCAAGCAAAGAGAGGGATTACT-3′. PCR products were also analyzed by agarose gel electrophoresis.
2.10. Western blot
Cells were washed with PBS, and nuclear proteins were isolated using nuclear and cytoplasmic extraction reagents (NE-PER Nuclear and Cytoplasmic Extraction Reagents, Thermo Scientific). Nuclear lysates were centrifuged at 13,000g for 10 min, and the protein concentration was measured in the supernatant using the Bradford protein assay (Bio-Rad, Richmond, CA, USA). The same amount of protein was loaded in each lane of a SDS-PAGE gel. After separation, proteins were transferred to nitrocellulose membranes (Amersham Life Sciences Inc., Arlington Hights, IL, USA) and incubated with Nor-1 (1:500, H7833, R&D Systems) antibody or Histone H3 (1:1000, 3H1, Cell Signaling Technologies Inc., Beverly, MA, USA) antibody. Histone H3 was used as housekeeping protein for normalization of Nor-1 protein content. After three washes, membranes were incubated with goat anti-rabbit IgG–Peroxidase antibody (Sigma-Aldrich). The proteins were detected using the ECL system (Amersham Life Sciences Inc.). For quantification, we used EasyWin32 Herolab (Wiesloch, Germany) Software.
2.11. Luciferase reporter gene assays
A 190-bp sequence encompassing the Nor-1 binding site of the human insulin gene was amplified from genomic DNA obtained from human myotubes. A 207-bp sequence encompassing the rat binding site II of Ins1 was amplified from genomic DNA from INS-1E cells. The following primers (Invitrogen) were used: 5′-GGAGACCCTCTCCCTGACC-3′ and 5′-GGGACCCCGACTCTGACTTA-3′ for the human sequence, 5′-GCCACTTTGCTGAAGTTGTTT-3′ and 5′-CTGCCCACTCTCTCCCTACTT-3′ for the rat sequence. Both fragments were cloned into the luciferase reporter vector pGL3-Promoter (Promega, Madison, WI, USA). The vector harboring the human Nr4a3 gene was purchased from Origene (Rockville, MD, USA). HEK293 cells were seeded at 5000 cells per well in a 96-well plate, transfected with Lipofectamine (Invitrogen) 24 h after seeding and assayed 48 h after transfection using Dual-Glo Luciferase Assay reagent (Promega) according to the manufacturer's instructions.
2.12. Lentiviral transduction
Control vector, vector coding for rat Nr4a3, 3rd Generation Packaging Mix, Lentifectin, Polybrene and ViralPlus Transduction enhancer were purchased from Applied Biological Materials Inc (Vancouver, Canada). Virus production in HEK293FT cells (Invitrogen) and infection of INS-1E cells was performed according to the manufacturer's instructions.
2.13. Human data
Data from 1454 non-diabetic participants of the Tübingen Family (TÜF) Study for type 2 diabetes were analyzed in this study [24]. Data from repeated sampling oral glucose tolerance tests (OGTT; 0, 30, 60, 90, and 120 min) and TCF7L2 rs7903146 and NR4A3 rs12686676 genotypes were available from earlier studies [4,25]. Details on genotyping as well as on associations of these variants with insulin secretion and other traits were reported recently [4,25]. Insulin secretion during OGTT was estimated by the insulinogenic index as described previously [26].
2.14. Statistical analysis
For all statistical analysis, the Software package JMP 8.0.2 (SAS Institute, Cary, NC, USA) was used. If not indicated otherwise, data are provided as relative values with control conditions chosen as reference point. Data are given as mean±SEM. Two-group comparisons were performed using matched pairs Student's t-test. With regard to human data, we stratified the cohort for the TCF7L2 SNP rs7903146 and NR4A3 SNP rs12686676 genotypes using dominant inheritance models. Differences in insulin secretion between the NR4A3 genotypes were tested using multiple linear regression analysis with gender, age, and BMI as covariates. For all analyses, p-values≤0.05 were considered statistically significant.
3. Results
3.1. Nor-1 is a novel regulator of insulin genes
All mechanistic investigations in this study were performed in INS-1E, a commonly used model of insulin-secreting cells, and important findings were validated in primary pancreatic islets. First, we determined the expression of Nr4a1, Nr4a2, and Nr4a3 using RT-PCR. Expression of Nr4a1 in INS-1E cells was 12-fold higher than that of Nr4a3 which in turn was expressed 2-fold higher than Nr4a2 (Figure 1A). The isoform with the strongest expression in rat and human islets was Nr4a1 as well (Figure 1B and C). In rat islets, Nr4a2 expression was 5.5-fold higher than that of Nr4a3 (Figure 1B). Human islets expressed Nr4a2 and Nr4a3 at similar levels (Figure 1C).
Figure 1.
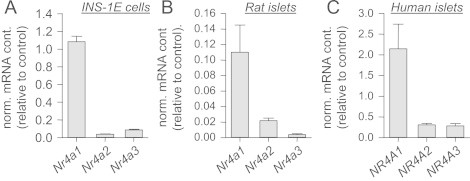
Expression of the Nr4a family members in INS-1E cells and primary islets. Expression of the indicated genes was measured in INS-1E cells (n=3), rat islets (n=8) and human islets (n=8). Data are given as mean±SEM.
Since the NR4A3 gene product Nor-1, but none of the other two family members, revealed an association with insulin secretion in humans [27], we intended to analyze whether Nor-1, as a transcription factor, affects insulin gene expression. Up to now, there are no physiological activators known for Nor-1. Therefore, we decided to use the pharmacological activator 6-MP. Activation of Nor-1 by 6-MP (48 h) induced a more than 2-fold increase in rat Ins1 and Ins2 gene expression (Figure 2A). To verify whether this gene induction is reflected at the protein level, we measured the intracellular insulin content in 6-MP-treated cells. Upon 6-MP treatment, the intracellular insulin content doubled compared to control cells (Figure 2B). Treatment of isolated rat islets with 6-MP for 24 h resulted in increased expression of Ins1 and Ins2 as well (Figure 2C).
Figure 2.
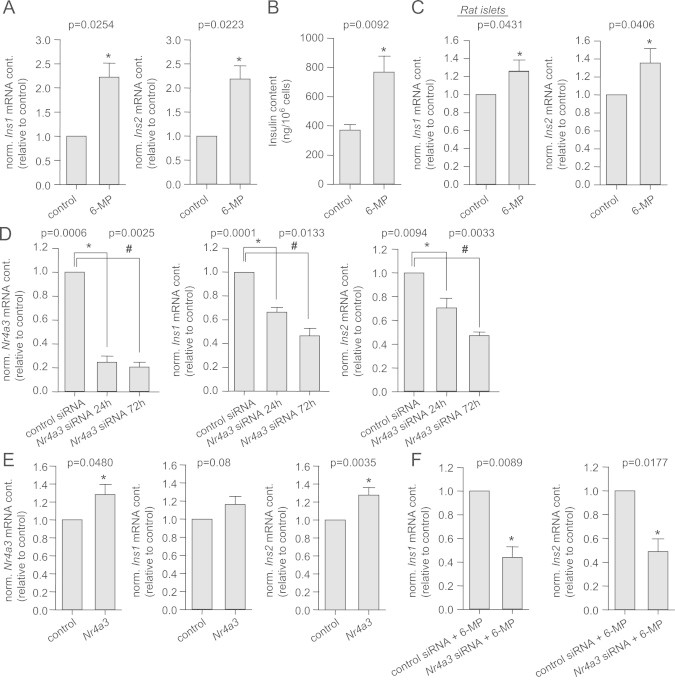
Nor-1-regulated expression of insulin genes. Expression of the rat insulin genes Ins1 and Ins2 (A) and intracellular insulin content (B) after incubation of INS-1E cells with 50 µM 6-MP for 48 h. Expression of the rat insulin genes Ins1 and Ins2 (C) after incubation of purified rat islets with 50 µM 6-MP for 24 h. After 24 h and 72 h treatment with Nr4a3 siRNA, the expression of Nr4a3, Ins1 and Ins2 (D) were measured in INS-1E cells. Expression of Nr4a3, Ins1 and Ins2 in Nr4a3-overexpressing INS-1E cells (E). Ins1 and Ins2 gene expression after Nr4A3 down-regulation with siRNA for 72 h and 6-MP (50 µM, 48 h) treatment of INS-1E cells (F). Data are given as mean±SEM. Two-group comparisons were performed using matched pairs Student's t-test (n≥3).
