Abstract
The emergence and convergence of cancer genomics, targeted therapies, and network oncology have significantly expanded the landscape of protein-protein interaction (PPI) networks in cancer for therapeutic discovery. Extensive biological and clinical investigations have led to the identification of protein interaction hubs and nodes that are critical for the acquisition and maintaining characteristics of cancer essential for cell transformation. Such cancer enabling PPIs have become promising therapeutic targets. With technological advances in PPI modulator discovery and validation of PPI-targeting agents in clinical settings, targeting PPI interfaces as an anticancer strategy has become a reality. Future research directed at genomics-based PPI target discovery, PPI interface characterization, PPI-focused chemical library design, and patient-genomic subpopulation-driven clinical studies is expected to accelerate the development of the next generation of PPI-based anticancer agents for personalized precision medicine. Here we briefly review prominent PPIs that mediate cancer-acquired properties, highlight recognized challenges and promising clinical results in targeting PPIs, and outline emerging opportunities.
Keywords: PPI, cancer genomics, signaling network, HTS, small molecule modulator, tumorigenesis
Rising interest in targeting PPIs
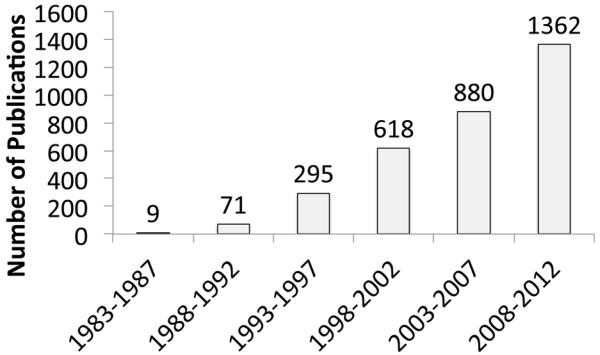
PPI interfaces represent a highly promising, although challenging, class of potential targets for therapeutic development [1]. In cancer, PPIs form signaling nodes and hubs to transmit pathophysiological cues along molecular networks to achieve an integrated biological output, thereby promoting tumorigenesis, tumor progression, invasion, and/or metastasis. Thus, pathway perturbation, through the disruption of PPIs critical for cancer, offers a novel and effective strategy to curtail the transmission of oncogenic signals. As our understanding of cancer biology has significantly increased in recent years, interest in targeting PPIs as anticancer strategies has increased as well (Figure 1).
Figure 1. Rising number of Publications in the field of cancer-related PPIs.
PubMed database was searched with the following keywords: protein-protein interaction, tumor, cancer, and inflammation.
PPI interfaces constitute basic units in oncogenic signaling networks
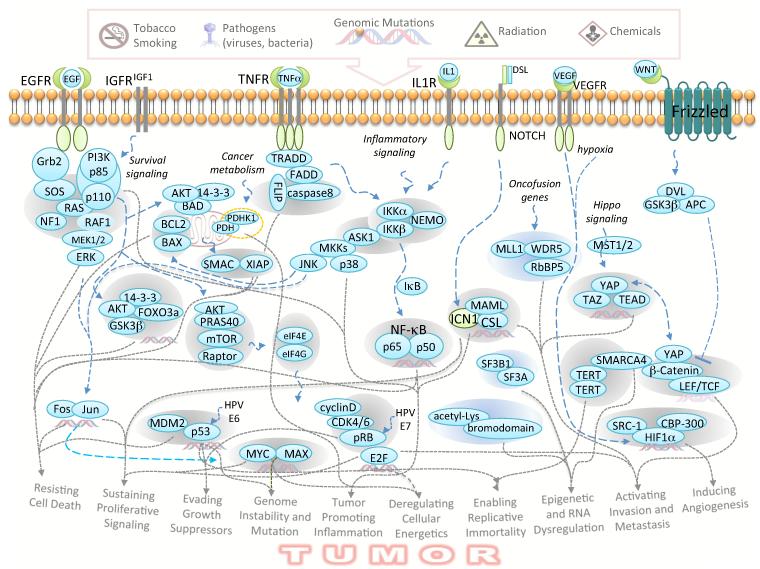
A variety of environmental, genetic, and epigenetic factors induce the re-programming of cancer initiating cells and the acquisition of physical and molecular features that promote tumorigenesis and provide resistance to therapeutics. These characteristics, including sustaining proliferative signaling and evading growth suppressors, permit the development and progression of cancer and have been recognized as distinctive hallmarks of cancer [2] (Figure 2). These hallmarks provide a molecular framework for our understanding of cancer, linking molecular signaling events to pathological outcomes. Indeed, the oncogenic potential of cells is determined by a combination of genetic and epigenetic alterations through the operation of well-orchestrated signaling networks. Importantly, PPIs represent the basic units within such vital networks.
Figure 2. Representative PPIs in oncogenic signaling networks that drive the acquisition and development of hallmarks of cancer.
Grey dotted arrows connect PPIs with corresponding cancer hallmarks. Some PPIs contribute to multiple features of cancer. It is cautioned that some PPIs may impact global processes of cell growth and their precise connections to cancer remain to be established.
Upon oncogenic stimulation, PPIs play essential roles in linking networks that relay oncogenic signals, enable the acquisition of hallmark features of cancer, and serve diverse roles in driving and maintaining the growth of cancer cells (Figure 2). From the engagement of receptors with dysregulated growth factors to dimerization of receptor tyrosine kinases triggered by gene amplification or mutations, PPIs initiate a cascade of reactions to promote uncontrolled cell proliferation [3]. Activated Ras, due to perturbations such as Epidermal Growth Factor Receptor (EGFR) activation, Neurofibromin 1 (NF1) deletion, or intrinsic mutation, assumes a conformation that allows binding to multiple regulatory proteins and results in enforced proliferation and survival. Survival signaling, activated by proteins such as Insulin-like Growth Factor 1 (IGF1) and Phosphoinositide-3-kinase (PI3K), or disabled negative regulator PTEN, enables tumors to resist cell death through a number of different mechanisms. For example, the Akt/FOXO3a/14-3-3 complex mediates a transcription-dependent mechanism, whereas the Akt/Bad/14-3-3 interaction mediates a transcription-independent anti-apoptotic mechanism [3]. In addition to providing resistance to cell death, Akt also regulates the mTOR complex to control cap-dependent translation, through the eIF4E/eIF4G PPI, of a large number of growth promoting genes, including c-Myc. In turn, amplified c-Myc favors binding to Max over Mad and thereby drives transcription of growth-promoting genes, such as cyclin D that modulate cell cycle progression [4].
