Abstract
Genome-wide association studies revealed that common non-coding variants in MTNR1B (encoding melatonin receptor 1B, also known as MT2) increase type 2 diabetes (T2D) risk1,2. Although the strongest association signal was highly significant (P<10−20), its contribution to T2D risk was modest (odds ratio, OR~1.10-1.15)1-3. We performed large-scale exon resequencing in 7,632 Europeans including 2,186 T2D patients and identified 40 non-synonymous variants, including 36 very rare variants (minor allele frequency, MAF<0.1%) associated with T2D (OR=3.31[1.78;6.18]95%); P=1.64×10−4. A four-tier functional investigation of all 40 mutants revealed that 14 were non-functional and rare (MAF<1%); four were very rare with complete loss of melatonin binding and signaling capabilities. Among the very rare variants, the partial or total loss-of-function variants, but not the neutral ones, contributed to T2D (OR=5.67[2.17;14.82]95%; P=4.09×10−4). Genotyping the four complete loss-of-function variants in 11,854 additional individuals revealed their association with T2D risk (Ncases=8,153/Ncontrols=10,100; OR=3.88[1.49;10.07]95%; P=5.37×10−3). This study establishes a firm functional link between MTNR1B and T2D risk.
Disruption of central and peripheral circadian rhythms, including the pancreatic clock, may lead to metabolic disorders and type 2 diabetes (T2D)4, 5. The neurohormone melatonin (MLT) is mainly secreted from the pineal gland in a circadian pattern with higher levels being observed during the night. MLT targets two high-affinity G protein-coupled receptors (GPCRs): melatonin receptor 1A (also known as MT1, encoded by MTNR1A) and MT2 (encoded by MTNR1B) that modulate both Gi protein/adenylyl cyclase and ERK1/2 pathways6, 7. Genome-wide association studies (GWAS) recently revealed an association between single nucleotide polymorphisms (SNPs) in MTNR1B and T2D risk in humans1, 2. However, the contribution of these SNPs to T2D risk was typical for common alleles1-3, and it explains less than 10% of T2D heritability so far3, 8. The missing heritability of T2D might be partially explained by numerous rare mutations which have a stronger functional effect9. However, rare variant effects cannot easily be identified via GWAS because of statistical power issues. Such rare variant effects have been discovered in type 1 diabetes10, 11 and hypertriglyceridemia12 GWAS-identified genes by large-scale resequencing. Recently, Coventry et al. sequenced two GWAS-associated T2D genes (HHEX and KCNJ11) in 13,715 subjects and found an excess of rare recent variants consistent with explosive population growth13. They suggested that increased disease [T2D] risk in contemporary populations might be heavily influenced by the distribution of rare variants. However, they had not tested for either T2D association or function of any of the variants13.
Based on the strong evidence for the association between the MTNR1B gene and T2D risk, together with the fact that MT2 belongs to the superfamily of GPCRs which are privileged drug targets, we wished to identify rare non-synonymous variants with putative effect on the function of MT2 that may associate with T2D. We sequenced the two exons of MTNR1B in 7,632 unrelated European individuals with known glycemic status, including 2,186 T2D patients (Supplementary Table 1). We identified 40 non-synonymous variants including two common SNPs (MAF>1% - p.Gly24Glu/rs8192552 and p.Lys243Arg/rs61747139); two less common variants (MAF between 0.1 and 1% - p.Arg138Cys/rs61746674 and p.Arg231His/rs8192553); and 36 very rare variants (MAF<0.1%) which were not previously listed in public SNP databases (Fig.1, Table 1). No association was found between T2D risk and either of the common SNPs (rs8192552 or rs61747139) (Table 1). The rarer variants were analysed by pooling them according to their MAF: we analysed one pool of two variants with MAF between 0.1 and 1% and a second pool of 36 variants with MAF<0.1%. By using the kernel-based adaptive cluster (KBAC) method14 embedded in a logistic regression model, we found that the variants with intermediate MAF did not associate with T2D risk (Ncontrols=4,804/Ncases=2,186; OR=1.00[0.40;3.20]95%; P=0.81; Table 1) whereas the rarest variants (with MAF<0.1%) contributed strongly to an increased T2D risk (OR=3.31[1.78;6.18]95%; P=1.64×10−4; Table 1).
Figure 1. Distribution of the 40 non-synonymous MTNR1B variants identified by exon resequencing.

Minor allele frequency (MAF) calculation was based on the whole sequencing dataset (7,632 individuals, including 2,186 T2D patients). Non-synonymous variants are colored as follows: blue, with MAF≥1%; green, with MAF between 0.1% and 1%; and red, with MAF<0.1%. Mutants devoid of any melatonin binding (and associated downstream signaling) are highlighted with a red flash. Mutants with impaired Gi protein-dependent signaling only are highlighted with a white flash. Mutants with impaired Gi protein-dependent signaling and ERK1/2 activation are highlighted with a black flash. Nomenclature for variants refers to functional protein sequences.
