Abstract
Serum metabolite concentrations provide a direct readout of biological processes in the human body, and are associated with disorders such as cardiovascular and metabolic diseases. Here we present a genome-wide association study with 163 metabolic traits using 1809 participants from the KORA population, followed up in the TwinsUK cohort with 422 participants. In eight out of nine replicated loci (FADS1, ELOVL2, ACADS, ACADM, ACADL, SPTLC3, ETFDH, SLC16A9) the genetic variant is located in or near enzyme or solute carrier coding genes, where the associating metabolic traits match the proteins’ function. Many of these loci are located in rate limiting steps of important enzymatic reactions. Use of metabolite concentration ratios as proxies for enzymatic reaction rates reduces the variance and yields robust statistical associations with p-values between 3×10−24 and 6.5×10−179. These loci explained 5.6% to 36.3% of the observed variance. For several loci, associations with clinically relevant parameters have previously been reported.
We have previously identified frequent genetic polymorphisms with large effects sizes that alter an individual’s metabolic capacities 1. In that study we described genetic variants in metabolism-related genes that lead to specific and clearly differentiated metabolic phenotypes, which we call “genetically determined metabotypes”. We argue that knowledge of these genetically determined metabotypes in human populations is key to identify the contributions and interactions of genetic and environmental factors in the etiology of complex diseases, providing a new paradigm for the study of gene-environment interactions. However, our original genome-wide association (GWA) study was limited in power due to its modest number of participants (n=284). In an effort to identify new major genetically determined metabotypes of biomedical relevance, we conducted a GWA study with metabolic traits in human serum in a much larger population using eight times the number of subjects.
Genotyping of the KORA samples was performed using the Affymetrix 6.0 GeneChip array and of TwinsUK using the Illumina Hap317K chip. Fasting serum concentrations of 163 metabolites, covering a biologically relevant panel of amino acids, sugars, acylcarnitines, and phospholipids, were determined by electrospray ionization tandem mass spectrometry (ESI-MS/MS) using the Biocrates AbsoluteIDQ™ targeted metabolomics technology. A full list of the measured metabolites and abbreviations used in this paper and their biological role is presented in the Online Methods section. Motivated by our previous finding that use of metabolite concentration ratios as proxies for enzymatic reaction rates reduces the variance and yields robust statistical associations 1,2, we tested all of the 163 metabolite concentrations and also all possible metabolite concentration ratios (163*162 = 26,406 traits) with a linear additive model for association with all SNPs that passed our selection criteria. The corresponding estimated genome-wide significance level after correction for testing 517,480 SNPs (MAF > 10%) and 26,406 multiple metabolic trait combinations is p = 3.64×10−12 (see Online Methods). This hypothesis-free approach allows the genetics to highlight pairs of metabolites that are more likely to be coupled either biochemically or physiologically.
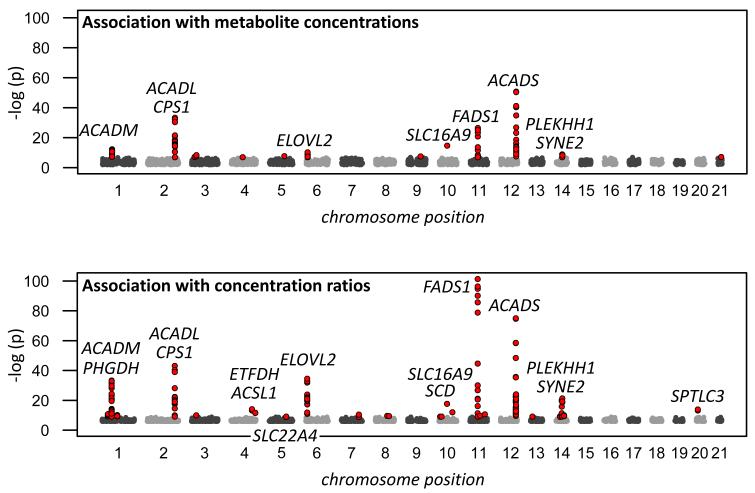
We applied a two-step discovery design in the KORA F4 population, followed by a replication in the TwinsUK cohort. Starting with a first discovery step based on samples of 1,029 male and female individuals of Southern German origin from the KORA F4 population, we selected all loci with p-values of association <10−7 for metabolite concentrations and <10−9 for concentration ratios in a genome-wide association screen (Fig. 1). 32 loci satisfy these criteria. One SNP for each locus was then tested in a second step in an independent sample of 780 participants selected from the remaining KORA F4 population. Identical genotyping and metabolomics techniques as in the first step were used. The metabolomics and genotyping experiments for this second step were conducted independently and after completion of the initial study at several months interval. Using data from all 1,809 KORA individuals, joint p-values of association were computed. Although this approach is less well powered than a full genome-wide joint analysis, it reflects the historical way in which we selected SNPs for follow-up. The top joint p-values from all 1,809 KORA samples are presented in Supplementary Table 1 and a full list is available on request.
Figure 1.
Manhattan plot of the strength of association with metabolite concentrations (top; data points with p<10−7 are plotted in red) and concentration ratios (bottom; data points with p<10−9 are plotted in red); based on association with 1029 samples (step 1 of discovery stage); for each SNP only the metabolic trait with the lowest p-value of association is shown; multiple dots thus indicate that several SNPs support the association at that locus (data provided as Supplementary Table 1).
