Abstract
Background
The progressive myoclonus epilepsies (PMEs) comprise a group of clinically and genetically heterogeneous disorders characterized by myoclonus, epilepsy, and neurological deterioration. We aimed to identify the underlying gene(s) in childhood-onset PME patients with unknown molecular genetic background.
Methods
Homozygosity mapping was applied on genome-wide SNP data of 18 Turkish patients. The potassium channel tetramerization domain-containing 7 (KCTD7) gene, previously associated with PME in a single inbred family, was screened for mutations. The spatiotemporal expression of KCTD7 was assessed in cellular cultures and mouse brain tissue.
Results
Overlapping homozygosity in 8/18 patients defined a 1.5 Mb segment on 7q11.21 as the major candidate locus. Screening of the positional candidate gene KCTD7 revealed homozygous missense mutations in two of the eight cases. Screening of KCTD7 in further 132 PME patients revealed four additional mutations (two missense, one in-frame deletion and one frameshift-causing) in five families. Eight patients presented with myoclonus and epilepsy and one with ataxia, the mean age of onset being 19 months. Within two years after onset progressive loss of mental and motor skills ensued leading to severe dementia and motor handicap. KCTD7 showed cytosolic localization and predominant neuronal expression, with widespread expression throughout the brain. None of three polypeptides carrying patient missense mutations affected the subcellular distribution of KCTD7.
Discussion
Our data confirm the causality of KCTD7 defects in PME, and imply that KCTD7 mutation screening should be considered in PME patients with onset around 2 years of age followed by rapid mental and motor deterioration.
Keywords: myoclonus, mutation, neurodegenerative disorders, mental retardation, pediatric epilepsy
INTRODUCTION
The progressive myoclonus epilepsies (PMEs) are a heterogeneous group of mainly autosomal recessively inherited symptomatic generalized epilepsies characterized by myoclonus, tonic-clonic seizures, and progressive neurological deterioration manifesting as psychomotor decline, cerebellar ataxia and/or early death.1, 2 Several clinically, pathologically and molecularly defined PME entities have been defined during the past two decades.2 Of these, for example Lafora disease (LD) and neuronal ceroid lipofuscinoses (NCLs) are associated with significant mental decline whereas in Unverricht-Lundborg disease mental decline is not prominent and the clinical features are dominated by myoclonus, epileptic seizures and ataxia.
In patients with disease onset in childhood, the most likely diagnosis is that of NCLs, which comprise the most common cause of neurodegeneration in children.3 The hallmark of NCLs is autofluorescent lipopigment of characteristic ultrastructural pathology that accumulates in cells, notably in neurons, of patients.3 Nine human NCL-causing genes have been identified thus far.4, 5 Despite progress in identification of NCL genes and those for other major forms of PME, a significant number of patients still remain without a molecular genetic diagnosis, among them a group of patients in which the manifestation is in early childhood.
The autosomal recessive mode of inheritance, characteristic of most PME entities,1 can be utilized to facilitate disease gene identification via the use of homozygosity mapping in consanguineous families. This powerful strategy requires only very small sample sizes to map the disease loci, which are identical by descent in the inbred families.6, 7 On the other hand, a pitfall of using inbred families is that the offspring can be homozygous over a significant portion of their genome, making it difficult to distinguish the true disease loci.8 Homozygosity mapping has been utilized successfully in PMEs to identify several novel genes such as PRICKLE1 for PME type 1B (EPM1B),9 KCTD7 for PME type 3 (EPM3)10 and MFSD8 for CLN7 disease, late infantile variant NCL.11
We here employed homozygosity mapping in genome-wide single nucleotide polymorphism (SNP) data in patients having onset of PME in early childhood to identify the underlying gene and to delineate the associated phenotype.
MATERIALS AND METHODS
Subjects and families
Eighteen unrelated Turkish patients with early childhood onset PME were genotyped in a genome-wide SNP scan. The onset was under 5 years of age. The patients presented with seizures and/or progressive mental and motor impairment leading to severe mental and motor handicap. Metabolic studies had shown normal results and the patients were negative for mutations in the eight known NCL genes. A panel of 108 unresolved Turkish PME patients and 22 NCL patients confirmed with electron microscopy (EM) analysis were screened for mutations in KCTD7. Clinical data were collected from hospital charts. All patients had been excluded for eight known NCL genes (CTSD, PPT1, TPP1, CLN3, CLN5, CLN6, MFSD8, and CLN8) either by sequencing or by enzyme activity analyses. One Pakistani family (Pak) was evaluated for 265 epilepsy-associated genes present on the NGS panel v1 (CeGaT GmbH, Tübingen, Germany). Parental samples in five families were available for segregation analysis of the identified mutation. The control panel consisted of 75 Turkish individuals. DNA was extracted from leukocytes using standard techniques. All samples were collected after an informed consent was obtained, according to the Declaration of Helsinki. An institutional review board of the Helsinki University Central Hospital approved the study.
Genome-wide SNP scan and homozygosity analysis
Genotyping was done using the Illumina Human610-Quad SNP chip (Illumina, San Diego, CA, USA) in the FIMM Technology Center, Helsinki, Finland. Only samples and markers with success rates >95% were included in the analysis, and minor allele frequency was limited to <1%.
Homozygosity analysis was performed using the PLINK analysis toolset v1.06.12 Genotype data was analyzed in windows of 500 kb, allowing for five missing calls and two heterozygotes per window per sample. Only regions of homozygosity greater than 1,000 kb in length and spanning over at least 100 consecutive SNPs were considered. Patients with overlapping homozygosity segments were grouped together, and regions showing most overlap between patients were designated for follow-up.
Mutation screening
The coding exons and exon-intron junctions of KCTD7 from the 1.5 Mb critical interval on 7q11.21 (chr7: 64.563.264-66.111.815; http://genome.ucsc.edu/cgi-bin/hgGateway) were screened for mutations from genomic DNA. Polymerase chain reaction (PCR) conditions and primer sequences are available from the authors upon request. The BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and an ABI 3730 DNA Analyzer (Applied Biosystems) were used to sequence the purified PCR products. Sequence analysis was done with Sequencher 4.8 program (Genes Codes Corporation, Ann Arbor, MI, USA).