Table 1.
Contribution of non-synonymous MTNR1B variants to T2D risk. Non-synonymous MTNR1B variants were identified by exon resequencing in 7,632 European individuals.
| Variants | MAF2 (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Position on Chr11 |
Amino acid change |
Allele change |
MAF1 (%) |
T2D (cases) N=2,186 |
IFG N=642 |
NG (controls) N=4,804 |
OR (95%CI) | P | |
| p.Gly24Glu | 92342610 | G>E | G>A | ≥1 | 6.43 | 5.45 | 6.96 | 0.87 (0.75;1.00) | 0.06 |
|
| |||||||||
| p.Lys243Arg | 92354765 | K>R | A>G | ≥1 | 2.42 | 2.26 | 2.76 | 0.86 (0.67;1.11) | 0.25 |
|
| |||||||||
| p. Arg138Cys | 92354449 | R>C | C>T | 0.1≤x<1 | 0.16 | 0.23 | 0.14 | 1.00 (0.40;3.20) | 0.81 |
| p. Arg231His | 92354729 | R>H | G>A | 0.1≤x<1 | 0.66 | 0.93 | 0.76 | ||
|
| |||||||||
| p.Ala8Ser | 92342561 | A>S | G>T | <0.1 | 0 | 0 | 0.01 | ||
| p.Ala13Val | 92342577 | A>V | C>T | <0.1 | 0 | 0 | 0.01 | ||
| p.Gly21Ser | 92342600 | G>S | G>A | <0.1 | 0 | 0 | 0.01 | ||
| p.Trp22Leu | 92342604 | W>L | G>T | <0.1 | 0.02 | 0 | 0 | ||
| p.Ala25Thr | 92342612 | A>T | G>A | <0.1 | 0 | 0 | 0.01 | ||
| p.Pro36Ser | 92342645 | P>S | C>T | <0.1 | 0.02 | 0 | 0 | ||
| p.Ala42Pro | 92342663 | A>P | G>C | <0.1 | 0.02 | 0 | 0 | ||
| p.Ala52Thr | 92342693 | A>T | G>A | <0.1 | 0.02 | 0 | 0 | ||
| p.Leu60Arg | 92342718 | L>R | T>G | <0.1 | 0.11 | 0.08 | 0.06 | ||
| p.Ala74Thr | 92342759 | A>T | G>A | <0.1 | 0.02 | 0 | 0.01 | ||
| p.Pro95Leu | 92354321 | P>L | C>T | <0.1 | 0.02 | 0 | 0 | ||
| p.Gly109Ala | 92354363 | G>A | G>C | <0.1 | 0 | 0 | 0.01 | ||
| p.Met120Val | 92354395 | M>V | A>G | <0.1 | 0 | 0 | 0.02 | ||
| p.Met120Ile | 92354397 | M>I | G>A | <0.1 | 0 | 0.08 | 0.01 | ||
| p.Ser123Arg | 92354406 | S>R | C>G | <0.1 | 0 | 0.08 | 0.01 | ||
| p.Val124Ile | 92354407 | V>I | G>A | <0.1 | 0.11 | 0.16 | 0.08 | ||
| p.Arg138Leu | 92354450 | R>L | G>T | <0.1 | 0 | 0 | 0.02 | ||
| p.Arg138His | 92354450 | R>H | G>A | >0.1 | 0 | 0 | 0.01 | 3.31 (1.78;6.18) | 1.64×10−4 |
| p.Tyr141Phe | 92354459 | Y>F | A>T | <0.1 | 0.02 | 0 | 0 | ||
| p.Met146Val | 92354473 | M>V | A>G | <0.1 | 0 | 0 | 0.01 | ||
| p.Arg154His | 92354498 | R>H | G>A | <0.1 | 0.02 | 0 | 0.01 | ||
| p.Leu166Ile | 92354533 | L>I | C>A | <0.1 | 0 | 0 | 0.01 | ||
| p.Thr201Met | 92354639 | T>M | C>T | <0.1 | 0.02 | 0 | 0 | ||
| p.Arg222His | 92354702 | R>H | G>A | <0.1 | 0.02 | 0 | 0 | ||
| p.Ile223Thr | 92354705 | I>T | T>C | <0.1 | 0.02 | 0 | 0 | ||
| p.Ala234Thr | 92354737 | A>T | G>A | <0.1 | 0 | 0 | 0.03 | ||
| p.Glu237Lys | 92354746 | E>K | G>A | <0.1 | 0 | 0 | 0.01 | ||
| p.Ser238Gly | 92354750 | S>G | G>A | <0.1 | 0.02 | 0 | 0 | ||
| p.Asp246Asn | 92354774 | D>N | G>A | <0.1 | 0.02 | 0 | 0 | ||
| p.Phe250Val | 92354786 | F>V | T>G | <0.1 | 0.02 | 0 | 0 | ||
| p.Tyr308Ser | 92354963 | Y>S | A>C | <0.1 | 0.02 | 0 | 0 | ||
| p.Arg316His | 92354984 | R>H | G>A | <0.1 | 0 | 0 | 0.010 | ||
| p.Arg330Trp | 92355025 | R>W | C>T | <0.1 | 0.02 | 0 | 0 | ||
| p.Ala342Val | 92355062 | A>V | C>T | <0.1 | 0.07 | 0 | 0 | ||
| p.Ile353Thr | 92355095 | I>T | T>C | <0.1 | 0.05 | 0 | 0.02 | ||
| p.Ala359Gln | 92355113 | A>Q | C>A | <0.1 | 0 | 0 | 0.01 | ||
Mutation position was indicated according to the human genome build NCBI36/hg18. The protein sequence reference used was NP_005950.1._MAF1 calculation was based on the whole sequencing dataset (7,632 individuals). Chr, Chromosome; MAF, minor allele frequency;T2D, type 2 diabetes; IFG, impaired fasting glucose; NG, individuals presenting normal fasting glucose; OR, odds ratio; CI, confidence interval; P, p-value.
