Abstract
Meta-analysis is becoming an increasingly popular and powerful tool to integrate findings across studies and OMIC dimensions. But there is the danger that hidden dependencies between putatively “independent” studies can cause inflation of type I error, due to reinforcement of the evidence from false-positive findings. We present here a simple method for conducting meta-analyses that automatically estimates the degree of any such non-independence between OMIC scans and corrects the inference for it, retaining the proper type I error structure. The method does not require the original data from the source studies, but operates only on summary analysis results from these in OMIC scans. The method is applicable in a wide variety of situations including combining GWAS and or sequencing scan results across studies with dependencies due to overlapping subjects, as well as to scans of correlated traits, in a meta-analysis scan for pleiotropic genetic effects. The method correctly detects which scans are actually independent in which case it yields the traditional meta-analysis, so it may safely be used in all cases, when there is even a suspicion of correlation amongst scans.
1. Introduction
Meta-analysis is becoming a common tool for integrating findings across multiple OMIC scans (e.g. Hsu et al., 2010; Moutselos et al., 2010). The advantages are most obvious when investigators do not have access to all of the source data, but only to summary results from each study. But such a meta analysis strategy is sometimes analytically preferred even when all of the individual level data are available, in situations where there is enough potential for study heterogeneity that a combined supermodel, mega-analysis would require estimation of many cross-study-by-covariate interaction terms (e.g. Ioannidis et al., 2002).
However, one potential problem with such meta-analyses is the danger of hidden non-independencies between elements of the scans that can occur when data are generated with overlapping subjects, related subjects, or other information. For example, there are overlapping subjects in several large scale NIH sponsored genetic epidemiology studies, such as the Framingham Heart Study, the NHLBI Family Heart, the HyperGen study, ARIC study, etc. Even if subjects are distinct across studies, if there are closely related subjects (e.g. siblings) across studies, this can cause non-independence of the observations and potentially inflate type I error in meta-analyses. The reason is that such non-independence violates the basic i.i.d. (independent and identically distributed) random variables assumptions of most statistical tests and models, including traditional meta-analysis ones, so that if a type I error (false positive) occurs in one study, and there is overlapping information to another study, then the other study is more likely to reflect this same false-positive trend in its corresponding result. A meta-analysis which ignores this fact, will take the reinforcement of “signals” between the two studies as a sign of independent replication, and overstate the significance of the meta-findings.
Conneely and Boehnke (2010) provide a method of conducting meta-analysis of correlated SNPs within an LD region, on multiple correlated traits, but they assume that the multiple studies that are being meta-analyzed are strongly independent, and they do not consider the possibility of overlapping subjects or correlated information between the OMIC scans. Riley et al. (2007) discuss the properties a bivariate random effects meta analysis model, in which they estimate what they call “between study correlation,” but it is clear from their hierarchical model that they are in fact making the assumption that studies are strongly independent of one another. Their “between study correlation” is actually the correlation between the true parameter values within a study, as distinct from what they call the “within study correlation” which is the correlation between the pair of estimates of the parameters for each study. So they are not modeling the kinds of across scans correlations that would arise from overlapping subjects or any of the other reasons that we consider in our correlated meta-analysis model. Lin and Sullivan (2009) provide an efficient method for analyzing overlapping subjects in multiple GWAS to avoid inflation of type I error, but their approach is only applicable to case/control data, not quantitative traits, and it requires either having access to all individual level data or at least having a complete census accounting of the exact numbers of overlapping cases and controls. Sometimes such information is not easily shared amongst studies, due to IRB concerns, and sometimes, such overlaps may not even be known to the investigators, as subjects may not always volunteer that they are participating in multiple studies. Turchin and Hirschhorn (2012) have provided a clever way to forensically detect overlapping subjects using cryptographic hashes on GWAS data that preserves confidentiality of subjects. But as stated above, overlapping subjects is not the only cause for non-independence.