To enable cancer progression, cells must acquire mechanisms to evade growth suppression. Several PPI complexes, including MDM2/p53 and CDK4/pRB, play key roles in neutralizing such tumor suppressive functions [2]. These tumor suppressor mechanisms are often hijacked by viral oncoproteins, such as Human papillomavirus E7 protein that binds to pRb and E6 protein that binds to p53, which allow the virus to induce tumors. Such PPIs offer tumor-specific targets. In addition to the examples given above, a large number of PPIs dictate signaling networks that enable the acquisition of or maintain other hallmarks of cancer. For instance, the VEGF/VEGFR and HIF1α/CBP PPIs mediate signals for inducing angiogenesis, the catalytic activity of TERT dimers enables replicative immortality, and a variety of re-programmed enzyme/substrate interactions, such as the onco-fusion gene regulated PDHK1/PDHA1 PPI, play integral roles in dysregulated cellular metabolism by controlling a metabolic switch between glycolysis and oxidative phosphorylation [5]. In addition, mutated p53 and Myc also play key roles in the regulation of cancer metabolism. The IKK/NEMO/ASK1 complex integrates the proinflammation function with stress response signaling initiated by reactive oxygen species [6]. Furthermore, It has recently been shown that epigenomic re-programming is a critical part of cancer development [7], and PPIs involved in epigenomic dysregulation, such as SMARCA4 interactions, have been described [8].
As a result of oncogenic network re-programming, some PPIs contribute to distinct features of cancer, whereas other PPIs are vital for multiple characteristics of cancer. For example, the MDM2/p53 and Myc/Max PPIs play key roles in evading growth suppression and cell death, as well as promoting genomic instability and cancer metabolism. Thus, it is expected that interception of certain critical PPIs may disable multiple mechanisms that cancer cells rely on for survival. As a large number of PPIs are involved in driving tumorigenesis through the regulation of oncogenic networks, these PPI interfaces represent a fertile ground for anticancer therapeutic discovery and development.
Overcoming challenges and current strategies for targeting PPIs
Challenges in discovering PPI modulators
A number of challenges and concerns exist regarding targeting PPIs, some of which include: (i) large PPI interface areas, (ii) lack of deep pockets, (iii) presence of non-contiguous binding sites, and (iv)the general lack of natural ligands. In addition, protein-protein interface surfaces are also differ from the small molecule binding sites in the shape and amino acid residue composition. In contrast to the well-defined and normally hydrophilic ligand binding cavities observed in the crystal structures of enzymes and GPCRs, the interface surfaces of the many protein-protein complexes are typically hydrophobic and relatively flat, often lacking deep grooves where a small molecule can dock. Recent studies have addressed some of these concerns as detailed in several publications [1, 9, 10]. Although the PPI interface generally covers an average area of 1150 to 10,000 Å2, which is larger than small molecule binding sites of 100 to 600 Å2, the presence of “hot spots” (i.e. small subsets of amino acid residues that contribute the most to free energy of binding) makes PPIs amenable for small molecule perturbations [10, 11]. Also, PPIs often are mediated by posttranslational modifications, such as the binding of 14-3-3 to phosphorylated Ser/Thr motifs, and the interaction of bromodomain proteins to acetylated lysine, which simplifies the definition of the targeting interfaces [12-14]. The typical lack of natural ligands for PPIs as starting points poses a significant challenge for structure-based drug design. However, promising examples of natural products that can act on PPIs do exist, such as rapamycin for mTOR and taxol for tubulin. Additionally, there are significant differences in the chemical space between PPI modulators (PPIMs) and conventional drug-like compounds [9, 15, 16]. In general, PPIMs have higher MW (> 400 Da) than that of typical drug-like compounds (200-500 Da), and PPIMs often violate the “Rule-of-Five” [9]. Therefore, the application of commonly used high-throughput screening (HTS) methods for PPIMs has also been limited due to the biased chemical composition towards classical target classes in current chemical libraries.
Current approaches for the discovery of PPI modulators
Structure-based design
Structural studies allow for the identification of peptide fragments and amino acid residues that are critical for PPI. This information, combined with that from functional assays, provides a basis for the rational design of PPIMs. Not surprisingly, mimicking the structure of binding peptides is one of the widely used approaches to design novel PPIMs [17]. Application of this approach led to the identification of potent inhibitors of BCL2, XIAP, NOTCH, and MDM2 [18-22]. General structure-based approaches include computational molecular modeling [23, 24], peptide engineering with display technologies, such as the phage display [25], design of small-molecules based on α-helix and β-sheet scaffolds [26, 27], and synthesis of conformationally constrained “stapled” peptides with a stabilized α-helical structure [28-31].
Small molecule screening methods
In contrast to the structure-based design, the screening approach allows discovery of small molecule PPIMs even if the structural information is very limited or unavailable. Indeed, various screening appoarches, including HTS, has been used to identify compounds that target “hot spots” of PPI interfaces [1]. Furthermore, HTS can be used to reveal inducible pockets in PPI interfaces as well as allosteric modulators. In many cases, the screening approach is combined with structure-based design to further enhance the physico-chemical and pharmacological properties of identified PPI modulators.