In the next step, Nr4a3 expression was down-regulated via RNA interference. Using small interfering RNA (siRNA), its expression was reduced by 75% after 24 h and by 80% after 72 h compared to control (Figure 2D). Nr4a3 down-regulation reduced the expression of Ins1 by 35% after 24 h and by 55% after 72 h, while the expression of Ins2 was reduced by 30% and 50%, respectively (Figure 2D). Nr4a2 down-regulation did not affect Ins1 or Ins2 expression (p≥0.06; n=4) excluding an involvement of this transcription factor in the regulation of insulin genes. However, Nr4a1 down-regulation reduced the expression of Ins1 and Ins2 by 15% (p=0.0151 and p=0.0076, respectively; n=4) after 24 h. Since the effect of Nr4a1 down-regulation was much lower compared to that of Nr4a3 down-regulation and no associations between common genetic variation in the NR4A1 locus and insulin secretion was detected in humans [27], we focused on NR4a3 in all subsequent experiments. At 72 h after Nr4a3 siRNA transfection, the effect on insulin gene expression was strongest. Therefore, we analyzed whether the intracellular insulin content was changed at this time point. However, we were not able to show a reduction in intracellular insulin content (p=0.8; n=4). Moderate lentiviral overexpression of Nr4a3 resulted in a trend for increased Ins1 expression and in significantly increased expression of Ins2 (Figure 2E).
Since 6-MP is an activator of all three NR4A members, we down-regulated Nr4a3 prior to 6-MP treatment. In these cells, 6-MP-induced Ins1 and Ins2 expression levels were markedly decreased (Figure 2F) excluding compensation of lost Nr4a3 expression by Nr4a1 or Nr4a2 and further strengthening the prominent role of Nr4a3 in the regulation of Ins1 and Ins2 expression.
Upon their expression, NR4A members bind to NBRE sites. In silico analysis of the rat insulin loci for putative NR4A binding sites displayed three NBREs in the Ins1 locus and two in Ins2 (Figure 3A). The human gene locus harbors one NBRE that represents a perfect consensus sequence (Figure 3A). The binding of Nor-1 to the rat insulin genes was analyzed by chromatin immunoprecipitation. Nor-1 preferentially binds to NBREII (Cp-value: 21.08±1.99) and weaker to NBREI (Cp-value: 34.74±2.09) and III (Cp-value: 33.76±2.10) in Ins1, while it shows comparable binding to both sites in Ins2 (Cp-values: 32.39±1.65 and 34.08±1.83, respectively). Qualitative data are shown in Figure 3B. Overexpression of a luciferase reporter vector containing either the human NBRE or the NBRE II of rat Ins1 resulted in an increased luciferase activity compared to control (Figure 3C). Nr4a3 co-transfection in both cases further increased luciferase activity (Figure 3C). These data indicate a direct role of Nor-1 in the transcriptional regulation of insulin genes.
Figure 3.
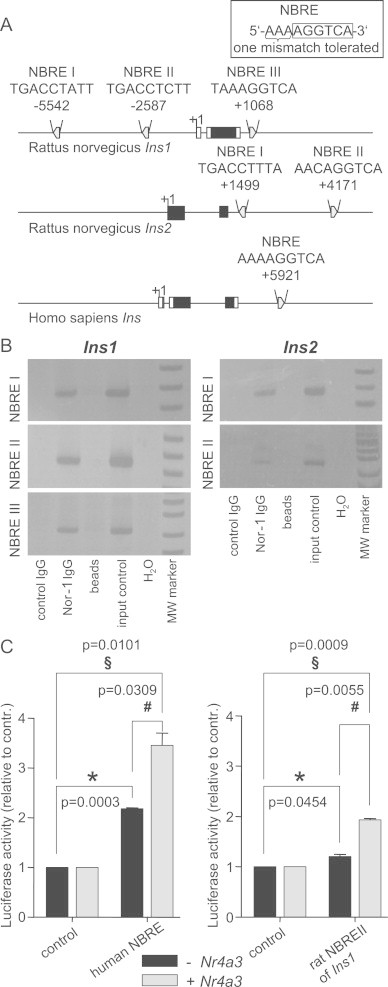
Nor-1 binding to insulin genes. In silico analysis of putative Nor-1 binding sites in the rat insulin genes Ins1 and Ins2 and in the human insulin gene Ins (A). Qualitative chromatin immunoprecipitation: Nor-1 binding to NBREs in Ins1 and Ins2. PCR reaction was stopped after 36 cycles for Ins1 NBRE I, 25 cycles for Ins1 NBRE II, 36 cycles for Ins1 NBRE III, 35 cycles for Ins2 NBRE I, and 34 cycles for Ins2 NBRE II, respectively (B). HEK293 cells were transiently transfected with luciferase reporter vectors containing the human NBRE or rat NBRE II of Ins1 with or without Nr4a3 co-transfection. Luciferase activity was measured 48 h after transfection. Data are given as mean±SEM. Two-group comparisons were performed using matched pairs Student's t-test (n=3).
3.2. Nor-1 regulates insulin secretion
Since we demonstrated an association between NR4A3 SNP rs12686676 and oral glucose-stimulated insulin secretion in humans [4], we analyzed GSIS in cells treated with Nr4a3 siRNA. Down-regulation of Nr4a3 did not affect basal insulin secretion (p=0.4), however, it reduced GSIS by 13% compared to cells treated with control siRNA (Figure 4A). Furthermore, exendin-4-enhanced GSIS was reduced by 18% compared to control cells (Figure 4A). Activation of Nr4a3 by 6-MP for 48 h resulted in a 2-fold increased insulin level in the supernatant (Figure 4B). 6-MP treatment of INS-1E cells and isolated rat islets resulted in increased basal insulin secretion as well (Figure 4C).
Figure 4.
