Abstract
The etiology and pathogenesis of ocular adnexal extranodal marginal zone lymphoma (OAEMZL) are still unknown and the association with Chlamydophila psittaci (C. psittaci) has been shown in only some geographic regions. Herein we comprehensively examined the frequency of chromosomal translocations as well as CARD11, MYD88 (L265P) and A20 mutations /deletions in 45 C. psittaci negative OAEMZLs. t(14;18)(q32;q21) IGH-MALT1 and t(11;18)(q21;q21) API2-MALT1 were not detected in any of the analyzed tumors while 3 tumors harbored IGH translocations to an unidentified partner. CARD11 mutations were not found in all the analyzed tumors while MYD88 L265P mutation was detected in 3 (6.7%) tumors. A20 mutations and deletions were each detected in 7(15.6%) and 6(13.3%) of the tumors, respectively. Therefore, the observed genetic aberrations could account for the activation of NF-kB signaling pathway in only a minority of the cases. Further studies are needed to identify the molecular mechanisms underlying the pathogenesis of OAEMZL.
Keywords: MALT lymphoma, ocular adnexa, translocations, mutations, NF-kB
Extranodal marginal zone lymphomas (also referred as mucosa-associated lymphoid tissue (MALT) lymphomas) are the third most common subtype of B-cell non-Hodgkin’s lymphomas (NHL)[1]. The majority of MALT lymphomas occur in the stomach, but they may also arise in additional extranodal sites devoid of significant lymphoid tissue such as the intestine, thyroid and salivary glands, lungs, liver, breast, skin and the ocular adnexa (lacrimal gland, orbit, conjunctiva and eyelid). While extranodal MALT lymphomas from different sites of origin are commonly characterized by an indolent clinical course and often remain localized for many years, they display variations in morphologic features, cytogenetic abnormalities and associations with underlying infections and inflammatory disorders[2–8]. These observations suggest that the process of lymphomagenesis may vary depending on differences in the anatomical site of origin of these tumors. Indeed, the underlying inflammatory disorders leading to lymphomagenesis vary widely between the distinct anatomic sites, being attributed to chronic Helicobacter pylori infection in the stomach[6], Berrelia Burgdoferi infection in the skin[7], Campylobacter Jejuni infection in the intestine[8] and Hashimoto’s thyroiditis[9] and Sjogren’s syndrome[10] in the thyroid and salivary glands, respectively. Furthermore, the frequency of chromosomal translocations t(11;18)(q21;q21)/API2-MALT1, t(1;14)(p22;q32)/BCL10-IGH, t(14;18)(q32;q21)/IGH-MALT1 and t(3;14)(p13;q32)/FOXP1-IGH, implicated in the pathogenesis of extranodal MALT lymphomas, vary markedly between different anatomic locations[2, 3, 11], further pointing to possible differences in the mechanism of lymphomagenesis.
While the pathogenesis of gastric MALT lymphoma was extensively studied, the etiology and pathogenesis of ocular adnexal extranodal marginal zone lymphomas (OAEMZLs), the second most common extranodal MALT lymphoma, are still controversial[12]. Chlamydophila psittaci (C. psittaci) infection has been implicated in the development of OAEMZLs in only some geographic regions, while no association with bacterial infections was found in most studies originating from North American institutions[4, 13–17]. Trisomies of chromosomes 3 and 18 have been detected by most investigators in OAEMZL tumors at frequencies ranging from 16–38%[2, 4, 18]. However, recurrent chromosomal translocations characteristic of MALT lymphomas were analyzed only in a small number of studies from different geographic regions and led to highly variable results. For example, t(11;18)(q21;q21)/API2-MALT1 was reported to be detected in between 0 to 16% of OAEMZL tumors[2, 3, 19, 20]. Whether the marked differences in the frequency of chromosomal translocations might be related to geographic variability or association with C. psittaci was not examined. Moreover, concomitant and comprehensive analyses of translocations, trisomies and mutations affecting genes reported to deregulate the nuclear factor (NF)-kB pathway, known to be implicated in MALT lymphoma pathogenesis in other anatomical sites[21], was not performed in large cohorts of OAEMZL patients. Consequently, herein we sought to comprehensively examine the frequency of chromosomal translocations, CARD11, A20 (also called TNFAIP3) and MYD88 (L265P) mutations and A20 deletions in a large cohort of C. psittaci negative OAEMZLs.