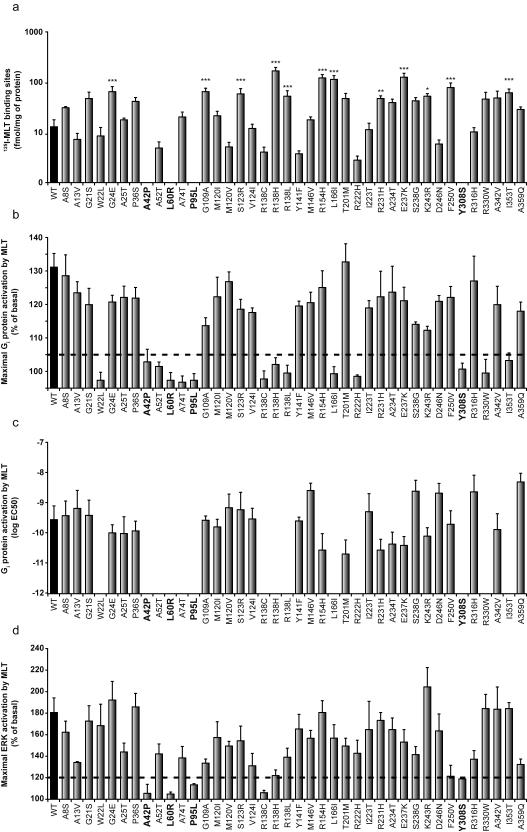
Subsequently, we wished to determine which non-synonymous MTNR1B variants altered MT2 function. Thus, we assessed the effects of all 40 non-synonymous MTNR1B variants on MT2 cell surface expression and 2(125I)-iodomelatonin (125I-MLT) binding, as well as on MLT-dependent Gi protein and ERK1/2 activation in human HEK293 cells. We found that cell surface expression of all 40 MT2 mutants was indistinguishable from that of the MT2 wild-type receptor when measured using immunohistochemical techniques (Supplementary Fig.1a). 125I-MLT saturation binding experiments revealed a marginal (four-fold) variation of the dissociation constant (Kd) for most mutants (Supplementary Fig.1b) and a ten-fold variation in the number of 125I-MLT binding sites (Fig.2a). Notably, four MT2 mutants (p.Ala42Pro, p.Leu60Arg, p.Pro95Leu and p.Tyr308Ser) showed no 125I-MLT binding capacity (Fig.2a, Supplementary Fig.1b). These same mutants did not activate downstream Gi protein-dependent (Fig.2b, Fig.2c) or ERK1/2 (Fig.2d) signaling pathways. Ten additional mutants with intact MLT binding (p.Trp22Leu, p.Ala52Thr, p.Ala74Thr, p.Arg138Cys, p.Arg138His, p.Arg138Leu, p.Leu166Ile, p.Arg222His, p.Arg330Trp and p.Ile353Thr) showed impaired Gi protein-dependent signaling, while the other mutants were similar to wild-type MT2 receptor (Fig.2b, Fig.2c). Out of the 10 mutants with impaired Gi protein signaling, only p.Arg138Cys was unable to activate the ERK1/2 pathway indicating the pathway-specific behaviour of the other nine mutants (Fig.2d). It is noteworthy that all of the variants that ablate MLT binding and all but one of the variants that inhibit signaling downstream from MT2 are very rare and have MAF<0.1%.
Figure 2. Functional characterization of wild-type and mutant MT2 receptors.