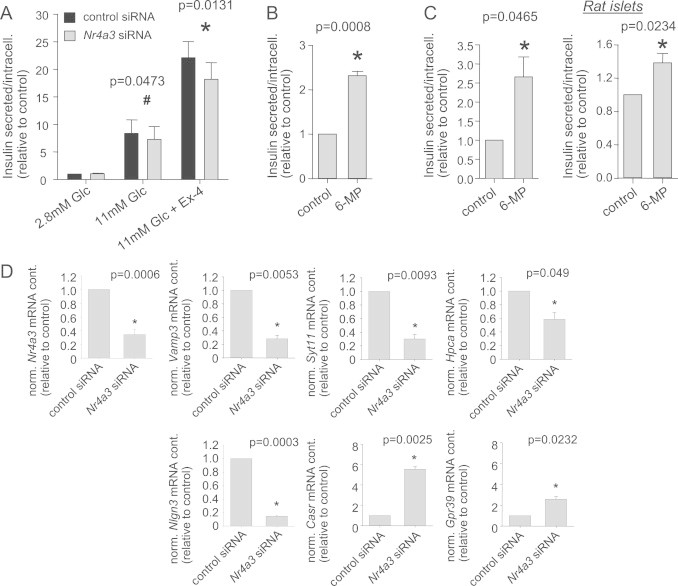
Nor-1 and insulin secretion. INS-1E cells were transfected with Nr4a3 siRNA 72 h prior to treatment. Basal, glucose- and incretin-stimulated insulin secretions as well as intracellular insulin content were measured. Secreted insulin was normalized for intracellular insulin content. Cells were pre-incubated with 2.8 mM glucose (Glc) for 3 h prior to incubation with indicated glucose concentrations in the presence or absence of exendin-4 (Ex-4) (A). Insulin concentration was measured in cell culture supernatant after incubation of INS-1E cells with 50 µM 6-MP for 48 h (B). Basal insulin secretion at 2.8 mM glucose after incubation of purified rat islets or INS1E cells with 50 µM 6-MP for 24 h (C). Regulation of several genes related to insulin exocytosis by Nr4A3 down-regulation for 72 h (D). Data are given as mean±SEM. Two-group comparisons were performed using matched pairs Student's t-test (n≥3).
72 h after transfection with Nr4a3 siRNA the intracellular insulin content was unchanged. Therefore, we hypothesized that Nor-1, as a transcription factor, may regulate the expression of additional genes involved in insulin secretion. To this end, we performed exploratory microarray analysis using mRNA from control cells and from cells treated with Nr4a3 siRNA. We found 313 genes to be differently regulated by Nr4a3 down-regulation (Supplementary Table S1, fold-change >2 or <0.5, respectively). At least six of these genes are potentially related to insulin secretion according to current literature. Therefore, we verified their altered expression by RT-PCR. Four of the six genes were down-regulated: Vamp3 by 70%, Syt11 by 70%, Hpca by 40%, and Nlgn3 by 85% (Figure 4D). Two of the six genes were up-regulated by Nr4a3 down-regulation: the expression of Casr was induced 5.5-fold, that of Gpr39 approximately 2.5-fold (Figure 4D). By in silico analysis of the 5′- and 3′-flanking regions of all these genes, we identified putative NBREs in each of these genes.
3.3. Nor-1 expression is regulated by the PKA pathway in β-cells
One of the major second messengers in β-cells is cAMP. It is produced upon activation of adenylyl cyclase after stimulation of the cells with diverse stimuli, such as glucose or insulinotropic enterohormones, i.e., incretins [28,29]. In the β-cell, cAMP has been shown to induce insulin expression via CREB [29]. Pharmacological activation of the PKA pathway via db-cAMP induced an 8-fold increase in Nr4a3 expression, while forskolin, an adenylyl cyclase activator, induced a 70-fold increase (Figure 5A). The forskolin effect was reflected at the protein level: treatment for 90 min or 3 h induced 4-fold and 3-fold increments in nuclear Nor-1 protein content, respectively (Figure 5B). In primary rat islets, forskolin treatment induced a 50-fold increase in Nr4a3 expression (Figure 5C). Glucose induced a modest, but not significant, increase in Nr4a3 expression (p=0.12), while exendin-4 alone induced a 4-fold increase in INS-1E cells (Figure 5D). The strongest effect was seen with a combination of exendin-4 and glucose (5.5-fold, Figure 5D). Similar effects were seen in isolated rat islets. Exendin-4 alone induced a 2.5-fold induction in Nr4a3 expression, while co-incubation of cells with exendin-4 and glucose induced a 12-fold increase (Figure 5E). Down-regulation of glucagon-like peptide 1 receptor (Glp1r) in INS-1E cells resulted in a blunted induction of Nr4a3 expression induced by glucose and exendin-4 (Figure 5F). Inhibition of the PKA pathway via the specific PKA inhibitor H89 reduced exendin-4/glucose-induced Nr4a3 expression by 45% in INS-1E cells (Figure 5G) and by 30% in rat islets (Figure 5H). Next, the impact of Creb1 on PKA-induced stimulation of Nr4a3 expression was examined. The expression of Creb1 mRNA was reduced by 90% via RNA interference (Figure 5I). Upon Creb1 knock-down, Nr4a3 expression was reduced by 40% (Figure 5I). Hence, via activation and inhibition of the PKA pathway, we were able to demonstrate the involvement of this pathway in Nr4a3 gene induction in β-cells and islets.
Figure 5.
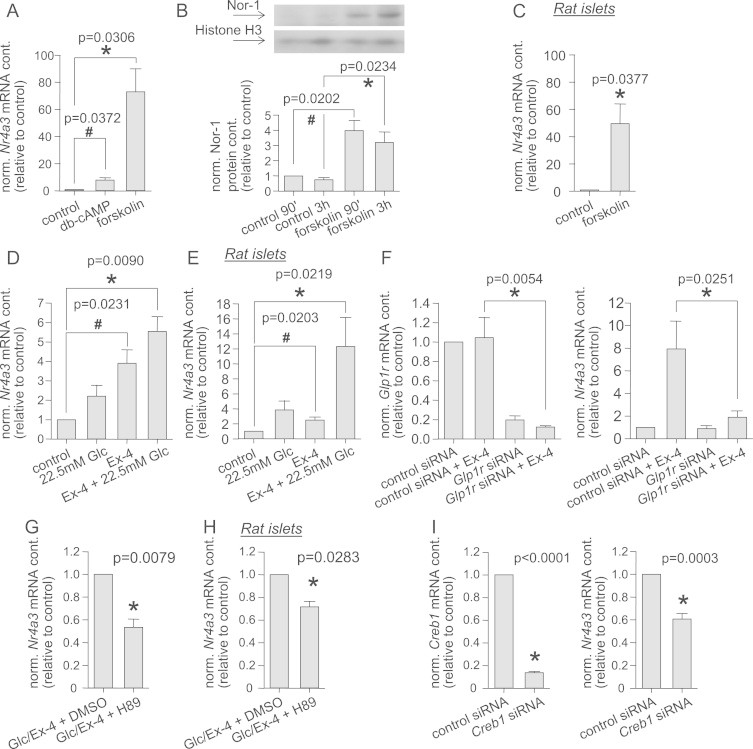
PKA pathway-regulated Nr4a3 expression. Nr4a3 expression after incubation of INS-1E cells for 90 min with 1 mM db-cAMP or 10 µM forskolin (A). Nor-1 and Histon H3 protein content was visualized by Western blotting after 90 min and 3 h of forskolin treatment. Nor-1 protein content was normalized by the housekeeping protein Histone H3. (B). Nr4a3 expression after incubation of rat islets with 1 µM forskolin for 90 min (C). Nr4a3 expression was measured after treatment of INS-1E cells (D) or purified rat islets (E) for 90 min with 10 nM exendin-4 (Ex-4) in the absence or presence of 22.5 mM glucose (Glc). INS-1E treated for 24 h with Glp1r siRNA and for 90 min with 10 nM exendin-4 in the presence of 22.5 mM glucose were lysed, and Glp1r and Nr4a3 expression were measured (F). Nr4a3 expression after pre-incubation with 10 µM H89 for 30 min prior to incubation with 10 nM exendin-4 in the presence of 22.5 mM glucose for 90 min: INS-1E cells (G) and purified rat islets (H). Nr4a3 and Creb1 expression were determined after down-regulation of Creb1 by siRNA for 24 h in INS-1E cells (I). Data are given as mean±SEM. Two-group comparisons were performed using matched pairs Student's t-test (n≥3).