Materials and methods
Patients and OAEMZL tumors
A total of 45 OAEMZL (median age 64 years, range 35–92; 23 females and 22 males) originating in the orbit (21), conjunctiva (18) and lacrimal gland (6) were used for analysis. All OAEMZL tumors were classified according to the WHO 2008 classification[1] on the basis of the morphologic features observed on routinely prepared hematoxylin and eosin–stained slides of formalin-fixed, paraffin-embedded tissues along with immunophenotypic (CD20+, cyclin D1-, CD5-, CD10- and BCL6-) and genotypic results. Flow cytometry immunophenotyping was performed in most cases. All cases were reviewed by two or more certified hematopathologists that agreed on the final diagnosis. Morphologic criteria for diagnosis included the finding of significant cytologic atypia and / or monocytoid features with an abnormally expanded CD20+ B cell population which lacked CD5, CD10 and / or BCL6 and cyclinD1. The interpretation of lymphoplasmacytic lymphoma was excluded based on the lack of the classically associated plasmacytoid lymphocytic population in combination with clinical findings. Other low grade lymphomas were eliminated based on cytologic features and immunoprofile, as described above. Clonality was confirmed by either Southern blot analysis or polymerase chain reaction (PCR) for immunoglobulin heavy or light chains in accordance with the BIOMED-2 recommendations[22]. Furthermore, immunoglobulin heavy chain clonality was also independently demonstrated by polymerase chain reactions using family specific leader primers followed by direct sequencing or cloning and sequencing, as previously reported by us[23]. This study was approved by the University of Miami Institutional Review Board (IRB) and written informed consent was obtained according to the approved protocol.
DNA extraction
DNA was previously extracted from a total of 65 fresh patient biopsy samples at the time of OAEMZL diagnosis (between 1991 and 2011) using a commercially available kit (QIAamp; Qiagen, Valencia, CA, USA), as previously reported by us[15, 23]. DNA from 45 tumor biopsies was available for the analyses performed in this study. No differences were present in the clinical characteristics and tumor locations between patients whose DNA was used in the analyses presented herein and those whose DNA was exhausted in our previous studies (not shown).
In addition, DNA was also extracted from peripheral blood mononuclear cells (PBMC) from three healthy volunteers (1 woman and 2 men), OCI-LY8 and Jeko-1 lymphoma cell lines using QIAamp DNA Blood Midi Kit (Qiagen, Valencia, CA) and quantified by NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific Inc). The cell lines were maintained in RPMI1640 medium with 10% fetal calf serum, penicillin (50 U/mL), streptomycin (50 g/mL) at 37°C in 5% of carbon dioxide.
Detection of C. psittaci DNA
Touchdown enzyme time-release polymerase chain reaction (PCR) for detection of C. psittaci DNA was performed as previously reported by us[15]. Blank reactions were always run concomitantly with the DNA from patients’ specimens to monitor for possible contamination of PCR reagents and to rule out false-positive results. All reactions were repeated twice.
A20, MYD88 and CARD11 mutation analysis
All the A20 coding exons were amplified by PCR using the primers and conditions reported by Novak et al[24]as follows: A20-E2-F, 5′-CCGGGAGTAGAGGTGCTAA-3′; A20-E2-R, 5′-GTCTGCTATTATCACATACCCC-3′; A20-E3-F, 5′-TCAGTTTGCCCTTGACTAGGA-3′; A20-E3-R, 5′-TGAGTCCCACTGGAGGTTTC-3′; A20-E4-F, 5′-CTCCCCAACTTTTGAGTTTGC-3′; A20-E5-R, 5′-AACCAAGCAAGTCACAGAACA-3′; A20-E6-F, 5′-CACCTCCAGGCTGGTTAATG-3′; A20-E6-R, 5′-TGTTTTGATTTGAAACCCAAGT-3′; A20-E7-F, 5′-GTTGCGTGAAAAGTGTGAGC-3′; A20-E7-R, 5′-CAGTGCCTTTTGCCTCCAT-3′; A20-E8-F, 5′-GATTGGTAAAGCCAAAGATGT-3′; A20-E8-R, 5′-GGAGGTAGCATTTCGGACC-3′; A20-E9-F, 5′-GCTTGGCGGTTTTCCTCAG-3′; A20-E9-R, 5′-CTTTGCTTTCTAAGGCCACCT-3′. Exon 5 of the MYD88 gene harboring the most commonly reported L265P mutation observed in diffuse large B cell and MALT lymphomas was amplified with the following primers: MYD88-Seq-E5F, 5′-GTTGTTAACCCTGGGGTTGAAG-3′; MYD88-Full-R, 5′-GCAGAAGTACATGGACAGGCAGACAGATAC-3′. The coiled-coil domain of CARD11 gene, harboring all the observed mutations in diffuse large B cell lymphomas was amplified using the following specific primers: CARD11-E5-1-F, 5′-GTGCCCCCTCTC CACAGT-3′; CARD11-E5-1-R, 5′-AGTACCGCTCCTGGAAGGTT-3′; CARD11-E5-2-F, 5′-GAAGAAGCAGATG- ACGCTGA-3′; CARD11-E5-2-R, 5′-GTCACCCTGGCGGAGTAG-3′; CARD11-E6-F, 5′-CACCCTTGGGGTATTTCAGA-3′; CARD11-E6-R, 5′-CAGGCCCTCACCTGGATG-3′; CARD11-E7-F, 5′-CCTGACCCTCTGAAACCTCCT CARD11-E7-R, 5′-GCGATCCCCACTCCCAC-3′; CARD11-E8-F, 5′-TCGATGCGCATATTGATTTC-3′; CARD11-E8-R, 5′-CTGCAGGTGGTGCCTGTA-3′; CARD11-E9-F, 5′-CCCAAAGCAG CCTTCGTC-3′; CARD11-E9-R, 5′-CCTGGTCCAGGTTGTTGCTGTCC3′. The integrity of DNA was verified by actin PCR amplification using specific primers yielding a 597 base pair (bp) amplicon as described previously[15, 23].