The most widely used HTS techniques for PPIs include fluorescence polarization (FP) and Föster/Fluorescence resonance energy transfer (FRET). FP measures the change in emitted polarization signals in solution upon association of a small fluorescent molecule (such as a peptide) with a relatively large binding partner. FRET is a nonradioactive, photophysical effect in which energy that is absorbed by a donor fluorophore is transferred to an acceptor fluorophore. By coupling the donor and acceptor fluorophores with appropriate spectral properties to two interacting molecules, the fluorophores may be brought into close proximity (10–100 Å) and induce a FRET signal. Both FP and FRET methods are extensively used in HTS campaigns for the discovery of PPIMs [32]. Other HTS methods include ELISA, flow cytometry, surface plasma resonance (SPR), and label-free platforms [33]. In addition to these biochemical HTS assays, intracellular PPIs can be coupled to readouts for cell-based reporter assays, which incorporate a more physiological cellular context and identify compounds that are cell permeable. For example, the p53/MDM2 PPI has been linked to a reporter assay based on cytoplasm-nuclear redistribution [34]. Various biosensors, such as protein complementation assays, can also be used to monitor PPIs in a HTS format.
In addition to biochemical and cell-based HTS assays, Fragment-based screening (FBS) is another commonly used approach for discovery of PPIMs. FBS aims to identify molecular fragments with binding activity for a target protein. Once the fragments have been identified, they are, or the interactions they identify, are built into a drug-like compound [35]. The main advantage of the fragment-based approach is that a large chemical space can be targeted with approximately 103 fragments. The ligand efficiency (LE) of fragment hits is high. Moreover, FBS can be successfully utilized for many targets that were found to be challenging using traditional HTS [36]. Due to a small molecular weight (MW ≈ 200 Da), and thus a limited contact area with a protein, the binding affinity of the fragments is relatively low (often in millimolar range). Therefore, to detect a binding event, sensitive biophysical methods are required, including X-ray crystallography, nuclear magnetic resonance (NMR), and SPR [35, 37, 38]. Key advantages of NMR include automated sampling, high sensitivity, quantitative data on binding affinity, and that ability to obtain structural information about the binding site [39]. Examples of successful application of NMR in FBS include discovery of XIAP [40], BCL2 [41], ZipA/FtsZ [42], and K-RAS/SOS [43] PPI inhibitors. Often, a combination of FBS assays is employed for a screening campaign. For instance, a combination of X-ray and NMR FBHTS led to identification of the Hsp90 inhibitor AT13387, which recently entered clinical trials [44, 45]. Another commonly used assay for FBS is SPR, which detects changes in the refractive index near a sensor surface. SPR fragment-based screening has been utilized to identify novel inhibitors of Hsp90 interactions [46, 47]. In contrast to methods for non-covalent binding, a “tethering” approach is used to detect reversible covalent bonds formed between the cysteine of a target protein and fragment molecules containing a disulfide bond [48]. Tethering FBS has been employed to identify PPI modulators such as IL2/IL2-αR inhibitors [1].
Two general approaches are used to perform computational screening of a 3D compound library: the ligand-based (also known as the pharmacophore-based) approach and structure-based virtual screening. A pharmacophore model represents the chemical features of a set of compounds critical for efficient protein binding [49]. These features or pharmacophore points (such as H-bond donors or acceptors, aromatic rings, and charges) have certain coordinates in a 3D space. This procedure is aimed at identifying compounds with a certain conformation and chemical composition that matches the requirements of a pharmacophore model. The pharmacophore-based approach has been successfully utilized to identify novel inhibitors of the MDM2/p53 interaction [50, 51], PPIs of BCL2 family proteins [52], and 14-3-3 inhibitors [53]. Conversely, the structure-based approach relies on the structural information of binding site on the target protein. For this type of screening, each compound in a chemical library has to be computationally docked to the binding site, and a binding affinity is estimated in terms of energy scoring functions. Application of the structure-based virtual screening has led to the discovery of PPI inhibitors of Ubc13-Uev1 [54], MDM2/p53 [55], and TCF4/β-catenin [56].
Clinical validation of targeting PPIs in cancer
Thousands of compounds have already been tested as potential inhibitors of various PPIs, and the results are promising. Titrobifan, a glycoprotein IIb/IIIa inhibitor, and Maraviroc, an inhibitor of the CCR5/gp120 interaction, are currently available on the market as cardiovascular and anti HIV drugs, respectively. These drugs demonstrate the feasibility of targeting PPIs for treatment of various diseases. Additionally, several anti-cancer compounds have entered clinical trials, highlighting the potential of the PPI targeting approach in cancer.
Inhibitors of the MDM2/p53 interaction – a breakthrough in PPI targeting
p53 plays a critical role in cell cycle regulation, DNA repair, angiogenesis, and apoptosis [57, 58]. Activation of p53 increases the expression of human protein double minute 2 (HDM2, MDM2 in mouse), which in turn directly binds to p53 and inhibits its tumor suppressive activity (Figure 2) [59, 60]. Structurally, the N-terminal domain of MDM2 binds a short 15-residue α-helical peptide of p53 [61]. Three hydrophobic residues of p53 (Phe19, Trp23, and Leu26) occupy a well-defined hydrophobic pocket of MDM2 (Figure 3). These structural features enable a strategy to target the MDM2/p53 PPI. To identify drug-like inhibitors of MDM2/p53 interactions, various approaches were used, including design of peptidomimetics, HTS, and computational drug design. As a result, several MDM2/p53 PPI inhibitors have entered clinical trials [60, 62]. For example, a series of cis-imidazoline analogs named Nutlins were identified by screening of compound libraries [59]. Nutlins employ the same binding mode as the p53 Phe19, Trp23, and Leu26 residues in the MDM2 binding pocket (Figure 3). Further chemical optimization of Nutlin-3 led to RG7112, the first MDM2 inhibitor to enter clinical trials in patients with advanced solid tumors in 2007 [63]. Another MDM2/p53 PPI inhibitor, RO5503781 is currently in a Phase I trial in patients with advanced malignancies [64]. The exciting success of potent MDM2/p53 PPI inhibitors has significantly accelerated studies to target other PPIs with small chemical compounds as anticancer drugs.