For patient Pak4, analyzed with the CeGaT platform, genomic DNA was fragmented, adaptor ligated, hybridized to complementary RNA baits from 265 epilepsy associated genes (NGS epilepsy panel v1, CeGaT GmbH, Tübingen, Germany), and followed by massive parallel sequencing on the SOLiD 4 platform according to the manufacturer’s instructions (Life Technologies, CA, USA). Sequencing of patient Pak4 generated 17.5 million mappable 50 bp reads, leading to an average coverage of 292/bp. All reads were mapped to the human genome and analyzed using the Bioscope software (Applied Biosystems, Foster City, CA, USA). The mutation identified using this method was further validated by Sanger sequencing, in all family members.
The impact of the identified mutations on the proteins was assessed with PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/). Mutation descriptions have been checked using the Mutalyzer program (http://www.LOVD.nl/mutalyzer/). The control chromosomes were screened for the identified mutations from genomic DNA. The reference sequence used for the KCTD7 mutation nomenclature was NM_153033.4.
Expression plasmids and site-directed mutagenesis
The cDNA of the human KCTD7 (NM_153033.4) was PCR-amplified and cloned into the hemagglutinin (HA) tag containing pAHC expression vector, a derivative of the pCIneo expression vector (Promega) (kindly provided by Prof. T. Mäkelä, University of Helsinki, Finland). Two constructs carrying an HA tag N-terminally (HAKCTD7wt) and C-terminally (KCTD7HAwt) were generated. Three nucleotide changes (c.280C>T, c.343G>T, and c.818A>T) corresponding to patient missense mutations were introduced into the HAKCTD7wt construct using the QuickChange® Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and verified by sequencing. The resultant constructs were HAKCTD7p.R94W, HAKCTD7p.D115Y, and HAKCTD7p.N273I. Detailed methods on cell cultures, transfections and immunofluorescence analysis performed with these constructs can be found in the data supplements.
Immunohistochemistry
Sagittal paraffin sections were obtained from postnatal (P) day 5, 7, 10, 14 as well as 1, 2, and 4 month-old 4% PFA-perfused mice (SV129-J, Jackson Laboratories). Paraffin was removed by incubation in xylene and decreasing graded series of ethanol solutions. Antigens were exposed by incubating the sections in 10 mM citrate buffer pH 6.0 at 95°C for 10 min. Unspecific antibody binding was blocked by incubating the slides in 10% FCS in PBS for 1 h at RT. The primary and secondary antibodies were diluted in 1% FCS in PBS, applied on the sections and incubated at +4°C for 18 h and 1 h, respectively. The sections were mounted with Gel Mount (Sigma-Aldrich) and visualized using an Axioplan 2 microscope. The images were obtained with AxioVision 3.1 and processed with Adobe Photoshop CS4 software.
Experiments in animals were in compliance with national laws and policies and approved by the Animal Ethics Committee of the State Provincial Office of Southern Finland (decision #STH524A).
Western blotting
Mouse hippocampal and cerebellar neurons, as well as astrocytes, microglia, untransfected and transfected COS-1 cells were lysed 18 h post transfection in lysis buffer (0.5% NP40, 50mM Tris pH 8.0, 10% glycerol, 0.1 mM EDTA, 0.25M NaCl, 1 mM Na3VO4, 50 mM NaF, 10 mM DTT, 0.04% 25x PIC). The cell lysates were collected, left to rotate at +4°C for 5 min, centrifuged at 10000 rpm for 10 min at +4°C, and the supernatants were collected. Cerebella from P 5, P14, 1 month and 2 month-old mice (SV129-J) were dissected and homogenized in lysis buffer with a FastPrep (FP120, ThermoSavant). After a 30 min incubation at +4°C the cell lysates were collected, centrifuged at 13000 rpm for 15 min at +4°C, and the supernatants were collected. Protein concentrations were measured with the Bradford assay (BioRad) and equalized protein concentrations were run on 14% SDS-PAGE gels. The proteins were transferred onto PVDF membranes (Millipore) by blotting. The antigens were detected with enhanced chemiluminescence (ECL) Western blotting detection reagents (Amersham Biosciences).
RESULTS
Identification of disease-associated mutations in KCTD7
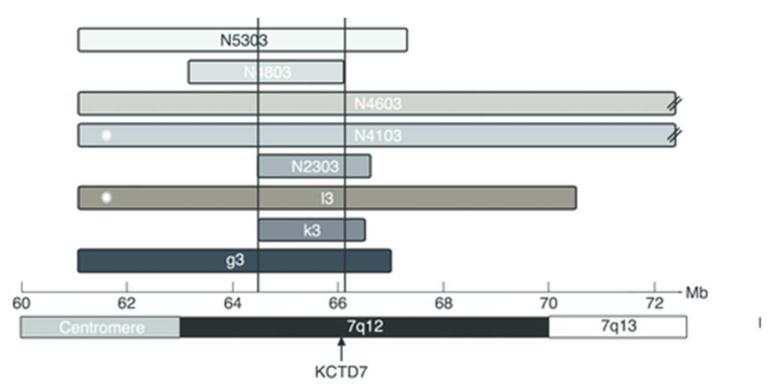
Analysis of the genome-wide SNP data for shared regions of homozygosity in 18 unrelated patients revealed a region on chromosome 7q11.21 as the strongest candidate locus. Eight patients shared homozygosity over a 1.5 Mb region extending between rs1812771 and rs4717338 (chr.7: 64.563.264-66.111.815), which contained 12 known genes (figure 1). The SNP haplotype was different in each of the eight patients, suggesting no common ancestry among them.
Figure 1. Results of homozygosity analysis at the candidate locus on 7q11.1-q13.