A total of 10–200 ng of DNA were amplified by GoTaq Green Master Mix (Promega, Madison, WI) in a final volume of 50 μl containing 10 pmol of specific 5′ primer and 10 pmol of antisense primer. PCR conditions for each gene and primer pair are shown in Supplementary Table 1. For each PCR, a control with no added template was used to check for contamination. PCR products (5μl) were analyzed by electrophoresis on 1–1.5% agarose gels stained with ethidium bromide. PCR products were purified using the QIAquick PCR Purification Kit(Qiagen, Valencia, CA, USA) and sequenced directly from both strands using the ABI Prism Big Dye Terminator Kit (Perkin Elmer, Foster City, CA) on a373 automatic DNA sequencer (Applied Biosystems, Foster City, CA), as recommended by the manufacturer. Sequences were compared with corresponding germ-line sequences (A20 – NM006290.2, MYD88-NM_002468.4, CARD11-NM_032415.3). All mutations were confirmed on independent PCR products and germ-line polymorphisms, including changes listed in the NCBI SNP database were excluded.
A20 copy number genotyping
A20 gene copy number variation was analyzed by TaqMan Copy Number Assay (assay ID Hs06775497_cn for Exon2 and Hs01790322_cn for Exon9; Applied Biosystems; Supplementary Table 2). TaqMan copy number assay amplifies the A20 gene and a reference gene (the RNase P H1 RNA gene) known to exist in two copies in a diploid genome simultaneously in a duplex real-time PCR. Genomic DNA from 3 healthy volunteers (1 woman and 2 men) was used as a calibrator. DNA from OCI-LY8, Jeko-1 lymphoma cell lines, known to harbor A20 monoallelic deletions, was selected as positive controls. Standard curves were generated using twofold serial dilutions of healthy adult genomic DNA to determine the real-time PCR amplification efficiency for each assay.
DNA concentrations were equalized before copy number genotyping of candidate genes. PCR was performed in 384-well plates using a PCR system (7900 Real-Time PCR System; Applied Biosystems). Thermal cycling conditions were described in the manufacturer’s instructions. Data were collected (Absolute Quantification Method, 7900 System SDS Software version 2.0; Applied Biosystems), and samples were assayed using triplicate wells for each gene of interest. Copy numbers were estimated using the ΔΔCt relative quantification method (CopyCaller Software version 2.0; Applied Biosystems). The copy number assays of deletion cases were repeated 3 times to confirm the reproducibility. Results of comparative genomic hybridization performed on 7 cases and previously reported by us were also reviewed.
FISH studies
Slides prepared from formalin-fixed paraffin-embedded tissues with three micron-thick sections were available for 25 OAEMZL tumors. FISH was performed according to protocols recommended by the manufacturer in a CLIA certified cyogenetics laboratory. The major steps consisted of slide deparaffinization, codenaturation of DNA probes and tissue materials on slides, hybridization, post hybridization washes and counterstaining with DAPI. Hybridization signals were analyzed with a fluorescence microscope equipped with a Cytovision system for image capturing and storage (Applied Imaging Corp, USA). All procedures were validated according the guidelines of College of American Pathologists (CAP), with cutoff values established for each type of probe used. Dual color fusion probes for t(14;18)(q32;q21) IGH-MALT1 and t(11;18)(q21;q21) API2-MALT1 and dual color break apart probes for MALT1, BCL6 and CEP18 (chromosome 18 centromere) were used for all the analyzed tumors. A dual color break-apart probe for IGH was used for 27 tumors. In addition to specific chromosome translocations or gene rearrangements, these probes also detect copy number variations caused by numerical or structural chromosomal abnormalities. For each probe, hybridization signals were counted in 200 cells and described using the International System for Human Cytogenetic Nomenclature(ISCN 2009)[25].
Long-distance inverse PCR
The long-distance inverse PCR (LDI-PCR) for the joining and switch region was done as described previously [26, 27]. Briefly, genomic DNA was digested with restriction enzymes (BglII, HindIII, PstI, and XbaI) overnight at 37°C. After column purification (Qiagen), 0.1–0.5 μg of digested DNA was ligated at 16°C overnight with DNA Ligation Kit (Takara). After purification, the ligated DNA was directly amplified in nested PCRs. PCR products were run on agarose gels. Bands were excised from the gels, column purified (Qiagen) and directly sequenced from both strands using a 373 automatic DNA sequencer (Applied Biosystems). Sequences were compared with the Genbank database using BLAST program. DNA obtained from JEKO-1 mantle cell lymphoma cell line harboring t(11;14)(q13;q32) and two cases of follicular lymphoma harboring t(14;18)(q32;q21) were used as positive control to establish the long-distance inverse PCR methodology in our laboratory.