3.4. Nr4a3 plays a role in PKA-dependent insulin gene induction
To understand the role of Nr4a3 in the PKA pathway, we down-regulated its expression for 72 h and, in parallel, activated the pathway for 24 h with forskolin. Forskolin treatment significantly induced Ins1 expression, and this was blunted in cells treated with Nr4a3 siRNA (Figure 6). Even though we did not observe forskolin-induced Ins2 expression, down-regulation of Nr4a3 resulted in a significant reduction of Ins2 expression in the presence or absence of forskolin (Figure 6). Similar results were seen after 48 h of forskolin treatment. These data further underline the role of Nor-1 as a novel important link between the PKA pathway and insulin gene expression.
Figure 6.
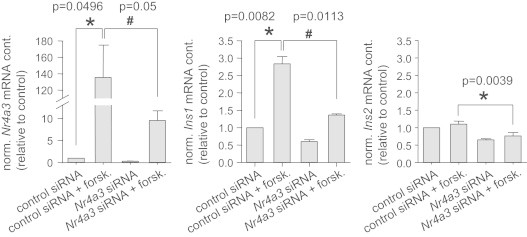
Role of NR4A3 in PKA-dependent insulin gene expression. INS-1E cells were treated for 72 h with Nr4a3 siRNA. Cells were incubated with forskolin (forsk., 10 µM) 24 h prior to lysis. Nr4a3, Ins1 and Ins2 expression were measured. Data are given as mean±SEM. Two-group comparisons were performed using matched pairs Student's t-test (n=3).
3.5. NR4A3 expression in healthy versus diabetic donors
Islets from diabetic donors showed somewhat lower NR4A3 and INS expression. This, however, did not reach significance in this small cohort (Supplementary Figure S1).
3.6. Genetic interaction between NR4A3 and TCF7L2 in humans
Recent genome-wide association studies revealed more than 50 type 2 diabetes risk genes [30–33] with TCF7L2 variant rs7903146 being the strongest predictor for the onset of the disease [34]. We and others have shown resistance to the incretin glucagon-like peptide-1 in subjects carrying the TCF7L2 risk allele [25,35]. Since we provide evidence of a new mechanism for incretin-dependent induction of insulin expression and secretion, we analyzed whether the aforementioned variant in the NR4A3 gene (SNP rs12686676) is able to aggravate or compensate for the secretion-compromising effect of the TCF7L2 risk allele. We therefore tested for gene–gene interactions between the TCF7L2 SNP rs7903146 and NR4A3 SNP rs12686676 with regards to insulin secretion during an OGTT. NR4A3 minor allele carriers show increased insulin secretion (as reported earlier [4] and shown in Figure 7A in a somewhat smaller cohort additionally genotyped for rs7903146). The well-described effect of the TCF7L2 risk allele on insulin secretion is also evident in our cohort (Figure 7B, [25]). The minor NR4A3 allele has no effect in TCF7L2 wildtype allele carriers that do not display compromised insulin secretion, but compensates for incretin resistance induced by the TCF7L2 risk allele (Figure 7C).
Figure 7.
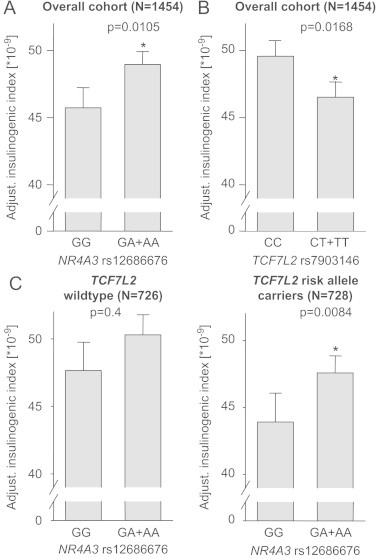
Gene–gene interaction between TCF7L2 and NR4A3. The effects of NR4A3 SNP rs12686676 and TCF7L2 SNP rs7903146 in our overall cohort are shown in (A) and (B), respectively. Thereafter, we stratified our cohort according to TCF7L2 SNP rs7903146 in order to assess the effect of NR4A3 SNP rs12686676 in both subgroups separately (C). Both SNPs, TCF7L2 SNP rs7903146 and NR4A3 SNP rs12686676, are presented in the dominant inheritance model. We adjusted for possible confounders (gender, age, and BMI). Data are given as mean±SEM.
4. Discussion
The impact of dysregulated insulin secretion gets more and more important for understanding the mechanisms leading to T2D. The majority of known T2D risk genes is associated with β-cell function suggesting that β-cell dysfunction is predominantly determined by genetics, while insulin resistance seems more prone to environmental influences [3]. We previously linked the polymorphism rs12686676 in the human NR4A3 gene, encoding for Nor-1, to increased insulin secretion in humans [4] revealing it as a beneficial minor allele. In the present study, we investigated the molecular function of Nor-1 in β-cells and introduce it as a novel transcriptional regulator of insulin genes and insulin secretion. Additionally, we provide a link to the best described T2D risk allele in TCF7L2 which is associated with reduced insulin secretion and reduced insulin gene expression [36].
Nor-1 and its isoforms Nur77 and Nurr1 are expressed in INS-1E cells, primary rat and human islets. Pharmacological activation of Nor-1 by 6-MP resulted in a strong induction of both insulin genes, Ins1 and Ins2, in rat insulin-secreting cells, while down-regulation of Nr4a3 resulted in reduced insulin expression showing that Nor-1 regulates the expression of these genes. The 6-MP effect on Ins1 and Ins2 expression was verified in isolated rat islets. Moderate overexpression of Nor-1 via lentiviral infection induced Ins2 expression significantly and showed a tendency towards increased Ins1 expression. Since down-regulation of Nr4a3 prior to 6-MP treatment prevented the expression of the insulin genes, we can assume that loss of Nor-1 is not compensated by either Nur77 or Nurr1, which are also activated by 6-MP [17]. Nevertheless, down-regulation of Nr4a1 also resulted in a weak reduction of Ins1 and Ins2 gene expression. This shows that Nur77, which is encoded by Nr4a1, might have a minor impact on insulin gene expression. The physiological relevance of this finding however appears questionable since we were not able to show an association between common genetic variation in NR4A1 and insulin secretion in humans [27]. The 6-MP-induced increase in insulin expression is reflected at the protein level. Acute down-regulation of Nr4a3 (72 h) did not result in reduced intracellular insulin content, probably due to the long half-life of 3–5 days of insulin granules [37]. The intracellular insulin content might however be reduced under chronic impairment of Nr4a3 expression. This could be the case in T2D, as it was shown that obese or diabetic rodent models express lower levels of Nr4a3 and Nr4a1 in muscle and adipose tissue [13]. Under the same conditions, the levels of all NR4A members are elevated in liver [10]. A human study with morbidly obese patients showed a high expression of all NR4A members that was normalized after weight loss [38]. These contradictory findings may be due to species- or tissue-specific differences in NR4A3 expression and therefore need further clarification. We tried to investigate NR4A3 expression in human islets from healthy versus diabetic donors, but did not obtain a significant difference.
