Abstract
Hematopoietic stem cells (HSCs) can either self-renew or differentiate into various types of cells of the blood lineage. Signaling pathways that regulate this choice of self-renewal versus differentiation are currently under extensive investigation. Here we report that deregulation of Notch signaling skews HSC differentiation in mouse models of Fanconi anemia (FA), a genetic disorder associated with bone marrow failure and progression to leukemia and other cancers. In mice expressing a transgenic Notch reporter, deletion of the Fanca or Fancc gene enhances Notch signaling in multipotential progenitors (MPPs), which is correlated with decreased phenotypic long-term HSCs and increased formation of MPP1 progenitors. Furthermore, we found an inverse correlation between Notch signaling and self-renewal capacity in FA hematopoietic stem and progenitor cells. Significantly, FA deficiency in MPPs deregulates a complex network of genes in the Notch and canonical NF-κB pathways. Genetic ablation or pharmacologic inhibition of NF-κB reduces Notch signaling in FA MPPs to near wide-type level, and blocking either NF-κB or Notch signaling partially restores FA HSC quiescence and self-renewal capacity. Taken together, these results suggest a functional crosstalk between Notch signaling and NF-κB pathway in regulation of HSC differentiation.
Introduction
Hematopoietic stem cells (HSCs) are rare cell population which can either self-renew or differentiate into various types of mature blood cells. Delineating the signaling pathways that regulate this choice of self-renewal versus differentiation remains an enduring challenge of foremost importance. One of the key regulators is Notch, an extracellular signal plays a critical role in hematopoietic homeostasis (1). Gain or loss of Notch signaling components has been directly linked to multiple human disorders including hematologic malignancies (2, 3). NF-κB is another known factor that controls cell fate, survival and differentiation (4, 5). NF-κB is indicated in various physiological processes, including immunity, inflammation, development and differentiation (4, 6). It has been shown that crosstalk between the canonical NF-κB and Notch signaling pathways may influence tissue homeostasis in certain cell types including hematopoietic progenitor cells (7, 8).
Fanconi anemia (FA) is an inherited disorder characterized by genome instability and an extremely high cancer predisposition (9–11). FA is genetically heterogeneous, with 15 complementation groups identified thus far. The genes encoding the groups A (FANCA), B (FANCB), C (FANCC), D1 (FANCD1/BRCA2), D2 (FANCD2), E (FANCE), F (FANCF), G (FANCG), I (FANCI/KIAA1794), J (FANCJ/BRIP1), L (FANCL), M (FANCM), N (FANCN/PALB2), O(FANCO/RAD51C) and P(FANCP/SLX4) have been cloned (9–13). The biological function of these FA proteins has been subjected to intensive investigation. The studies demonstrate that eight of the FA proteins (FANCA, B, C, E, F, G, L, and M) form a core complex, which functions as an ubiquitin ligase. In response to DNA damage or DNA replication stress, the FA core complex monoubiquitinates two downstream FA proteins, FANCD2 and FANCI, which then recruit other downstream FA proteins including FANCD1 (which is the breast cancer protein BRCA2), FANCJ, FANCN and other DNA repair factors, to nuclear loci containing damaged DNA and consequently influence important cellular processes such as DNA replication, cell-cycle control, and DNA damage response and repair (12, 13).
Two of the most important clinical hallmarks of FA are bone marrow (BM) failure and progression to leukemia caused by HSC depletion and malignant transformation (14). Indeed, there are fewer HSCs in FA patients and knockout mice compared to normal controls (11, 15). In fact, HSCs from FA patients or knockout mice display severe defects in in vitro survival and in vivo repopulating (14, 16–18). Although these hematologic phenotypes suggest that the FA pathway likely plays specific roles in hematopoiesis, the mechanisms by which FA proteins regulate HSC self-renewal and differentiation is unknown. In the present study, we show that FA deficiency enhances Notch signaling in multipotential progenitors (MPPs), which is correlated with decreased phenotypic long-term HSCs and increased formation of MPP1 progenitors. Mechanistically, we show that FA deficiency in MPPs deregulates genes controlled by the NF-κB pathway leading to enhanced Notch signaling. The study thus identifies a functional cross-talk between the NF-κB pathway and Notch signaling in HSC differentiation and establishes a role of FA proteins in the control of balance between renewal and lineage commitment. In addition, our data also lends support to the recent report that several members of FA core complex, including FANCA, FANCF, FANCG and FANCL functionally interact with HES1, a key player in the Notch signaling pathway (19).
Materials and Methods
Mice and treatment
Notch-eGFP;Fanca−/− or Notch-eGFP;Fancc−/− mice were generated by interbreeding the heterozygous Fanca+/− or Fancc+/− mice (20, 21) with Notch-eGFP transgenic mice (22). Notch-eGFP;Fanca−/−;p65f/f mice were generated by interbreeding the RelA/p65 mice (23) with Notch-eGFP-Fanca+/−. p65 gene deletion was accomplished by Cre-mediated deletion of floxed alleles by crossing the Notch-eGFP;Fanca+/−;p65f/f mice with a Cre-ERT2 strain (24) and intraperitoneally (i.p.) injecting 100 µl of Tamoxifen (20 mg/ml, Sigma- Aldrich, St Louis, MO) daily for 3 days. Animals were maintained in the animal barrier facility at Cincinnati Children’s Hospital Medical Center.
For in vivo treatment, mice were subcutaneously injected with γ-secretase inhibitor, DAPT in a range of doses (0, 10, 50, 100 and 200 mg/kg, Tocris, R&D, Minneapolis, MN) daily for 2 days followed by bone marrow cell isolation on day 3 (25–27). For TNF-α treatment, a single injection (100 µg/kg) of mouse recombinant TNF-α(Peprotech, Rocky Hill, NJ) was given by intraperitoneally (i.p.) injection 10 days before sacrifice (28). BAY 11–7082 (Calbiochem- Novabiochem Corporation, La Jolla, CA) was administered (i.p.) to mice at doses of 20 mg/kg/day for 20 days (29). Mice were then sacrificed for bone marrow cell isolation 12 hours after the final injection.
Flow Cytometry analysis and Cell Cycle analysis
The lineage marker (Lin) mixture (BD Biosciences, San Jose, CA) for BM cells from Notch-eGFP-WT and Notch-eGFP-Fanca−/− or Notch-eGFP-Fancc−/− mice included the following biotinylated antibodies: CD3ε (145-2C11), CD11b (M1/70), CD45R/B220 (RA3-6B2), mouse erythroid cells Ly-76 (Ter119), Ly6G and Ly-6C (RB6–8C5). Other conjugated antibodies (BD Biosciences, San Jose, CA) used for surface staining included: CD45.1 (A20), CD45.2 (A104), Sca1 (D7), c-kit (2B8), CD34 (RAM34), Flt3 (A2F10.1), CD48 (HM48-1), CD150 (9D1), IL-7Rα (HIL-7R-M21). Biotinylated primary antibodies were detected by incubation of antibody coated cells with streptavidin-PerCP Cy5.5 (BD Biosciences, San Jose, CA) in a two-step staining procedure.
Intracellular staining for the active Notch1 (NICD) was conducted as previously described (30). Briefly, surface marker stained CD150+CD48− LSK cells (LT-HSCs) from Notch-eGFP-Fanca+/+ or Notch-eGFP-Fanca−/− mice pretreated with different doses of DAPT were first fixed and permeablized with a Phosflow kit (BD Pharmingen, San Jose, CA) and then stained with Notch1 antibody (mN1A) according to the manufacturer’s instructions (BD sciences, San Jose, CA).
For cell cycle analysis, cells were surface stained, fixed and permeabilized in BD Cytofix/Cytoperm Buffer (BD Biosciences, San Jose, CA), then stained with 5 µg/ml Hoechst 33342 and 150 ng/ml Pyronin Y (Sigma-Aldrich, St Louis, MO). Cells were then subjected to Flow Cytometric analysis (BD Biosciences, San Jose, CA).
In vitro T cell differentiation assay
4000 LSK cells isolated from Notch-eGFP-Fanca+/+ or Notch-eGFP-Fanca−/− were seeded to 24-well plate pre-coated with OP9-DL1 stromal cells (20,000 cells plated the day before the co-culture). The co-culture was conducted in α-MEM medium (Invitrogen, Grand Island, NY) supplemented with 20% fetal bovine serum and the following cytokines: 5ng/ ml IL-7, 5 ng/ml Flt3L (Peprotech, Rocky Hill, NJ).Suspension cells were subjected to Flow cytometric analysis using antibodies specific for CD3 (BD Pharmingen, San Jose, CA), CD4 (eBioscience, San Diego, CA), CD8, TCRαβ(BD Pharmingen, San Jose, CA) and TCRγδ (Beckman Coulter, Brea, CA) on day 14 and day 21.
CFU assay
The upper 30% of Notch-eGFP Lin−Sca1+c-kit+ (LSK) cells or lower 30% of Notch-eGFP LSK cells were sorted from Notch-eGFP-WT, Notch-eGFP-Fanca−/− or Notch-eGFP-Fancc−/− mice. 100 LSK cells were seeded in duplicate and cultured in cytokine-supplemented methylcellulose medium (MethoCult 3434; Stem Cell Technologies, Vancouver, Canada) in 35 mm-gridded dishes. For CFU assay using Notch-eGFP− or Notch-eGFP+ LT-HSCs or MMP1 cells, GFP positive and negative cells from LT-HSC or MPP1 gated cell compartment were sorted using FACS AriaII (BD Biosciences, San Jose, CA). Subsequently, 20 LT-HSCs or 200 MMP1 were plated. Colonies were counted on day 7, then isolated, replated and cultured for another 7 days.