Results
MYD88 and CARD11 mutations and A20 mutations and deletions
A20 mutations and deletions were previously reported in MALT lymphomas as well as in small cohorts of OAEMZL patients without specific reference to C. psittaci infection[18, 28–31]. Mutations of CARD11 and MYD88 were reported in DLBCL and MYD88 mutations were also detected in 9% of gastric MALT but were not comprehensively analyzed in OAEMZL[32, 33], except a recent study published while our study was prepared for publication[34]. In this study MYD88 and CARD11 mutations were detected in 6(5.7%) and 1 (0.9%) of 105 OAEMZL patients without specific reference to C. psittaci infection[34]. Herein, we comprehensively analyzed CARD11 and MYD88 mutations and A20 mutations and deletions in 45 C. psittaci negative OAEMZL tumors. CARD11 coiled-coil domain mutations were not found in any of the analyzed tumors. The MYD88 L265P mutation was detected in 3 (6.7%) tumors. A total of 9 A20 mutations were discovered in 7 (15.6%) tumors. One tumor harbored 3 distinct mutations (F149C; 1bp deletion in exon 3 and 2bp deletion in exon 5). Among the 9 detected A20 mutations, the majority (89%) would produce truncated proteins due to out-of-frame insertion/deletion (7), while one of these bp deletions was located at a known splice site. Only 2 missense mutations were observed, including one in a tumor in which 2 concomitant deletions were also present (as described above). Overall the detected mutations (Supplementary Figures 1 and 2) were of similar nature to those previously reported in other tumors and most probably would impair protein function[28].
A20 heterozygous deletions were detected in 6 (13.3%) tumors (Supplementary Figure 3). There was no association between A20 mutations and heterozygous deletion in any of the analyzed tumors. A total of 13 (28.9%) OAEMZL tumors harbored either A20 mutations or heterozygous deletions. None of the tumors concomitantly harbored A20 mutation/deletion and a MYD88 L265P mutation. Overall, we detected MYD88 and A20 mutations / deletions that might deregulate the NF-kB signaling pathway in 16 (35.6%) OAEMZLs (Supplementary Table 3).
Interphase FISH analysis
A total of 25 OAEMZL tumors with available material were analyzed by interphase FISH using probes as described in the Materials and Methods. Chromosomal alterations were detected in 7 tumors but t(14;18)(q32;q21) IGH-MALT1 and t(11;18)(q21;q21) API2-MALT1 were not detected in any of the analyzed tumors. A total of 2 tumors harbored an additional copy of BCL6, most probably due to trisomy 3 in 38 and 50% of cells, respectively. The following aberrations were each detected in one tumor in 38 and 77 % of analyzed cells: an additional copy of the IGH gene and extra copies of both IGH and MALT1 genes without translocations. Extra copy of MALT1 most likely indicates trisomy 18. Comparative genomic hybridization (CGH) array analysis was previously performed and reported on 2 of the cases analyzed by FISH and also detected trisomy 18 in the same case in which it was detected by FISH. In addition, these CGH studies revealed 7p14.1 and 19p13.11 deletions and trisomy 15 in one tumor each, respectively[35]. In 3 additional tumors dual color IGH break-apart probe also detected IGH rearrangement to an unidentified partner. Overall, translocations involving IGH gene to an unidentified partner were detected in 3(12%) of the analyzed OAEMZL tumors (Supplementary Table 3).
None of the tumors with the MYD88 L265P mutation harbored FISH-detectable chromosomal alterations. A20 mutations/deletions were detected concomitantly with additional chromosomal aberrations in 2 tumors: A20 mutations were detected in a tumor with extra signals of both IGH and MALT1 genes as well as in one tumor with an additional copy of BCL6.
Identification of translocation partners by long-distance inverse PCR
FISH analysis using a dual color IGH break-apart probe detected IGH rearrangement to an unidentified partner in 3 tumors, in two of which (cases 7806 and 11652) we had sufficient amounts of high molecular weight DNA to perform LDI-PCR. In case 7806 we identified t(6;14)(p21.1;q32.33), with the breakpoint located in a non-coding region (Figure 1A and Supplementary Figure 4). The gene nearest to this translocation is ectonucleotide pyrophosphatase/phosphodiesterase 5 (ENPP5). In case 11652 we identified t(14;17)(q32.33;p13.1), with the breakpoint also located in a non-coding region (Figure 1B and Supplementary Figure 4). The gene nearest to this translocation is myosin-10 isoform 1 (MYH10). Similar translocations were previously not reported in OAEMZL tumors. Unfortunately, pathological material was exhausted and we could not confirm the presence of these translocations by an independent method (e.g. FISH).
Figure 1. Translocation sequences detected by long-distance inverse PCR.