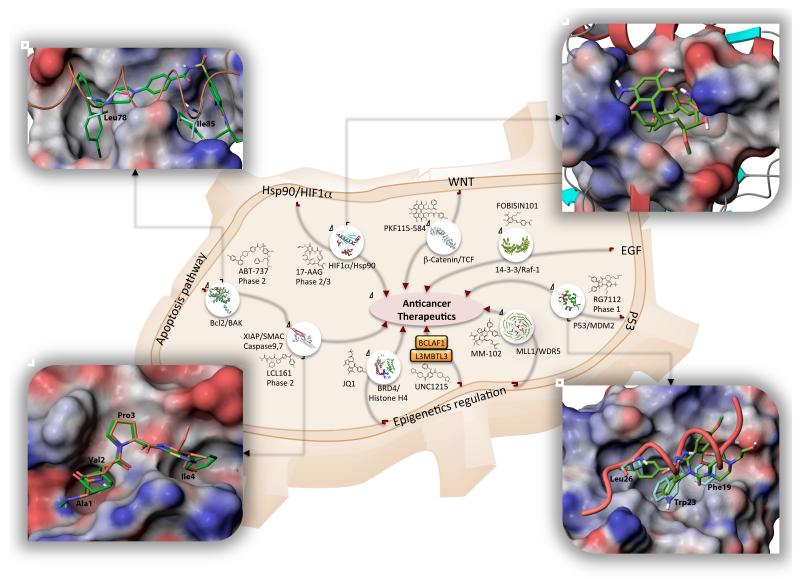
Figure 3. Examples of PPI inhibitors entered clinical trials and emerging agents.
Inhibitors of MDM2/p53, Bcl2, XIAP, and Hsp90 PPIs are in Phase 1-3. Examples of promising PPI targets with recently identified novel inhibitors include MLL1/WDR5, β-Catenin/TCF, BCLAF1/L3MBTL3, BRD4/Histone H4, and 14-3-3 interactions. Chemical structures of representative inhibitors are shown along with available crystal structures of protein-protein complexes. Shown in details are: a superimposition of Bak peptide (orange, carbon atoms are colored cyan) and ABT-737 (carbon atoms are colored green) bound at BCL-XL (PDB ID: 1BXL, 2YXJ). ABT-737 occupies the same hydrophobic pocket on Bcl-XL surface as the peptide, overlapping with Leu78 and Ile85 Bak residues critical for the peptide binding (top-left); a crystal structure of Hsp90 in a complex with Geldanamycin (PDB ID: 1YET), the first Hsp90 inhibitor entered clinical trials (top-right); a superimposition of p53 peptide and Nutline-2 (one of the first identified potent MDM2/p53 inhibitors) bound to N-terminal domain of MDM2 (PDB ID: 1YCR, 1RV1). Phe19, Trp23, and Leu26 residues of p53 peptide occupy hydrophobic pocket of MDM2. The ethoxy- and chlorophenyl groups of Nutlin-2 match the positions of Phe19, Trp23, and Leu26, respectively (bottom-right); a superimposition of SMAC AVPI peptide (the carbon atoms are colored orange) and GDC-0152 the first SMAC mimetic entered clinical trials (the carbon atoms colored green) (bottom-left, PDB ID: 3UW5, 1G73). The molecular surfaces of Hsp90, Bcl-XL, MDM2, and XIAP are colored by electrostatic potential (blue – positive, white – neutral, red – negative).
Mimicking the structure of Smac peptides resulted in new XIAP antagonists in the clinic
One widely used strategy to identify lead PPIM compounds is to mimic the structure of binding peptides [26, 27]. For example, the BIR3 domain of the X-linked Inhibitor of Apoptosis protein (XIAP) binds and inhibits pro-apoptotic Caspase-9. In turn, the anti-apoptotic activity of XIAP can be neutralized by Smac, which is released from mitochondria during apoptosis (Figure 2). The XIAP/Caspase-9 interaction can be disrupted by a Smac tetrapeptide (AVPI) [65, 66], which provides a novel PPI target. A combination of structure-based design and targeted compound library generation led to the identification of GDC-0152, the first Smac mimetic to enter clinical trials in patients with locally advanced or metastatic malignancies [19]. GDC-0152 binds to BIR domains with low nM affinity at the same binding site on IAPs as the SMAC AVPI peptide (Figure 3). A Phase I trial with another Smac mimetic, GDC-0917 (CUDC-427), has been completed in patients with advanced solid tumors and lymphomas. The structurally related LCL161 Smac mimetic has entered Phase II trials in patients with triple negative breast cancer (Figure 3) [20, 67]. Other non-peptide XIAP/Smac inhibitors include an orally available derivative, AT-406, and two bivalent Smac mimetics, TL32711 and HGS1029, all of which are currently in clinical trials for various cancers [68, 69].
FBS-based discovery of mitochondrial apoptosis pathway modulators
BH3-containing pro-apoptotic proteins, such as Bax, bind to the hydrophobic pocket of anti-apoptotic Bcl-2 proteins through a single α-helix (the BH3 domain). Mimicking the BH3 domain with small-molecule compounds has shown significant therapeutic potential [18]. Several BH3-mimetics have been identified using NMR-based FBS combined with structure-based optimization. For instance, ABT-737 has a high binding affinity (nM range) to Bcl-2 and Bcl-XL [70]. This compound occupies the same hydrophobic pocket on Bcl-XL as a Bak-derived peptide, overlapping with the Leu78 and Ile85 of Bak, which are critical residues for peptide binding (Figure 3) [70-72]. ABT-737 was further improved to generate orally bioavailable ABT-263 (Navitoclax, Phase I) and ABT-199 (Phase I) with enhanced water solubility [73]. Another oral BH3-mimetic, Obatoclax (GS-01570), discovered by screening natural product libraries, is currently in Phase II clinical trials in patients with small cell lung cancer [18, 74, 75].