Interestingly, altered Nr4a3 gene expression also affected insulin secretion. This cannot be explained by changes in intracellular insulin content, since the insulin content was not changed by Nr4a3 down-regulation. Down-regulation of Nr4a3 reduced glucose- and incretin-induced insulin secretion, while basal insulin secretion was unchanged. On the other hand, 6-MP-dependent activation of Nor-1 induced insulin secretion, and 6-MP treatment of isolated rat islets resulted in increased basal insulin secretion as well. Since Nor-1 is a transcription factor, we analyzed whether the expression of proteins involved in insulin exocytosis is affected by Nr4a3 down-regulation. We found four proteins to be down-regulated (Vamp3, Nlgn3, Syt11 and Hpca) while two others were induced (Gpr39 and Casr). Cellubrevin (encoded by Vamp3) is attached to the insulin granules by a C-terminal transmembrane domain and is, as a part of the SNARE complex, involved in Ca2+-dependent insulin secretion [39,40]. Neuroligin-3 (encoded by Nlgn3) has been shown to be expressed in human and rat islets, and overexpression of the protein increased basal insulin secretion. However, an influence on GSIS was not analyzed [41]. Also involved in Ca2+-dependent insulin secretion are members of the synaptotagmin (Syt) family. The expression of several Syt isoforms has been shown in different insulin-secreting cells, and two isoforms, Syt III and Syt VII, have been shown to co-localize with insulin granules and to play a role in Ca2+-dependent insulin release [42]. Consequently, synaptotagmin VII null mutant mice revealed impaired insulin secretion [43]. Even though up to now, there are no data suggesting an involvement of Syt XI (encoded by Syt11) in insulin secretion, our data indirectly suggest that Syt XI is a novel member of this protein family being involved in the exocytosis of insulin. Hippocalcin (encoded by Hpca) is a member of the Ca2+ sensor protein subfamily of visinin-like proteins. Visinin-like protein-1, another member of the same family, has been shown to regulate insulin secretion in primary mouse β-cells [44]. Surprisingly, the down-regulation of Nr4a3 also induced the expression of two genes related to insulin secretion suggesting that Nor-1 may also function as a gene repressor. GPR39 (encoded by Gpr39) seems to play a role in insulin secretion in knock-out mice under increased insulin demand, i.e., in the state of insulin resistance [45]. In human islets, activation of the calcium-sensing receptor (CasR, encoded by Casr) induced a rapid but relatively transient stimulation of insulin secretion in the absence of glucose suggesting that CasR is linked to the secretory process [46]. The results of our analysis indicate that several proteins related to insulin secretion are regulated by Nor-1, and changes in their expression could possibly explain our results showing reduced insulin secretion after Nr4a3 down-regulation as well as increased insulin secretion in subjects carrying a SNP in this gene. Further studies are however needed to corroborate this.
In the β-cell, cAMP is one of the major second messengers and regulates insulin secretion in a PKA-dependent as well as a PKA-independent manner (for review, see [47]), and cAMP has also been shown to induce insulin expression via CREB [48]. Here, we demonstrate a novel cAMP/PKA/CREB-dependent player inducing insulin expression. B-cell specific activation of the PKA pathway by the incretin mimetic exendin-4 induced Nr4a3 expression in INS-1E cells as well as in isolated rat islets. The strongest effect was seen with a combination of exendin-4 and glucose reflecting the amplifying effect of glucose on the incretin effect in β-cells [49]. The expression of Nr4a3 was reduced by pharmacological inhibition of this pathway in INS-1E cells as well as in primary rat islets. Additionally, we found Nr4a3 to be a CREB target gene in insulin-secreting cells. Here, we show that Nor-1 directly binds to regulatory sequences in the insulin genes. Insulin gene expression is induced upon Nor-1 activation via 6-MP and reduced upon down-regulation of Nr4a3 expression. Therefore, elevated cAMP levels not only regulate insulin gene expression directly via CREB binding to insulin genes but also via CREB-dependent Nr4a3 induction. Therefore, Nor-1 represents a novel link in the PKA pathway inducing insulin genes. Since down-regulation of Nr4a3 had such a strong effect on insulin gene expression, it is conceivable that Nor-1 plays an important role in the PKA/CREB pathway leading to insulin gene expression. Since it is possible to activate Nor-1 by external stimuli (6-MP), it represents a potential therapeutic target for the treatment of incretin resistance. Therefore, it would be worthwhile to develop new Nor-1 activators with fewer side effects than 6-MP.
Genetic variation in TCF7L2 is the most important genetic risk factor for type 2 diabetes known up to now and is involved in incretin- and glucose-mediated insulin secretion [25,50,51]. In the present study, we introduce a novel incretin-dependent mechanism of insulin secretion. Therefore, we intended to analyze whether the effects of the NR4A3 and TCF7L2 SNPs, with regard to insulin secretion in humans are influencing each other or if they are even counteracting each other. By stratification of our cohort according to TCF7L2 SNP rs7903146, we indeed were able to show that the SNP in NR4A3 is counteracting the effect of the TCF7L2 SNP. Based on our findings of Nor-1's function, we hypothesize that the SNP in NR4A3 that associates with improved insulin secretion can circumvent and compensate for the impaired incretin response of TCF7L2 risk allele carriers, e.g., by directly increasing the expression of insulin genes and insulin secretion-related genes (Figure 8). The effect of NR4A3 SNP rs12686676 is only visible in subjects carrying the TCF7L2 risk allele since incretin signaling is intact in TCF7L2 non-risk allele carriers. These genetic findings underline the importance of Nor-1's connection to incretin signaling in humans and render Nor-1 a promising new target for pharmacological intervention. Moreover, this finding may help to create personalized therapies for type 2 diabetes and support the efforts which have been started with regard to TCF7L2 [52]. Nevertheless, our understanding of the role of Nor-1 in human insulin gene expression and insulin secretion is still limited and needs further clarification.
Figure 8.
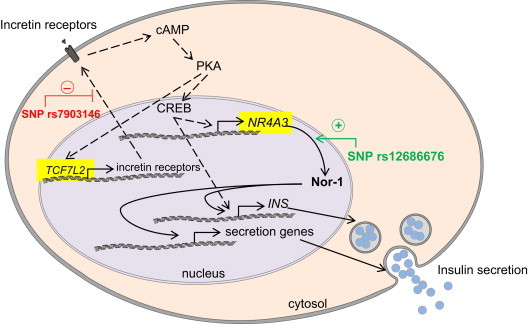
Role of Nor-1 in incretin-induced insulin expression and secretion and hypothetical localization of the TCF7L2 and NR4A3 SNP effects. The well-known TCF7L2 SNP rs7903146 leads to impaired incretin response, inhibited PKA pathway, and reduced insulin secretion (dashed lines). Based on our molecular data, we propose a gain-of-function mechanism in NR4A3 SNP rs12686676 carriers independent of incretin action. Increased Nor-1 expression enhances insulin secretion in these subjects probably by induction of genes encoding for exocytosis proteins and maybe by increasing intracellular insulin content thereby bypassing the defect caused by the TCF7L2 SNP (solid lines).
5. Conclusions
In this study, we introduce the novel transcriptional regulator of insulin genes, Nor-1. Its expression is induced via the PKA pathway in insulin-secreting cells and primary pancreatic islets in an incretin- and glucose-dependent manner. It directly regulates the expression of insulin genes and insulin secretion. Since Nor-1 has also been shown to play a major role in metabolism in other tissues, it might be an interesting new pharmacological target to fight type 2 diabetes. Especially patients with impaired insulin gene expression or impaired insulin secretion, for example TCF7L2 SNP carriers, might profit from pharmacological activation of Nor-1.