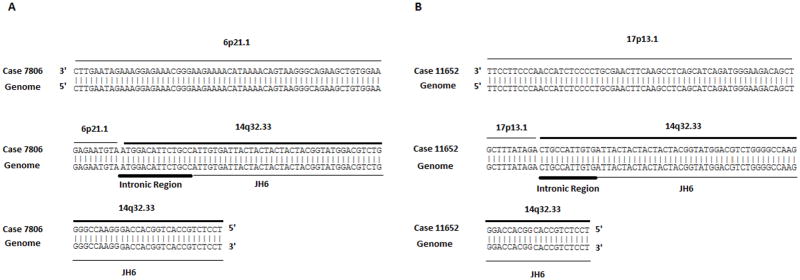
A. Translocation t(6;14)(p21.1;q32.33). B Translocation (14;17)(q32.33;p13.1).
Discussion
The etiology and pathogenesis of OAEMZLs are still controversial, not well established and may vary in different geographic regions[12]. Initial studies from Italy demonstrated the frequent presence of C. psittaci in OAEMZL tumors. However, subsequent studies suggested marked variation in the association between the infection and OAEMZL in different geographic regions, being frequent in Italy, Germany and Korea but relatively uncommon or absent in the United Kingdom, Japan, China and certain areas of the USA[14]. Similarly, genetic aberrations implicated in the activation of the canonical NF-kB signaling pathway are commonly observed in gastric EMZL but are reported at variable frequencies in OAEMZL (Table 1). Only one of the previously reported studies concomitantly analyzed genetic aberrations and the presence of the C. psittaci in OAEMZL tumors[4]. Furthermore, no direct evidence for activation of the NF-kB pathway in OAEMZL was previously provided. Herein we examined the frequency of chromosomal translocations, CARD11, A20 and MYD88 (L265P) mutations and A20 deletions in a large cohort of C. psittaci negative OAEMZLs.
Table 1.
Genetic aberrations reported in OAEMZL
| Reference | Country | N | C.psittaci | t(11;18) (q21;q21)* | t(14;18) (q32;q21)* | t(1;14) (p22;q32)* | t(3;14) (p13;q32)* | t(14;?) (q32;?)* | A20 mutations* | A20 deletions* | MYD88 mutations* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Streubel[2] | Austria and Germany | 37 | NA | 2.7 | 24.3 | 0 | NA | NA | NA | NA | NA |
| Streubel[36] | Austria and Germany | 8 | NA | NA | 38 | NA | NA | NA | NA | NA | NA |
| Streubel[11] | Austria | 20 | NA | 0 | 0 | 0 | 20 | NA | NA | NA | NA |
| Adachi[19] | Japan | 50 | NA | 0 | NA | NA | NA | NA | NA | NA | NA |
| Ye[3] | Multi- country | 55 | NA | 16.4 | NA | NA | NA | NA | NA | NA | NA |
| Ye[20] | Multi- country | 73 | NA | 13.7 | 9.6 | 0 | NA | NA | NA | NA | NA |
| Liu[39] and Kim[18] | Korea and Japan | 24 | NA | 0 | 0 | NA | NA | NA | NA | 38 | 0 |
| Chanudet[31] | Multi- country | 33 | NA | 3 | 0 | 3 | 0 | NA | NA | 19.0** | NA |
| Chanudet[30] | UK and USA | 13 | NA | NA | NA | NA | NA | NA | 23.1 | 23.1 | NA |
| Kato[28] | Japan | 13 | NA | NA | NA | NA | NA | NA | 16.6 | 26.2 | NA |
| Bi[29] | China | 105 | NA | 1.1 | 1.1 | 0 | 0 | 3.4 | 28.6 | 8.6 | NA |
| Ruiz[4] | USA | 34 | Negative | 0 | 0 | 0 | 0 | 10.4 | NA | NA | NA |
| Yan[34] | China | 105 | NA | NA | NA | NA | NA | NA | NA | NA | 5.7 |
| Current | USA | 45 | Negative | 0 | 0 | NA | NA | 12 | 15.6 | 13.3 | 6.7 |
indicates percentage of positive cases;
42 cases analyzed; NA- not analyzed
In contrast to EMZL originating in other anatomical locations[2] and concordant with some of the previous OAEMZL studies[4, 19], all the tested C. psittaci negative OAEMZLs were negative for MALT1 translocations. However, several studies reported the presence of the t(14;18)(q32;q21) IGH-MALT1 and t(11;18)(q21;q21) API2-MALT1 translocations in 9.6 –38% and 2.7–16.4% of OAEMZL tumors, respectively, with unknown C. psittaci status[2, 3, 20, 36] (Table 1). Notably, almost all the studies detecting t(14;18)(q32;q21) IGH-MALT1 and t(11;18)(q21;q21) API2-MALT1 translocations in OAEMZL were performed in European institutions, while these translocations were rarely detected in studies originating from Asia and the US (Table 1). In our cohort only 3 of the analyzed tumors (12%) harbored IGH translocations, in two of which we identified novel previously unreported partners. Similarly, Ruiz et al. reported the presence of IGH translocations with non-MALT1, non-BCL10 and non-FOXP1 partners in 10% of C. psittaci negative OAEMZLs[4]. Translocations in these cases as well as in the additional case from our cohort that could not be analyzed by LDI-PCR may involve either novel partners or the CNN3(1p), ODZ2(5q), JMJD2C (9p) and GPR34(X) genes recently reported to rearrange with IGH in several EMZLs, including OAEMZLs[37].