We identified fifteen loci where the strength of association (indicated by decreasing p-values) increases when additional data is added and selected only these for further investigation. All fifteen loci display genome-wide significant p-values of association that are smaller than 3.64×10−12 in this joint analysis (Table 1; local association plots, boxplots by genotype, and QQ-plots are presented in Supplementary Fig. 1-3). In a third step, as a replication in an independent population, we used metabolomics data measured on our platform from serum samples of 422 female participants of the TwinsUK cohort. Nine of the fifteen loci are replicated (p<0.05) after Bonferroni correction for 15 tests. Five loci displayed signals of association with similar effect size estimates but were above this significance threshold and should at present be considered as non-replicated. However, as we show in the discussion of the individual loci, most of these five “suggestive loci” are supported by biological evidence (CPS1, SCD, SLC22A4, PHGDH) and two of them represent indirect replications of previous studies (CPS1 and SLC22A4). Note also that the TwinsUK study had only limited power due to its smaller sample size. Moreover, all but one SNP were imputed, and the signal to noise ratio in the TwinsUK metabolomics data was about 20% higher than in KORA. We therefore believe that these five suggestive loci are worthy of analysis in further studies and report relevant biological information on these loci in the supplementary material.
Table 1.
Summary of the 15 loci that were identified in the in the KORA population. Loci that were then replicated in the TwinsUK cohort (p<0.05after correction for 15 tests) are indicated by a ‘*’. Beta’ is the estimation of the effect per allele copy, normalized by the mean of the metabolic trait. Eta2 is the proportion of the variance that can be explained by the genotype in the linear model. Gene variants are reported on the positive strand of NCBI build 36. Additional information on the 50 strongest associating metabolic traits for each locus is provided as Supplementary Table 2. Additional p-values computed using log-scaled metabolite concentration ratios and also from the first discovery step alone are given in Supplementary Table 3. Abbreviations for metabolite names are as follows: phophatidylcholine (PC), acyl-acyl-PC (aa), acyl-alkyl-PC (ae), lyso-PC (a), sphingomyelin (SM), acylcarnitine (C), composition of fatty acid side chains (x:y, x=number of carbons, y=number of double bonds), with decarboxyl group (DC), with hydroxyl group (OH), with methyl group (M). A full list of metabolites determined in this study is provided in the Online Methods section.
| rs-number | SNP type | Locus | Chr | Coded/ noncoded allele |
Position | Minor allele frequency |
Strongest association |
N KORA |
beta’ KORA |
eta2 KORA |
p-value KORA |
N TwinsUK |
beta’ TwinsUK |
p-value TwinsUK |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs174547 | intronic | FADS1 | 11 | T/C | 61,327,359 | 30.4% | PC aa C36:3 / PC aa C36:4 |
1806 | 0.151 | 36.3% | 6.5×10−179 | 422 | 0.156 | 2.0×10−26 * |
| rs2014355 | intronic | ACADS | 12 | T/C | 119,659,907 | 27.7% | C3 / C4 | 1790 | −0.218 | 21.5% | 5.1×10−96 | 416 | −0.229 | 2.1×10−24 * |
| rs211718 | upstream | ACADM | 1 | C/T | 75,879,263 | 30.5% | C12 / C10 | 1804 | 0.120 | 14.6% | 1.3×10−63 | 299 | 0.133 | 2.8×10−11 * |
| rs2286963 | coding | ACADL | 2 | T/G | 210,768,295 | 36.5% | C9 / C10:2 | 1806 | 0.219 | 13.8% | 3.1×10−60 | 421 | 0.312 | 2.4×10−19 * |
| rs9393903 | intronic | ELOVL2 | 6 | G/A | 11,150,895 | 24.6% | PC aa C40:3 / PC aa C42:5 |
1803 | 0.087 | 9.8% | 2.3×10−42 | 419 | 0.076 | 4.0×10−5 * |
| rs2216405 | downstream | CPS1 | 2 | A/G | 211,325,139 | 18.5% | Glycine / PC ae C38:2 |
1792 | 0.129 | 7.1% | 1.9×10−30 | 420 | 0.094 | 0.014 |
| rs7156144 | upstream | PLEKHH1 | 14 | G/A | 67,049,466 | 41.4% | PC ae C32:1 / PC ae C34:1 |
1799 | −0.042 | 6.6% | 1.7×10−28 | 405 | −0.023 | 0.0024 * |
| rs11158519 | intronic | SYNE2 | 14 | G/A | 63,434,338 | 14.5% | PC ae C38:1 / PC aa C28:1 |
1763 | −0.083 | 6.5% | 1.5×10−27 | 394 | −0.098 | 0.0075 |
| rs168622 | upstream | SPTLC3 | 20 | G/T | 12,914,089 | 37.5% | SM (OH) C24:1 / SM C16:0 |
1796 | 0.061 | 5.8% | 5.2×10−25 | 421 | 0.061 | 2.9×10−4 * |
| rs8396 | downstream | ETFDH | 4 | T/C | 159,850,267 | 29.8% | C14:1-OH / C10 | 1778 | 0.102 | 5.6% | 3.5×10−24 | 421 | 0.114 | 3.1×10−7 * |
| rs7094971 | intronic | SLC16A9 | 10 | A/G | 61,119,570 | 13.5% | C0 | 1786 | −0.091 | 4.6% | 3.8×10−20 | 421 | −0.089 | 9.3×10−4 * |
| rs2046813 | upstream | ACSL1 | 4 | T/C | 186,006,153 | 32.2% | PC ae C44:5 / PC ae C42:5 |
1804 | 0.033 | 4.1% | 3.6×10−18 | 409 | 0.002 | 0.91 |
| rs603424 | upstream | SCD | 10 | G/A | 102,065,469 | 19.4% | C14 / C16:1 | 1805 | 0.054 | 4.0% | 1.5×10−17 | 422 | 0.023 | 0.10 |
| rs272889 | intronic | SLC22A4 | 5 | G/A | 131,693,277 | 38.5% | Valine / C5 | 1809 | −0.075 | 3.3% | 7.9×10−15 | 422 | −0.052 | 0.0098 |
| rs541503 | upstream | PHGDH | 1 | T/C | 120,009,820 | 37.9% | Ornithine / Serine |
1809 | 0.058 | 2.7% | 3.0×10−12 | 419 | 0.029 | 0.087 |
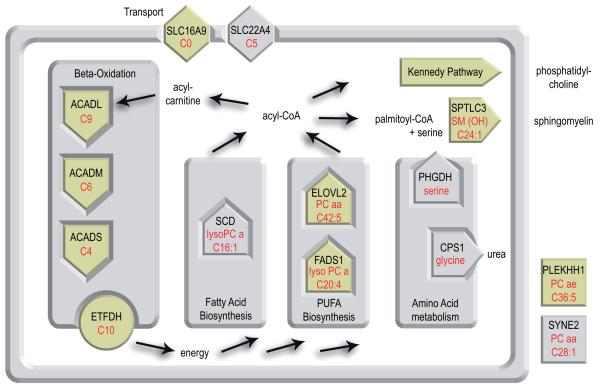