The x-axis shows the genomic position in Megabases (Mb) in NCBI build 37. The horizontal bars indicate the lengths and positions of the homozygous segments found in each of the eight patients. Asterisks at the left end of patients’ N4103 and L3 haplotype blocks indicate that these patients carry mutations in KCTD7. The segments for patients N4103 and N4603 extend beyond the illustrated region (designated by the two diagonal lines). The minimum overlap interval of 1.5 Mb is marked by the vertical black lines. The position of KCTD7 is indicated with an arrow.
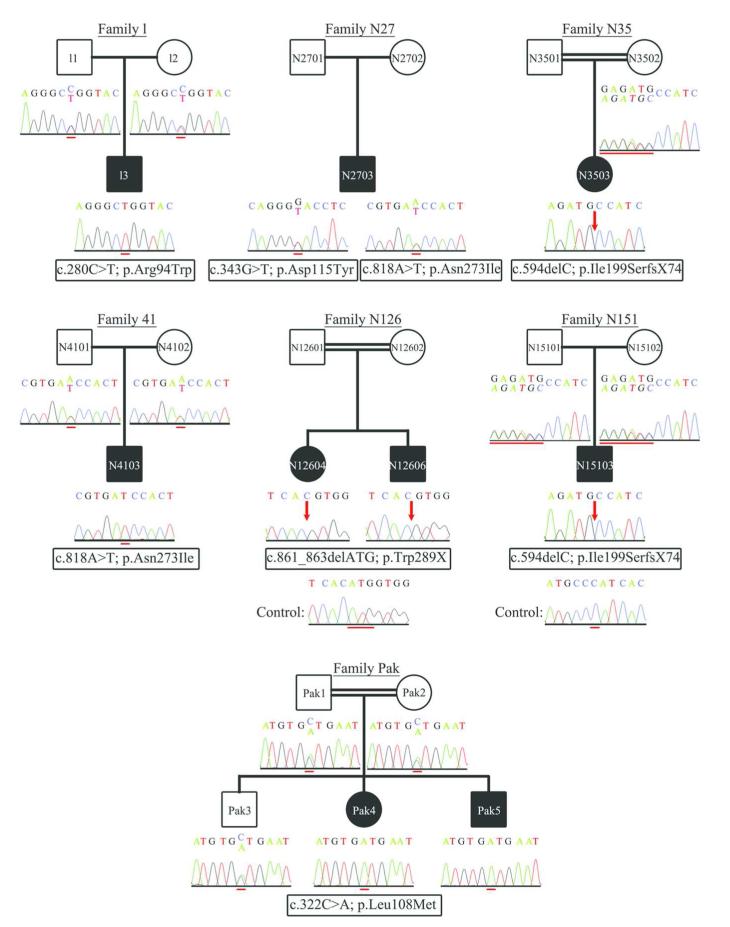
The KCTD7 gene was screened as the primary positional candidate due to its previous association to a PME phenotype in an inbred Moroccan family.10 Two out of the eight patients homozygous over the critical region harbored novel mutations in KCTD7 (figure 2). Screening of KCTD7 in the cohort of 108 additional Turkish PME patients and one Pakistani family revealed four additional novel mutations (figure 2). Of the identified six mutations four were missense, one was an in-frame deletion of a single amino acid and one resulted in a frameshift. A homozygous transition of C to T in exon 2 (c.280C>T; p.Arg94Trp) was identified in patient L3, and a homozygous transversion of A to T in exon 4 (c.818A>T; p.Asn273Ile) in patient N4103. Both p.Arg94Trp and p.Asn273Ile mutations affect highly conserved amino acid residues among vertebrates and were predicted to be probably damaging by PolyPhen-2. Patient N2703 was compound heterozygous for the mutations c.818A>T (p.Asn273Ile) and c.343G>T (p.Asp115Tyr) in exon 3. The asparagine at position 115 is not well conserved across vertebrates and the p.Asp115Tyr change was predicted to be benign by PolyPhen-2. The affected siblings Pak4 and Pak5 were homozygous for c.322C>A (p.Leu108Met) in exon 3 affecting a highly conserved residue and thus predicted to be probably damaging. Patients N3503 and N15103 were homozygous for c.594delC in exon 4, resulting in a frameshift, that introduces 74 novel amino acids before terminating the protein prematurely by 16 residues (p.Ile199SerfsX74). Finally, two affected siblings were homozygous for an in-frame deletion of three bases (c.861_863delATG), resulting in the deletion of the last amino acid residue (tryptophan) of KCTD7 (p.Trp289X). Parents from whom DNA was available were heterozygous carriers of the respective mutations (figure 2). None of the 22 NCL patients carried mutations in KCTD7. None of the identified changes were present in 150 Turkish control chromosomes or in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Figure 2. KCTD7 gene mutations.
The seven families in which mutations in KCTD7 have been identified are illustrated. For each family member the chromatogram showing the change identified is given below the corresponding pedigree symbol. For the deletion mutations identified in families N126 and N151 the control sequence is shown below the mutation sequence in the affected offspring (N12604, N12606 and N15103). A red line is used to highlight the mutations, except for mutations c.861_863delATG and c.594delC identified in families N126 and N151 respectively, where the red line shows the deleted bases in control sequence. At the mutation sites the reference nucleotides are given as superscript while the mutant nucleotides as subscript. The positions at which nucleotide deletions have been detected, are indicated by red arrows. All sequence chromatograms are shown in the forward orientation.
Phenotype associated with KCTD7 mutations
The patients’ clinical symptoms are summarized in table 1 and the EEG and MRI findings in table 2. More detailed clinical descriptions can be found in the data supplements.
Table 1.