Allosteric regulation of PPI: Hsp90 inhibitors
The Hsp90 chaperone protein regulates the activity and stability of numerous client proteins. Inhibition of Hsp90 can simultaneously shut down multiple oncogenic pathways, which has sparked interest in targeting Hsp90 PPIs for cancer treatment [76-78]. The natural product geldanamycin (GM) inhibits Hsp90/Src complex formation by binding to a 15 Å deep ATP-binding site in the N-terminal domain of Hsp90 (Figure 3). A 17-allylamino,17-demetoxy-substituted GM derivative, 17-AAG, was later developed as a clinical candidate, and is currently in Phase I and II trials in patients with multiple myeloma, lymphoma, stage IV pancreatic cancer, non-small cell lung cancer, and solid tumors [79]. Other Hsp90 inhibitors in clinical testing include IPI-504 (Phase II), BIIB021 (Phase I and II), PU-H71 (Phase I), NVP-AUY922 (Phase II), AT13387 (Phase I/II), and KW-2478 (Phase I/II) [78].
Emerging opportunities for targeting PPIs
Although validated PPIs remain active targets for therapeutic development, new concepts and promising PPIs have emerged for anticancer drug discovery (Figure 2). For example, increased knowledge of cancer genomics and PPI-mediated epigenetic mechanisms and identification of cancer-specific onco-fusion proteins expose a large number of new PPIs that are directly associated with pathology of cancer. Recent insight into the consequences of various cancer therapeutics and the induced therapeutic resistance offers unanticipated PPIs as potential cancer targets to enhance therapeutic efficacy.
Cancer genomics
Large scale genomics initiatives, such as The Cancer Genome Atlas (http://cancergenome.nih.gov) and The International Cancer Genome Consortium (http://icgc.org/icgc/cgp), have led to the discovery of a plethora of genomic alternations that drive tumorigenesis and/or progression [80, 81]. It is not hard to imagine that such changes will lead to alteration of protein interaction networks that regulate cell growth. For example, Akt-activating mutations often re-wire downstream phospho-relay systems via altered PPIs, such as enhanced 14-3-3 interactions with FOXO3a, Bad, and PRAS40 (Figure 2). To systematically examine PPI network changes in cancer, we have conducted large scale experiments to establish cancer-associated PPI network maps based on genomic information from Glioblastoma multiforme [82] and other tumor types. Such studies, along with predicted new PPIs [83], have revealed novel PPIs that act as major drivers of cancer and thus, are potential targets for therapeutic exploration.
PPIs that regulate epigenetic mechanisms
Cancer genomics studies have not only validated the importance of classical hallmarks of cancer, but also revealed new characteristics that are intricately associated with cancer, such as epigenetic dysregulation and RNA splicing [8, 80, 81]. Recent advances outlining the contribution of dysregulated epigenetic mechanisms to cancer offer new opportunities for targeting PPIs. For instance, dysregulated histone methylation and acetylation have been found to be associated with tumorigenesis. These changes in turn dictate the specific recognition of modified histones by methyllysine-binding proteins and by acetlylysine-binding bromodomains (Figure 2) [7, 13]. Recently, a potent and selective compound, UNC1215, has been identified that effectively disrupts the interaction of methylated histone with the L3MBTL3 methyllysine binding protein [84]. UNC1215 demonstrated significant selectivity against more than 200 other analogous methyllysine-recognition domains, making it a highly promising agent for probing L3MBTL3 function in cancer. For the interaction of acetylated histone with bromodomain-containing proteins, two small molecules, JQ1 and I-BET, have been developed, which are pan-bromodomain and extraterminal domain (BET) family inhibitors [85]. Antitumor activity has been observed for JQ1 in a patient-derived xenograft animal model. It is particularly valuable for Myc-driven tumors [86]. I-BET-151 exhibited promising efficacy against onco-fusion driven leukemia [87]. Cancer-associated mutations in the RNA-splicing machinery indicate the importance of PPIs in the regulation of RNA processing in cancer, such as the association of frequently mutated splicing factor 3b (SF3B1) with 3a (SF3A) in the U2 small nuclear ribonucleoproteins complex[81].
Onco-fusion PPIs offer cancer selectivity
PPIs are important for the catalytic activity of many enzymes, including epigenetic modifying enzymes, which can also be targeted. One example is the development of high-affinity peptidomimetics that antagonize the interaction of the histone methyltransferase, mixed lineage leukemia (MLL1), and its activator WDR5. Dysregulated MLL1 is associated with various leukemias. Disruption of the MLL1/WDR5 PPI with peptidomimetics effectively decreased MLL1-fusion mediated leukemogenesis [88]. Similarly, targeting the MLL1/menin PPI has led to the development of a series of lead compounds with therapeutic potential [89]. Importantly, fusion proteins, such as MLL1, offer tumor-selective targets; thus, future efforts targeting onco-fusion protein-specific PPIs are not only warranted, but much needed [90].
PPIs in protein complexes
As indicated for MLL1 and many other hub proteins that mediate oncogenic signaling, PPIs often involve multiprotein complexes. Selective inhibition of a particular PPI in the complex for a desired therapeutic effect is challenging. However, selective disruption of MLL1 with WDR5 gave rise to promising antileukemogenesis activity [88]. Inhibition of MAML with ICN1/CSL by a stapled peptide in NOTCH1 signaling is another example, which offers a novel strategy for treating NOTCH1-dependent cancer [22]. Another challenge is experimentally identifying selective agents with HTS. For example, 14-3-3 proteins interact with multiple partners, such as Raf-1, Bad, and FOXO [14]. Although these interactions engage a common binding groove, some partner-selective residues have been suggested. Technologies that can identify pan and specific modulators are expected to greatly accelerate the development of selective PPI inhibitors.
PPIs in combination therapies
Another emerging opportunity for targeting PPIs in cancer is re-wired PPIs in oncogenic signaling networks triggered by therapeutic insults. For example, inhibition of mTOR induces a paradoxal activation of Akt [91] . Activated Akt is known to trigger phosphorylation-dependent PPIs, such as 14-3-3-mediated PPIs [3, 12]. Such induced PPIs may yield new cancer dependency and serve as new targets to overcome pharmacologically-induced drug resistance. Interestingly, cancer cells treated with a MEK inhibitor renders them sensitive to the Bcl-2 PPI inhibitor, ABT-263 [92]. PPI modulation is expected to have broad and important roles in future mechanism-based combination therapies.