When the functional roles of the genes in these loci are considered, we can draw the most comprehensive to date view of genetic variation in human metabolism. The connections between these genes in the human metabolism are outlined in Fig. 2. As each locus and association has its particularities and may be of different relevance to the individual reader, we provide a full discussion of all loci from their specific perspectives as a Supplementary Note, while focusing here on the overall picture. Table 2 summarizes the relevant information from that discussion for all loci. For eight of nine fully replicated genetic polymorphisms and also four of the five suggestive loci, the genetic variant is located in or near enzyme or solute carrier coding genes, where the associating metabolic traits match the proteins’ function. Many of these polymorphisms are located in rate limiting steps of important enzymatic reactions. Four loci replicate associations from previous studies (FADS1, ACADS, ACADM, ELOVL2), including our own. Two of the suggestive loci replicate previous studies on related metabolic traits (SLC22A4 and CPS1). For some loci, new hypotheses on the genes’ function can be derived from the associating metabolite pattern (e.g. SLC16A9 and PLEKHH1). For three loci, an association with clinical endpoints has previously been reported (SLC22A4 with Crohn’s disease, FADS1 with hyperactivity and cholesterol/triglyceride levels, ACADS as susceptibility locus for ethylmalonic aciduria).
Figure 2.
A systemic view of genetic variation in human metabolism as identified by this study. Eight of nine fully replicated genetic polymorphisms (beige) and also four of five suggestive loci (grey) are located in or near enzyme-coding genes that are central to the different processes in human lipid metabolism, that is, the beta-oxidation (ACADS, ACADM, ACADL), the polyunsaturated fatty acid biosynthesis (FADS1, ELOVL2), the fatty acid synthesis (SCD), the breakdown of fats and proteins to energy (ETFDH), and the biosynthesis of phospholipids (SPTLC3). Two SNPs are located in or near genes coding for carrier proteins (SLC22A4, SLC16A9) and two SNPs involve enzymes that are related to amino acid metabolism (PHGDH, CPS1). Only for two genetic variants does the attribution of a metabolic function remain elusive (PLEKHH1, SYNE2). For each locus the most strongly associating single metabolite is indicated in red.
Table 2.
Summary of findings for the loci discussed in this paper; top: fully replicated loci, middle: loci from discovery phase that were not replicated in TwinsUK; bottom: already described risk loci that associate here with metabolic traits but that do not reach the genome-wide significance level. Metabolites with the lowest p-values of association for these loci with respect to metabolite concentrations are reported; note that concentration ratios with the lowest p-values of association are given in Table 1 and data for the 50 strongest associations is provided as Supplementary Table 2. Loci that have already been published elsewhere are indicated as replication. The term “indirect replication” is used when the study in question did not measure the same, but related metabolites. Commonly used but non-standard gene names are mentioned in parentheses.
| Locus | Gene function | Metabolites | Notes | Reference |
|---|---|---|---|---|
| Fully replicated loci | ||||
| FADS1 | fatty acid desaturase 1 | lysoPC a C20:4, PC aa C38:4, PC aa C36:4 |
Risk locus for high cholesterol 6-8; linked to hyperactivity 23; covariate in association breastfeeding-IQ 24; C20:4 fatty acids are products of FADS1. |
Replication of KORA 1 and indirect replication of INCHIANTI 25, who measured selected PUFAs |
|
ACADS
(SCAD) |
acyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain |
C4 | Predisposition allele that can cause ACADS deficiency if additional factors are present 26; C4 fatty acids are substrates of ACADS. |
Replication of KORA 1 and a target gene study in a US population 27 |
|
ACADM
(MCAD) |
acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain |
C6*, C8, C10, C10:1 | C4 to C12 fatty acids are substrates of ACADM | Replication of KORA 1 |
|
ACADL
(LCAD) |
acyl-Coenzyme A dehydrogenase, long chain |
C9 | Coding SNP; patented in a method for diagnosing the risk of thermo-labile phenotype in influenza-associated encephalopathy 14; long chain fatty acids (incl. C9) are substrates of ACADL. |
- |
| ELOVL2 | elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 |
PC aa C42:5, PC aa C42:6 | PUFA products and/or substrates of ELOVL2 are incorporated into phosphatidylcholines, such as PC aa C42:5 and PC aa C42:6. |
Indirect replication of INCHIANTI 25, who measured selected PUFAs |
| PLEKHH1 | pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
PC ae C36:5, PC ae C32:2 | Gene function unknown; our results suggest that this locus may be functionally related to the metabolism of plasmalogens, such as PC ae C36:5 and PC ae C32:2. |
- |
| SPTLC3 | serine palmitoyltransferase, long chain base subunit 3 |
SM (OH) C24:l | The SPT complex catalyses the rate limiting step of SM biosynthesis; SPTLC3 is the regulatory subunit; association with ratios between sphingolipids indicates that this polymorphism may modify the cellular mechanism to adjust SPT activity to tissue-specific requirements of sphingolipid synthesis. |
- |
| ETFDH | electron-transferring- flavoprotein dehydrogenase |
C10, C8 | Breakdown of fats and proteins to energy (beta-oxidation of substrates such as C8 and C10 fatty acids); genetic variant in this locus also involves changes in acylcarnitine hydroxylation and carboxylation. |
- |
|
SLC16A9
(MCT9) |
solute carrier family 16, member 9 (monocarboxylic acid transporter 9) |