Clinical symptoms and findings in KCTD7 mutation positive patients. Manifestation of a particular symptom in each patient is indicated with a plus (+).
| Patient code | Gender | Age of onset | Present age | Presenting symptom |
Seizure type | Mental regression |
Motor impairment |
Retinal findings | Scoliosis | Ataxia | Treatment | Pathology (tissue) |
Mutation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L3 | M | 1-2 y | 8 y | Seizures | Myoclonic Atonic |
+ | + | Normal | No | + | VPA, CLZ, TPM, LEV (No seizure control) |
EM: no storage material (skin) |
c.280C>T; p.Arg94Trp |
| N2703 | M | 1-2 y | 9.5 y | Seizures | Myoclonic GTCS Hypomotor |
+ | + | Normal | + | + | VPA, LMT, VGB, DZM, PH, LEV, CLZ, steroid, ACTH, PYRP (Seizures controlled) |
No Lafora bodies (skin); Normal (Muscle biopsy) |
c.343G>T; p.Asp115Tyr and c.818A>T; p.Asn273Ile |
| N3503 | F | 6-12 mo | 14 y | Seizures | Myoclonic | + | + | Normal | + | + | VPA, CLZ (Seizures controlled) |
Not performed | c.594delC; p.Ile199SerfsX74 |
| N4103 | M | 1-2 y | 8 y | Seizures | Myoclonic GTCS |
+ | + (never walked) |
Normal | + | Not present |
PH, VPA, CLZ, CLB, LEV (Seizures controlled partially) |
Not performed | c.818A>T; p.Asn273Ile |
| N12604 | F | >2 y | 8 y | Ataxia | Myoclonic GTCS |
+ | + | Normal | + | + | VPA, CLZ, LEV (Seizures controlled) |
Not performed | c.861_863delATG ; p.Trp289X |
| N12606 | M | 1-2 y | 3.5 y | Seizures | Myoclonic GTCS |
+ | + | Normal | + | + | VPA (Seizures controlled) |
Not performed | c.861_863delATG ; p.Trp289X |
| N15103 | M | 1-2 y | 4 y | Seizures | GTCS Myoclonic Hypomotor |
+ | + | Normal | No | + | VPA, LEV (Seizures controlled at 75%) |
EM: no storage material (skin) |
c.594delC; p.Ile199SerfsX74 |
| Pak4 | F | 1-2 y | 4 y 11 mo | Seizures (initially febrile) |
Myoclonic | + | + | Normal | No | + | VPA, CLB (Seizure control for 6 mo), ETX, LEV, LMT, PDN, RFM, STP (No seizure control) |
EM: no storage material (skin) |
c.322C>A; p.Leu108Met |
| Pak5 | M | 1-2 y | 3 y 3 mo |
Seizures | Myoclonic Atonic |
+ | + | Normal | No | + | VPA, CLB, ETX, LEV, LMT, PDN, RFM, STP, MSM (No seizure control) |
Not performed | c.322C>A; p.Leu108Met |
Abbreviations used: male (M), female (F), not available (n.a.), generalized tonic clonic seizures (GTCS), valproate (VPA), clonazepam (CLZ), topiramate (TPM), levetiracetam (LEV), lamotrigine (LMT), vigabatrin (VGB), diazepam (DZM), phenobarbital (PH), pyridoxal phosphate (PYRP), clobazam (CLB), ethosuximide (ETX), prednisolone (PDN), rufinamide (RFM), stiripentol (STP), mesuximide (MSM)
Table 2.
EEG and MRI findings in KCTD7 mutation positive patients. The time at which the EEG or MRI examination was performed is given in the table next to the corresponding findings.
| EEG | MRI | |||
|---|---|---|---|---|
| Patient | Time of examination |
Findings | Time of examination |
Findings |
| L3 | At onset | Spikes from posterior areas of both hemispheres. No high amplitude waves on photic stimulation |
At onset | Normal |
| N2703 | 5 years after onset |
Generalized sharp wave activity beginning from the left fronto-temporo-occipital and spreading to the whole hemisphere |
6 months after onset |
Ventricular dilatation |
| N3503 | At onset | Slow background activity. Epileptic discharges start from the left frontotemporal and right temporal regions |
3 years after onset |
Cerebral, cerebellar, and hippocampal atrophy |
| N4103 | 2 years after onset |
Multifocal spike wave activity that is prominent in the posterior cranial region |
2 years after onset |
Volume loss in bilateral posterior sylvian fissure and frontotemporal cerebral region. Small nonspecific focal lesion on the left frontal posterior white matter |
| N12604 | 5 years after onset |
Sharp spike activity on parieto-occipital and parieto-temporal lobes. Background activity is composed of irregular delta activity |
5 years after onset |
Hyper-intensity on posterior periventricular region on T2 weighted images |
| N12606 | At onset | Epileptiform activity starts from the fronto- central region of the right hemisphere |
At onset | Hyper-intensity on posterior periventricular region on T2 weighted images |
| N15103 | 2 years after onset |
Epileptiform activity in the left posterior hemisphere |
2.5 years after onset |
Frontotemporal cortical atrophy |
| Pak4 | 1 year after onset |
Sequences of rhythmic high amplitude delta waves with superimposed spikes predominant in posterior regions. No abnormal photic stimulation |
2 years after onset |
Discrete non-specific focal subcortical white matter lesions |
| Pak5 | 2 months after onset |
Sequences of rhythmic high amplitude delta waves with superimposed spikes predominant in posterior regions. No abnormal photic stimulation |
2 months after onset |
Normal |
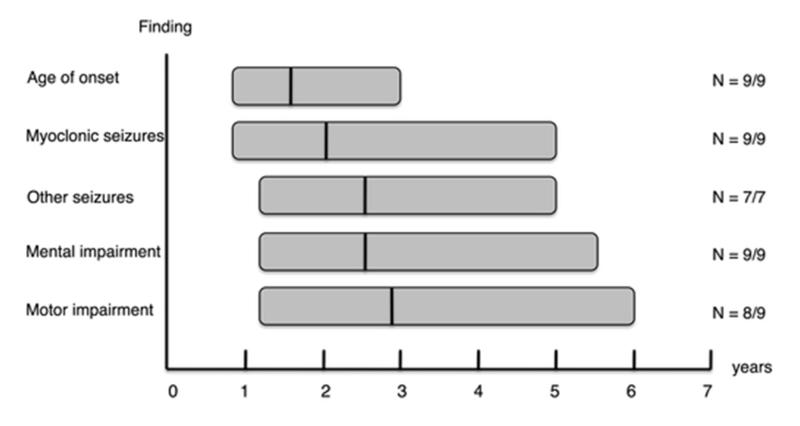
The mean age of presentation of the KCTD7 mutation positive patients was 19 months (range 10 months–3 years) (table 1; figure 3). The presenting symptoms in eight patients were myoclonic and/or tonic-clonic seizures and in one ataxia. Antiepileptic drug (AED) treatment resulted in complete or partial epilepsy control in the majority of patients (N=6/9). Three patients had intractable seizures (L3, Pak4, Pak5). In all patients but one more than one AED was used (table 1). Psychomotor decline became evident 0-22 months after onset of seizures and resulted in severe motor and mental retardation in all patients. Ataxia developed within 28 months after onset in all patients but N4103. Retinal findings were normal in all patients evaluated. Routine biochemical and pathological examinations performed were normal. No lysosomal storage material compatible with NCL was detected in EM analysis of skin biopsies obtained from three patients (table 1). MRI was normal near disease onset in two patients, with atrophic changes in one patient and non-specific focal lesions in the second present only in more advanced disease stages (table 2; supplementary figure S3). EEG revealed prominent epileptiform activity with predominance in the posterior region (table 2; supplementary figure S4). All patients are alive and their present age varies between 3.2-14 years.