A20 is a negative regulator of the canonical Nf-kB pathway, inactivating several proteins necessary for NF-kB signaling such as TRAF6, NEMO, TAK1 and receptor interacting proteins (RIP) 1/2[38]. A20 mutations and deletion are frequently observed in various non-Hodgkin lymphomas[28]. We have detected A20 mutations or deletions in 28.9% C. psittaci negative OAEMZLs. The observed frequencies of A20 mutations (15.6%) and deletions (13.3) were in the lower frequency range of previously reported A20 mutations (16.6–28.6%) and deletions (8.6–38%) (Table 1). The wide frequency range of A20 mutations and deletions in OAEMZLs may reflect geographic differences and usage of different methodologies to detect A20 deletions. A20 functions as a molecular “brake” or rheostat that determines responsiveness to stimuli activating NF-kB; thus its mutations and deletions should not be sufficient to initiate activation of this signaling pathway. In almost all the reported cases A20 genetic abnormalities are usually mutually exclusive from chromosomal translocations activating NF-kB signaling (Table 1). However, the frequency of these translocations in the analyzed cohorts is low and A20 aberrations were not examined in studies concomitantly demonstrating high frequencies of chromosomal translocations (Table 1). However, overall these observations suggest the presence of additional genetic changes potentially activating NF-kB signaling. While CARD11 mutations were not observed in any of the analyzed tumors, we detected the presence of MYD88 mutations in 6.7% of C. psittaci negative OAEMZLs. During the preparation of this report, Yan et all reported similar incidence of CARD11 and MYD88 mutations in OAEMZL patients without specific reference to C. psittaci presence in the tumors[34]. MYD88 mutations were previously reported in 8% of gastric EMZLs[32], but were not found in 24 OAEMZL cases analyzed by Liu et al[39]. Recent studies also detected only single OAEMZL tumors with mutations in BIRC3 and PRDM1 genes and failed to detect mutations in CD79A[34, 39]. Mutations affecting other components of the NF-kB pathway, while rare in DLBCL (e.g. TNFRF11A, TRAF2, TRAF5, TAK1)[40], may be more common in OAEMZL, and need to be examined by unbiased whole genome or exome sequencing in future studies. Overall, the currently identified relatively low frequency of these mutations as well as the newly identified IGH translocation partners cannot account for NF-kB activation in the majority of OAEMZLs.
Cell extrinsic stimulation, such as B-cell receptor (BCR) stimulation, may also activate the canonical NF-kB signaling pathway. We have previously examined for evidence of antigen selection based on analysis of somatic mutations in the clonal immunoglobulin heavy and light chains as a potential surrogate for BCR activation[23, 41]. Evidence for immunoglobulin antigen selection was present in the majority of OAEMZL tumors, suggesting that extrinsic BCR activation may contribute to the activation of the canonical NF-kB signaling pathway. Identifying antigens to which tumor BCR can bind may explain NF-kB signaling activation in these tumors and these studies are in progress in our laboratory. In addition, studies examining tonic or chronic active BCR signaling as well as alternative extracellular stimulations leading to the activation of canonical NF-kB pathway in these tumors are needed, but restricted by the absence of appropriate cell lines originating from OAEMZL.
To the best of our knowledge, NF-kB signaling activation was not previously directly examined in primary OAEMZL tumors by biochemical or immunohistochemical methods, but was implicated in gastric EMZL based on association with chromosomal translocations targeting genes regulating this pathway, gene expression analyses of primary gastric tumors and in vitro studies following overexpression of the translocation constructs[21, 42, 43]. Consequently, without direct demonstration of the NF-kB signaling activation in OAEMZL tumors, the role of this pathway in pathogenesis of these tumors is proposed but not confirmed. We have attempted to analyze presence of nuclear p50 and p52 by immunohistochemistry, as was previously reported[40]. Despite usage of the reported methodology and antibodies, the observed immunohistochemical staining was not reproducible in control and tumor tissues (not shown), thus preventing direct demonstration of the NF-kB activation on OAEMZL tumors. Development of better antibodies for immunohistochemistry or other techniques that can be applied to small tumor biopsies will be needed to confirm the role of the NF-kB signaling activation in pathogenesis of OAEMZL tumors.
In summary, herein we demonstrate that while deletion of A20 and mutations of A20 and MYD88 are observed in C. psittaci negative OAEMZLs, they most probably contribute to NF-kB activation in only a minority of cases. Therefore, further studies are needed to demonstrate NF-kB signaling activation and its underling molecular mechanisms in C. psittaci negative OAEMZLs.
Supplementary Material
Acknowledgments
I.S.L. is supported by National Institutes of Health (NIH) grants NIH CA109335, NIH CA122105 and the Dwoskin Family, Recio Family and Anthony Rizzo Family Foundations.