C0 | Hypothesis: carnitine (C0) is a monocarboxylic acid and may therefore be the substrate of this “orphan” transporter. |
- |
|
| ||||
| “Suggestive loci“ from discovery stage that were not replicated in TwinsUK | ||||
| CPS1 | carbamoyl-phosphate synthetase 1, mitochondrial |
Glycine | Locus associates with glycine to arginine and glutamine ratios; these amino acids are linked to carbamoyl phosphate in the urea cycle. |
Indirect replication of Paré et al.28, who show an association with the related metabolite homocysteine. |
| SYNE2 | spectrin repeat containing, nuclear envelope 2 |
PC aa C28:l | Gene function unknown; causative SNP may possibly be linked to a modification in the nearby sphingosine-l-phosphate phosphatase 1 (SGPP1) activity. |
- |
|
SCD
(FADS5) |
stearoyl-CoA desaturase (delta-9-desaturase) |
lysoPC a C16:l | Fatty acid C16:l is the direct product of SCD; SCD activity is linked to adiposity and plasma lipid profiles 29; C16:l palmitoleate regulates insulin signaling and glucose metabolism 30 |
- |
|
SLC22A4
(OCTN1) |
solute carrier family 22 (organic cation/ ergothioneine transporter), member 4 |
C5 | Locus associates with Crohn’s disease; association with C5 carnitine suggests role of modified transport of this metabolite in disease etiology; the causative SNP may also be located in neighboring SLC22A5 (OCTN2) gene 31. |
Indirect replication of Peltekova et .al31, who show that this genetic variation at this locus associates with carnitine concentrations (Fig. 3a in Peltekova et al. 31); |
| PHGDH | phosphoglycerate dehydrogenase |
Serine |
PHGDH catalyses the rate limiting step of serine biosynthesis; PHGDH deficiency leads to congenital microcephaly, seizures, and severe psychomotor retardation 32. |
- |
|
| ||||
| Already described risk loci that associate here with metabolic traits above the genome-wide significance level | ||||
|
| ||||
|
APO-
cluster |
apolipoprotein | PC aa C36:2/PC aa C38:1 (p=1.8×10−11) |
SNP rs964184 in the APOA1-APOC3-APOA4-APOA5 cluster strongly associates with blood triglyceride levels (p<10−60) 4 |
- |
| GCKR | glucokinase (hexokinase 4) regulator |
PC ae C34:2/PC aa C32:2 (p=3.2×10−8) |
SNP rs1260326 (P446L polymorphism) in GCKR inversely modulates fasting glucose (p=8×10−13) and triglyceride (p=1×10−4) levels and reduces type 2 diabetes risk in the DESIR prospective general French population 10 |
- |
| MTNR1B | melatonin receptor 1B | tryptophan/phenylalanine (p=5.7×10−6) |
SNP rs10830963 in the melatonin-receptor (MTNR1B) associates with fasting glucose 11; phenylalanine is a precursor of melatonin. |
- |
Note that C6 cannot be discerned from C4:1-DC by the tandem mass spectrometry technique used here.
For several other loci, loss of function of the corresponding gene leads to severe disorders (e.g. ACADM, ACADL, ETFDH) indicating that the genetic variants we identify here or variants in linkage disequilibrium may induce a related but likely more moderate phenotype. This is in line with findings of a recent GWA study on kidney function which identified UMOD to be associated with glomerular filtration rate 3. Rare mutations in the UMOD gene are known to be the cause of monogenic autosomal dominant kidney diseases. Common mutations in the same gene region can be the cause of disease-related phenotypes of less severity on the population level. As discussed in Gieger et al. 1, a ratio between the concentrations of two metabolites that are linked to a substrate/product pair of some enzymatic reaction may constitute an approximation of the conversion rate of that reaction. From the effect size of the association (beta’ in the linear model), one can therefore derive the per-allele difference in metabolic capacities of an individual with respect to the considered enzymatic reaction. For instance, beta’ of the association of rs211718 (ACADM) with C12/C10 is 0.12. Assuming an additive-per-copy effect (see Supplementary Fig. 2), this implies that individuals who are homozygotes of the major allele of ACADM burn fatty acids with a chain length of 12 carbons about 24% faster than carriers of two copies of the minor allele. Similar arguments hold for the other loci (beta’ for all loci is reported in Supplementary Table 2).
The SNPs identified in this study can now be used for the identification of true positives in GWA studies with clinical parameters. As an example, in our previous study 1 we suggested FADS1 to be a risk locus for perturbed blood lipid parameters. This was supported by the observed association with different phospholipids and the fact that two published GWA studies with lipid levels reported p-values of association for the FADS1 locus with LDL, HDL, and total cholesterol levels that ranged between 1.89×10−4 and 6.07×10−5 4,5. These associations had not been included in the list of potential candidates for replication in those studies, as their p-values taken alone were not sufficiently small in the context of a “classical” GWA study. Three more well powered GWA studies with lipid parameters have only recently confirmed this prediction 6-8, thereby proving that a combination of a GWA study using metabolomic phenotypic traits with data from previous GWA studies can identify new candidate SNPs associated to known phenotypes of clinical relevance. To facilitate further studies we provide a list of all high scoring associations of this study as supplemental data set for a similar use by other consortia (Supplementary Table 1).