Concluding remarks
Future efforts aimed at targeting PPIs will be greatly accelerated by a number of recent advances. Understanding the nature of PPI interfaces and successful PPIMs may provide rationale design strategies for PPI-focused libraries. PPI assay technologies that closely reflect physiological conditions and address multiprotein complex issues are likely to shorten the process of lead discovery. PPI target discovery coupled with functional validation in genetically defined model systems are vital to move PPIMs into the pipeline for clinical evaluation. These activities are fueled by a new U.S. national initiative, the Cancer Target Discovery and Development (CTD2) network (http://ocg.cancer.gov/programs/ctdd.asp). The CTD2 aims to bridge the gap between the vast amount of cancer genomics information and limited therapeutics by accelerating the discovery of new promising targets, including PPIs. Emphasizing collaborative interactions among members with complementary and unique expertise, the CTD2 network focuses on rapid identification and characterization of potential targets for the development of cancer therapeutics. These scientific efforts will significantly accelerate the expansion of the PPI target landscape, which we hope will lead to a paradigm shift in targeting the once “undruggable” for personalized cancer therapy and precision medicine.
Acknowledgement
Work in authors’ laboratory was supported in part by the US National Institutes of Health grants P01 CA116676 and U01 CA168449. FRK and HF are Georgia Cancer Coalition Distinguished Scholars.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- [2].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [3].Hennessy BT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- [4].Prochownik EV, Vogt PK. Therapeutic Targeting of Myc. Genes & cancer. 2010;1:650–659. doi: 10.1177/1947601910377494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hitosugi T, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44:864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Puckett MC, et al. Integration of the Apoptosis signal-regulating kinase 1-mediated stress signaling with the Akt/PKB-IkappaB kinase cascade. Molecular and cellular biology. 2013 doi: 10.1128/MCB.00047-13. DOI: 10.1128/MCB.00047-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kelly TK, et al. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Buchwald P. Small-molecule protein-protein interaction inhibitors: therapeutic potential in light of molecular size, chemical space, and ligand binding efficiency considerations. IUBMB Life. 2010;62:724–731. doi: 10.1002/iub.383. [DOI] [PubMed] [Google Scholar]
- [10].Moreira IS, et al. Hot spots--a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- [11].Perot S, et al. Druggable pockets and binding site centric chemical space: a paradigm shift in drug discovery. Drug Discov Today. 2010;15:656–667. doi: 10.1016/j.drudis.2010.05.015. [DOI] [PubMed] [Google Scholar]
- [12].Watanabe N, Osada H. Phosphorylation-dependent protein-protein interaction modules as potential molecular targets for cancer therapy. Curr Drug Targets. 2012;13:1654–1658. doi: 10.2174/138945012803530035. [DOI] [PubMed] [Google Scholar]
- [13].Muller S, Brown PJ. Epigenetic chemical probes. Clin Pharmacol Ther. 2012;92:689–693. doi: 10.1038/clpt.2012.154. [DOI] [PubMed] [Google Scholar]
- [14].Fu H, et al. 14-3-3 proteins: structure, function, and regulation. Annual review of pharmacology and toxicology. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- [15].Sperandio O, et al. Rationalizing the chemical space of protein-protein interaction inhibitors. Drug Discov Today. 2010;15:220–229. doi: 10.1016/j.drudis.2009.11.007. [DOI] [PubMed] [Google Scholar]
- [16].Barker A, et al. Expanding medicinal chemistry space. Drug Discov Today. 2012;18:298–304. doi: 10.1016/j.drudis.2012.10.008. [DOI] [PubMed] [Google Scholar]
- [17].Mason JM. Design and development of peptides and peptide mimetics as antagonists for therapeutic intervention. Future Med Chem. 2010;2:1813–1822. doi: 10.4155/fmc.10.259. [DOI] [PubMed] [Google Scholar]
- [18].Billard C. Design of novel BH3 mimetics for the treatment of chronic lymphocytic leukemia. Leukemia. 2012;26:2032–2038. doi: 10.1038/leu.2012.88. [DOI] [PubMed] [Google Scholar]
- [19].Flygare JA, et al. Discovery of a potent small-molecule antagonist of inhibitor of apoptosis (IAP) proteins and clinical candidate for the treatment of cancer (GDC-0152) J Med Chem. 2012;55:4101–4113. doi: 10.1021/jm300060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Houghton PJ, et al. Initial testing (stage 1) of LCL161, a SMAC mimetic, by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer. 2012;58:636–639. doi: 10.1002/pbc.23167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rew Y, et al. Structure-based design of novel inhibitors of the MDM2-p53 interaction. J Med Chem. 2012;55:4936–4954. doi: 10.1021/jm300354j. [DOI] [PubMed] [Google Scholar]
- [22].Moellering RE, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rubinstein M, Niv MY. Peptidic modulators of protein-protein interactions: progress and challenges in computational design. Biopolymers. 2009;91:505–513. doi: 10.1002/bip.21164. [DOI] [PubMed] [Google Scholar]
- [24].Vanhee P, et al. Computational design of peptide ligands. Trends Biotechnol. 2011;29:231–239. doi: 10.1016/j.tibtech.2011.01.004. [DOI] [PubMed] [Google Scholar]
- [25].Rothe A, et al. In vitro display technologies reveal novel biopharmaceutics. FASEB J. 2006;20:1599–1610. doi: 10.1096/fj.05-5650rev. [DOI] [PubMed] [Google Scholar]
- [26].Cummings CG, Hamilton AD. Disrupting protein-protein interactions with non-peptidic, small molecule alpha-helix mimetics. Curr Opin Chem Biol. 2010;14:341–346. doi: 10.1016/j.cbpa.2010.04.001. [DOI] [PubMed] [Google Scholar]