Footnotes
Author contributions
Daxing Zhu-designed the study, performed experiments and analyzed the data
Offiong F Ikpatt- designed the study, performed experiments and analyzed the data
Sander R Dubovy- provided valuable materials and analyzed the data
Chen Lossos-analyzed the data
Yasodha Natkunam- performed experiments and analyzed the data
Jennifer R. Chapman-Fredricks- designed the study, performed experiments and analyzed the data
Yao-Shan Fan- designed the study, performed experiments and analyzed the data
Izidore S. Lossos- designed the study, analyzed the data and wrote the paper
All the authors reviewed the manuscripts and agree with its content and do not have conflicts of interest relevant to this manuscript.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Geneva, Switzerland: World Health Organization; 2008. [Google Scholar]
- 2.Streubel B, Simonitsch-Klupp I, Mullauer L, et al. Variable frequencies of MALT lymphoma-associated genetic aberrations in MALT lymphomas of different sites. Leukemia. 2004;18:1722–1726. doi: 10.1038/sj.leu.2403501. [DOI] [PubMed] [Google Scholar]
- 3.Ye H, Liu H, Attygalle A, et al. Variable frequencies of t(11;18)(q21;q21) in MALT lymphomas of different sites: significant association with CagA strains of H pylori in gastric MALT lymphoma. Blood. 2003;102:1012–1018. doi: 10.1182/blood-2002-11-3502. [DOI] [PubMed] [Google Scholar]
- 4.Ruiz A, Reischl U, Swerdlow SH, et al. Extranodal marginal zone B-cell lymphomas of the ocular adnexa: multiparameter analysis of 34 cases including interphase molecular cytogenetics and PCR for Chlamydia psittaci. Am J Surg Pathol. 2007;31:792–802. doi: 10.1097/01.pas.0000249445.28713.88. [DOI] [PubMed] [Google Scholar]
- 5.Isaacson PG, Du MQ. MALT lymphoma: from morphology to molecules. Nat Rev Cancer. 2004;4:644–653. doi: 10.1038/nrc1409. [DOI] [PubMed] [Google Scholar]
- 6.Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, et al. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338:1175–1176. doi: 10.1016/0140-6736(91)92035-z. [DOI] [PubMed] [Google Scholar]
- 7.Roggero E, Zucca E, Mainetti C, et al. Eradication of Borrelia burgdorferi infection in primary marginal zone B-cell lymphoma of the skin. Hum Pathol. 2000;31:263–268. doi: 10.1016/s0046-8177(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 8.Lecuit M, Abachin E, Martin A, et al. Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N Engl J Med. 2004;350:239–248. doi: 10.1056/NEJMoa031887. [DOI] [PubMed] [Google Scholar]
- 9.Hyjek E, Isaacson PG. Primary B cell lymphoma of the thyroid and its relationship to Hashimoto’s thyroiditis. Hum Pathol. 1988;19:1315–1326. doi: 10.1016/s0046-8177(88)80287-9. [DOI] [PubMed] [Google Scholar]
- 10.Hyjek E, Smith WJ, Isaacson PG. Primary B-cell lymphoma of salivary glands and its relationship to myoepithelial sialadenitis. Hum Pathol. 1988;19:766–776. doi: 10.1016/s0046-8177(88)80259-4. [DOI] [PubMed] [Google Scholar]
- 11.Streubel B, Vinatzer U, Lamprecht A, et al. T(3;14)(p14. 1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia. 2005;19:652–658. doi: 10.1038/sj.leu.2403644. [DOI] [PubMed] [Google Scholar]
- 12.Stefanovic A, Lossos IS. Extranodal marginal zone lymphoma of the ocular adnexa. Blood. 2009;114:501–510. doi: 10.1182/blood-2008-12-195453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husain A, Roberts D, Pro B, et al. Meta-analyses of the association between Chlamydia psittaci and ocular adnexal lymphoma and the response of ocular adnexal lymphoma to antibiotics. Cancer. 2007;110:809–815. doi: 10.1002/cncr.22843. [DOI] [PubMed] [Google Scholar]
- 14.Decaudin D, Dolcetti R, de Cremoux P, et al. Variable association between Chlamydophila psittaci infection and ocular adnexal lymphomas: methodological biases or true geographical variations? Anticancer Drugs. 2008;19:761–765. doi: 10.1097/CAD.0b013e32830b58c4. [DOI] [PubMed] [Google Scholar]
- 15.Rosado MF, Byrne GE, Jr, Ding F, et al. Ocular adnexal lymphoma: a clinicopathologic study of a large cohort of patients with no evidence for an association with Chlamydia psittaci. Blood. 2006;107:467–472. doi: 10.1182/blood-2005-06-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang GS, Winter JN, Variakojis D, et al. Lack of an association between Chlamydia psittaci and ocular adnexal lymphoma. Leuk Lymphoma. 2007;48:577–583. doi: 10.1080/10428190601132105. [DOI] [PubMed] [Google Scholar]
- 17.Matthews JM, Moreno LI, Dennis J, et al. Ocular Adnexal Lymphoma: no evidence for bacterial DNA associated with lymphoma pathogenesis. Br J Haematol. 2008;142:246–249. doi: 10.1111/j.1365-2141.2008.07112.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim WS, Honma K, Karnan S, et al. Genome-wide array-based comparative genomic hybridization of ocular marginal zone B cell lymphoma: comparison with pulmonary and nodal marginal zone B cell lymphoma. Genes, chromosomes & cancer. 2007;46:776–783. doi: 10.1002/gcc.20463. [DOI] [PubMed] [Google Scholar]