Figure 3. Schematic representation of the disease course in the nine patients carrying KCTD7 mutations.
The x axis shows the age in years. The y axis shows the symptoms. Each bar represents one symptom. The length of each bar represents the age range between which the symptom develops. The black vertical lines on the bars show the mean age of onset of individual symptoms/findings. On the right side of the graph the number of patients (N) that showed the symptom is given.
Subcellular localization of wild-type and mutant KCTD7
Specificity of the KCTD7 antibody was confirmed in immunofluorescence microscopy, where the staining obtained with the rabbit anti-HA and the rabbit anti-KCTD7 antibodies showed full overlap (supplementary figure S1A-C) and in western blot analyses where the signals detected by both antibodies corresponded to the expected molecular masses (supplementary figure S1D, E).
A wide intracellular distribution of KCTD7 across the cell was detected in COS-1, BHK, and HeLa cells in which HAKCTD7wt was transiently overexpressed (figure 4A). The distribution of KCTD7 remained unaltered when constructs carrying changes representing patient mutations (HAKCTD7p.R94W, HAKCTD7p.D115Y, and HAKCTD7p.N273I) were transiently overexpressed (figure 4B, C, and data not shown). To test whether the diffuse pattern of KCTD7 across the cell was compatible with cytoplasmic or plasma membrane localization, saponin was used to remove the cytosol in BHK cells. Following this treatment a significant decrease in KCTD7 staining was observed, suggesting that KCTD7 is a cytosolic protein (figure 4A versus 4D). The strong nuclear staining (figure 4D, F) observed in the cells with removed cytosol reflects most probably unspecific staining that might have occurred from partial permeabilization of the nuclear membrane. No co-localization was observed with markers for endosomes, ER, Golgi, lysosomes, and cytoskeleton (figure 4I, L, and data not shown).
Figure 4. Intracellular distribution and expression of wild-type and mutant KCTD7.
(A-C) In BHK cells transfected with HAKCTD7wt, HAKCTD7p.R94W, and HAKCTD7p.D115Y the wild-type and mutant polypeptides showed a similar widespread distribution across the cell, compatible with cytosolic localization. (D-F) The signal obtained for KCTD7 when HA-tagged KCTD7 (D) constructs were co-transfected with the plasma membrane marker wtCD8 (E) in BHK cells and the cytosol was removed by saponin treatment is much weaker, suggesting that KCTD7 is a soluble cytoplasmic protein (D versus A). (D, F) The nuclear staining observed upon cytoplasm removal is considered to be unspecific and to have occurred through permeabilization of also the nuclear membrane. (G-L) Double-stainings in BHK cells for KCTD7 (G, J) and markers for endosomes (H), or ER (K) did not reveal co-localization. (I, L). The nucleus was stained blue with Hoechst. (M) In western blot analyses wild-type (HAKCTD7wt and KCTD7HAwt) and mutant KCTD7 (HAKCTD7p.R94W, HAKCTD7p.D115Y, and HAKCTD7p.N273I) was visualized as an apparently 37 kDa band of which 2 kDa correspond to the HA-tag. Comparison of the mutant constructs to the wild-type protein did not reveal differences in the levels of protein expression. A β–tubulin (load) was used to affirm that equal amounts of protein were loaded into each lane. The molecular weight of the wild-type and mutant KCTD7 proteins is given on the right. The scale bars correspond to 20 μm.
Endogenous KCTD7 was not detected in HeLa or COS1 cells (figure 4M, and data not shown). In COS1 cells transiently expressing HAKCTD7wt, the tagged protein was visualized as a sharp band of approximately 37 kDa. The protein expression level and molecular weight were not affected by the patient missense mutations, as determined by western blot analysis (figure 4M).
Expression of KCTD7 in mouse neuronal cultures and brain
Cultured mouse hippocampal neurons showed intense KCTD7 immunoreactivity in the cell soma (figure 5A), the neuritic varicosities along the developing neuronal extensions (figure 5B, C), and the neurite growth cones (figure 5D), but not in the nucleus. No co-localization of neuronal KCTD7 with markers for pre- or post-synaptic vesicles (figure 5B-F) or lysosomes (figure 5G) was observed.
Figure 5. Expression of endogenous KCTD7 in mouse embryonic neuronal cultures and brain tissue.
