Abstract
Brain diseases including Alzheimer’s and Parkinson’s involve the cellular ‘unfolded protein’ (UPR) stress response. Psychiatric illnesses such as depressive disorders are thought to involve brain stress-response pathways. The XBP1 gene encodes a key transcription factor in the UPR stress response and therefore could be involved in the pathophysiology of depressive disorders. A functional polymorphism (−116C→G) in the XBP1 promoter was linked in some studies to bipolar disorder. Among 132 adults (mean age 39 yr) who presented with a major depressive episode, this polymorphism was found to be associated with a worse course during 1-yr prospective follow-up. In a subgroup (n=22), the polymorphism was associated with higher plasma levels of the stress hormone cortisol. The results suggest that hypothalamic–pituitary–adrenocortical and cellular stress pathways involving the XBP1 gene may be involved in the pathophysiology of major depressive disorder. These relationships merit further study.
Keywords: Cortisol, endoplasmic reticulum stress, genetics, major depressive disorder, unfolded protein response
Introduction
The XBP1 gene encodes a transcription factor with a key role in the ‘unfolded protein response’ (UPR) to endoplasmic reticulum (ER) stress (Yoshida et al. 2001). The ER is a cell organelle involved in quality control of secretory proteins. ER stress occurs when a build-up of unfolded proteins overloads the quality-control equipment resulting in UPR (Kaufman, 2002). ER function has been linked to bipolar disorder (reviewed by So et al. 2007). Compared to cells from healthy controls, lymphoblasts from persons with bipolar disorder had lower XBP1 transcription after laboratory-induced cellular stress (So et al. 2007). A polymorphism (−116C→G) in the XBP1 promoter that is associated with lower gene transcription was associated with bipolar disorder in a Japanese case-control study (Kakiuchi et al. 2003), but not in European (Cichon et al. 2004) or Chinese (Hou et al. 2004) samples. Here, we evaluate an association of the higher expressing −116C polymorphism with higher baseline cortisol and worse 1-yr course after an index major depressive episode (MDE).
Methods
Subjects
The sample included adults (n=132) who presented to our research clinic for evaluation and treatment of a MDE and who met DSM-IV criteria for major depressive disorder (MDD). The mean age was 39 yr (S.D.=12.3 yr) and 59% were female. For inclusion, subjects had to score at least 16 on the 17-item Hamilton Depression Rating Scale (HAMD-17). Exclusion criteria included unstable medical problems, significant neurological illness or past head injury, current substance dependence, anorexia or bulimia within 1 yr, and electroconvulsive therapy within 6 months. After complete description of the study, participants gave written informed consent as approved by the Institutional Review Board.
Blood samples for DNA extraction and PCR amplification were collected at baseline and processed as reported (Huang et al. 2003). The PCR fragment was digested with the BsaAI restriction enzyme which cuts allele C(−116) (ACGT core sequence) but not allele G(−116) (AGGT core sequence) of the XBP1 gene (Y. Y. Huang et al., unpublished protocol). At 09:00 hours a subgroup (n=22) had plasma cortisol measured by radioimmunoassay.
After a comprehensive baseline evaluation, subjects received follow-up assessments at 3 months and 1 yr. These included the Structured Clinical Interview for DSM-IV to ascertain whether they had met criteria for MDE at any point during the time since the last research assessment. Treatment of the baseline MDE was open, determined clinically by treating psychiatrists. Follow-up interviews inventoried the highly varied treatment regimens.
Results
During 1-yr follow-up, 28% (n=37) of subjects did not meet MDE criteria and 72% (n=95) did at some point. Presence of the XBP1 C allele (CC or CG genotype) was associated with meeting MDE criteria at some point during follow-up vs. not (χ2=4.75, d.f.=1, p=0.029). Because of the unequal follow-up times, percentage of time spent in a MDE during follow-up was calculated by dividing the number of days spent in a MDE by the total number of days in follow-up with available data for that patient; for those who were not depressed during follow-up the measurement was coded as 0. The C allele (CG or CC genotype) was associated with a greater percentage of follow-up time spent in a MDE (Mann–Whitney U test z=−2.19, p=0.028). Median percentage of follow-up time spent in MDE, by genotype, was 2% (GG) vs. 25% (CG or CC). Mean percentage of follow-up time spent in MDE by genotype, was 19.5% (S.D.=28.3%) (GG) vs. 34.4% (S.D.=32.6) (CG or CC).