Competitive transplantation
Competitive repopulation experiments were conducted by transplanting 500 Notch-eGFPlo or Notch-eGFPhi LSK cells from Notch-eGFP-WT, Notch-eGFP-Fanca−/− or Notch-eGFP-Fancc−/− mice along with 0.3 million of congenic BM cells as competitors into lethally irradiated BoyJ recipients as previously described (31). For BMT using Notch-eGFP− or Notch-eGFP+ LT-HSCs or MMP1 cells, 50 LT-HSCs or 1000 MPP1 (CD45.2+) cells plus 0.3 million CD45.1+ cells were used. Donor-derived chimerism (CD45.2+) in recipient at 4, 8, 16 and 24 weeks post transplantation were determined by CD45.1-PE and CD45.2-FITC markers staining followed by Flow Cytometry FACSCanto I (BD Biosciences, San Jose, CA) analysis.
RNA isolation, Pathway-specific gene array analysis and Real time-PCR
Total RNA of purified MPP1 subsets from Notch-eGFP-WT, Notch-eGFP-Fanca−/− and Notch-eGFP-Fancc−/− mice was prepared with RNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s procedure. To address the inhibitory effect of DAPT, total RNA was extracted from MPP cells isolated from Notch-eGFP-Fanca+/+ or Notch-eGFP-Fanca−/− mice treated with or without DAPT (100 mg/kg body weight). Reverse transcription was performed with random hexamers and Superscript II RT (Invitrogen, Grand Island, NY) and was carried out at 42 °C for 60 min and stopped at 95 °C for 5 min. First-strand cDNA was used for pathway-specific arrays (SABiosciences, Valencia, CA). Genes involved in DNA repair, Cell cycle control, oxidative stress, inflammation, apoptosis and Notch signaling pathways were analyzed. Genes with folder changes>3 were picked up for confirmation by real time-PCR.
For real time-PCR, 1st cDNA described above were employed for PCR amplification using the primers listed in Table 1. Samples were normalized to the level of GAPDH mRNA, and the relative expression levels were determined by the standard curve method.
Table 1.
Primers used for Real-time PCR
| Symbol | Forward primer | Reverse primer |
|---|---|---|
| Gadd45b | CTCCTGGTCACGAACTGTCA | GGGTAGGGTAGCCTTTGAGG |
| Rela | GCGTACACATTCTGGGGAGT | ACCGAAGCAGGAGCTATCAA |
| Sod2 | GCCCCCTGAGTTGTTGAATA | AGACAGGCAAGGCTCTACCA |
| Tnfrsf1b | TGGCAGAGGAGCCTAGTTGT | CACACCCAGGAACAGTCCTT |
| Stat1 | TGGTGAAATTGCAAGAGCTG | CAGACTTCCGTTGGTGGATT |
| Xiap | TTGGAACATGGACATCCTCA | TGCCCCTTCTCATCCAATAG |
| Hes1 | CCCACCTCTCTCTTCTGACG | AGGCGCAATCCAATATGAAC |
| Hey1 | AGCAGTGAGGTGAAGGGAGA | AACGGTGAAATCCGTGAGAC |
| Hoxb4 | CTGGATCACAGCCTGGATTT | TGGAAGGGAGTGAGCAGTCT |
| Fzd1 | CAAGGTTTACGGGCTCATGT | GTAACAGCCGGACAGGAAAA |
| Notch2 | CCTGAACGGGCAGTACATTT | GCGTAGCCCTTCAGACACTC |
| Numb | CGGGAAAGAAAGCAGTGAAG | AGTGGTGCCATCACGACATA |
| GAPDH | AACI I IGGCATTGTGGAAGG | ACACATTGGGGGTAGGAACA |
Preparation of cell extracts, Immunoblotting
To prepare whole cell extracts, Lin− cells from Notch-eGFP-Fanca−/− mice treated with indicated doses of DAPT were washed with ice-cold PBS, and resuspended in ice-cold lysis buffer containing 50mM Tris-HCL (pH 7.4), 0.1% NP40, and 1M NaCl supplemented with protease and phosphatase inhibitors (10µg/ml aprotinin, 25µg/ml leupeptin, 10µg/ml pepstatin A, 2mM PMSF, 0.1M NaP2O4, 25mM NaF and 2mM sodium orthovandate) for 30 min on ice. Cell lysates were resolved on SDS-PAGE, and immunoblots were analyzed with antibodies for the active form of Notch1 (NICD), Stat1 (Cell Signaling, Boston, MA), or β-actin (Sigma-Aldrich, St Louis, MO). Each lane contains protein from 50,000 Lineage depleted cells (Miltenyi Biotec, Auburn, CA). Signals were visualized by incubation with anti-mouse or anti-rabbit secondary antibodies followed by ECL chemiluminescence (Amersham Biosciences, Piscataway, NJ).
Statistical analyses
Paired or unpaired student’s t-test was used for two-group comparison, and one-way ANOVA for more than two-group comparison. Values of p less than 0.05 were considered statistically significant. Results are presented as mean ± SD. * indicates p<0.05; ** indicates p<0.01; *** indicates p<0.001.
Results
Enhanced Notch signaling in FA MPPs
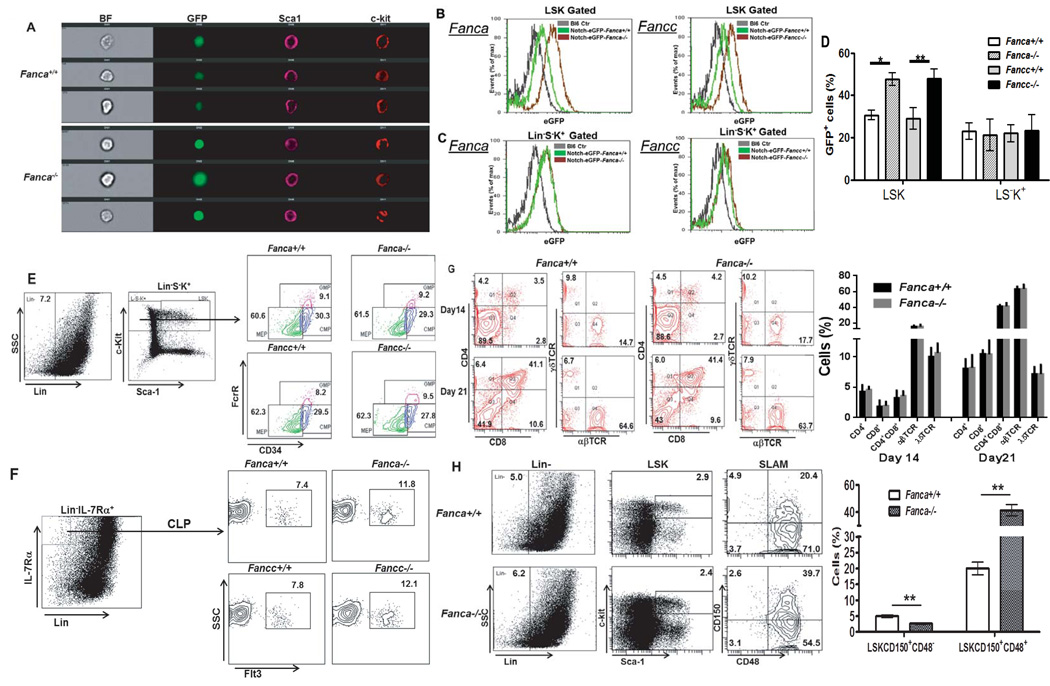
To examine whether FA murine HSCs can balance self-renewal with differentiation, we crossed two FA mouse models (Fanca+/− and Fancc+/−) with a reporter strain, in which Notch-driven eGFP expression acts as a sensor for HSC differentiation (22). We first used ImageStream to analyze Notch signaling in Notch-eGFP expression in Lin−Sca1+c-kit+ (LSK) cells, a population containing HSCs and multipotential progenitors. We observed higher Notch-eGFP expression in LSK cells from FA mice compared to WT littermates (Fig 1A). Consistent with this, flow cytometric analysis showed that FA LSK cells expressed higher eGFP than their WT counterparts (Fig 1B, D). It appeared that Notch-eGFP was also expressed in Lin−S−K+ cells (Fig 1C), a population consisting mostly of myeloid progenitors, from mice of either genotype, albeit at lower levels than more primitive LSK cells. However, no difference in Notch-eGFP expression was observed between WT and FA Lin−S−K+ cells (Fig 1C, D).
Figure 1. Increased Notch-eGFP expression in FA HSPCs.