Author contributions
A.-M.O. performed the in vitro experiments, analyzed the data, and wrote the manuscript. O.R., C.H., I.T. provided technical support and analyzed the data. R.G., M.H., S.H.-S., A.B., F.M. collected and analyzed the human data. S.U. contributed to the design of the in vitro experiments. N.S., A.F. and H.-U.H. designed and carried out the human study. H.S. designed the in vitro experiments, analyzed the data, and edited the manuscript. All authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank all the participants of the TÜF Study for their cooperation. We thank Alke Guirguis and Roman Werner for technical assistance. We thank Reiner Lammers and Stephan Huber for methodological support. This study was supported in part by a grant from the German Federal Ministry of Education and Research (BMBF) to the German Centre for Diabetes Research (DZD e.V.).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.molmet.2013.06.003.
Appendix A. Supplementary materials
Figure S1.
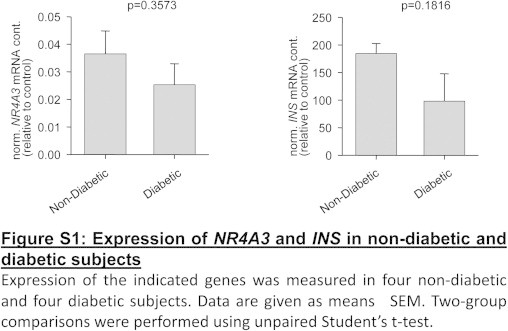
Expression of the indicated genes was measured in four non-diabetic and four diabetic subjects. Data are given as mean±SEM. Two-group comparisons were performed using unpaired Student’s t-test.
Supplementary data
References
- 1.Donath M.Y., Halban P.A. Decreased beta-cell mass in diabetes: significance, mechanisms and therapeutic implications. Diabetologia. 2004;47:581–589. doi: 10.1007/s00125-004-1336-4. [DOI] [PubMed] [Google Scholar]
- 2.Folli F., Okada T., Perego C., Gunton J., Liew C.W., Akiyama M., D'Amico A., La Rosa S., Placidi C., Lupi R., Marchetti P., Sesti G., Hellerstein M., Perego L., Kulkarni R.N. Altered insulin receptor signalling and beta-cell cycle dynamics in type 2 diabetes mellitus. PLoS One. 2011;6:e28050. doi: 10.1371/journal.pone.0028050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staiger H., Machicao F., Fritsche A., Haring H.U. Pathomechanisms of type 2 diabetes genes. Endocrine Reviews. 2009;30:557–585. doi: 10.1210/er.2009-0017. [DOI] [PubMed] [Google Scholar]
- 4.Weyrich P., Staiger H., Stancakova A., Schafer S.A., Kirchhoff K., Ullrich S., Ranta F., Gallwitz B., Stefan N., Machicao F., Kuusisto J., Laakso M., Fritsche A., Haring H.U. Common polymorphisms within the NR4A3 locus, encoding the orphan nuclear receptor Nor-1, are associated with enhanced beta-cell function in non-diabetic subjects. BMC Medical Genetics. 2009;10 doi: 10.1186/1471-2350-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milbrandt J. Nerve growth-factor induces a gene homologous to the glucocorticoid receptor gene. Neuron. 1988;1:183–188. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- 6.Law S.W., Conneely O.M., Demayo F.J., Omalley B.W. Identification of a new brain-specific transcription factor, Nurr1. Molecular Endocrinology. 1992;6:2129–2135. doi: 10.1210/mend.6.12.1491694. [DOI] [PubMed] [Google Scholar]
- 7.Ohkura N., Ito M., Tsukada T., Sasaki K., Yamaguchi K., Miki K. Structure, mapping and expression of a human NOR-1 gene, the third member of the Nur77/NGFI-B family. Biochimica et Biophysica Acta—Gene Structure and Expression. 1996;1308:205–214. doi: 10.1016/0167-4781(96)00101-7. [DOI] [PubMed] [Google Scholar]
- 8.Pols T.W., Ottenhoff R., Vos M., Levels J.H., Quax P.H., Meijers J.C., Pannekoek H., Groen A.K., de Vries C.J. Nur77 modulates hepatic lipid metabolism through suppression of SREBP1c activity. Biochemical and Biophysical Research Communications. 2008;366:910–916. doi: 10.1016/j.bbrc.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 9.Myers S.A., Eriksson N., Burow R., Wang S.C., Muscat G.E. Beta-adrenergic signaling regulates NR4A nuclear receptor and metabolic gene expression in multiple tissues. Molecular and Cellular Endocrinology. 2009;309:101–108. doi: 10.1016/j.mce.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Pei L., Waki H., Vaitheesvaran B., Wilpitz D.C., Kurland I.J., Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nature Medicine. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 11.Chao L.C., Zhang Z., Pei L., Saito T., Tontonoz P., Pilch P.F. Nur77 coordinately regulates expression of genes linked to glucose metabolism in skeletal muscle. Molecular Endocrinology. 2007;21:2152–2163. doi: 10.1210/me.2007-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pearen M.A., Myers S.A., Raichur S., Ryall J.G., Lynch G.S., Muscat G.E. The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 2008;149:2853–2865. doi: 10.1210/en.2007-1202. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y., Luo L., Luo N., Zhu X., Garvey W.T. NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: potential role in insulin resistance. Journal of Biological Chemistry. 2007;282:31525–31533. doi: 10.1074/jbc.M701132200. [DOI] [PubMed] [Google Scholar]
- 14.Lynch G.S., Ryall J.G. Role of beta-adrenoceptor signaling in skeletal muscle: implications for muscle wasting and disease. Physiological Reviews. 2008;88:729–767. doi: 10.1152/physrev.00028.2007. [DOI] [PubMed] [Google Scholar]
- 15.Paulsen R.F., Granas K., Johnsen H., Rolseth V., Sterri S. Three related brain nuclear receptors, NGFI-B, Nurr1, and NOR-1, as transcriptional activators. Journal of Molecular Neuroscience. 1995;6:249–255. doi: 10.1007/BF02736784. [DOI] [PubMed] [Google Scholar]
- 16.Philips A., Lesage S., Gingras R., Maira M.H., Gauthier Y., Hugo P., Drouin J. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Molecular and Cellular Biology. 1997;17:5946–5951. doi: 10.1128/mcb.17.10.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wansa K.D., Harris J.M., Yan G., Ordentlich P., Muscat G.E. The AF-1 domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine anti-metabolite 6-mercaptopurine. Journal of Biological Chemistry. 2003;278:24776–24790. doi: 10.1074/jbc.M300088200. [DOI] [PubMed] [Google Scholar]
- 18.Flaig R., Greschik H., Peluso-Iltis C., Moras D. Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. Journal of Biological Chemistry. 2005;280:19250–19258. doi: 10.1074/jbc.M413175200. [DOI] [PubMed] [Google Scholar]
- 19.Susini S., Roche E., Prentki M., Schlegel W. Glucose and glucoincretin peptides synergize to induce c-fos, c-jun, junB, zif-268, and nur-77 gene expression in pancreatic beta(INS-1) cells. FASEB Journal. 1998;12:1173–1182. [PubMed] [Google Scholar]