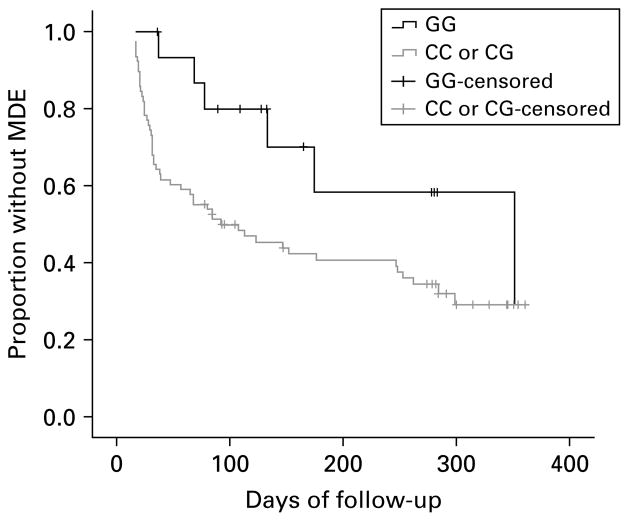
We performed non-parametric survival analysis of time to MDE among subjects who did not meet criteria for MDE for at least the first 14 d of follow-up (n=94, 71% of sample). In these subjects, XBP1 genotype (GG vs. CG/CC) was not associated with demographics: age (t=0.51, d.f.=92, p=0.609), sex (χ2=2.0, d.f.=1, p=0.157), race (Non-Hispanic white/other: Fisher’s exact test p=0.369), or Hispanic ethnicity (Fisher’s exact test p=0.450). XBP1 was not associated with clinical severity: baseline HAMD-17 score (t=−0.67, d.f.=92, p=0.502), length in weeks of the baseline MDE (z=−0.37, p=0.710), number of prior MDEs (z=−0.31, p=0.757), or lifetime substance use disorder (χ2=1.54, d.f.=1, p=0.215). XBP1 genotype was not associated with number of follow-up days on which subjects took antidepressant medication (z=−0.28, p=0.783). The log rank test showed a trend towards longer survival time until a MDE in the GG genotype subgroup (median time for subjects with GG genotype: 352 d vs. 92 d for others; χ2=3.80, d.f.=1, p=0.051) (Fig. 1).
Fig. 1.
Survival plot of proportion without major depressive episode (MDE) among subjects (n=94) who did not have MDE for at least the first 14 d of 1-year follow-up by XBP1 genotype (log rank χ2=3.8, d.f.=1, p=0.051).
Given the putative relationship of XBP1 to stress-modulating pathways, we performed an exploratory test for association of C allele presence to baseline plasma cortisol (n=22). The C allele was associated with higher morning plasma cortisol [CC or CG mean plasma cortisol: 15.3 μg/dl (S.D.=5.4) vs. GG mean plasma cortisol: 10.6 μg/dl (S.D.=2.8); z=−2.03, p= 0.043].
Discussion
Intracellular stress and environmental stress experienced by persons with mood disorders are at very different levels of organization. However, in a rodent model, ER stress and increased XBP1 activity were triggered by environmental stress (foot shock) and also by pharmacologically induced serotonin release (Toda et al. 2006). This suggests transduction of environmental stress to intracellular systems involving XBP1 as well as a link between depression and stress-related biochemical pathways. Perturbations in XBP1 expression, such as through genetic variation, may render the cell more vulnerable to environmental stress. The associations we find between XBP1 genotype, plasma cortisol, and clinical depression are consistent with the hypothesis that higher cortisol levels are related to the XBP1 gene and perhaps associated with impaired UPR function. This may also relate to evidence that hypothalamic–pituitary–adrenocortical dysfunction may be associated with a poorer clinical course of major depression (Belmaker & Agam, 2008).