(A) eGFP+ LSK cells from Fanca−/− mice are brighter than those from WT. Whole bone marrow cells (WBMCs) from Notch-eGFP-Fanca+/+or Notch-eGFP-Fanca−/− mice were isolated followed by Lineage+ cell depletion. LinSca1+c-kit+ (LSK) cells were labeled and subjected to ImageStream analysis. Representative images were shown (n=5-7). (B) Enhanced Notch signaling in FA LSK cells. Whole bone marrow cells (WBMCs) from Notch-eGFP-Fanca−/−, Notch-eGFP-Fancc−/−, or WT littermates were isolated for cell surface marker staining. LSK cells were gated for eGFP expression analysis by Flow Cytometry. (C) No difference in Notch expression between FA and WT LinK+S cells. Cells described in (A) were gated for Linc-kit+Sca1 (LinK+S) population followed by Flow Cytometry analysis for eGFP expression. (D) Quantification of eGFP expression in LSK and LinK+S cells. Results are means plus or minus SD of 3 independent experiments (n=12 per group). (E–F) FA deficiency does not alter myeloid differentiation (E) or lymphoid differentiation (F). Cells described in (B) were subjected to Flow Cytometry analysis for myeloid or lymphoid progenitors. (G) FA deficiency does not affect T cell differentiation in vitro. 4000 LSK cells isolated from either Fanca+/+ or Fanca−/− mice were seeded to 24-well plate pre-coated with OP9-DL1 cells. Suspension cells were collected on day 14 and day 21 followed by Flow cytometric analysis for CD3, CD4, CD8, αβTCR and γδTCR. Representative image (Upper) and quantification (Lower) are shown. Results are means plus or minus SD of 3 independent experiments. (H) FA deficiency results in the reduction of LT-HSC and increased MPP frequency. Cells described in (A) were stained for SLAM markers followed by Flow Cytometry analysis. Representative image (Left) and quantification (Right) are shown. Results are means plus or minus SD of 3 independent experiments (n=5–7 per group).
Since Notch signaling favors lymphoid while interferes with myeloid differentiation (32–35), we next determined if increased Notch expression in FA LSK cells was associated with alteration in lineage differentiation. Surprisingly, we did not see reduced myeloid differentiation in FA mice. Specifically, when we looked further into the subsets in the stem/progenitor compartment, that is, granulocyte-macrophage progenitors (GMP), common myeloid progenitors (CMP) and megakaryocyte-erythrocyte progenitors (MEP), we saw no difference between WT and FA mice (Fig1E) although a slight increased portion of common lymphoid progenitors (CLP) was observed in FA mice (Fig 1F). Consistent with this, the frequencies of CD4+ single positive (SP) T cells, CD8+ SP T cells, CD4+CD8+ double positive (DP) T cells, as well as T cells bearing αβTCR+ or γδRCT+ cells derived from HSPCs at different stages of co-culture with OP9-DL1 feeder cells in vitro (36) were similar between the genotypes (Fig 1G). HSC differentiate through intermediate stages, generating at least two subsets of multipotential progenitors (MPP1 and MPP2; 37). Only LT-HSCs have the ability to self-renew. Therefore, we subdivided the LSK fraction using signaling lymphocytic-activation molecule markers (CD150 and CD48; SLAM; 38). We observed lower LT-HSC (LSKCD150+CD48−) number but higher MPP1 (LSKCD150+CD48+) cells in both Fanca−/− and Fancc−/− mice than in WT mice (Fig 1H). Therefore, reduction in LT-HSCs is associated with an increase in the number of MPP1 cells in the BM of FA mice.
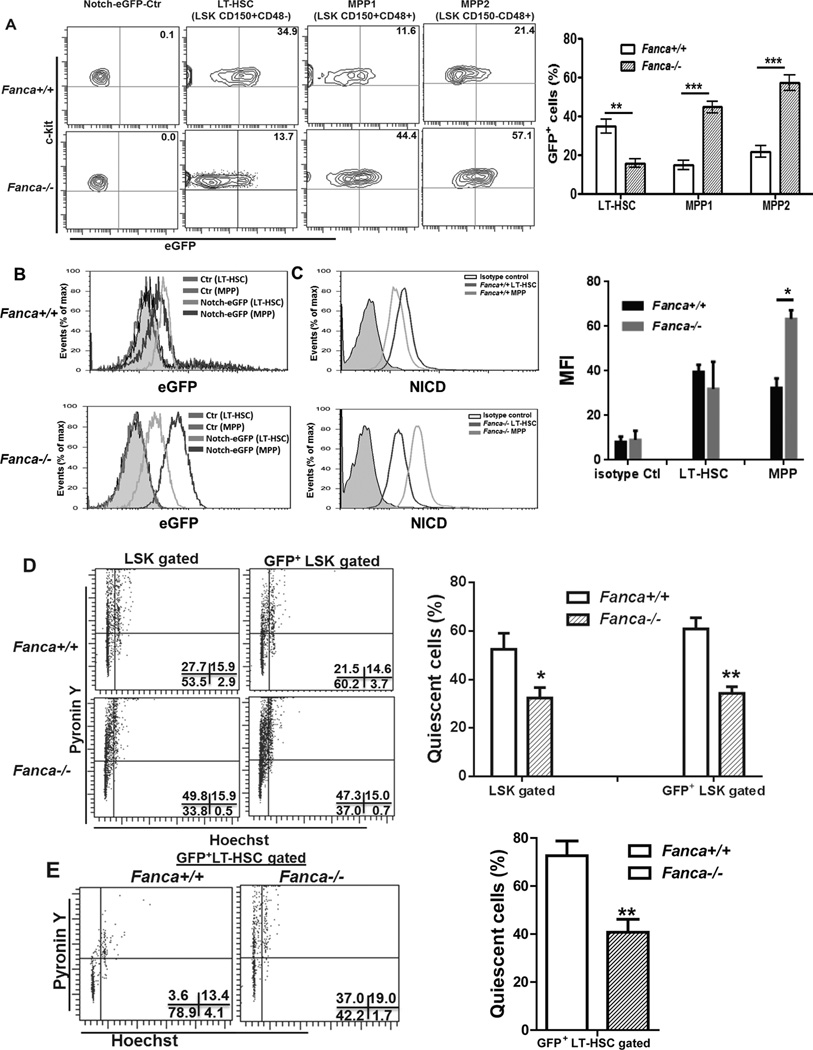
Since we observed higher Notch-eGFP expression in FA LSK cells than in WT LSK cells, we next analyzed Notch signaling in more defined HSPC populations. Surprisingly, WT and FA mice showed strikingly different patterns of Notch-eGFP expression in LT-HSCs vs MPPs. Specifically, the percentage of WT LT-HSCs expressing Notch-eGFP was significantly higher than that of Fanca−/− LT-HSCs (Fig 2A). In contrast, FA MPPs expressed much higher Notch-eGFP than WT MPPs (Fig 2A). This was confirmed by analyzing the intracellular level of Notch-eGFP expression in single cells from these subsets (Fig 2B). Further, intracellular staining using antibody specific for the active Notch1 (NICD; 30, 39, 40) showed positive correlation between eGFP expression level and active Notch signal in vivo (Fig 2C). Thus, these results link Notch signaling to specific stages of HSPC frequencies.
Fig 2. Enhanced Notch signaling in Fanca−/− MPPs.
(A) Percentage of GFP+ cells in LT-HSC and MPP populations. WBMCs from Notch-eGFP-Fanca+/+ or Notch-eGFP-Fanca−/− mice were isolated and stained for SLAM markers followed by Flow Cytometry analysis. Representative GFP expression (Upper) from three cell populations: LT-HSC (LinSca1+c-kit+CD150+CD48); MPP1 (LinSca1+c-kit+CD150+CD48+); or MPP2 ((Lin-Sca1+c-kit+CD150−CD48+) and quantification (Lower) were shown. Results are means plus or minus SD of 3 independent experiments (n=9 per group). (B) Intracellular expression levels of GFP in LT-HSC and MPP subsets. The levels of GFP expression from cell populations described in (A) were plotted for histogram display. (C) eGFP expression is positively correlated with active Notch1 in HSPCs. WBMCs from Notch-eGFP-Fanca+/+ or Notch-eGFP-Fanca−/− mice were subjected to intracellular staining for active Notch1 (NICD). LT-HSC or MPP cell portions were gated for histogram display. (D) Decreased quiescence in Fanca−/− HSPCs. Cell described in (A) were subjected to cell cycle analysis by Hochest 33342/Pyronin Y staining gated on total or eGFP+ LSK cells respectively. (E) Decreased quiescence of Fanca−/− LT-HSCs. eGFP+ LT-HSCs from cells described in (A) were gated for cell cycle analysis. Representative flow graph (Left) and quantification (Right) were shown. Results are means plus or minus SD of 3 independent experiments (n=9 per group).
Because loss of cellular quiescence contributes to HSC exhaustion (41), we next determined the cell cycle status of FA HSCs by PY/Hochest staining. We noticed that both total and eGFP-positive LSK cells from FA mice showed significantly decreased G0 quiescent cells compared to those from WT mice (Fig 2D). To examine further the potential linkage between Notch signaling and HSC quiescence, we analyzed cell-cycle status of eGFP-positive LT-HSCs. Similar to the result obtained with the heterogeneous LSK cells, we observed significantly reduced quiescent Notch-eGFP+ LT-HSCs in FA mice compared to WT Notch-eGFP+ LT-HSCs (Fig 2E). Therefore, the data indicate that Notch signaling fails to rescue loss of quiescence in FA HSCs.