(a) Expression levels in Flp-In HEK 293 cells determined by 125I-MLT saturation binding experiments. (b) Maximal Gi protein-dependent signaling. (c) EC50 values of Gi protein-dependent signaling determined in MLT dose-response curves. (d) ERK activations in the presence of 100nM MLT. The threshold line in panels (b) and (d) was defined according to the known variability of each assay. Ligand binding deficient MT2 mutants are highlighted in bold. Data are means ± standard error of at least three independent experiments, each performed in duplicate (*P<0.05; **P<0.01; ***P<0.001 significantly different from wild-type MT2). WT, wild-type; EC50, half maximal effective concentration; MLT, melatonin; 125I-MLT, 2(125I)-iodomelatonin.
The four MLT binding-deficient MT2 mutants contain amino-acid substitutions in the predicted transmembrane domains I (p.Ala42Pro, p.Leu60Arg), II (p.Pro95Leu) and VII (p.Tyr308Ser) (Fig.1). The ten MT2 mutants with impaired Gi protein signaling are located in the predicted N-terminus (p.Trp22Leu), transmembrane domains (p.Ala52Thr, p.Ala74Thr, p.Arg138Cys, p.Arg138His, p.Arg138Leu, p.Leu166Ile, p.Arg222His) and the C-terminus (p.Arg330Trp and p.Ile353Thr) (Fig.1). Four of these variants affect conserved motifs in MT2: p.Tyr308Ser alters a conserved ‘NP(A)XXY’ motif, while p.Arg138Cys, p.Arg138Leu and p.Arg138His alter the ‘D(N)RY’ motif, both of which are involved in GPCR activation15.
To further assess the observed deficits in MT2 function, we compared our functional data with sequence- and structure-based predictions made by the PolyPhen-2 software (Polymorphism Phenotyping version v2; Supplementary Table 2)16. By considering variants predicted ‘possibly’ or ‘probably’ damaging by PolyPhen-2 as ‘loss-of-function’, we found 60% concordance between our functional data and prediction by PolyPhen-2 (Supplementary Table 3). Furthermore, the software predicted 30% false-positive and 7.5% false-negative loss-of-function variants compared to our own data. It is noteworthy that the most recent version (v2.0.23; Dec 9th 2010) is more sensitive but shows more ‘potentially damaging’ variants that were not biologically confirmed than the previous version (v2.0.22; Supplementary Table 3).
We then assessed whether the four total loss-of-function MT2 variants had a significant effect on increased T2D risk, confirming or not what was previously observed for all rare variants with MAF<0.1%. We genotyped the p.Ala42Pro, p.Leu60Arg, p.Pro95Leu and p.Tyr308Ser variants in 11,854 additional French subjects, including 5,967 T2D patients. By analysing these variants as a pool using the KBAC method embedded in a logistic regression model, we found that the p.Ala42Pro, p.Leu60Arg, p.Pro95Leu and p.Tyr308Ser variants had a strong and significant effect on increased T2D risk (Ncontrols=10,100/Ncases=8,153; OR=3.88[1.49;10.07]95%; P=5.37×10−3; Table 2), confirming the association of total loss-of-function of MT2 variants with T2D. A study has described impairment of G protein-dependent MT2 function caused by the p.Leu60Arg mutation17. No association with T2D was found, probably due to statistical power issues17. This emphasizes the need for extensive sequencing/genotyping/biological studies in very large populations.
Table 2.
Contribution of non-synonymous MTNR1B variants to increased T2D risk according to their consequences on MT2 function.
| Consequences on MT2 function |
Variants |
NT2D (% of variant carriers) |
NNG (% of variant carriers) |
OR (95% CI) |
P | ||
|---|---|---|---|---|---|---|---|
|
No melatonin binding
(and downstream signaling) |
A42P | L60R P95L |
Y308S | 8,153 (0.22) | 10,100 (0.13) | 3.88 (1.49;10.07) | 5.37×10 −3 |
|
| |||||||
|
No GI protein-dependent
signaling |
W22L A52T A74T |
R138C R138H R138L L166I |
R222H R330W I353T |
2,186 (0.64) | 4,804 (0.42) | 2.66 (1.01;7.00) | 0.047 |
|
| |||||||
| No ERK activation | R138C | 2,186 (0.32) | 4,804 (0.27) | 1.24 (0.44;3.51) | 0.68 | ||
|
| |||||||
|
Loss-of-function variants
with MAF<0.1% |
W22L A42P A52T L60R |
A74T P95L R138H R138L L166I |
R222H Y308S R330W I353T |
2,186 (0.69) | 4,804 (0.27) | 5.67 (2.17;14.82) | 4.09×10 −4 |
|
| |||||||
|
Neutral variants
with MAF<0.1% |
A8S A13V G21S A25T P36S G109A M120V M120I |
S123R V124I Y141F M146V R154H T201M I223T |
A234T E237K S238G D246N F250V R316H A342V A359Q |
2,186 (0.73) | 4,804 (0.52) | 2.15 (0.93;4.95) | 0.072 |
T2D, type 2 diabetes; NG, normal fasting glucose; OR, odds ratio; CI, confidence interval; P,p-value; MAF, minor allele frequency; Loss-of-function variants are variants which impair MT2 function according to our four-tier functional investigation.