The initial report of an association of bipolar disorder to the XBP1 −116C→G polymorphism found that the lower transcription activity G allele conferred greater risk. In our MDD sample, we found the higher-expressing C allele to be associated with a worse course during 1-yr follow-up after MDE. The divergence in results may be partly related to the different mood disorder subtypes. In a bipolar disorder sample (n=58) from our clinic, while CC/CG vs. GG genotype frequencies did not differ statistically from our MDD sample, XBP1 genotype was not associated with MDE during follow-up (Fisher’s exact test p=0.24). An association of risk with the opposite allele vs. prior reports – the ‘flip-flop’ phenomenon – does not necessarily indicate a false positive, but may be due to association of the putative risk allele with other contributory loci in genetically complex disease (Lin et al. 2007).
ER stress response and UPR function are involved in the pathophysiology of neurodegenerative diseases including Alzheimer’s and Parkinson’s disease (Lindholm et al. 2006). Early ER stress and UPR mechanisms appear to be neuroprotective while prolongation may trigger apoptotic cell death (Kaufman, 2002; Lindholm et al. 2006). It is unclear if a lower- vs. higher-expressing polymorphism in the XBP1 gene would be advantageous. Further investigation of this gene in relation to the HPA axis, sensitivity to stressful life events, and psychiatric illness is warranted.
Acknowledgments
Supported by PHS grants MH48514 and MH62185 (Conte Neuroscience Center); Stanley Medical Research Institute; NIMH K23 MH076049 (M. F. Grunebaum); and NIMH K25 MH074068 (H. C. Galfalvy).
Footnotes
Statement of Interest
Dr Grunebaum received donated medication from GlaxoSmithKline for a federally funded (K23 award) clinical trial study. Dr Mann receives research grant support from GlaxoSmithKline and Novartis.
References
- Belmaker RH, Agam G. Major depressive disorder. New England Journal of Medicine. 2008;358:55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- Cichon S, Buervenich S, Kirov G, Nirmala A, Dimitrova A, Green E, Schumacher J, Klopp N, Becker T, Ohlraun S, et al. Lack of support for a genetic association of the XBP1 promoter polymorphism with bipolar disorder in probands of European origin. Nature Genetics. 2004;36:783–784. doi: 10.1038/ng0804-783. [DOI] [PubMed] [Google Scholar]
- Hou SJ, Feng-Chang Y, Cheng CY, Tsai SJ, Hong CJ. X-box binding protein 1 (XBP1) C-116G polymorphisms in bipolar disorders and age of onset. Neuroscience Letters. 2004;367:232–234. doi: 10.1016/j.neulet.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Huang YY, Oquendo MA, Friedman JM, Greenhill LL, Brodsky B, Malone KM, Khait V, Mann JJ. Substance abuse disorder and major depression are associated with the human 5-HT1B receptor gene (HTR1B) G861C polymorphism. Neuropsychopharmacology. 2003;28:163–169. doi: 10.1038/sj.npp.1300000. [DOI] [PubMed] [Google Scholar]
- Kakiuchi C, Iwamoto K, Ishiwata M, Bundo M, Kasahara T, Kusumi I, Tsujita T, Okazaki Y, Nanko S, Kunugi H, et al. Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nature Genetics. 2003;35:171–174. doi: 10.1038/ng1235. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Orchestrating the unfolded protein response in health and disease. Journal of Clinical Investigation. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. American Journal of Human Genetics. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death and Differentiation. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- So J, Warsh JJ, Li PP. Impaired endoplasmic reticulum stress response in B-lymphoblasts from patients with bipolar-I disorder. Biological Psychiatry. 2007;62:141–147. doi: 10.1016/j.biopsych.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Toda H, Suzuki G, Nibuya M, Shioda K, Nishijima K, Wakizono T, Kanda Y, Watanabe Y, Shimizu K, Nomura S. Behavioral stress and activated serotonergic neurotransmission induce XBP-1 splicing in the rat brain. Brain Research. 2006;1112:26–32. doi: 10.1016/j.brainres.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]