Inverse correlation between Notch signaling and self-renew capacity in FA HSCs
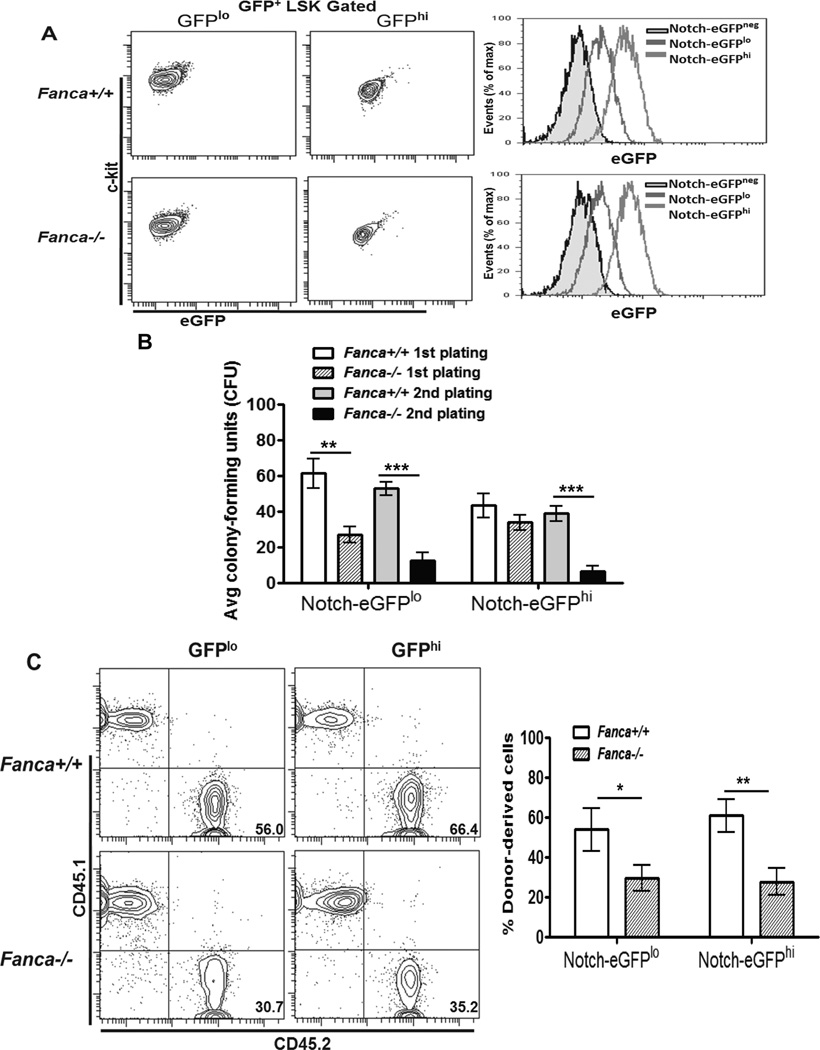
The observations that Notch-eGFP expression was low in FA LT-HSCs but high in FA MPP cells suggested that alteration in Notch signaling in FA LT-HSCs and MPP cells could influence their capacity to self-renew and differentiate. To test this hypothesis, we used FACS to divide the Notch-eGFP+ LSK population into Notch-eGFPhi (MFI values: 49.41± 3.29 for Fanca+/+ and 68.59 ± 3.07 for Fanca−/−) and Notch-eGFPlo (MFI values: 20.25 ± 2.68 for Fanca+/+ and 26.4 ± 4.72 for Fanca−/− ) fractions (Fig 3A) and determined the self-renewal capacity of these two populations. Colony-forming unit (CFU) assay showed that although the first plating showed that both WT and the FA Notch-eGFPhi fractions had similar capacity to generate colonies; replating of the FA Notch-eGFPhi LSK cells yielded much fewer colonies (Fig 3B), suggesting a possible loss of in vitro self-renewing capacity. To complement these in vitro studies, we did competitive BM transplantation with 500 LSK cells purified from both fractions (expressing the marker CD45.2) along with 0.3 million CD45.1+ competitor cells. Interestingly, we found that FA Notch-eGFPhi LSK cells still displayed defects in hematopoietic repopulation compared to the same number of WT Notch-eGFPhi LSK cells (Fig 3C).
Fig 3. Self-renewal defect in Fanca−/− Notch-eGFPhi HSPCs.
(A) Flow-based purification of GFPlo and GFPhi subsets. The upper 30% of Notch-eGFP LSK cells (GFPhi) or lower 30% of Notch-eGFP LSK cells (GFPlo) were sorted from mice with the indicated genotypes. Representative flow images (Left) and histogram overlay (Right) are shown. (B) Replating defects of Notch-eGFPhi LSK cells. 100 GFPlo or GFPhi LSK cells were seeded in duplicate and cultured in cytokine-supplemented methycellulose medium for 7 days. Colonies were then counted, isolated, replated and cultured for another 7 days. Error bars indicate standard deviation (std) of three experiments (n=9). (C) Hematopoietic reconstitution defects of Fanca−/− Notch-eGFPhi LSKs. 500 GFPlo or GFPhi LSK (CD45.2+) cells plus 0.3 million cells (CD45.1+) were tail vein injected to lethally irradiated recipient (BoyJ) mice. Donor-derived chimera was assessed by Flow Cytometry analysis 16 weeks post BMT. Representative dot graph (Left) and quantification (Right) are shown. Results are means plus or minus SD of 3 independent experiments (n=9 per group).
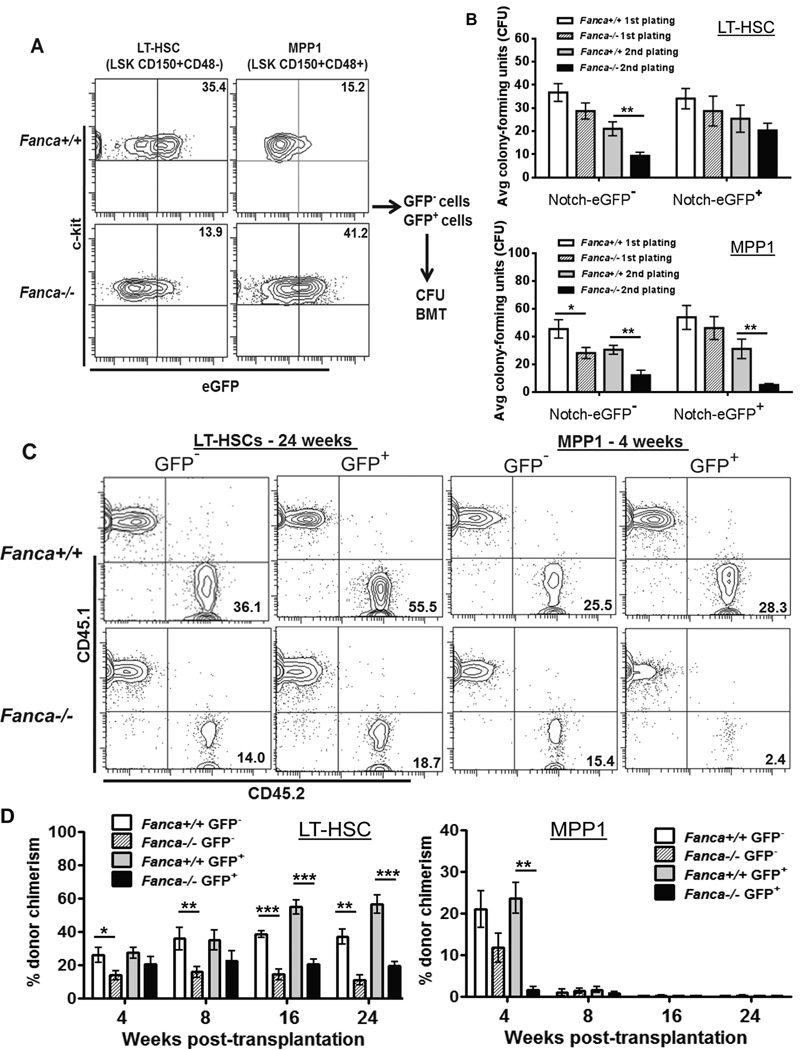
This result prompted us to reevaluate the LSK population used to identify engrafting HSCs by dividing the LT-HSCs and MPP1 population into Notch-eGFP− and Notch-eGFP+ cells (Fig 4A). As shown in Figure 4B, eGFP− LT-HSCs from Fanca−/− mice showed reduced colony forming units compared to WT eGFP− LT-HSC in the second plating, indicating an impaired in vitro self-renewal capacity. In contrast, Fanca−/− eGFP+ LT-HSCs show comparable colony-forming ability to their WT counterparts in both platings (Fig 4B). On the other hand, both eGFP+ and eGFP− MPP1 cells from Fanca−/− mice showed defective colony-forming capacity in the second plating (Fig 4B). These results suggest that Notch signaling may play distinct roles in self-renewal of LT-HSCs and MPP cells in vitro.
Fig 4. Inverse correlation between Notch signaling and self-renewal capacity of Fanca−/− HSPCs.
(A) Flow-based purification of GFP− and GFP+ subsets. GFP− and GFP+ cells from LT-HSC or MPP1 compartments of Notch-eGFP-Fanca−/− or Notch-eGFP-WT mice were sorted. Representative images (Left) and histogram overlay (Right) were shown. GFP− and GFP+ subsets were then subjected to CFU assay (B) and BMT (C). (B) Increased Notch signaling results in replating defect in Fanca−/− MPPs. 20 LT-HSCs or 200 MPP1 cells were seeded in duplicated and were cultured in cytokine-supplemented methylcellulose medium (MethoCult 3434). Subsequently, colonies were counted on day 7, then were isolated, replated and cultured for another 7 days. Error bars indicate standard deviation (STD) of three experiments (n=9). (C) Increased Notch signaling leads to self-renew defects in Fanca−/− HSCs. 50 LT-HSCs or 1000 MPP1 (CD45.2+) cells plus 0.3 million cells (CD45.1+) were tail vein injected to lethally irradiated recipient (BoyJ) mice. Donor-derived chimera was assessed by Flow Cytometry analysis at 4, 8, 16 and 24 weeks after transplantation. (D) Quantification of self-renewal capacity of Fanca−/− HSCs. Experiments described in (C) were quantified. Results are means plus or minus SD of 3 independent experiments (n=9 per group).