- [27].Whitby LR, Boger DL. Comprehensive peptidomimetic libraries targeting protein-protein interactions. Acc Chem Res. 2012;45:1698–1709. doi: 10.1021/ar300025n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Brown CJ, et al. Stapled Peptides with Improved Potency and Specificity That Activate p53. ACS Chem Biol. 2013;8:506–512. doi: 10.1021/cb3005148. [DOI] [PubMed] [Google Scholar]
- [29].Henchey LK, et al. Contemporary strategies for the stabilization of peptides in the alpha-helical conformation. Curr Opin Chem Biol. 2008;12:692–697. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kawamoto SA, et al. Design of triazole-stapled BCL9 alpha-helical peptides to target the beta-catenin/B-cell CLL/lymphoma 9 (BCL9) protein-protein interaction. J Med Chem. 2012;55:1137–1146. doi: 10.1021/jm201125d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:7264–7270. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- [32].Du Y, et al. A dual-readout F2 assay that combines fluorescence resonance energy transfer and fluorescence polarization for monitoring bimolecular interactions. Assay Drug Dev Technol. 2011;9:382–393. doi: 10.1089/adt.2010.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Arkin MR, et al. Inhibition of Protein-Protein Interactions: Non-Cellular Assay Formats. In: Sittampalam GS, et al., editors. Assay Guidance Manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2012. [PubMed] [Google Scholar]
- [34].Dudgeon DD, et al. Characterization and optimization of a novel protein-protein interaction biosensor high-content screening assay to identify disruptors of the interactions between p53 and hDM2. Assay Drug Dev Technol. 2010;8:437–458. doi: 10.1089/adt.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Valkov E, et al. Targeting protein-protein interactions and fragment-based drug discovery. Top Curr Chem. 2012;317:145–179. doi: 10.1007/128_2011_265. [DOI] [PubMed] [Google Scholar]
- [36].Scott DE, et al. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51:4990–5003. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- [37].Meireles LM, Mustata G. Discovery of modulators of protein-protein interactions: current approaches and limitations. Curr Top Med Chem. 2011;11:248–257. doi: 10.2174/156802611794072632. [DOI] [PubMed] [Google Scholar]
- [38].Winter A, et al. Biophysical and computational fragment-based approaches to targeting protein-protein interactions: applications in structure-guided drug discovery. Q Rev Biophys. 2012;45:383–426. doi: 10.1017/S0033583512000108. [DOI] [PubMed] [Google Scholar]
- [39].Maurer T. Advancing fragment binders to lead-like compounds using ligand and protein-based NMR spectroscopy. Methods Enzymol. 2011;493:469–485. doi: 10.1016/B978-0-12-381274-2.00018-2. [DOI] [PubMed] [Google Scholar]
- [40].Wu B, et al. HTS by NMR of Combinatorial Libraries: A Fragment-Based Approach to Ligand Discovery. Chem Biol. 2013;20:19–33. doi: 10.1016/j.chembiol.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lugovskoy AA, et al. A novel approach for characterizing protein ligand complexes: molecular basis for specificity of small-molecule Bcl-2 inhibitors. J Am Chem Soc. 2002;124:1234–1240. doi: 10.1021/ja011239y. [DOI] [PubMed] [Google Scholar]
- [42].Tsao DH, et al. Discovery of novel inhibitors of the ZipA/FtsZ complex by NMR fragment screening coupled with structure-based design. Bioorg Med Chem. 2006;14:7953–7961. doi: 10.1016/j.bmc.2006.07.050. [DOI] [PubMed] [Google Scholar]
- [43].Sun Q, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Murray CW, et al. Fragment-based drug discovery applied to Hsp90. Discovery of two lead series with high ligand efficiency. J Med Chem. 2010;53:5942–5955. doi: 10.1021/jm100059d. [DOI] [PubMed] [Google Scholar]
- [45].Woodhead AJ, et al. Discovery of (2,4-dihydroxy-5-isopropylphenyl)-[5-(4-methylpiperazin-1-ylmethyl)-1,3-dihydrois oindol-2-yl]methanone (AT13387), a novel inhibitor of the molecular chaperone Hsp90 by fragment based drug design. J Med Chem. 2010;53:5956–5969. doi: 10.1021/jm100060b. [DOI] [PubMed] [Google Scholar]
- [46].Miura T, et al. Lead generation of heat shock protein 90 inhibitors by a combination of fragment-based approach, virtual screening, and structure-based drug design. Bioorganic & Medicinal Chemistry Letters. 2011;21:5778–5783. doi: 10.1016/j.bmcl.2011.08.001. [DOI] [PubMed] [Google Scholar]
- [47].Suda A, et al. Design and synthesis of novel macrocyclic 2-amino-6-arylpyrimidine Hsp90 inhibitors. Bioorganic & Medicinal Chemistry Letters. 2012;22:1136–1141. doi: 10.1016/j.bmcl.2011.11.100. [DOI] [PubMed] [Google Scholar]
- [48].Erlanson Da Fau - Wells JA, et al. Tethering: fragment-based drug discovery. [DOI] [PubMed] [Google Scholar]
- [49].Yang SY. Pharmacophore modeling and applications in drug discovery: challenges and recent advances. Drug Discov Today. 2010;15:444–450. doi: 10.1016/j.drudis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- [50].Lu Y, et al. Discovery of a nanomolar inhibitor of the human murine double minute 2 (MDM2)-p53 interaction through an integrated, virtual database screening strategy. J Med Chem. 2006;49:3759–3762. doi: 10.1021/jm060023+. [DOI] [PubMed] [Google Scholar]
- [51].Zhuang C, et al. Discovery, Synthesis, and Biological Evaluation of Orally Active Pyrrolidone Derivatives as Novel Inhibitors of p53-MDM2 Protein-Protein Interaction. J Med Chem. 2012;55:9630–9642. doi: 10.1021/jm300969t. [DOI] [PubMed] [Google Scholar]
- [52].Mukherjee P, et al. Targeting the BH3 domain mediated protein-protein interaction of Bcl-xL through virtual screening. J Chem Inf Model. 2010;50:906–923. doi: 10.1021/ci1000373. [DOI] [PubMed] [Google Scholar]