Endogenous KCTD7 was detected with the anti-KCTD7 antibody. (A-G) No co-localization was observed with either β–tubulin (A), pre- (B-D) or post-synaptic (E, F), or lysosomal markers (G). (C, D) Magnified images of the neurite varicosities (C, white arrows) and the tip of a growth cone (D, white arrows). (H-J) In brain sections from P5, P14 and 4 month-old mice, KCTD7 was prominently expressed in the cerebellar Purkinje cells. (H-J) The neuronal progenitor cells detected with Cdc47 (H), the astrocytes with GFAP (I), and some neuronal populations such as the PV-immunopositive interneurons of the cerebellar molecular layer (J), are not immunopositive for KCTD7. (H-J) KCTD7 resides in the soma of the neurons although expression in the neurites of Purkinje cells can also be seen (J). (H-J) White arrowheads point to Purkinje cells (H-J), and white arrows to cerebellar granule cells (H, I) or cerebellar interneurons (J). (K) Cell lysates were evaluated for expression of KCTD7 from embryonic hippocampal neurons, cerebellar neurons, astrocyte and microglia cell cultures, and cell lysates from homogenized cerebella of different postnatal time points (P5, P14, 1mo and 2mo). Endogenous KCTD7 is detected as a single band with a molecular weight of 35 kDa. KCTD7 expression is already seen in embryonic hippocampal as well as in postnatal cerebellar granular neurons (lanes 1-2). In contrast with the neuronal cells, KCTD7 does not seem to be expressed in the microglia (lane 3) or astrocytes (lane 4). The faint band detected in the astrocyte lysate is likely to be due to contamination of the astrocyte culture from neuronal cells. The KCTD7 expression levels in cerebellar lysates from P5, P14, 1mo, and 2mo-old mice remained unaltered across the evaluated time-points, suggesting constant and unaffected protein expression throughout the maturation (lanes 5-8). Immunostaining with β–actin was used as a control for protein loading. The molecular weight of KCTD7 is given on the left. The scale bar is corresponding to 20 μm.
Immunohistochemistry from mouse brain demonstrated that KCTD7 is widely expressed in post-mitotic neurons throughout the brain (figure 5H-J and supplementary figure S2). Prominent expression was for example observed in the cortical neurons (supplementary figure S2A-C), in the granular and pyramidal cell layers of the hippocampus (supplementary figure S2D-F), as well as in the cerebellar Purkinje cells (figure 5H-J) from P7 onwards. However, not all neuronal cell populations were immunopositive for KCTD7, exemplified by the lack of KCTD7 immunostaining in the PV-positive interneurons of the cerebellar molecular layer (figure 5J). Additionally, no obvious KCTD7 staining was seen in Cdc47 immunopositive neural progenitor cells (figure 5H) or GFAP-positive astrocytes (figure 5I).
Western blot analyses showed expression of KCTD7 in cultured embryonic and post-natal neurons but not in astrocytes or microglial cells (figure 5K, lanes 1-4) further supporting the neuron-specific expression of KCTD7. In mouse cerebellar lysates expression was constant from P5 to 2 months (figure 5K, lanes 5-8).
DISCUSSION
In this study we confirm that KCTD7 mutations cause an early childhood onset PME phenotype, delineate the resulting clinical phenotype, and provide a primary characterization of the defective protein. Six different mutations were identified in KCTD7 in six Turkish and one Pakistani family. Previously a nonsense mutation (p.Arg99X) in KCTD7 exon 2 had been reported to segregate with a PME phenotype in a consanguineous Moroccan family with three affected children.10 Detection in this study of a further seven KCTD7 positive families establishes the causal role of KCTD7 defects in PME patients with onset in early childhood.
All patients except one in our study presented at below two years of age with seizures, similar to the Moroccan patients in whom seizure onset was at 16-24 months of age.10 In one patient the presenting symptom at 3 years of age was ataxia, seizures developing only two years later. The seizures associated with KCTD7 mutations are most often myoclonic and tonic-clonic, but also atonic and/or hypomotor seizures are frequently reported.this study, 10 The epileptic phenotype was ameliorated in some of the patients, usually via a combination of two or more drugs, although this was not feasible in all patients. We did not notice improvement of the neurological condition with better epilepsy control as seen in the Moroccan patients.10 On the contrary, despite better seizure control in some patients, the disease progressed rapidly with progressive loss of mental and motor skills and development of ataxia ultimately leading to severe motor handicap and dementia. Lack of retinopathy and the characteristic storage materialthis study, 10 are features that distinguish the KCTD7-associated disorder in our patients from NCLs, the most common cause of neurodegeneration in children.
Of the mutations characterized here, four were missense, one created a frameshift and one deletes a single amino acid. Of the missense mutations, three were predicted to be damaging, while in one the resulting amino acid change (p.Asp115Tyr) was predicted to be benign. We, however, argue that also this mutation is likely to be disease-causing, since it is found in compound heterozygosity with a predicted damaging mutation and it was absent from the control chromosomes. The KCTD7-associated missense defects are likely to mediate their deleterious effect through disturbed functional properties, as none of the three tested missense mutations resulted in altered intracellular protein distribution or changes in the levels of protein expression. The frameshift-causing c.594delC, on the other hand, is expected to result in an mRNA that will be targeted for nonsense-mediated decay13 and thus also to result in loss of function.
The c.861_863delATG (p.Trp289X) mutation is predicted to remove only the last tryptophan residue prior to the stop codon from isoform 1 of KCTD7, the resulting protein product predicted to be identical to KCTD7 isoform 2 (UniProt accession number ID: Q96MP8-2). In KCTD7 isoform 2 the effect of the c.861_863delATG mutation is either deletion of the last amino acid or altered splicing of the transcript. Segregation of c.861_863delATG with the disease phenotype, absence of it in controls and the consequent predicted total lack of KCTD7 isoform 1 argue in favor of a disease-causing role of this variant. Interestingly, the c.861_863delATG mutation was identified in two affected sibs, one of them being the only KCTD7 mutation positive patient with onset remarkably later and the presenting symptom being ataxia, and not seizures. This implies that additional genetic factors rather than less deleterious functional properties of the c.861_863delATG mutation per se, are likely to account for this intra-familial variation in phenotype.
Interestingly, only two of the eight patients homozygous over the locus 7q11.21 carried mutations in KCTD7. It is possible that some of these patients carry KCTD7 mutations that escaped identification, such as copy number variations, as only exons and exon-intron boundaries were screened from genomic DNA. The 7q11 region has previously been shown to be prone to chromosomal rearrangements such as duplications and deletions in other neurodevelopmental disorders, like the Williams-Beuren syndrome.14 Moreover, it is possible that the remaining patients have mutations in another gene that resides in close proximity to KCTD7. Finally, as the clinical presentation was rather unspecific and as five of the six remaining patients were from inbred families having more than 7% of their genome homozygous, the observed homozygosity over the 7q locus in these patients may have occurred by chance with the homozygous region not indicating the true disease-associated locus in the family.