The association data presented in this study can be used to learn more about already known risk loci. Using the catalog of published GWA studies 9 we identified three such loci that also associate with metabolic traits in our study (Table 2). For instance, SNP rs964184 in the apolipoprotein APOA1-APOC3-APOA4-APOA5 cluster strongly associates with blood triglyceride levels (p<10−60) 4. We find that the same SNP associates with ratios between different phosphatidylcholines (e.g. PC aa C36:2/PC aa 38:1, p=1.8×10−10), which are biochemically connected to triglycerides by the intermediary of only a few enzymatic reaction steps. SNP rs1260326 (P446L polymorphism) in the glucose kinase regulator protein (GCKR) inversely modulates fasting glucose (p=8×10−13) and triglyceride levels (p=1×10−4) and reduces type 2 diabetes risk in the DESIR prospective general French population 10. This locus associates with different ratios between plasmalogens and phosphatidylcholines (e.g. PC ae C34:2/PC aa C32:2, p=3.2×10−8), thereby providing new avenues for further investigation on the functional background of this association. SNP rs10830963 in the melatonin-receptor (MTNR1B) associates with fasting glucose 11. The same SNP associates in this study with tryptophan/phenylalanine ratios (p=5.7×10−6). This is of particular interest since phenylalanine is a precursor of melatonin, indicating a functional relationship between this pathway and the regulation of glucose homeostasis. We expect the list of loci with parallel association of clinically relevant parameters and metabolic traits to grow as new GWA studies will become available, and therefore provide our association data as supplemental material for such use.
The SNPs identified in this study can now be used in clinical studies for association with response to drug treatment. One published example is a common polymorphism in the dihydropyrimidine dehydrogenase (DPYP) gene that associates strongly with fluoropyrimidine-related toxicity in cancer patients 12. Carriers of this variant could benefit from individual dose adjustment of the fluoropyrimidine drug or alternate therapies. It is now possible to use the here identified SNPs in association studies with phenotypes that are specific to a disease, such as the development of particular complications during the course of a disease or treatment. One published example is a SNP in carnitine palmitoyltransferase II (CPT-II) that is a predisposing factor for influenza-associated encephalopathy (thermo-labile phenotype) 13. Indeed, the SNP that we identified here in the ACADL gene has been patented by others 14 along with the CPT-II polymorphism as a method of diagnosing risk of a thermo-labile phenotype.
In summary, this study allowed us to draw a systemic perspective of the genetic variation that is found in human metabolism. In contrast to most GWA studies with clinically relevant endpoints, it appears that for metabolic traits most of the associations are linked to genetic variants in genes with a matching metabolic function (Fig. 2). The use of metabolite concentration ratios demonstrates a pronounced sharpening of the association with dramatically decreased p-values when compared to an analysis of single metabolites. Moreover, as we show with examples (FADS1, APO-cluster, MTNR1B, GCKR), it allows the deriving of new functional information from GWA studies with clinically relevant endpoints, and this based on a relatively small number of samples. Our study shows the exciting potential of metabolomics to unravel the genetics of human metabolism. The presented genome-wide perspective of genetic variation in human metabolism will continue to improve as more extensive metabolite panels become available for use at a genome-wide scale, including additional studies with nutritional challenges of the participants. We believe that the introduction of metabolomics into the field of molecular epidemiology provides a new and more hypothesis-driven approach to GWA studies.
ONLINE METHODS
This study is based on genotyping and metabolic profiling efforts in the German KORA and the British TwinsUK population for which we report the essentials here.
Study population
The KORA S4 survey, an independent population-based sample from the general population living in the region of Augsburg, Southern Germany, was conducted in 1999/2001. The standardized examinations applied in the survey (4261 participants, response 67%) have been described in detail 15 and references therein. A total of 3,080 subjects participated in a follow-up examination of S4 in 2006-08 (KORA F4), comprising individuals who, at that time, were aged 32–81 years. Informed consent has been given. The study has been approved by the local ethical committee. For the first genome-wide screening step, 1048 blood samples of KORA F4 participants were metabolically characterized. For 1029 samples of this group genome-wide genotype data were also available (509 males, 520 females). In a second step, 972 samples were metabolically characterized in an independent experimental batch. For 780 of these samples genome-wide genotype data were available (374 males, 406 females). For the joint analysis in KORA, metabolomics and genotype data for a total of 1809 individuals were available. No evidence of population stratification has been found in multiple published analyses using the KORA cohort.
Genotyping and imputation
In KORA F4 genotyping was done using the Affymetrix 6.0 GeneChip array. Analysis in the discovery stage was performed solely on genotyped SNPs. As is current standard for GWA studies, we excluded all X-chromosome-linked SNPs owing to the following reasons: (i) the X chromosome has to be treated differently from the autosomes; (ii) it cannot be predicted which allele is active, (iii) testing males separately results in different sample sizes and power. Imputation of SNPs in the HapMap CEU population was performed using IMPUTE 16 for use in the regional association plots. In the discovery stage we limited our analysis to SNPs with a moderate to high minor allele frequency (MAF>10%), high genotyping quality (call rate >95%), and with respect to Hardy-Weinberg equilibrium (p(HWE)>0.001). A total of 517,480 SNPs satisfy all of these criteria.
Blood sampling
Blood samples for metabolic analysis were collected between 2006 and 2008 in parallel with the KORA F4 examinations. To avoid variation due to circadian rhythm, blood was drawn in the morning between 8 and 10 am after a period of overnight fasting. Material was drawn into serum gel tubes, gently inverted two times, followed by 30 min resting at room temperature to obtain complete coagulation. The material was then centrifuged for 10 minutes (2750g; 15°C). Serum was aliquoted and kept for max. 6 hours at 4°C, after which it was deep frozen to −80°C until analysis.