To examine the in vivo role of Notch signaling in self-renewal of LT-HSCs and MPP cells, we transplanted 50 LT-HSCs or 1000 MPP1 cells from each of the two fractions into lethally irradiated recipients to investigate their repopulating potential. Fanca−/− Notch-eGFP− LT-HSCs showed typical repopulating deficit at each time point post-transplantation (Fig 4C, D). Consistent with previous report (20), WT Notch-eGFP+ LT-HSCs exhibited enhanced long-term repopulating capacity. However, Fanca−/− Notch-eGFP+ LT-HSCs displayed quantitatively defective long-term repopulating activity with less than 20% donor-derived cells compared to more than 50% repopulated by WT Notch-eGFP+ LT-HSCs at 16 and 24 weeks post-transplant (Fig 4C, D). All LT-HSCs, regardless of genotype status or Notch-eGFP expression, generate HSPCs as well as all mature lineages 24 weeks after transplantation (Fig S1). Neither WT nor FA MPP1 cells, regardless of Notch-eGFP expression, provided long-term reconstitution (Fig 4C, D). In fact, Notch-eGFP+ FA MPP1 progenies were almost undetectable at 4 weeks after transplantation (Fig 4C, D). Taken together, these results indicate that Notch signaling fails to rescue repopulating defect of FA LT-HSCs and impairs the short-term repopulating ability of FA MPPs.
FA deficiency in MPPs deregulates the gene network of Notch and NF-κB pathways
To further understand the effect of enhanced Notch signaling mechanistically in FA MPPs, we purified MPP1 subsets and used pathway-specific gene array analysis to define the gene-expression signatures of each population (Fig S2A). Since FA cells have defects in DNA repair, Cell-cycle control, anti-oxidant defense, inflammatory response, and apoptotic signaling, we focused on these pathways as well as the Notch signaling pathway (Fig S2A). A substantial increase in the expression of genes involved in the NF-κB and Notch signaling pathways was observed in the Fanca−/− or Fancc−/− MPP1 cells (Table 2, Fig S2B, S2C). These include NF-κB target genes Gadd45b, Icam1, Irf1, Rela (p65) Sod2, Stat1, Tnfrsf1b (TNF receptor II) and Xiap, as well as Notch signature genes Hes1, Hey1, Hoxb4, Fzd1, Fzd2, Nfkb1, Notch1, Notch2 and Numb (Fig S2B, S2C).
Table 2.
Increased expression of genes involved in the NF-κB pathway and Notch signaling pathway.
| Pathway specific Array | Symbol | Accession # | Fold Up-Regulation |
|---|---|---|---|
| NF-κB signaling Pathway |
Ccr5 | NM_009917 | 3.04 |
| Gadd45b | NM_008655 | 8.79 | |
| Icam1 | NM_010493 | 6.26 | |
| Irf1 | NM_008390 | 4.84 | |
| Rela | NM_009045 | 6.17 | |
| Sod2 | NM_013671 | 9.10 | |
| Stat1 | NM_009283 | 9.96 | |
| Tnfrsf1b | NM_011610 | 5.37 | |
| Xiap | NM_009688 | 8.91 | |
| Notch Signaling Pathway |
Fzd1 | NM_021457 | 5.71 |
| Fzd2 | NM_020510 | 4.27 | |
| Hes1 | NM_00823S | 5.79 | |
| Hey1 | NM_010423 | 5.79 | |
| Hoxb4 | NM_010459 | 3.79 | |
| Nfkb1 | NM_008689 | 3.59 | |
| Notch1 | NM_008714 | 6.52 | |
| Notch2 | NM_010928 | 9.81 | |
| Numb | NM_010949 | 3.28 | |
| Pdpk1 | NM_011062 | 3.52 |
Inflammatory stress-activated Notch signaling compromises HSC self-renewal
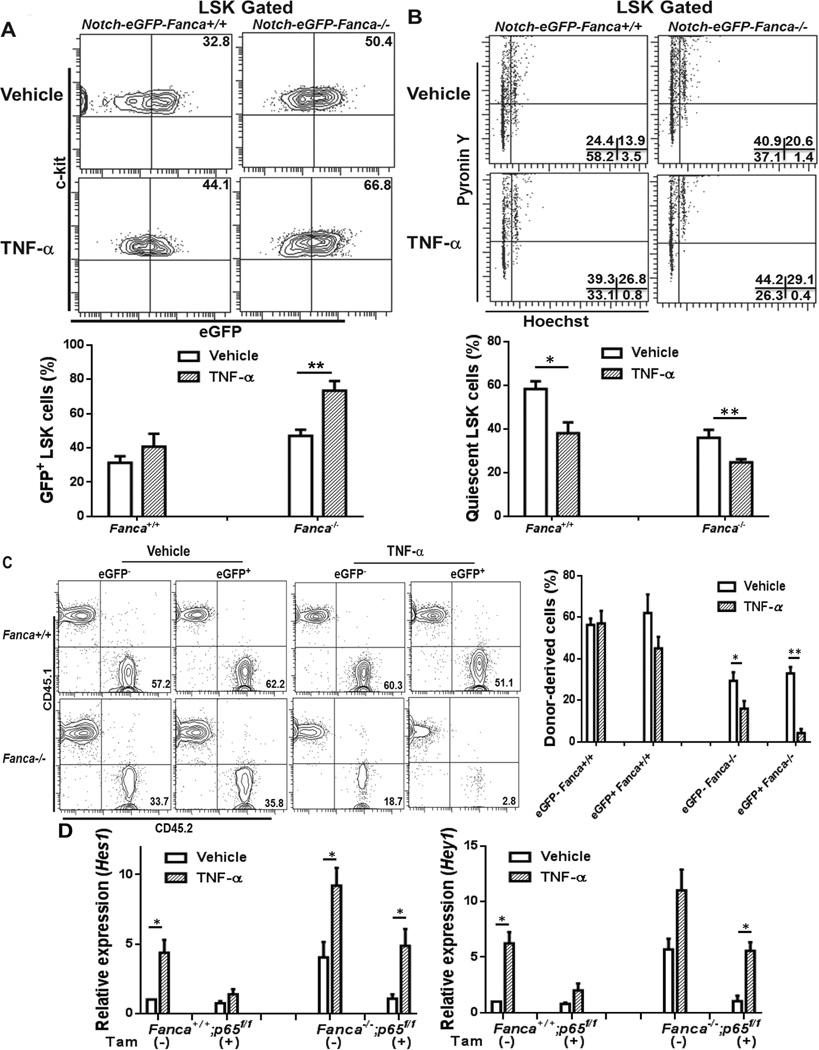
Because our gene expression analysis suggests a potential NF-κB-Notch crosstalk in FA HSPC cells and because FA HSPC cells are hypersensitive to inflammatory stress (42), we next determined whether TNF-α, a NF-κB activator and a major inflammation mediator in FA (43), would enhance Notch signaling in FA HSPCs. To this end, we treated Notch-eGFP-Fanca−/− mice and Notch-eGFP-WT littermates with TNF-α and examined Notch-eGFP expression in BM LSK cells. Interestingly, we found that TNF-α affected eGFP-positive HSPCs in both WT and FA mice. However, the effect of TNF-α was significantly exacerbated in FA mice compared to WT mice. Specifically, the frequency of GFP+ LSK cells was further increased in TNF-α-treated Notch-eGFP-Fanca−/− mice (Fig 5A). This was associated with a further decrease in HSC quiescence (Fig 5B). To substantiate these phenotypic analysis, we performed competitive BM transplantation by injecting 500 GFP− LSK or GFP+ LSK cells isolated from the mice treated with TNF-α along with 0.3 million congenic WT BM cells into lethally irradiated recipients and determined donor-derived chimera 24 weeks post transplantation. We observed dramatically further impairment in hematopoietic repopulation of GFP+ LSK cells from TNF-α-treated Notch-eGFP-Fanca−/− mice (Fig 5C). In fact, donor-derived cells from recipients transplanted with TNF-α-treated FA eGFP+LSK cells were almost undetectable at 24 weeks after transplantation (Fig 5C). It is also noteworthy that Notch-eGFP+ HSPCs, regardless of genotypes, were more vulnerable to TNF-α treatment than Notch-eGFP− HSPCs in the context of long-term hematopoietic repopulation (Fig 5C). To assess whether the enhanced expression of Notch-eGFP in TNF-α-treated Fanca−/− mice required canonical NF-κB signaling, we crossed Notch-eGFP-Fanca+/− mice with mice carrying a conditional gene encoding the p65 subunit of NF-κB (p65f/f) (23). Inducible deletion of p65 was accomplished by Cre-mediated deletion of floxed p65 alleles using a Cre-ERT2 strain (24) and Tamoxifen (Fig S3). We found that inactivation of p65 resulted in attenuation of Hes1 and Hey1 expression in LSK cells from TNF-α-treated Notch-eGFP-positive WT and Fanca−/− mice (Fig 5D). Furthermore, p65 deletion caused a significant reduction in Notch target gene expression in untreated Notch-eGFP-positive Fanca−/− LSK cells (Fig 5D). These results suggest that inflammatory stress activates Notch signaling and compromises hematopoietic repopulating capacity of HSPCs.