- 19.Adachi A, Tamaru J, Kaneko K, et al. No evidence of a correlation between BCL10 expression and API2-MALT1 gene rearrangement in ocular adnexal MALT lymphoma. Pathol Int. 2004;54:16–25. doi: 10.1111/j.1440-1827.2004.01580.x. [DOI] [PubMed] [Google Scholar]
- 20.Ye H, Gong L, Liu H, et al. MALT lymphoma with t(14;18)(q32;q21)/IGH-MALT1 is characterized by strong cytoplasmic MALT1 and BCL10 expression. J Pathol. 2005;205:293–301. doi: 10.1002/path.1715. [DOI] [PubMed] [Google Scholar]
- 21.Du MQ. MALT lymphoma: many roads lead to nuclear factor-kappab activation. Histopathology. 2011;58:26–38. doi: 10.1111/j.1365-2559.2010.03699.x. [DOI] [PubMed] [Google Scholar]
- 22.van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 23.Zhu D, Lossos C, Chapman-Fredricks JR, et al. Biased use of the IGHV4 family and evidence for antigen selection in Chlamydophila psittaci-negative ocular adnexal extranodal marginal zone lymphomas. PloS one. 2011;6:e29114. doi: 10.1371/journal.pone.0029114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novak U, Rinaldi A, Kwee I, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113:4918–4921. doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaffer LG, Slovak ML, Campbell LJ. S Karger Ag. 2009. An International System for Human Cytogenetic Nomenclature (2009): Recommendations of the International Standing Committee on Human Cytogenetic Nomenclature. [Google Scholar]
- 26.Willis TG, Jadayel DM, Coignet LJ, et al. Rapid molecular cloning of rearrangements of the IGHJ locus using long-distance inverse polymerase chain reaction. Blood. 1997;90:2456–2464. [PubMed] [Google Scholar]
- 27.Sonoki T, Willis TG, Oscier DG, et al. Rapid amplification of immunoglobulin heavy chain switch (IGHS) translocation breakpoints using long-distance inverse PCR. Leukemia. 2004;18:2026–2031. doi: 10.1038/sj.leu.2403500. [DOI] [PubMed] [Google Scholar]
- 28.Kato M, Sanada M, Kato I, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 29.Bi YW, Zeng NY, Chanudet E, et al. A20 inactivation in ocular adnexal MALT lymphoma. Haematol-Hematol J. 2012;97:926–930. doi: 10.3324/haematol.2010.036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chanudet E, Huang Y, Ichimura K, et al. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma (vol 24, pg 483, 2010) Leukemia. 2010;24:488–489. doi: 10.1038/leu.2009.234. [DOI] [PubMed] [Google Scholar]
- 31.Chanudet E, Ye H, Ferry J, et al. A20 deletion is associated with copy number gain at the TNFA/B/C locus and occurs preferentially in translocation-negative MALT lymphoma of the ocular adnexa and salivary glands. Journal of Pathology. 2009;217:420–430. doi: 10.1002/path.2466. [DOI] [PubMed] [Google Scholar]
- 32.Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 34.Yan Q, Wang M, Moody S, et al. Distinct involvement of NF-kappaB regulators by somatic mutation in ocular adnexal malt lymphoma. Br J Haematol. 2013;160:851–854. doi: 10.1111/bjh.12162. [DOI] [PubMed] [Google Scholar]
- 35.Braggio E, Dogan A, Keats JJ, et al. Genomic analysis of marginal zone and lymphoplasmacytic lymphomas identified common and disease-specific abnormalities. Mod Pathol. 2012;25:651–660. doi: 10.1038/modpathol.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streubel B, Lamprecht A, Dierlamm J, et al. T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood. 2003;101:2335–2339. doi: 10.1182/blood-2002-09-2963. [DOI] [PubMed] [Google Scholar]
- 37.Vinatzer U, Gollinger M, Mullauer L, et al. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:6426–6431. doi: 10.1158/1078-0432.CCR-08-0702. [DOI] [PubMed] [Google Scholar]
- 38.Shembade N, Harhaj EW. Regulation of NF-kappaB signaling by the A20 deubiquitinase. Cellular & molecular immunology. 2012;9:123–130. doi: 10.1038/cmi.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F, Karube K, Kato H, et al. Mutation analysis of NF-kappaB signal pathway-related genes in ocular MALT lymphoma. Int J Clin Exp Pathol. 2012;5:436–441. [PMC free article] [PubMed] [Google Scholar]
- 40.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu D, Lossos C, Chapman-Fredricks JR, et al. Biased immunoglobulin light chain use in the Chlamydophila psittaci negative ocular adnexal marginal zone lymphomas. Am J Hematol. 2013 doi: 10.1002/ajh.23416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamoudi RA, Appert A, Ye H, et al. Differential expression of NF-kappaB target genes in MALT lymphoma with and without chromosome translocation: insights into molecular mechanism. Leukemia. 2010;24:1487–1497. doi: 10.1038/leu.2010.118. [DOI] [PubMed] [Google Scholar]
- 43.Zhou H, Du MQ, Dixit VM. Constitutive NF-kappaB activation by the t(11;18)(q21;q21) product in MALT lymphoma is linked to deregulated ubiquitin ligase activity. Cancer cell. 2005;7:425–431. doi: 10.1016/j.ccr.2005.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