Metabolite measurements
Liquid handling of serum samples (100 μl) was performed with Hamilton Star (Hamilton Bonaduz AG, Bonaduz, Switzerland) robot and prepared for quantification using the AbsoluteIDQ kit (BIOCRATES Life Sciences AG, Innsbruck, Austria). Sample analyses were done on API 4000 Q TRAP LC/MS/MS System (Applied Biosystems, Darmstadt, Germany) equipped with a Schimadzu Prominence LC20AD pump and a SIL-20AC auto sampler. The complete analytical process was performed using the MetIQ™ software package, which is an integral part of the AbsoluteIDQ™ kit. We did not apply any data correction, nor were any data points removed. The experimental metabolomics measurement technique is described in detail by patent US 2007/0004044 (accessible online at http://www.freepatentsonline.com/20070004044.html) and in the manufacturer’s manuals. A summary of the method can be found in 17,18 and a comprehensive overview of the field and the related technologies is given in the review paper by Wenk 19. Briefly, a targeted profiling scheme is used to quantitatively screen for known small molecule metabolites using multiple reaction monitoring, neutral loss and precursor ion scans. Quantification of the metabolites of the biological sample is achieved by reference to appropriate internal standards. The method has been proven to be in conformance with 21CFR (Code of Federal Regulations) Part 11, which implies proof of reproducibility within a given error range (see Supplementary Table 4 for data). It has been applied in different academic and industrial applications 1,20,21. Concentrations of all analyzed metabolites are reported in μM.
Metabolite panel
In total, 163 different metabolites were detected (Table 3 in Online Methods). The metabolomics dataset contains 14 amino acids, hexose (H1), free carnitine (C0), 40 acylcarnitines (Cx:y), hydroxylacylcarnitines (C(OH)x:y), and dicarboxylacylcarnitines (Cx:y-DC), 15 sphingomyelins (SMx:y) and N-hydroxylacyloylsphingosyl-phosphocholine (SM (OH)x:y), 77 phosphatidylcholines (PC, aa=diacyl, ae=acyl-alkyl) and 15 lysophosphatidylcholines. Lipid side chain composition is abbreviated as Cx:y, where x denotes the number of carbons in the side chain and y the number of double bonds. E.g. “PC ae C33:1” denotes an acyl-alkyl phosphatidylcholine with 33 carbons in the two fatty acid side chains and a single double bond in one of them. Full biochemical names are provided in Supplementary Table 4. The precise position of the double bonds and the distribution of the carbon atoms in different fatty acid side chains cannot be determined with this technology. In some cases, the mapping of metabolite names to individual masses can be ambiguous. For example, stereo-chemical differences are not always discernible, neither are isobaric fragments. In such cases, possible alternative assignments are indicated.
Table 3.
Metabolite concentrations determined using the Biocrates AbsolutelDQ™ kit. Full biochemical names are provided in Supplementary Table 4.
| Metabolite class | N | Metabolite name or abbreviation | Biological relevance (selected examples) |
|---|---|---|---|
| Amino acids | 14 | Arginine, Glutamine, Glycine, Histidine, Methionine, Ornithine, Phenylalanine, Proline, Serine, Threonine, Tryptophan, Tyrosine, Valine, (Iso)Leucine |
Amino acid metabolism, urea-cycle, activity of gluconeogenesis and glycolysis, insulin sensitivity, neurotransmitter metabolism, oxidative stress |
| Sum of hexoses | 1 | H1 | Carbohydrate metabolism |
| Carnitine | 1 | C0 | |
| Acylcarnitines | 26 | C2, C3, C3:1, C4, C4:1, C5, C5:1, C6 (or C4:1-DC), C6:1, C8, C8:1, C9, C10, C10:1, C10:2, C12, C12:1, C14, C14:1, C14:2, C16, C16:1, C16:2, C18, C18:1, C18:2 |
Energy metabolism, fatty acid transport and , mitochondrial fatty acid oxidation, ketosis, oxidative stress, mitochondrial membrane damage |
| Hydroxy- and dicarboxy- acylcarnitines |
14 | C3-OH, C4-OH (or C3-DC), C5-DC (or C6-OH), C5-OH (or C3-DC-M), C5:1-DC, C5-M-DC, C7-DC, C12-DC, C14:1-OH, C14:2-OH, C16:1-OH, C16:2-OH, C16-OH, C18:1-OH , |
|
| Sphingomyelins | 10 | SM C16:0, SM C16:1, SM C18:0, SM C18:1, SM C20:2, SM C22:3, SM C24:0, SM C24:1, SM C26:0, SM C26:1 |
Signaling cascades, membrane damage (e.g. neurodegeneration) |
| Hydroxysphingomyelins | 5 | SM (OH) C14:1, SM (OH) C16:1, SM (OH) C22:1, SM (OH) C22:2, SM (OH) C24:1 |
|
| Diacyl-phosphatidyl- cholines |
38 | PC aa C24:0/C26:0/C28:1/C30:0/C30:2/C32:0/C32:1/C32:2/C32:3/ C34:1/C34:2/C34:3/C34:4/C36:0/C36:1/C36:2/C36:3/C36:4/C36:5/ C36:6/C38:0/C38:1/C38:3/C38:4/C38:5/C38:6/C40:1/C40:2/C40:3/ C40:4/C40:5/C40:6/C42:0/C42:1/C42:2/C42:4/C42:5/C42:6 |
Dyslipidemia, membrane composition and damage, fatty acid profile, activity of desaturases |
| Acyl-alkyl-phosphatidyl- cholines |
39 | PC ae C30:0/C30:1/C30:2/C32:1/C32:2/C34:0/C34:1/C34:2/C34:3/C36:0/ C36:1/C36:2/C36:3/C36:4/C36:5/C38:0/C38:1/C38:2/C38:3/C38:4/ C38:5/C38:6/C40:0/C40:1/C40:2/C40:3/C40:4/C40:5/C40:6/C42:0/ C42:1/C42:2/C42:3/C42:4/C42:5/C44:3/C44:4/C44:5/C44:6 |
|
| Lyso-phosphatidyl- cholines |
15 | lysoPC a C6:0/C14:0/C16:0/C16:1/C17:0/C18:0/C18:1/C18:2/C20:3/ C20:4/C24:0/C26:0/C26:1/C28:0/C28:1 |
Degradation of phospholipids, membrane damage, signaling cascades, fatty acid profile |
|
| |||
| total | 163 | ||
Statistical analysis
In the statistical analysis only SNPs with a minor allele frequency of at least 10% were included in order to avoid spurious associations due to small numbers. Additive genetic models assuming a trend per copy of the minor allele were used to specify the association between genotype categories and each of the 163 metabolite concentrations as well as all possible metabolite concentration ratios (163*162 = 26.406 traits). No further adjustment was performed. The linear regression algorithm implemented in the statistical analysis system R (http://www.r-project.org/) was used in the genome wide association study and SPSS for Windows (Version 17.0, Chicago: SPSS Inc.) was used for statistical analysis on a case-by-case level. Motivated by our previous observation that the use of ratios may lead to a strong reduction in the overall variance and a corresponding improvement in the p-values of association 20, we also computed all possible pairs of metabolite concentration ratios for those cases and used those ratios as quantitative traits. We present in the main text the results for the untransformed ratios for ease of interpretation. We used a conservative estimate of a genome-wide significance level (using a Bonferroni correction) based on a nominal level of 0.05 is 5.93×10−10 (0.05 / (163*517,480). To estimate whether deviation from normality of metabolite ratios may have biased our results we tested associations for both untransformed and log-scaled ratios, not detecting significant differences (Supplementary Table 3). The reported p-values of step 1 of the discovery stage were not corrected for genomic inflation, as the genomic control inflation factors (lambda) are small, ranging from 1.00 to 1.03, both in KORA and in TwinsUK.