The spatiotemporal expression of KCTD7 in the brain tissue, its ubiquitous expression in cultured neurons as well as the absence of KCTD7 expression from microglia and astrocytes imply that the function of KCTD7 is critical for a large subset of neuronal populations. Compatible with recently reported findings15 we found KCTD7 to be localized in the cytoplasm in both neurons and transfected non-neuronal cells, with no overlap with any of the organelle markers tested. The cytosolic distribution was further supported by the dramatic loss of KCTD7 immunoreactivity upon cytosol removal from BHK cells. In the cultured neurons the diffuse KCTD7 staining was found not to be restricted only to the cell soma, but to extend to the neurites and even the growth cones.
The molecular function of KCTD7 remains unknown. The sequence homology of the KCTD7 N-terminal domain to the T1 domain of voltage-gated channels makes it tempting to speculate that KCTD7 would be involved in ion channel function, as most epilepsy-associated genes are.16 Recently overexpression of KCTD7 was shown to lead to hyperpolarization of neuronal resting membrane potential and to decreased neuronal excitability in in vitro patch clamp experiments.15 Loss of KCTD7 function was predicted to be associated with depolarized resting membrane potential and increased excitability with consequent susceptibility to epileptic seizures. However, a direct association of KCTD7 to ion channels on the plasma membrane is not supported by its diffuse cytosolic localization. Indeed, KCTD7 was suggested to comprise a part of the E3 ubiquitin ligase multi-protein complex, via its interaction with Cullin-3,15 its effect on ion channel function thus being possibly indirectly mediated with an as yet unknown mechanism. Similarly to KCTD7, another KCTD member, KCTD5 has been shown to interact with Cullin-3.17 Furthermore KCTD5 also localizes to cytoplasm, and has been explicitly shown not to participate in the formation and function of potassium channels (Kv4.2, Kv3.4, Kv2.1, or Kv1.2).17 Taken together these data imply that KCTD5 and KCTD7 could affect the neuronal electrophysiology indirectly via mediating the ubiquitination and proteasome-degradation of proteins required for the correct ion channel function, or through other mechanisms that await characterization.15, 17 Also KCTD8, KCTD12 and KCTD16 have been associated with neuronal function, acting as auxiliary subunits of the GABAB receptors.18, 19 Other KCTD members, however, have been associated with mainly non-neurological phenotypes.20-23
This study consolidates KCTD7 as a gene underlying early childhood onset and rapidly progressing PME leading to severe motor handicap and dementia. The possibility of a KCTD7 defect should be considered in such patients in the absence of any specific metabolic or pathologic hallmarks. Although KCTD7 mutations have so far been limited to Moroccan, Pakistani and Turkish patients, it is anticipated that the geographic distribution of KCTD7 defects will expand once molecular diagnosis of this disorder is more widely carried out.
Supplementary Material
Supplementary figure S1. Specificity of the antibody recognizing KCTD7.
Supplementary figure S2. Spatiotemporal expression of KCTD7 in the postnatal mouse brain.
Supplementary figure S3. Cranial magnetic resonance imaging (MRI) samples of patients N15103 and Pak4.
Supplementary figure S4. Electroencephalographic recording of patient Pak4.
ACKNOWLEDGMENTS
We would like to thank the families that have contributed samples for the purposes of this study. We would also like to thank Saara Tegelberg for providing the cerebellar lysates, Inken Körber for the lysates from astrocyte and microglial cultures and Paula Hakala, Hanna Hellgren, and Svetlana Zueva for the technical assistance in the immunohistochemistry, cloning experiments, and preparation for EM samples, respectively.
FUNDING This work was supported by the Center of Excellence in Complex Disease Genetics of the Academy of Finland, the Folkhälsan Research Foundation, the Epilepsy Research Foundation, the Biomedicum Helsinki Foundation and the Emil Aaltonen Foundation. VA was supported by the Häme Student Foundation.
Footnotes
COMPETING INTERESTS None
PATIENT CONSENT Consents to carry out the study were received from all patients. Separate consents were not received though, as the data presented preserve the patient anonymity.
ETHICS APPROVAL This study was approved by an institutional review board of the Helsinki University Central Hospital.