Fig 5. TNF-α activates Notch signaling in HSCs.
(A) Enhanced Notch signaling in Fanca−/− HSPCs by TNF-α. Notch-eGFP-Fanca−/− or Notch-eGFP-WT mice were treated with vehicle or TNF-α(single injection; 100 µg/kg). Twenty-four hours later, WBMCs (whole bone marrow cells) were isolated and subjected to Flow Cytometry analysis for GFP expression in gated LSK cells. Representative flow graphs (Left) and quantification (Right) were shown. Results are means plus or minus SD of three experiments (n=9 per group). (B) TNF-α further decreases HSC quiescence in Fanca−/− mice. Cells described in (A) were subjected to cell cycle analysis using Hochest 33342/Pyronin Y staining. Representative flow graphs (Left) and quantification (Right) were shown. Results are means plus or minus SD of three independent experiments (n=9 per group). (C) Stress-activated Notch signaling compromises HSC self-renew. 500 GFP− or GFP+ LSK cells (CD45.2+) isolated from the mice treated with TNF-α along with 0.3 million cogenic WT BM cells (CD45.1+) were transplanted into lethally irradiated recipients and determined donor-derived chimera 24 weeks post BMT. Representative flow graphs (Upper) and quantification (Lower) were shown. Results are means plus or minus SD of three independent experiments (n=6 per group). (D) Inactivation of p65 resulted in attenuation of Hes1 and Hey1 expression in TNF-α-treated Fanca−/− mice. LSK cells from TNF-α-treated Fanca−/−p65f/f or Fanca+/+p65f/fCreER mice (plus or minus Tamoxifen injection for 3 days) were isolated for RNA extraction followed by 1st cDNA synthesis. cDNA were then subjected for Real time-PCR using primers for Hes1 or Hey1. The results were plotted after normalization with GAPDH as internal control. Data are shown as mean plus or minus SD of 3 independent experiments.
Inhibition NF-κB or Notch partially restores FA HSPC function
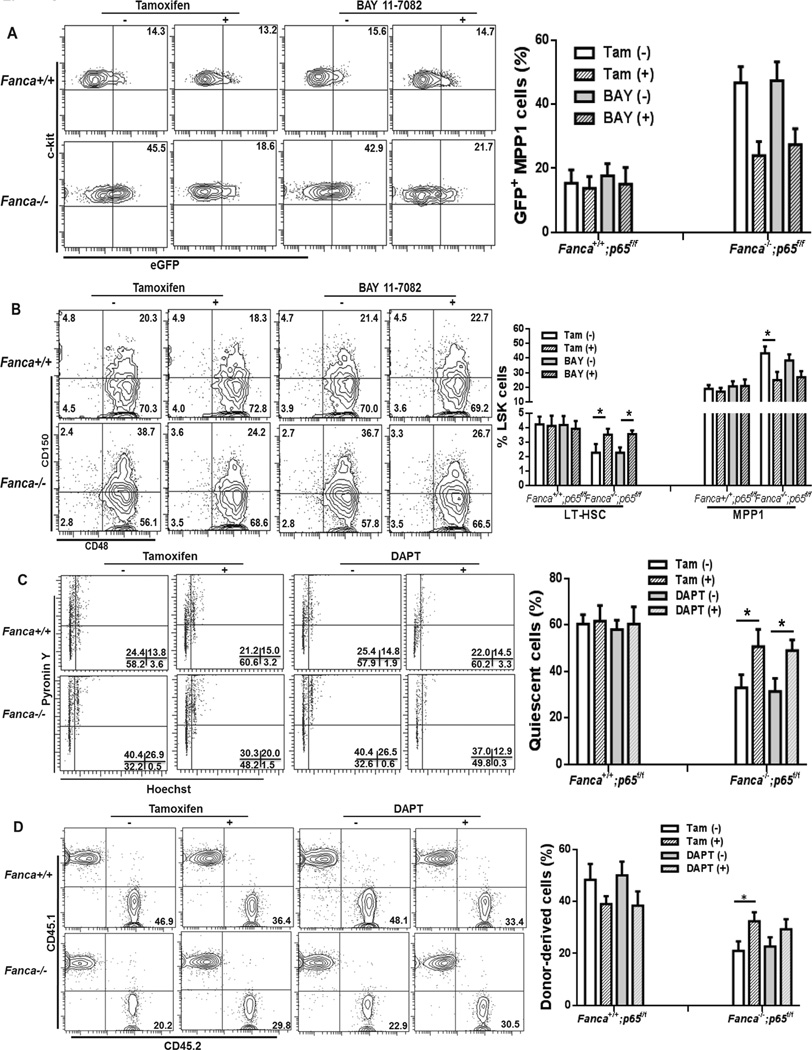
To further determine the functional relevance of Notch-NF-κB cross-talk_in FA HSPCs, we sought to test whether inhibition of NF-κB or Notch signaling could restore FA HSPC function. We took two approaches on NF-κB inhibition: genetic ablation of p65, a subunit of NF-κB transcription complex (23), and pharmacologic inhibition of NF-κB. We found that p65 deletion in MPP1 cells of Notch-eGFP-Fanca−/− mice reduced Notch expression to nearly WT level (Fig 6A). To further substantiate this finding, we made use of BAY11–7082, an inhibitor of cytokine-induced IκBα phosphorylation (29). Consistent with p65 knockout, BAY11–7082 treatment also significantly reduced Notch-eGFP expression in FA MPP1 cells (Fig 6A). Phenotypic analysis of the HSPC subsets in the LSK compartment revealed that inhibition of NF-κB by either p65 deletion or BAY11–7082 treatment increased LT-HSCs and decreased MPP1 cells in Fanca−/− mice to near WT levels (Fig 6B). Thus, these data provide molecular evidence that links NF-κB-Notch crosstalk to regulation of FA HSC differentiation.
Fig 6. Inhibition of NF-KB partially rescues FA HSC phenotype.
(A) Inhibition of NF-κB reduces Notch-eGFP expression in Fanca−/− MPP1 to nearly WT level. WBMCs from genetic ablation (3 days vehicle or Tamoxifen-injected Notch-eGFP-Fanca+/+p65f/f-CreER or Notch-eGFP-Fanca−/−p65f/fCreER mice) or pharmacologic inhibition (BAY 11-7082-treated Notch-eGFP-Fanca+/+ or Notch-eGFP-Fanca−/− mice) of NF-κB were isolated for Flow cytometry analysis. MPP1 cells were gated for GFP expression. Representative flow graphs (Upper) and quantification (Lower) were shown. Results are means plus or minus SD of 3 independent experiments (n=9 per group). (B) Inhibition of NF-κB increases LT-HSCs and decreases MPP1 in Fanca−/− mice. Cells described in (A) were subjected to SLAM analysis using Flow Cytometry. Representative flow graphs (Lower) and quantification (Upper) were shown. Results are means plus or minus SD of 3 independent experiments (n=9 per group). (C) Inhibition of NF-κB or Notch partially restores HSPC quiescence. BMCs from genetic ablation (3 days vehicle or Tamoxifen-injected Notch-eGFP-Fanca+/+ p65f/fCreER or Notch-eGFP-Fanca−/− p65f/fCreER mice) or DAPT-treated Notch-eGFP-Fanca+/+ or Notch-eGFP-Fanca−/− mice were isolated for cell cycle analysis. Representative flow graphs (Upper) and quantification (Lower) were shown. Results are means plus or minus SD of 3 independent experiments (n=9 per group). (D) Inhibition of NF-κB or Notch partially restores HSPC self-renewal capacity. 500 LSK cells (CD45.2+) from mice described in (C) along with 0.3 million cogeneic WT BM cells (CD45.1+) were transplanted to lethally irradiated recipients. Donor-derived chimerism was determined 24 weeks after BMT. Representative flow graphs (Lower) and quantification (Upper) were shown. Results are means plus or minus SD of 3 independent experiments (n=9 per group).
To determine the functional consequence of inhibition of NF-κB and Notch signaling in FA HSPCs, we blocked either NF-κB or Notch signaling by conditional deletion of p65 in Fanca−/− mice or treated the mice with the γ-secretase inhibitor, DAPT that inhibits Notch signaling (25, 26), respectively, and assessed cell cycle status and repopulating capacity of HSPCs. Deletion of p65 partially rescued the loss of quiescence of Fanca−/− but not WT HSPCs (Fig 6C). For DAPT treatment, we found a dose-dependent (up to 100 mg/kg body weight) inhibition of Notch signaling evidenced by a decreased generation of the active form of Notch 1 (NICD) without affecting non-Notch signaling protein Stat1, which paralleled a decreased level of eGFP-positive MPP cells in Fanca−/− mice (Fig S4A–B). Consistent with this, mice treated with DAPT at the dose of 100 mg/kg body showed effective inhibition of the expression of Notch target genes without affecting NF-κB transactivation in MPP cells (Fig S4C). Consequently, cell-cycle analysis demonstrated that DAPT treatment restored quiescence of LSK cells from Fanca−/− mice (Fig 6C). Furthermore, LSK cells from Fanca−/− mice subjected to p65 deletion exhibited significant increase in long-term hematopoietic repopulation when transplanted to lethally irradiated recipients compared to untreated controls (Fig 6D ). A notable increase, albeit not statistically significant, in hematopoietic repopulation was also observed in DAPT- treated Fanca−/− LSK cells. These results indicate that blocking NF-κB or Notch signal can partially correct the functional defect of FA HSPCs.