The pool of ten partial loss-of-function MT2 variants (which cause impaired Gi protein signaling without MLT binding deficiency) showed a trend for association with T2D risk (Ncontrols=4,804/Ncases=2,186; OR=2.66[1.01;7.00]95%; P=0.047; Table 2). This suggests that Gi protein-dependent signaling might contribute to T2D risk. Interestingly, the original group of 36 non-synonymous variants with MAF<0.1% that associated with T2D according to Table 1, can be split into two categories after our current biological assessment: a group of 13 ‘loss-of-function’ variants that strongly associated with T2D risk (Ncontrols=4,804/Ncases=2,186; OR=5.67[2.17;14.82]95%; P=4.09×10−4; Table 2; Fig.3); and a second group of 23 apparently ‘neutral’ variants that did not associate with T2D (Table 2; Fig.3). Therefore, the association between rare non-synonymous MTNR1B variants and T2D risk is further supported by the mutants that show evidence for melatonin signaling impairment.
Figure 3. Odds ratio estimates of partial or total loss-of-function versus neutral very rare variants (MAF<0.1%) for T2D risk based on MTNR1B sequencing data.

For each pool of variants (purple: loss-of-function, cyan: neutral and black: altogether), we generated an estimate of the effect size and its associated standard error. The figure represents assumed normal distributions of the odds ratios (log scale) centered on the effect sizes and with standard deviations equal to the respective estimated standard error. The figure illustrates the relative impact of functional variants among all variants with MAF below 0.1%. MAF, minor allele frequency; Loss-of-function variants are variants which impair MT2 function according to our four-tier functional investigation.
Based on RNA expression data, Lyssenko et al. have reported that the T2D at-risk allele of the GWAS-identified SNP rs10830963 (located in the unique MTNR1B intron), was associated with increased MTNR1B transcript levels in human islets18. As pharmacological doses of MLT inhibit insulin secretion from murine beta-cell lines18, 19, the authors suggested that inhibiting MLT receptors could be a therapeutic avenue for T2D18. Our functional analyses at the protein level, showing an association between defective MT2 receptor function and T2D risk, suggests that the previously observed increased MTNR1B expression is not causal but could be rather due to the absence of negative feedback regulatory events under conditions of impaired MT2 receptor signaling. Plausible mechanisms are the loss of melatonin’s acute inhibitory effect on insulin secretion or the loss of melatonin effect on circadian rhythm entrainment. Hence, MLT receptor agonists which are currently prescribed for sleep and circadian rhythm disorders, as well as for depression20, may be beneficial for T2D therapy.
Our data confirm that resequencing GWAS-identified susceptibility genes for a complex disease (like T2D), followed by the careful biological evaluation of newly identified rare but potentially damaging variants, can contribute to a more complete picture of the genetic architecture of the disease risk.
METHODS
Our study design is summarized in Supplementary Fig.2.
Study participants
Clinical characteristics and phenotypic data collected from the study populations are reported in Supplementary Table 1. Fasting plasma glucose levels and type 2 diabetes (T2D) statuses were determined for all participants. All subjects in this study were unrelated and of European ancestry.
Sequencing of the two MTNR1B exons and genotyping of the four total loss-of-function MTNR1B mutations (p.Ala46Pro, p.Leu60Arg, p.Pro95Leu and p.Tyr308Ser) were performed in several general European cohort studies and T2D case-control studies (Supplementary Table 1). The non-diabetic subjects were obtained from five European studies: 1) the Data from the Epidemiological Study on the Insulin Resistance Syndrome (D.E.S.I.R.) cohort which is a longitudinal study in a French general population, fully described elsewhere22; 2) the Ely study (Cambridgeshire, UK) which was established in 1990 as a prospective population-based cohort study of the aetiology and pathogenesis of T2D23; 3) the Hertfordshire Cohort Study which consists of men and women born between 1931 and 1939 and still residing in the English county of Hertfordshire (UK), of whom almost 3,000 have been extensively characterised24; 4) obese probands from obesity French pedigrees who were recruited at the Centre National de la Recherche Scientifique (CNRS)-UMR8199 unit (Lille, France) through an ongoing national media campaign25; and 5) the French SU.VI.MAX study which is a randomized double-blind, placebo-controlled, primary prevention trial designed to assess the usefulness of daily supplementation with antioxidant vitamins and minerals in reducing the frequency of major health problems in industrialized countries26. In total, 685 T2D patients were obtained from the five previously described studies. The majority of T2D patients were recruited from: 1) the Endocrinology-Diabetology Department of the Corbeil-Essonnes Hospital in Corbeil, France27; 2) the French Diabhycar/Diab2-Néphrogène/Surdiagène study28; and 3) CNRS-UMR8199 unit in Lille, France27.