Replication in the TwinsUK study
The TwinsUK cohort (KCL, http://www.twinsuk.ac.uk) is an adult twin British registry. These unselected twins were recruited from the general population through national media campaigns in the UK and shown to be comparable to age-matched population singletons in terms of disease-related and lifestyle characteristics 22. Ethics approval was obtained from the Guy’s and St. Thomas’ Hospital Ethics Committee. Written informed consent was obtained from every participant to the study. A total of 2277 individuals of European ancestry (1073 singletons and 602 dizygotic twins (DZs)) from the TwinsUK registry were genotyped using the Illumina Hap317K chip (Illumina, San Diego, USA). We applied a strict quality control at both individual and SNP levels. We excluded 51 individuals due to non-European ancestry and 3366 SNPs due to MAF <1%, call rate < 95% if MAF < 5% or call rate <99% if MAF >5%, or PHWE <1×10−4 . After the quality control, 305,811 autosomal SNPs available on 2226 individuals (1046 singletons and 590 DZs) were available and used for imputation. The imputation was carried out using the IMPUTE software 16. NCBI build 36 was used for strand reference. 2.5 million autosomal imputed SNPs data are made available on these 2226 individuals. 422 unrelated individuals from these genotyped were selected for the metabolomics assay. For the TwinsUK study, blood samples were taken after at least 6 hours fasting. The samples were immediately inverted three times, followed by 40 min resting at 4°C to obtain complete coagulation. The samples were then centrifuged for 10 minutes at 3,000RPM. Serum is removed from the centrifuged brown-topped tubes as the top, yellow, clear layer of liquid. Aliquot in 4 × 1.5mls skirted microcentrifuge tubes was then stored in a −45°C freezer until sampling. Metabolite measurements were performed using the same metabolomics platform and following an identical protocol as for the KORA study at the Genome Analysis Centre of the Helmholtz Zentrum München. For the purpose of the replication for the KORA study, the data on the metabolites, the ratios of the concentration, and the genotypes were extracted from the available database, and the association between the concentration of the metabolites or the ratios and the corresponding SNPs were tested using a linear regression model in STATA version 10 (StatCorp LP, College Station, TX, USA).
Supplementary Material
ACKNOWLEDGEMENTS
The KORA research platform (KORA: Kooperative Gesundheitsforschung in der Region Augsburg) and the MONICA Augsburg studies (Monitoring trends and determinants on cardiovascular diseases) were initiated and financed by the Helmholtz Zentrum München - National Research Center for Environmental Health, which is funded by the German Federal Ministry of Education, Science, Research and Technology and by the State of Bavaria. Part of this work was financed by the German National Genome Research Network (NGFNPlus: 01GS0823) and by grants from the “Genomics of Lipid-associated Disorders – GOLD” of the “Austrian Genome Research Programme GEN-AU”. Computing resources have been made available by the Leibniz Supercomputing Centre of the Bavarian Academy of Sciences and Humanities (HLRB project h1231) and the DEISA Extreme Computing Initiative (project PHAGEDA). Part of this research was supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. The TwinsUK study was funded by the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F2-2008-201865-GEFOS and (FP7/2007-2013), ENGAGE project grant agreement HEALTH-F4-2007-201413 and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). The study also receives support from the Dept of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London. TDS is an NIHR senior Investigator. The project also received support from a Biotechnology and Biological Sciences Research Council (BBSRC) project grant (G20234) .The authors acknowledge the funding and support of the National Eye Institute via an NIH/CIDR genotyping project (PI: Terri Young). We gratefully acknowledge the contributions of P. Lichtner, G. Eckstein, Guido Fischer, T. Strom and all other members of the Helmholtz Zentrum München genotyping staff in generating the SNP dataset, those of Tamara Halex and Arsin Sabunchi to the metabolomics measurements, as well as the contribution of all members of field staffs who were involved in the planning and conduct of the MONICA/KORA Augsburg studies. The KORA group consists of H.E. Wichmann (speaker), A. Peters, C. Meisinger, T. Illig, R. Holle, J. John and their co-workers who are responsible for the design and conduct of the KORA studies. For TwinsUK we thank the staff from the Genotyping Facilities at the Wellcome Trust Sanger Institute for sample preparation, Quality Control and Genotyping led by Leena Peltonen and Panos Deloukas; Le Centre National de Génotypage, France, led by Mark Lathrop, for genotyping; Duke University, North Carolina, USA, led by David Goldstein, for genotyping; and the Finnish Institute of Molecular Medicine, Finnish Genome Center, University of Helsinki, led by Aarno Palotie. Genotyping was also performed by CIDR as part of an NEI/NIH project grant. Finally, we express our appreciation to all participants of the KORA and the TwinsUK studies for donating their blood and time.