- 20.Briand O., Helleboid-Chapman A., Ploton M., Hennuyer N., Carpentier R., Pattou F., Vandewalle B., Moerman E., Gmyr V., Kerr-Conte J., Eeckhoute J., Staels B., Lefebvre P. The nuclear orphan receptor Nur77 Is a lipotoxicity sensor regulating glucose-induced insulin secretion in pancreatic beta-cells. Molecular Endocrinology. 2012 doi: 10.1210/me.2011-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ranta F., Avram D., Berchtold S., Dufer M., Drews G., Lang F., Ullrich S. Dexamethasone induces cell death in insulin-secreting cells, an effect reversed by exendin-4. Diabetes. 2006;55:1380–1390. doi: 10.2337/db05-1220. [DOI] [PubMed] [Google Scholar]
- 22.Staiger H., Haas C., Machann J., Werner R., Weisser M., Schick F., Machicao F., Stefan N., Fritsche A., Haring H.U. Muscle-derived angiopoietin-like protein 4 is induced by fatty acids via peroxisome proliferator-activated receptor (PPAR)-delta and is of metabolic relevance in humans. Diabetes. 2009;58:579–589. doi: 10.2337/db07-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heni M., Hennige A.M., Peter A., Siegel-Axel D., Ordelheide A.M., Krebs N., Machicao F., Fritsche A., Haring H.U., Staiger H. Insulin promotes glycogen storage and cell proliferation in primary human astrocytes. PLoS One. 2011;6:e21594. doi: 10.1371/journal.pone.0021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schafer S., Kantartzis K., Machann J., Venter C., Niess A., Schick F., Machicao F., Haring H.U., Fritsche A., Stefan N. Lifestyle intervention in individuals with normal versus impaired glucose tolerance. European Journal of Clinical Investigation. 2007;37:535–543. doi: 10.1111/j.1365-2362.2007.01820.x. [DOI] [PubMed] [Google Scholar]
- 25.Schafer S.A., Tschritter O., Machicao F., Thamer C., Stefan N., Gallwitz B., Holst J.J., Dekker J.M., 't Hart L.M., Nijpels G., van Haeften T.W., Haring H.U., Fritsche A. Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia. 2007;50:2443–2450. doi: 10.1007/s00125-007-0753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herzberg-Schafer S.A., Staiger H., Heni M., Ketterer C., Guthoff M., Kantartzis K., Machicao F., Stefan N., Haring H.U., Fritsche A. Evaluation of fasting state-/oral glucose tolerance test-derived measures of insulin release for the detection of genetically impaired beta-cell function. PLoS One. 2010;5:e14194. doi: 10.1371/journal.pone.0014194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mussig K., Machicao F., Machann J., Schick F., Claussen C.D., Stefan N., Fritsche A., Haring H.U., Staiger H. No association between variation in the NR4A1 gene locus and metabolic traits in white subjects at increased risk for type 2 diabetes. BMC Medical Genetics. 2010;11:84. doi: 10.1186/1471-2350-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capito K., Hedeskov C.J. Effects of glucose, glucose metabolites and calcium ions on adenylate cyclase activity in homogenates of mouse pancreatic islets. Biochemical Journal. 1977;162:569–573. doi: 10.1042/bj1620569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moens K., Heimberg H., Flamez D., Huypens P., Quartier E., Ling Z., Pipeleers D., Gremlich S., Thorens B., Schuit F. Expression and functional activity of glucagon, glucagon-like peptide I, and glucose-dependent insulinotropic peptide receptors in rat pancreatic islet cells. Diabetes. 1996;45:257–261. doi: 10.2337/diab.45.2.257. [DOI] [PubMed] [Google Scholar]
- 30.Cho Y.S., Chen C.H., Hu C., Long J., Hee Ong R.T., Sim X., Takeuchi F., Wu Y., Go M.J., Yamauchi T., Chang Y.C., Kwak S.H., Ma R.C., Yamamoto K., Adair L.S., Aung T., Cai Q., Chang L.C., Chen Y.T., Gao Y., Hu F.B., Kim H.L., Kim S., Kim Y.J., Lee J.J., Lee N.R., Li Y., Liu J.J., Lu W., Nakamura J., Nakashima E., Ng D.P., Tay W.T., Tsai F.J., Wong T.Y., Yokota M., Zheng W., Zhang R., Wang C., So W.Y., Ohnaka K., Ikegami H., Hara K., Cho Y.M., Cho N.H., Chang T.J., Bao Y., Hedman A.K., Morris A.P., McCarthy M.I., Takayanagi R., Park K.S., Jia W., Chuang L.M., Chan J.C., Maeda S., Kadowaki T., Lee J.Y., Wu J.Y., Teo Y.Y., Tai E.S., Shu X.O., Mohlke K.L., Kato N., Han B.G., Seielstad M. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nature Genetics. 2011 doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., Balkau B., Heude B., Charpentier G., Hudson T.J., Montpetit A., Pshezhetsky A.V., Prentki M., Posner B.I., Balding D.J., Meyre D., Polychronakos C., Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 32.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., McCulloch L.J., Ferreira T., Grallert H., Amin N., Wu G., Willer C.J., Raychaudhuri S., McCarroll S.A., Langenberg C., Hofmann O.M., Dupuis J., Qi L., Segre A.V., van Hoek M., Navarro P., Ardlie K., Balkau B., Benediktsson R., Bennett A.J., Blagieva R., Boerwinkle E., Bonnycastle L.L., Bengtsson B.K., Bravenboer B., Bumpstead S., Burtt N.P., Charpentier G., Chines P.S., Cornelis M., Couper D.J., Crawford G., Doney A.S., Elliott K.S., Elliott A.L., Erdos M.R., Fox C.S., Franklin C.S., Ganser M., Gieger C., Grarup N., Green T., Griffin S., Groves C.J., Guiducci C., Hadjadj S., Hassanali N., Herder C., Isomaa B., Jackson A.U., Johnson P.R., Jorgensen T., Kao W.H., Klopp N., Kong A., Kraft P., Kuusisto J., Lauritzen T., Li M., Lieverse A., Lindgren C.M., Lyssenko V., Marre M., Meitinger T., Midthjell K., Morken M.A., Narisu N., Nilsson P., Owen K.R., Payne F., Perry J.R., Petersen A.K., Platou C., Proenca C., Prokopenko I., Rathmann W., Rayner N.W., Robertson N.R., Rocheleau G., Roden M., Sampson M.J., Saxena R., Shields B.M., Shrader P., Sigurdsson G., Sparso T., Strassburger K., Stringham H.M., Sun Q., Swift A.J., Thorand B., Tichet J., Tuomi T., van Dam R.M., van Haeften T.W., van Herpt T., Vliet-Ostaptchouk J.V., Walters G.B., Weedon M.N., Wijmenga C., Witteman J., Bergman R.N., Cauchi S., Collins F.S., Gloyn A.L., Gyllensten U., Hansen T., Hide W.A., Hitman G.A., Hofman A., Hunter D.J., Hveem K., Laakso M., Mohlke K.L., Morris A.D., Palmer C.N., Pramstaller P.P., Rudan I., Sijbrands E., Stein L.D., Tuomilehto J., Uitterlinden A., Walker M., Wareham N.J., Watanabe R.M., Abecasis G.R., Boehm B.O., Campbell H., Daly M.J., Hattersley A.T., Hu F.B., Meigs J.B., Pankow J.S., Pedersen O., Wichmann H.E., Barroso I., Florez J.C., Frayling T.M., Groop L., Sladek R., Thorsteinsdottir U., Wilson J.F., Illig T., Froguel P., van Duijn C.M., Stefansson K., Altshuler D., Boehnke M., McCarthy M.I. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., Ardlie K., Bostrom K.B., Bergman R.N., Bonnycastle L.L., Borch-Johnsen K., Burtt N.P., Chen H., Chines P.S., Daly M.J., Deodhar P., Ding C.J., Doney A.S., Duren W.L., Elliott K.S., Erdos M.R., Frayling T.M., Freathy R.M., Gianniny L., Grallert H., Grarup N., Groves C.J., Guiducci C., Hansen T., Herder C., Hitman G.A., Hughes T.E., Isomaa B., Jackson A.U., Jorgensen T., Kong A., Kubalanza K., Kuruvilla F.G., Kuusisto J., Langenberg C., Lango H., Lauritzen T., Li Y., Lindgren C.M., Lyssenko V., Marvelle A.F., Meisinger C., Midthjell K., Mohlke K.L., Morken M.A., Morris A.D., Narisu N., Nilsson P., Owen K.R., Palmer C.N., Payne F., Perry J.R., Pettersen E., Platou C., Prokopenko I., Qi L., Qin L., Rayner N.W., Rees M., Roix J.J., Sandbaek A., Shields B., Sjogren M., Steinthorsdottir V., Stringham H.M., Swift A.J., Thorleifsson G., Thorsteinsdottir U., Timpson N.J., Tuomi T., Tuomilehto J., Walker M., Watanabe R.M., Weedon M.N., Willer C.J., Illig T., Hveem K., Hu F.B., Laakso M., Stefansson K., Pedersen O., Wareham N.J., Barroso I., Hattersley A.T., Collins F.S., Groop L., McCarthy M.I., Boehnke M., Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genetics. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Florez J.C., Jablonski K.A., Bayley N., Pollin T.I., de Bakker P.I., Shuldiner A.R., Knowler W.C., Nathan D.M., Altshuler D. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. New England Journal of Medicine. 2006;355:241–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyssenko V., Lupi R., Marchetti P., Del Guerra S., Orho-Melander M., Almgren P., Sjogren M., Ling C., Eriksson K.F., Lethagen A.L., Mancarella R., Berglund G., Tuomi T., Nilsson P., Del Prato S., Groop L. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. Journal of Clinical Investigation. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.da Silva X., Mondragon A., Sun G., Chen L., McGinty J.A., French P.M., Rutter G.A. Abnormal glucose tolerance and insulin secretion in pancreas-specific Tcf7l2-null mice. Diabetologia. 2012;55:2667–2676. doi: 10.1007/s00125-012-2600-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halban P.A. Structural domains and molecular lifestyles of insulin and its precursors in the pancreatic beta cell. Diabetologia. 1991;34:767–778. doi: 10.1007/BF00408349. [DOI] [PubMed] [Google Scholar]
- 38.Veum V.L., Dankel S.N., Gjerde J., Nielsen H.J., Solsvik M.H., Haugen C., Christensen B.J., Hoang T., Fadnes D.J., Busch C., Vage V., Sagen J.V., Mellgren G. The nuclear receptors NUR77, NURR1 and NOR1 in obesity and during fat loss. International Journal of Obesity (Lond) 2012;36:1195–1202. doi: 10.1038/ijo.2011.240. http://dx.doi.org/10.1038/ijo.2011.240, in press. [DOI] [PubMed] [Google Scholar]
- 39.Jewell J.L., Oh E., Thurmond D.C. Exocytosis mechanisms underlying insulin release and glucose uptake: conserved roles for Munc18c and syntaxin 4. American Journal of Physiology Regulatory, Integrative and Comparative Physiology. 2010;298:R517–R531. doi: 10.1152/ajpregu.00597.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regazzi R., Wollheim C.B., Lang J., Theler J.M., Rossetto O., Montecucco C., Sadoul K., Weller U., Palmer M., Thorens B. VAMP-2 and cellubrevin are expressed in pancreatic beta-cells and are essential for Ca(2+)-but not for GTP gamma S-induced insulin secretion. EMBO Journal. 1995;14:2723–2730. doi: 10.1002/j.1460-2075.1995.tb07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suckow A.T., Comoletti D., Waldrop M.A., Mosedale M., Egodage S., Taylor P., Chessler S.D. Expression of neurexin, neuroligin, and their cytoplasmic binding partners in the pancreatic beta-cells and the involvement of neuroligin in insulin secretion. Endocrinology. 2008;149:6006–6017. doi: 10.1210/en.2008-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Z., Reavey-Cantwell J., Young R.A., Jegier P., Wolf B.A. Synaptotagmin III/VII isoforms mediate Ca2+-induced insulin secretion in pancreatic islet beta-cells. Journal of Biological Chemistry. 2000;275:36079–36085. doi: 10.1074/jbc.M004284200. [DOI] [PubMed] [Google Scholar]
- 43.Gustavsson N., Lao Y., Maximov A., Chuang J.C., Kostromina E., Repa J.J., Li C., Radda G.K., Sudhof T.C., Han W. Impaired insulin secretion and glucose intolerance in synaptotagmin-7 null mutant mice. Proceedings of the National Academy of United States of America. 2008;105:3992–3997. doi: 10.1073/pnas.0711700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai F.F., Zhang Y., Kang Y., Wang Q., Gaisano H.Y., Braunewell K.H., Chan C.B., Wheeler M.B. The neuronal Ca2+ sensor protein visinin-like protein-1 is expressed in pancreatic islets and regulates insulin secretion. Journal of Chemical Biology. 2006;281:21942–21953. doi: 10.1074/jbc.M512924200. [DOI] [PubMed] [Google Scholar]
- 45.Tremblay F., Richard A.M., Will S., Syed J., Stedman N., Perreault M., Gimeno R.E. Disruption of G protein-coupled receptor 39 impairs insulin secretion in vivo. Endocrinology. 2009;150:2586–2595. doi: 10.1210/en.2008-1251. [DOI] [PubMed] [Google Scholar]
- 46.Gray E., Muller D., Squires P.E., Asare-Anane H., Huang G.C., Amiel S., Persaud S.J., Jones P.M. Activation of the extracellular calcium-sensing receptor initiates insulin secretion from human islets of Langerhans: involvement of protein kinases. Journal of Endocrinology. 2006;190:703–710. doi: 10.1677/joe.1.06891. [DOI] [PubMed] [Google Scholar]
- 47.Seino S., Takahashi H., Fujimoto W., Shibasaki T. Roles of cAMP signalling in insulin granule exocytosis. Diabetes, Obesity and Metabolism. 2009;11(Suppl. 4):180–188. doi: 10.1111/j.1463-1326.2009.01108.x. [DOI] [PubMed] [Google Scholar]
- 48.Philippe J., Missotten M. Functional characterization of a cAMP-responsive element of the rat insulin I gene. Journal of Biological Chemistry. 1990;265:1465–1469. [PubMed] [Google Scholar]
- 49.Holst J.J., Deacon C.F., Vilsboll T., Krarup T., Madsbad S. Glucagon-like peptide-1, glucose homeostasis and diabetes. Trends in Molecular Medicine. 2008;14:161–168. doi: 10.1016/j.molmed.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Shu L., Sauter N.S., Schulthess F.T., Matveyenko A.V., Oberholzer J., Maedler K. Transcription factor 7-like 2 regulates beta-cell survival and function in human pancreatic islets. Diabetes. 2008;57:645–653. doi: 10.2337/db07-0847. [DOI] [PubMed] [Google Scholar]
- 51.Shu L., Matveyenko A.V., Kerr-Conte J., Cho J.H., McIntosh C.H., Maedler K. Decreased TCF7L2 protein levels in type 2 diabetes mellitus correlate with downregulation of GIP- and GLP-1 receptors and impaired beta-cell function. Human Molecular Genetics. 2009;18:2388–2399. doi: 10.1093/hmg/ddp178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson E.R., Donnelly L.A., Kimber C., Whitley A., Doney A.S., McCarthy M.I., Hattersley A.T., Morris A.D., Palmer C.N. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56:2178–2182. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