For each individual, glycemic status was defined according to 1997 American Diabetes Association criteria29: normal glucose was defined as fasting plasma glucose levels <6.1mmol/l without treatment with antidiabetic agents, impaired fasting glucose (IFG) was defined as fasting plasma glucose levels ≥6.1mmol/l and <7.0mmol/l without treatment with antidiabetic agents, and T2D was defined as fasting plasma glucose levels ≥7.0mmol/l or treatment with antidiabetic agents.
All samples used had been collected with appropriate informed consent consistent with their use in the present study.
Sequencing
Sanger sequencing of the MTNR1B exons was performed in 7,632 unrelated European subjects, including 2,186 T2D patients. MTNR1B is located on human chromosome 11q21-q22 and encodes a 362-amino-acid protein (NCBI NM_005959.3 and NP_005950.1). The two MTNR1B exons were analysed in five fragments using previously described primer sequences and PCR conditions1. Fragments were bidirectionally sequenced using the automated 3730×l DNA Analyzer (Applied Biosystems). Electrophoregram reads were assembled and analysed using the Variant Reporter software (Applied Biosystems). Each novel non-synonymous mutation was confirmed by another bidirectional Sanger sequencing. The location of each variant is displayed by base numbers counting from the ATG-translation initiation codon according to the Human Genome Variation Society nomenclature for the description of sequence variations. The position of mutations was indicated according to the human genome build NCBI36/hg18.
Genotyping
Genotyping of the four total loss-of-function MTNR1B mutations (p.Ala46Pro, p.Leu60Arg, p.Pro95Leu and p.Tyr308Ser) was performed in 14,909 additional unrelated European individuals. The DNA samples were pooled and screened by High Resolution Melting (HRM); samples that potentially contained sequence polymorphisms were studied individually using Sanger sequencing. We used the LightCycler 480 HRM Master kit (Roche Diagnostics) following the manufacturer’s protocol. Briefly, for each mutation genotyping, 10ng DNA per sample were used in a 5μl Master Mix (2×) with 1μl Primer Mix (4μM) and 1.2μl MgCl2 (25mM). Primers are described in Supplementary Table 4. PCR and HRM were performed on 384-well plates using the LightCycler 480 Real-Time PCR System (Roche Diagnostics) following the protocol: 1) Pre-incubation, one cycle, 95°C for 5min; 2) Touch Down, 10 cycles, 95°C for 10s _ 70°C to 63°C for 15s (−0.7°C per cycle) _ 72°C for 25s; 3) Amplification, 45 cycles, 95°C for 10s _ 63°C for 15s _ 72°C for 25s; 4) HRM, one cycle, 95°C for 1min _ 40°C for 1min _ temperature gradient from 63°C to 95°C for 1s each temperature; and 5) Cooling 40°C. We demonstrated that pools of four DNA samples per well provided accurate genotypes (Supplementary Fig.3), but conservatively, we chose to pool three DNA samples in each reaction. Each 384-well plate contained two positive controls: one well with a mutated DNA sample only and one well with a mutated sample and two wild-type DNA samples (Supplementary Fig.3). When a HRM profile matched the profile of a positive control, we resequenced the three DNA samples following a standard protocol. A total of 3,055 samples were both directly sequenced for the two MTNR1B exons and subsequently genotyped for the four mutations. We found 100% concordance (3,055/3,055) between the two protocols.
Statistical analyses
We independently assessed the effect of frequent variants (with MAF≥1%) on T2D risk using a logistic regression adjusted for age, gender, and body mass index (BMI), under an additive model. Rarer variants were analysed by pooling them based on their MAF (between 0.1 and 1%, or <0.1%) or their functional consequences. We assessed the effect of these pooled variants on T2D risk via the kernel-based adaptive cluster (KBAC) method14 embedded in a logistic regression model adjusted for age, gender and BMI. The KBAC method was developed to overcome the problems of detecting rare variant associations in the presence of misclassification. This method was extensively compared to three widely accepted methods (the weighted sum statistic [WSS], the combined multivariate and collapsing method [CMC], and the comparison of rare variants found exclusively in cases to those found only in controls [RVE]) and was demonstrated to be the most powerful under the assumption of phenotypic effects inversely correlated with MAF14. It is noteworthy that the age adjustment used in the assessment of the effect of total loss-of-function MTNR1B variants on T2D risk showed a positive impact on OR. To further assess this phenomenon, we performed a Mantel-Haenszel stratified analysis using three or four strata based on the quantiles of the age distribution without any adjustments for other confounders. Both the OR and the corresponding p-value for T2D risk measured for the three- and four-strata analyses had the same magnitude as those reported by the logistic regression model adjusted for age, gender and BMI, which shows the robustness of our analysis. With regard to a possible population stratification effect on our results, we used SNP arrays data (Illumina Metabochip or Illumina CNV370-Duo DNA arrays) which were available for carriers of partial or total loss-of-function MTNR1B variants. By using the population specific genotypes database released by the HapMap project, we found that 39,000 SNPs present on both chips were identified as good markers of ethnicity, since a primary principal component analysis (PCA) of those variants allowed us to clearly distinguish European-Caucasians (HapMap CEU), Africans (HapMap YRI) and Asians (HapMap JPT and CHB) on the first factorial plan. We then achieved a secondary PCA including the mutation carriers so as to detect potential admixture. The latter analysis revealed that it was highly unlikely that the mutation carriers did not have European ancestry (Supplementary Fig.4). All statistical analyses were performed with R (version 2.12) and SPSS software (version 14.0 for Windows).