Footnotes
DISCLOSURE STATEMENTS
The authors have no competing interests to disclose.
REFERENCES
- 1.Gieger C, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altmaier E, et al. Bioinformatics analysis of targeted metabolomics--uncovering old and new tales of diabetic mice under medication. Endocrinology. 2008;149:3478–89. doi: 10.1210/en.2007-1747. [DOI] [PubMed] [Google Scholar]
- 3.Kottgen A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009 doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hindorff LA, Junkins HA, Mehta JP, Manolio TA. [accessed 14 April 2009];A Catalog of Published Genome-Wide Association Studies. www.genome.gov/26525384.
- 10.Vaxillaire M, et al. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 2008;57:2253–7. doi: 10.2337/db07-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gross E, et al. Strong association of a common dihydropyrimidine dehydrogenase gene polymorphism with fluoropyrimidine-related toxicity in cancer patients. PLoS ONE. 2008;3:e4003. doi: 10.1371/journal.pone.0004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, et al. Thermolabile phenotype of carnitine palmitoyltransferase II variations as a predisposing factor for influenza-associated encephalopathy. FEBS Lett. 2005;579:2040–4. doi: 10.1016/j.febslet.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 14.KIDO H, KINOSHITA M, MIZUGUCHI H, TAKAHASHI N. METHOD OF DIAGNOSING THE RISK OF THERMOLABILE PHENOTYPE DISEASES BY USING GENE. OTSUKA PHARMACEUTICAL CO., LTD. & The University of Tokushima; 2007. [Google Scholar]
- 15.Wichmann HE, Gieger C, Illig T. KORA-gen--resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl 1):S26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 16.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 17.Weinberger KM. Metabolomics in diagnosing metabolic diseases. Ther Umsch. 2008;65:487–91. doi: 10.1024/0040-5930.65.9.487. [DOI] [PubMed] [Google Scholar]
- 18.Weinberger KM, Graber A. Using Comprehensive Metabolomics to Identify Novel Biomarkers. Screening Trends in Drug Discovery. 2005;6:42–45. [Google Scholar]
- 19.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 20.Altmaier E, et al. Bioinformatics analysis of targeted metabolomics - uncovering old and new tales of diabetic mice under medication. Endocrinology. 2008;149:34783489. doi: 10.1210/en.2007-1747. [DOI] [PubMed] [Google Scholar]
- 21.Wang-Sattler R, et al. Metabolic profiling reveals distinct variations linked to nicotine consumption in humans--first results from the KORA study. PLoS ONE. 2008;3:e3863. doi: 10.1371/journal.pone.0003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrew T, et al. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–77. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 23.Brookes KJ, Chen W, Xu X, Taylor E, Asherson P. Association of fatty acid desaturase genes with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2006;60:1053–61. doi: 10.1016/j.biopsych.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 24.Caspi A, et al. Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proc Natl Acad Sci U S A. 2007;104:18860–5. doi: 10.1073/pnas.0704292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregersen N, et al. Identification of four new mutations in the short-chain acyl-CoA dehydrogenase (SCAD) gene in two patients: one of the variant alleles, 511C-->T, is present at an unexpectedly high frequency in the general population, as was the case for 625G-->A, together conferring susceptibility to ethylmalonic aciduria. Hum Mol Genet. 1998;7:619–27. doi: 10.1093/hmg/7.4.619. [DOI] [PubMed] [Google Scholar]
- 27.Nagan N, et al. The frequency of short-chain acyl-CoA dehydrogenase gene variants in the US population and correlation with the C(4)-acylcarnitine concentration in newborn blood spots. Mol Genet Metab. 2003;78:239–46. doi: 10.1016/s1096-7192(03)00034-9. [DOI] [PubMed] [Google Scholar]
- 28.Pare G, et al. Novel Associations of CPS1, MUT, NOX4, and DPEP1 With Plasma Homocysteine in a Healthy Population: A Genome-Wide Evaluation of 13 974 Participants in the Women’s Genome Health Study. Circ Cardiovasc Genet. 2009;2:142–150. doi: 10.1161/CIRCGENETICS.108.829804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou YE, Egeland GM, Meltzer SJ, Kubow S. The association of desaturase 9 and plasma fatty acid composition with insulin resistance-associated factors in female adolescents. Metabolism. 2009;58:158–66. doi: 10.1016/j.metabol.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Cao H, et al. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–44. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peltekova VD, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–5. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 32.Pind S, et al. V490M, a common mutation in 3-phosphoglycerate dehydrogenase deficiency, causes enzyme deficiency by decreasing the yield of mature enzyme. J Biol Chem. 2002;277:7136–43. doi: 10.1074/jbc.M111419200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.