Discussion
The current study identifies a potential interaction between the FA pathway and Notch signaling in HSC differentiation and establishes a role of FA proteins in the control of balance between renewal and lineage commitment. There are several findings that highlight the significance of our study: (1) Loss of murine FA proteins results in enhanced Notch signaling in MPPs, which is correlated with decreased phenotypic and functional LT-HSCs and increased formation of MPP1 progenitors; (2) Deletion of Fanca or Fancc gene deregulates genes in the Notch signaling and the NF-κB pathway; (3) TNF-α stimulation enhances Notch signaling in Fanca−/− and Fancc−/− LSK cells, leading to decreased HSC quiescence and compromised HSC self-renewal; (4) Inflammation-activated Notch target gene expression in Fanca−/−and Fancc−/− MPP cells requires NF-κB; (5) Genetic ablation or pharmacologic inhibition of NF-κB reduces Notch signaling in FA MPPs to near wide-type level and significantly increases LT-HSCs and decreases MPP1 cells in FA mice; (5) Blocking either NF-κB or Notch signaling partially restored FA HSC quiescence and self-renewal capacity.
Several previous studies also suggest a crosstalk between the FA proteins and Notch targets. Tremblay et al. identified the interaction between components of the FA core complex and HES1, a Notch target that participates in many of the Notch-assigned functions including proliferation, differentiation, apoptosis, self-renewal, and asymmetric cell division regulation (19, 44). Another study demonstrates that FA core complex interacts with HES1 and antagonizes HES1-mediated transcriptional repression (45). Here, we show that enhanced Notch signaling in FA MPPs skews HSC differentiation, evidenced by exhaustion of LT-HSC pool and increase in MPP1 frequency. Deregulated Notch signaling was further demonstrated by a marked increase in the transcription of Notch target genes (including Hes1, Hey1, Notch1 and Notch2) in Fanca−/− and Fancc−/− MPP1 cells. Based on these results and the studies by others described above, we propose that one critical function of FA proteins in hematopoiesis is to regulate HSC differentiation, probably through modulating the integrity of the Notch signaling pathway.
Recent studies have identified the Notch pathway as a principal player on stem cell regulation and differentiation (46). While Notch plays a predominant role in the generation of HSCs in the embryo, it appears that the maintenance of adult HSCs does not require Notch (47). However, our present study reveals a plethora of Notch functions in HSPC cell populations that is dependent on the cell context and environment. For example, we found that enhanced Notch signaling in murine FA MPP cells was associated with the reduction of LT-HSC pool and the increase in MPP production in the BM (Fig 1G and Fig 2A). Functionally, FA Notch-eGFP+ HSPC cells show hematopoietic repopulation defect upon transplantation into lethally irradiated recipients (Fig 3C and Fig 4C), which might result from increased HSC differentiation demanded by the expanded Notch-eGFP+ MPPs. On the other hand, inflammatory stress generated by TNF-α stimulation enhanced Notch signaling of FA HSPC cells and led to decreased HSC quiescence and compromised HSC self-renewal (Fig 5). TNF-α is known to be a common activator for both Notch signaling and NF-κB pathways (47, 48). It has been shown that FA BM HSPC cells are hypersensitive to inflammatory cytokines including TNF-α(13). In addition, TNF-α is often found overproduced in FA patients (43, 49). Conceivably, increased TNF-α observed in FA patients could change the BM microenvironment and cause deregulation of the Notch and NF-κB signaling pathways. Our finding thus raises an important question as to whether deregulated Notch and NF-κB signaling caused by the elevated level of TNF-α in FA BM microenvironment is the mechanism that links to the defects in HSC self-renewal and differentiation.
Quiescence has been postulated to prevent HSC exhaustion (50). In the BM, HSCs are kept in a low proliferative, relatively quiescent state within the microenvironment (niche; 51). This dormancy is crucial to maintain the appropriate HSC pool. In this study, we demonstrate that HSPC (LSK) cells as well as LT-HSCs from Fanca−/− and Fancc−/− mice are less quiescent compared with WT controls, as evidenced by the reduced G0 population (Fig 2C and 2D), indicating that FA proteins may play a role in HSC maintenance by regulating cell cycle entry. Although loss of quiescence in FA HSCs is independent of Notch signaling in the steady state, enhanced Notch signaling in FA HSPCs leads to impaired long-term repopulation abilities of FA HSCs (Fig 4). Notably, inflammatory stress (TNF-α stimulation) activated Notch signaling in both WT and FA HSPC (LSK) cells, which was correlated with decreased quiescence and impaired hematopoietic repopulation (Fig 5). Moreover, blockage of Notch signaling partially restored FA HSC quiescence and repopulating capacity (Fig 6). Therefore, our results suggest that while Notch signaling is dispensable for the maintenance of the adult HSCs, deregulated Notch signaling under inflammatory stress could tip HSCs toward less quiescent state leading to impaired stem cell function.
Recent evidence of interaction between Notch and NF-κB suggests that the crosstalk between these two important signaling pathways may influence tissue hemeostasis (7, 8, 52). On the one hand, it has been shown that transcriptional repressor CBF1, the essential transcription factor downstream in Notch signaling, interacts with a dual NF-κB/CBF1-binding site (κB2) in the IκBα promoter and de-repress IκBα gene transcription leading to NF-κB activation in hematopoietic progenitor cells (8, 53). On the other hand, it was found that activation of the canonical NF-κB pathway by TNF-α stimulation induced optimal expression of Notch targets such as Hes1 and Hey1 through a mechanism involving epigenetic activation of the Notch gene promoters by TNF-α stimulation (7). In this study, we provide several lines of evidence demonstrating the crosstalk between NF-κB and Notch pathways in the regulation of HSC maintenance. First, we found that TNF-α stimulation enhanced Notch signaling in Fanca−/− and Fancc−/− HSCP (LSK) cells, leading to decreased HSC quiescence and compromised hematopoietic repopulation (Fig 5). Second, although further study is required to reveal the underlying molecular mechanism, it is intriguing that FA deficiency in MPPs deregulates a complex network of genes in the Notch and canonical NF-κB pathways (Fig S2), suggesting a potential link between these two pathways. To support this, we show that inflammation-activated transcription of Notch target genes in Fanca−/− MPP cells requires NF-κB (Fig 5D and data not shown). Third, genetic ablation or pharmacologic inhibition of NF-κB reduces Notch signaling in Fanca−/− MPPs to near wide-type level and significantly increases LT-HSCs and decreases MPP1 cells in Fanca−/− mice (Fig 6A–B). Finally, genetic ablation of NF-κB or pharmacologic inhibition of Notch signaling partially restored FA HSC quiescence and hematopoietic repopulation (Fig 6C–D). Thus our results favor the mechanism by which NF-κB regulates Notch pathway probably via influencing the expression of Notch target genes.
Supplementary Material
Acknowledgements
We thank Dr. Manuel Buchwald (Hospital for Sick Children, University of Toronto) for the Fancc+/− mice, Dr. Madeleine Carreau (Laval University) for the Fanca+/− mice, the Comprehensive Mouse and Cancer Core of the Cincinnati Children’s Research Foundation (Cincinnati Children’s Hospital Medical Center) for bone marrow transplantation service.
This work was supported by NIH grants R01 HL076712 and R01 CA157537. Q.P. is supported by a Leukemia & Lymphoma Society Scholar award. W.D. is supported by a NIH T32 training grant.
Footnotes
Author Contributions
W. D., designed research, performed research, analyzed data and wrote the paper; S. A., performed research and analyzed data; J. S., performed research; J. S., performed research; K. S., contributed vital new reagents; Q. P., designed research and wrote the paper.
Conflict of Interest
The authors declare no conflict of interest.