- [53].Corradi V, et al. Computational techniques are valuable tools for the discovery of protein-protein interaction inhibitors: the 14-3-3sigma case. Bioorg Med Chem Lett. 2011;21:6867–6871. doi: 10.1016/j.bmcl.2011.09.011. [DOI] [PubMed] [Google Scholar]
- [54].Scheper J, et al. Protein-protein interaction antagonists as novel inhibitors of non-canonical polyubiquitylation. PLoS One. 2010;5:e11403. doi: 10.1371/journal.pone.0011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lawrence HR, et al. Identification of a disruptor of the MDM2-p53 protein-protein interaction facilitated by high-throughput in silico docking. Bioorg Med Chem Lett. 2009;19:3756–3759. doi: 10.1016/j.bmcl.2009.04.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tian W, et al. Structure-based discovery of a novel inhibitor targeting the beta-catenin/Tcf4 interaction. Biochemistry. 2012;51:724–731. doi: 10.1021/bi201428h. [DOI] [PubMed] [Google Scholar]
- [57].Wang S, et al. Targeting the MDM2-p53 protein-protein interaction for new cancer therapeutics. Top Med. Chem. 2012;8:57–80. [Google Scholar]
- [58].Wang X, Jiang X. Mdm2 and MdmX partner to regulate p53. FEBS Lett. 2012;586:1390–1396. doi: 10.1016/j.febslet.2012.02.049. [DOI] [PubMed] [Google Scholar]
- [59].Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science (New York, N.Y.) 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- [60].Vu BT, Vassilev L. Small-molecule inhibitors of the p53-MDM2 interaction. Curr Top Microbiol Immunol. 2011;348:151–172. doi: 10.1007/82_2010_110. [DOI] [PubMed] [Google Scholar]
- [61].Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science (New York, N.Y.) 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- [62].Popowicz GM, et al. The structure-based design of Mdm2/Mdmx-p53 inhibitors gets serious. Angew Chem Int Ed Engl. 2011;50:2680–2688. doi: 10.1002/anie.201003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Andreeff M, et al. A Multi-Center, Open-Label, Phase I Study of Single Agent RG7112, A First In Class p53-MDM2 Antagonist, In Patients with Relapsed/Refractory Acute Myeloid and Lymphoid Leukemias (AML/ALL) and Refractory Chronic Lymphocytic Leukemia/Small Cell Lymphocytic Lymphomas (CLL/SCLL) Blood (ASH Annual Meeting Abstracts) 2010;116:657. [Google Scholar]
- [64]. http://clinicaltrials.gov/ct2/show/NCT01462175%3Fterm%3DRO5503781%26rank%3D1.
- [65].Franklin MC, et al. Structure and function analysis of peptide antagonists of melanoma inhibitor of apoptosis (ML-IAP) Biochemistry. 2003;42:8223–8231. doi: 10.1021/bi034227t. [DOI] [PubMed] [Google Scholar]
- [66].Srinivasula SM, et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- [67].Infante JR, et al. AACR 101st Annaul Meeting; Washingtin DC, USA. 2010; Abstract 2775. [Google Scholar]
- [68].Cai Q, et al. A potent and orally active antagonist (SM-406/AT-406) of multiple inhibitor of apoptosis proteins (IAPs) in clinical development for cancer treatment. J Med Chem. 2011;54:2714–2726. doi: 10.1021/jm101505d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Eckhardt SG, et al. ASCO Annaul Meeting; Chicago, IL, USA. 2010; Abstract 2580. [Google Scholar]
- [70].Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- [71].Lee EF, et al. Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 2007;14:1711–1713. doi: 10.1038/sj.cdd.4402178. [DOI] [PubMed] [Google Scholar]
- [72].Sattler M, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science (New York, N.Y.) 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- [73].Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- [74].Nguyen M, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci U S A. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Paik PK, et al. A phase II study of obatoclax mesylate, a Bcl-2 antagonist, plus topotecan in relapsed small cell lung cancer. Lung Cancer. 2011;74:481–485. doi: 10.1016/j.lungcan.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Maloney A, Workman P. HSP90 as a new therapeutic target for cancer therapy: the story unfolds. Expert Opin Biol Ther. 2002;2:3–24. doi: 10.1517/14712598.2.1.3. [DOI] [PubMed] [Google Scholar]
- [77].Porter JR, et al. Discovery and development of Hsp90 inhibitors: a promising pathway for cancer therapy. Curr Opin Chem Biol. 2010;14:412–420. doi: 10.1016/j.cbpa.2010.03.019. [DOI] [PubMed] [Google Scholar]
- [78].Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Schulte TW, Neckers LM. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol. 1998;42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- [80].Vogelstein B, et al. Cancer genome landscapes. Science (New York, N.Y.) 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- [82].Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zhang QC, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].James LI, et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat Chem Biol. 2013;9:184–191. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Karatas H, et al. High-Affinity, Small-Molecule Peptidomimetic Inhibitors of MLL1/WDR5 Protein-Protein Interaction. J Am Chem Soc. 2013;135:669–682. doi: 10.1021/ja306028q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhou H, et al. Structure-based design of high-affinity macrocyclic peptidomimetics to block the menin-mixed lineage leukemia 1 (MLL1) protein-protein interaction. J Med Chem. 2013;56:1113–1123. doi: 10.1021/jm3015298. [DOI] [PubMed] [Google Scholar]
- [90].Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sun SY, et al. Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 2005;65:7052–7058. doi: 10.1158/0008-5472.CAN-05-0917. [DOI] [PubMed] [Google Scholar]
- [92].Sale MJ, Cook SJ. The BH3 mimetic ABT-263 synergizes with the MEK1/2 inhibitor selumetinib/AZD6244 to promote BIM-dependent tumour cell death and inhibit acquired resistance. Biochem J. 2013;450:285–294. doi: 10.1042/BJ20121212. [DOI] [PubMed] [Google Scholar]