REFERENCES
- 1.Berkovic SF, Andermann F, Carpenter S, Wolfe LS. Progressive myoclonus epilepsies: specific causes and diagnosis. N Engl J Med. 1986 Jul 31;315(5):296–305. doi: 10.1056/NEJM198607313150506. [DOI] [PubMed] [Google Scholar]
- 2.Ramachandran N, Girard JM, Turnbull J, Minassian BA. The autosomal recessively inherited progressive myoclonus epilepsies and their genes. Epilepsia. 2009 May;50(Suppl 5):29–36. doi: 10.1111/j.1528-1167.2009.02117.x. [DOI] [PubMed] [Google Scholar]
- 3.Haltia M. The neuronal ceroid-lipofuscinoses. J Neuropathol Exp Neurol. 2003 Jan;62(1):1–13. doi: 10.1093/jnen/62.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Kousi M, Lehesjoki AE, Mole SE. Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum Mutat. 2011 Oct 11;33(1):42–63. doi: 10.1002/humu.21624. [DOI] [PubMed] [Google Scholar]
- 5.Noskova L, Stranecky V, Hartmannova H, Pristoupilova A, Baresova V, Ivanek R, Hulkova H, Jahnova H, van der Zee J, Staropoli JF, Sims KB, Tyynelä J, Van Broeckhoven C, Nijssen PC, Mole SE, Elleder M, Kmoch S. Mutations in DNAJC5, Encoding Cysteine-String Protein Alpha, Cause Autosomal-Dominant Adult-Onset Neuronal Ceroid Lipofuscinosis. Am J Hum Genet. 2011 Aug 12;89(2):241–252. doi: 10.1016/j.ajhg.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander ES, Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987 Jun 19;236(4808):1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 7.Kruglyak L, Daly MJ, Lander ES. Rapid multipoint linkage analysis of recessive traits in nuclear families, including homozygosity mapping. Am J Hum Genet. 1995 Feb;56(2):519–527. [PMC free article] [PubMed] [Google Scholar]
- 8.Woods CG, Cox J, Springell K, Hampshire DJ, Mohamed MD, McKibbin M, Stern R, Raymond FL, Sandford R, Malik Sharif S, Karbani G, Ahmed M, Bond J, Clayton D, Inglehearn CF. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am J Hum Genet. 2006 May;78(5):889–896. doi: 10.1086/503875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bassuk AG, Wallace RH, Buhr A, Buller AR, Afawi Z, Shimojo M, Miyata S, Chen S, Gonzalez-Alegre P, Griesbach HL, Wu S, Nashelsky M, Vladar EK, Antic D, Ferguson PJ, Cirak S, Voit T, Scott MP, Axelrod JD, Gurnett C, Daoud AS, Kivity S, Neufeld MY, Mazarib A, Straussberg R, Walid S, Korczyn AD, Slusarski DC, Berkovic SF, El-Shanti HI. A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy-ataxia syndrome. Am J Hum Genet. 2008 Nov;83(5):572–581. doi: 10.1016/j.ajhg.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Bogaert P, Azizieh R, Desir J, Aeby A, De Meirleir L, Laes JF, Christiaens F, Abramowicz MJ. Mutation of a potassium channel-related gene in progressive myoclonic epilepsy. Ann Neurol. 2007 Jun;61(6):579–586. doi: 10.1002/ana.21121. [DOI] [PubMed] [Google Scholar]
- 11.Siintola E, Topcu M, Aula N, Lohi H, Minassian BA, Paterson AD, Liu XQ, Wilson C, Lahtinen U, Anttonen AK, Lehesjoki AE. The novel neuronal ceroid lipofuscinosis gene MFSD8 encodes a putative lysosomal transporter. Am J Hum Genet. 2007 Jul;81(1):136–146. doi: 10.1086/518902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999 Feb 5;96(3):307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 14.Merla G, Brunetti-Pierri N, Micale L, Fusco C. Copy number variants at Williams-Beuren syndrome 7q11.23 region. Hum Genet. 2010 Jul;128(1):3–26. doi: 10.1007/s00439-010-0827-2. [DOI] [PubMed] [Google Scholar]
- 15.Azizieh R, Orduz D, Van Bogaert P, Bouschet T, Rodriguez W, Schiffmann SN, Pirson I, Abramowicz MJ. Progressive myoclonic epilepsy-associated gene KCTD7 is a regulator of potassium conductance in neurons. Mol Neurobiol. 2011 Aug;44(1):111–121. doi: 10.1007/s12035-011-8194-0. [DOI] [PubMed] [Google Scholar]
- 16.Baulac S, Baulac M. Advances on the genetics of Mendelian idiopathic epilepsies. Clin Lab Med. 2010 Dec;30(4):911–929. doi: 10.1016/j.cll.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Dementieva IS, Tereshko V, McCrossan ZA, Solomaha E, Araki D, Xu C, Grigorieff N, Goldstein SA. Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol. 2009 Mar 20;387(1):175–191. doi: 10.1016/j.jmb.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwenk J, Metz M, Zolles G, Turecek R, Fritzius T, Bildl W, Tarusawa E, Kulik A, Unger A, Ivankova K, Seddik R, Tiao JY, Rajalu M, Trojanova J, Rohde V, Gassmann M, Schulte U, Fakler B, Bettler B. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010 May 13;465(7295):231–235. doi: 10.1038/nature08964. [DOI] [PubMed] [Google Scholar]
- 19.Bartoi T, Rigbolt KT, Du D, Kohr G, Blagoev B, Kornau HC. GABAB receptor constituents revealed by tandem affinity purification from transgenic mice. J Biol Chem. 2010 Jul 2;285(27):20625–20633. doi: 10.1074/jbc.M109.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elks CE, Loos RJ, Sharp SJ, Langenberg C, Ring SM, Timpson NJ, Ness AR, Davey Smith G, Dunger DB, Wareham NJ, Ong KK. Genetic markers of adult obesity risk are associated with greater early infancy weight gain and growth. PLoS Med. 2010 May 25;7(5):e1000284. doi: 10.1371/journal.pmed.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Correale S, Pirone L, Di Marcotullio L, De Smaele E, Greco A, Mazza D, Moretti M, Alterio V, Vitagliano L, Di Gaetano S, Gulino A, Pedone EM. Molecular organization of the cullin E3 ligase adaptor KCTD11. Biochimie. 2011 Apr;93(4):715–724. doi: 10.1016/j.biochi.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Gallo R, Zazzeroni F, Alesse E, Mincione C, Borello U, Buanne P, D’Eugenio R, Mackay AR, Argenti B, Gradini R, Russo MA, Maroder M, Cossu G, Frati L, Screpanti I, Gulino A. REN: a novel, developmentally regulated gene that promotes neural cell differentiation. J Cell Biol. 2002 Aug 19;158(4):731–740. doi: 10.1083/jcb.200202024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argenti B, Gallo R, Di Marcotullio L, Ferretti E, Napolitano M, Canterini S, De Smaele E, Greco A, Fiorenza MT, Maroder M, Screpanti I, Alesse E, Gulino A. Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J Neurosci. 2005 Sep 7;25(36):8338–8346. doi: 10.1523/JNEUROSCI.2438-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure S1. Specificity of the antibody recognizing KCTD7.
Supplementary figure S2. Spatiotemporal expression of KCTD7 in the postnatal mouse brain.
Supplementary figure S3. Cranial magnetic resonance imaging (MRI) samples of patients N15103 and Pak4.
Supplementary figure S4. Electroencephalographic recording of patient Pak4.