Reference
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Gridley T. Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 2003;12:R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 3.Louvi A, Artavanis-Tsakonas S. Notch signaling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 4.Stanic AK, Bezbradica JS, Park JJ, Matsuki N, Mora AL, Van Kaer L, Boothby MR, Joyce S. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J. Immunol. 2004;172:2265–2273. doi: 10.4049/jimmunol.172.4.2265. [DOI] [PubMed] [Google Scholar]
- 5.Rizo A, Vellenga E, de Haan G, Schuringa JJ. Signaling pathways in self-renewing hematopoietic and leukemic stem cells: do all stem cells need a niche? Hum. Mol. Genet. 2006;15:210–219. doi: 10.1093/hmg/ddl175. [DOI] [PubMed] [Google Scholar]
- 6.Gerondakis S, Banerjee A, Grigoriadis G, Vasanthakumar A, Gugasyan R, Sidwell T, Grumont RJ. NF-κB subunit specificity in hemopoiesis. Immunol. Rev. 2012;246:272–285. doi: 10.1111/j.1600-065X.2011.01090.x. [DOI] [PubMed] [Google Scholar]
- 7.Maniati E, Bossard M, Cook N, Candido JB, Emami-Shahri N, Nedospasov SA, Balkwill FR, Tuveson DA, Hagemann T. Crosstalk between the canonical NF-κB and Notch signaling pathways inhibits Pparγ expression and promotes pancreatic cancer progression in mice. J. Clin. Invest. 2011;121:4685–4699. doi: 10.1172/JCI45797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng P, Zlobin A, Volgina V, Gottipati S, Osborne B, Simel EJ, Miele L, Gabrilovich DI. Notch-1 regulates NF-kappaB activity in hemopoietic progenitor cells. J. Immunol. 2001;167:4458–4467. doi: 10.4049/jimmunol.167.8.4458. [DOI] [PubMed] [Google Scholar]
- 9.Bagby GC., Jr Genetic basis of Fanconi anemia. Curr. Opin. Hematol. 2003;10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes. Dev. 2000;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 11.Green AM, Kupfer GM. Fanconi anemia. Hematol Oncol. Clin. North. Am. 2009;23:193–214. doi: 10.1016/j.hoc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes. Dev. 2012;26:1393–408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pang Q, Andreassen PR. Fanconi anemia proteins and endogenous stresses. Mutat. Res. 2009;668:42–53. doi: 10.1016/j.mrfmmm.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly PF, Radtke S, von Kalle C, Balcik B, Bohn K, Mueller R, Schuesler T, Haren M, Reeves L, Cancelas JA, Leemhuis T, Harris R, Auerbach AD, Smith FO, Davies SM, A.Williams D. Stem cell collection and gene transfer in Fanconi anemia. Mol. Ther. 2007;15:211–219. doi: 10.1038/sj.mt.6300033. [DOI] [PubMed] [Google Scholar]
- 17.Pulliam AC, Hobson MJ, Ciccone SL, Li Y, Chen S, Srour EF, Yang FC, Broxmeyer HE, Clapp DW. AMD3100 synergizes with G-CSF to mobilize repopulating stem cells in Fanconi anemia knockout mice. Exp. Hematol. 2008;36:1084–1090. doi: 10.1016/j.exphem.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haneline LS, Li X, Ciccone SL, Li Y, Chen S, Srour EF, Yang FC, Broxmeyer HE, Clapp DW. Retroviral-mediated expression of recombinant Fancc enhances the repopulating ability of Fancc−/− hematopoietic stem cells and decreases the risk of clonal evolution. Blood. 2003;101:1299–1307. doi: 10.1182/blood-2002-08-2404. [DOI] [PubMed] [Google Scholar]
- 19.Tremblay CS, Huang FF, Habi O, Huard CC, Godin C, Lévesque G, Carreau M. HES1 is a novel interactor of the Fanconi anemia core complex. Blood. 2008;112:2062–2070. doi: 10.1182/blood-2008-04-152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng NC, van de Vrugt HJ, van der Valk MA, Oostra AB, Krimpenfort P, de Vries Y, Joenje H, Berns A, Arwert F. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum. Mol. Genet. 2000;9:1805–1811. doi: 10.1093/hmg/9.12.1805. [DOI] [PubMed] [Google Scholar]
- 21.Chen M, Tomkins DJ, Auerbach W, McKerlie C, Youssoufian H, Liu L, Gan O, Carreau M, Auerbach A, Groves T, Guidos CJ, Freedman MH, Cross J, Percy DH, Dick JE, Joyner AL, Buchwald M. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat. Genet. 1996;112:448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- 22.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, Reya T. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 23.Steinbrecher KA, Harmel-Laws E, Sitcheran R, Baldwin AS. Loss of epithelial RelA results in deregulated intestinal proliferative/apoptotic homeostasis and susceptibility to inflammation. J. Immunol. 2008;180:2588–2599. doi: 10.4049/jimmunol.180.4.2588. [DOI] [PubMed] [Google Scholar]
- 24.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 25.Borghese L, Dolezalova D, Opitz T, Haupt S, Leinhaas A, Steinfarz B, Koch P, Edenhofer F, Hampl A, Brüstle O. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem. Cells. 2010;28:955–964. doi: 10.1002/stem.408. [DOI] [PubMed] [Google Scholar]
- 26.Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalén M, Gerhardt H, Betsholtz C. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 27.Cao Q, Li P, Lu J, Dheen ST, Kaur C, Ling EA. Nuclear factor-κB/p65 responds to changes in the Notch signaling pathway in murine BV-2 cells and in amoeboid microglia in postnatal rats treated with the γ-secretase complex blocker DAPT. J. Neurosci. Res. 2010;88:2701–2714. doi: 10.1002/jnr.22429. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Sejas DP, Zhang X, Qiu Y, Nattamai KJ, Rani R, Rathbun KR, Geiger H, Williams DA, Bagby GC, Pang Q. TNF-α induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J. Clin. Invest. 2007;117:3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dewan MZ, Terashima K, Taruishi M, Hasegawa H, Ito M, Tanaka Y, Mori N, Sata T, Koyanagi Y, Maeda M, Kubuki Y, Okayama A, Fujii M, Yamamoto N. Rapid tumor formation of human T-cell leukemia virus type 1-infected cell lines in novel NOD-SCID/gamma c (null) mice: suppression by an inhibitor against NF-kappaB. J. Virol. 2003;77:5286–5294. doi: 10.1128/JVI.77.9.5286-5294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rathinam C, Matesic LE, Flavell RA. The E3 ligase Itch is a negative regulator of the homeostasis and function of hematopoietic stem cells. Nat. Immunol. 2011;12:399–407. doi: 10.1038/ni.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Sipple J, Pang Q, Du W. Salidroside stimulates DNA repair enzyme Parp-1 activity in mouse HSC maintenance salidroside. Blood. 2012;119:4162–4173. doi: 10.1182/blood-2011-10-387332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat. Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 33.Wilson A, MacDonald HR, Radtke F. Notch 1- deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, Pear WS. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 35.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, Nakagami-Yamaguchi E, Hamada Y, Aizawa S, Hirai H. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt TM, de Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zúñiga-Pflücker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat. Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- 37.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, Lió P, Macdonald HR, Trumpp A. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 38.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 39.Maillard I, Koch U, Dumortier A, Shestova O, Xu L, Sai H, Pross SE, Aster JC, Bhandoola A, Radtke F, Pear WS. Canonical notch signaling is dispensable for the maintenance of adult hematopoietic stem cells. Cell. Stem. Cell. 2008;2:356–366. doi: 10.1016/j.stem.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao W, Sweeney C, Connolly M, Kennedy A, Ng CT, McCormick J, Veale DJ, Fearon U. Notch-1 mediates hypoxia-induced angiogenesis in rheumatoid arthritis. Arthritis. Rheum. 2012;64:2104–2113. doi: 10.1002/art.34397. [DOI] [PubMed] [Google Scholar]
- 41.Sato T, Onai N, Yoshihara H, Arai F, Suda T, Ohteki T. Interferon regulatory factor-2 protects quiescent hematopoietic stem cells from type I interferon-dependent exhaustion. Nat. Med. 2009;15:696–700. doi: 10.1038/nm.1973. [DOI] [PubMed] [Google Scholar]
- 42.Du W, Adam Z, Rani R, Zhang X, Pang Q. Oxidative stress in Fanconi anemia hematopoiesis and disease progression. Antioxid Redox Signal. 2008;10:1909–1921. doi: 10.1089/ars.2008.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Briot D, Macé-Aimé G, Subra F, Rosselli F. Aberrant activation of stress-response pathways leads to TNF-alpha oversecretion in Fanconi anemia. Blood. 2008;111:1913–1923. doi: 10.1182/blood-2007-07-099218. [DOI] [PubMed] [Google Scholar]
- 44.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 45.Tremblay CS, Huard CC, Huang FF, Habi O, Bourdages V, Lévesque G, Carreau M. The Fanconi Anemia Core Complex Acts as a Transcriptional Co-regulator in Hairy Enhancer of Split 1 Signaling. J. Biol. Chem. 2009;284:13384–13395. doi: 10.1074/jbc.M807921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bigas A, Robert-Moreno A, Espinosa L. The Notch pathway in the developing hematopoietic system. Int. J. Dev. Biol. 2010;54:1175–1188. doi: 10.1387/ijdb.093049ab. [DOI] [PubMed] [Google Scholar]
- 47.Lee SH, Hong HS, Liu ZX, Kim RH, Kang MK, Park NH, Shin KH. TNF-α enhances cancer stem cell-like phenotype via Notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem. Biophys. Res. Commun. 2012;424:58–64. doi: 10.1016/j.bbrc.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ang HL, Tergaonkar V. Notch and NF-kappa B signaling pathways: Do they collaborate in normal vertebrate brain development and function? Bioessays. 2007;29:1039–1047. doi: 10.1002/bies.20647. [DOI] [PubMed] [Google Scholar]
- 49.Dufour C, Corcione A, Svahn J, Haupt R, Poggi V, Béka’ssy AN, Scimè R, Pistorio A, Pistoia V. TNF-alpha and IFN-gamma are overexpressed in the bone marrow of Fanconi anemia patients and TNF-alpha suppresses erythropoiesis in vitro. Blood. 2003;102:2053–2059. doi: 10.1182/blood-2003-01-0114. [DOI] [PubMed] [Google Scholar]
- 50.Ho AD. Kinetics and symmetry of divisions of hematopoietic stem cells. Exp Hematol. 2005;33:1–8. doi: 10.1016/j.exphem.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 52.Guan E, Wang J, Laborda J, Norcross M, Baeuerle PA, Hoffman T. T cell leukemia-associated human Notch/translocation-associated Notch homologue has IκB-like activity and physically interacts with nuclear factor-κB proteins in T cells. J. Exp. Med. 1996;183:2025–2032. [Google Scholar]
- 53.Oakley F, Mann J, Ruddell RG, Pickford J, Weinmaster G, Mann DA. Basal expression of IkappaB alpha is controlled by the mammalian transcriptional repressor RBP-J (CBF1) and its activator Notch1. J. Biol. Chem. 2003;278:24359–24370. doi: 10.1074/jbc.M211051200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.