Functional characterization of the MT2 mutants
The human MT2 cDNA was fused at its 5′-end with a mouse myc epitope and subcloned into the Flp-In pcDNA5/FRT plasmid (Invitrogen). Each of the forty non-synonymous mutations was generated in this plasmid using the QuickChange site-directed mutagenesis kit (Stratagene) and was confirmed by Sanger sequencing of the full-length clone. We established 41 stable Flp-In HEK293 cell lines stably expressing the wild-type receptor or one of the 40 mutant MT2 receptors according to the manufacturer’s recommendations (Invitrogen). Briefly, Flp-In HEK293 cells were co-transfected with each MT2 containing Flp-In pcDNA5/FRT plasmid and the pOG44 vector encoding the Flp recombinase at a 1/9 ratio, using the FuGENE HD transfection reagent (Roche Diagnostics). 48h after transfection, cells were cultured in the presence of 100μg/ml hygromycin B (Invitrogen) for 2–3 weeks. Hygromycin B-resistant cells were collected and expression of MT2 mutants was assessed by 2(125I)-iodomelatonin (125I-MLT) binding (see below) and Western blotting (not shown). Use of Flp-In cells is attractive since only one copy of the mutated MTNR1B coding gene is integrated at a specific predefined insertion site into the cell genome. This renders expression levels independent of the number of inserted copies and the insertion site, and allows the comparison of the expression levels of the 40 mutants solely on the basis of the receptor’s intrinsic properties.
Surface expression of MT2 variants was evaluated by In-Cell Western experiments using human HEK293T cells transfected with each plasmid and seeded onto sterile polyL-lysine-coated 24-well plates one day following transfection. After 24h, cells were fixed with phosphate buffered saline (PBS)-paraformaldehyde (PFA) 4% for 15min. After a 10min permeabilization step in PBS-Triton X-100 0.1%, cells were blocked for 1h with 3% bovine serum albumin (BSA) in PBS. Cells were immunolabelled for 1h with primary antibodies (monoclonal anti-myc antibodies at 0.2μg/ml (sc-40, Santa Cruz)). Immunoreactivity was revealed using IRDye secondary antibodies and quantified using the LI-COR Odyssey infrared imaging system (Sciencetec).
Radioligand saturation binding experiments were performed in stable MT2 Flp-In 293 cell lines. As previously described21, saturation binding assays were carried out on crude membrane preparations with increasing concentrations of 125I-MLT (PerkinElmer) to determine dissociation constants (Kd) and expression levels (Bmax) of wild-type and mutant MT2 receptors with the PRISM program (GraphPad Software).
Gi protein activation was determined in HEK293T cells expressing each MT2 mutant and the chimeric Gαq/i9 protein, which links Gi-coupled receptors to the activation of the phospholipase C pathway and inositol phosphate (IP) production. Cells were incubated for 30min with different MLT concentrations (10−13 - 10−6 mol/l) and IP levels determined with the IP-one HTRF kit (Cisbio Bioassays) according to the manufacturer instructions. Half maximal effective concentrations (EC50) were calculated with the PRISM program (GraphPad Software).
ERK1/2 activation was monitored in Flp-In HEK293 cell lines stably expressing MT2 mutants. Cells were stimulated for 2, 5, 7.5 and 10min with 100nM MLT and the level of ERK1/2 phosphorylation was determined. Reactions were stopped in Laemmli sample buffer and total cell lysates separated by SDS-PAGE. Proteins were transferred to nitrocellulose membranes and ERK1/2 phosphorylation determined by immunoblotting using anti-pERK1/2 antibodies (sc-7383, Santa Cruz). Immunoreactivity was revealed using IRDye secondary antibodies and quantified using the LI-COR Odyssey infrared imaging system (Sciencetec). Levels of loaded proteins were verified by determining total ERK2 levels by immunoblotting (sc-154, Santa-Cruz). ERK1/2 phosphorylation kinetics were transient with peak levels at 2-5 min. No differences in kinetics were observed between MT2 mutants and wild-type receptors.
Supplementary Material
ACKNOWLEDGMENTS
We are sincerely indebted to all participants in the genetic study. We thank Marianne Deweirder and Frédéric Allegaert for their technical assistance and their precious management of DNA samples, and Béatrice Gardiola-Lemaître for her invaluable advice. This study was supported by: the French Agence Nationale de la Recherche (ANR-08-GENOPAT to P.F., ANR-11-blanc “MLT2D” and ANR-11-META “MELA-BETES” to R.J. and P.F.), the Contrat de Projets Etat-Région Nord-Pas-De-Calais (CPER 2007-2013 ‘Axe Cardio-Diabète’, to P.F.), the Société Francophone du Diabète (to A.B.), the Fondation Recherche Médicale (‘Equipe FRM’, to R.J.), Institut National de la Santé et de la Recherche Médicale (Inserm) and Centre National de la Recherche Scientifique (CNRS). I.B. acknowledges funding from Wellcome Trust grant 077016/Z/05/Z and from United Kingdom NIHR Cambridge Biomedical Research Centre and the MRC Centre for Obesity and Related Metabolic Diseases.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Bouatia-Naji N, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 2.Prokopenko I, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–54. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–31. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubocovich ML, et al. International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharmacol Rev. 2010;62:343–80. doi: 10.1124/pr.110.002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jockers R, Maurice P, Boutin JA, Delagrange P. Melatonin receptors, heterodimerization, signal transduction and binding sites: what’s new? Br J Pharmacol. 2008;154:1182–95. doi: 10.1038/bjp.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–9. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downes K, et al. Reduced expression of IFIH1 is protective for type 1 diabetes. PLoS One. 2010;9:e12646. doi: 10.1371/journal.pone.0012646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–7. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coventry A, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu DJ, Leal SM. A novel adaptive method for the analysis of next-generation sequencing data to detect complex trait associations with rare variants due to gene main effects and interactions. PLoS Genet. 2010;6:e1001156. doi: 10.1371/journal.pgen.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30:249–59. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersson EA, et al. MTNR1B G24E variant associates with BMI and fasting plasma glucose in the general population in studies of 22,142 Europeans. Diabetes. 2010;59:1539–48. doi: 10.2337/db09-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyssenko V, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–8. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peschke E, Muhlbauer E. New evidence for a role of melatonin in glucose regulation. Best Pract Res Clin Endocrinol Metab. 2010;24:829–41. doi: 10.1016/j.beem.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Dubovsky SL, Warren C. Agomelatine, a melatonin agonist with antidepressant properties. Expert Opin Investig Drugs. 2009;18:1533–40. doi: 10.1517/13543780903292634. [DOI] [PubMed] [Google Scholar]
- 21.Ayoub MA, et al. Monitoring of ligand-independent dimerization and ligand-induced conformational changes of melatonin receptors in living cells by bioluminescence resonance energy transfer. J Biol Chem. 2002;277:21522–8. doi: 10.1074/jbc.M200729200. [DOI] [PubMed] [Google Scholar]
- 22.Balkau B. [An epidemiologic survey from a network of French Health Examination Centres, (D.E.S.I.R.): epidemiologic data on the insulin resistance syndrome] Rev Epidemiol Sante Publique. 1996;44:373–5. [PubMed] [Google Scholar]
- 23.Williams DR, et al. Undiagnosed glucose intolerance in the community: the Isle of Ely Diabetes Project. Diabet Med. 1995;12:30–5. doi: 10.1111/j.1464-5491.1995.tb02058.x. [DOI] [PubMed] [Google Scholar]
- 24.Syddall HE, et al. Cohort profile: the Hertfordshire cohort study. Int J Epidemiol. 2005;34:1234–42. doi: 10.1093/ije/dyi127. [DOI] [PubMed] [Google Scholar]
- 25.Meyre D, et al. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31-q23.2. Diabetes. 2004;53:803–11. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]
- 26.Hercberg S, et al. A primary prevention trial using nutritional doses of antioxidant vitamins and minerals in cardiovascular diseases and cancers in a general population: the SU.VI.MAX study--design, methods, and participant characteristics. SUpplementation en VItamines et Mineraux AntioXydants. Control Clin Trials. 1998;19:336–51. doi: 10.1016/s0197-2456(98)00015-4. [DOI] [PubMed] [Google Scholar]
- 27.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 28.Hadjadj S, et al. Prognostic value of the insertion/deletion polymorphism of the ACE gene in type 2 diabetic subjects: results from the Non-insulin-dependent Diabetes, Hypertension, Microalbuminuria or Proteinuria, Cardiovascular Events, and Ramipril (DIABHYCAR), Diabete de type 2, Nephropathie et Genetique (DIAB2NEPHROGENE), and Survie, Diabete de type 2 et Genetique (SURDIAGENE) studies. Diabetes Care. 2008;31:1847–52. doi: 10.2337/dc07-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.AmericanDiabetesAssociation Standards of medical care in diabetes. Diabetes Care. 2008;31(Suppl 1):S12–54. doi: 10.2337/dc08-S012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.