Hartung (1999) proposed the first true correlated meta-analysis test, using an approach similar to ours based upon the inverse normal MVN. But his approach estimates a single dependency correlation amongst all scans, assuming they are all equally correlated, which can be problematic when some pairs of OMIC scans are more related than others. Additionally, his method estimates this single correlation for each hypothesis (or “OMIC unit”, below) separately. This works well under the null hypothesis OMIC units, avoiding accumulation of evidence from correlated false-positives which results in inflation of type I error. But this approach can overcorrect for those OMIC unit tests under either the complete alternative hypothesis or partial alternative (incomplete null), resulting in potential loss of power. There, we want correlated true positive evidence to accumulate—we do not want to correct it out Our approach, first proposed by Province in 2005 for combining linkage scans, estimates the complete M×M correlation matrix for M OMIC scans, and thus allows for different scans to be correlated at different levels. We also estimate the average study dependency correlation matrix across all OMIC units in a set of scans, exploiting the biological fact that most of the OMIC units will be under the null. Thus, our method is more likely to only be correcting for dependencies under the null, retaining power under the alternative (or partial alternative) OMIC units.
2. Methods
2.1. OMIC unit of inference
Definition
An “OMIC unit of inference” is the basic unit for which statistical testing is performed for a particular OMIC scan.
For example, in a micro-array experiment, the OMIC unit of inference would be the gene, since the scan would consist of one statistical test for each of the 20,000 genes on the array. In a proteomic scan, the OMIC unit would be “proteins”, since we have one statistical test for each measured protein. In a linkage scan, the OMIC unit of inference might be the linkage markers themselves, or it might be centimorgan locations equally spaced throughout the genome, at which Identity-By-Descent (IBD) estimates have been made for each relative pair. There, we would have one multipoint LOD-score for each cM location. In a Whole Exome Sequencing (WES) experiment where the goal is to find rare variants, the OMIC unit of inference could be the variants themselves if power is sufficient to support individual testing of rare variants. But often there is not enough power to detect rare variants at an individual variant level (unless the variants are unusually penetrant). More commonly a statistical burden test is applied, so that the OMIC unit inference would be the gene, not the variant. Even though the smallest unit of measurement in the WES is the variant, the statistical tests are conducted at the gene level not the variant level, by collapsing/weighting all exonic variants within the same gene into a single composite predictor for that gene. So the gene is considered the OMIC unit of inference in this case. In a GWAS, the most natural OMIC unit of inference would be SNPs genotyped on the GWAS chip. But this might be reduced to a gene-level OMIC unit of inference, by taking, say, only the most significant SNP for each gene. Or it might be expanded to include all SNPs catalogued in a standard reference panel, such as HapMap or 1000 Genomes, by first performing genetic imputation (estimating the unmeasured SNPs via haplotype matching) and then conducting statistical tests on each imputed as well as measured SNP.
2.2. Harmonization of OMIC units of inference and missing data patterns
In order to conduct any meta-analyses of multiple OMIC scans we must first put them all on a common OMIC unit of inference scale, so that we can see if the combined evidence reinforces or destroys the overall signal at that particular OMIC unit. Exactly how this is done depends very much on the scientific goals, the types of OMIC scans one wishes to meta-analyze, the granularity and extent of the available data, and many other factors which we will not address in this paper. It is important to note that the methods we propose here do not depend upon the details of the process by which harmonization of OMIC units of inference across scans is accomplished. Nor is it necessary that this is accomplished comprehensively with identical OMIC units of inference across all OMIC scans. We can in fact have quite complex patterns of missing data within and across the OMIC scans, and our method will still apply. We do not make the strong assumption that data are missing “at random”, but instead, we make the slightly weaker assumption that the missing data patterns are “ignorable.” For example, we may wish to meta-analyze “I” different expression array experiments along with “K” different GWAS scans in combination with “J” linkage scans, and “M” WESs. We can reduce each of these I+J+K+M scans at the gene OMIC unit of inference, by taking the “most significant” SNP/gene in the GWASes, the highest LOD score over each gene in the linkage scans, and use burden tests for each gene in the exome scans. But we may also be interested in going beyond the genes into the intergenic regions, leaving out the expression arrays, and meta-analyzing the J+K+M GWAS+linkage+exome scans. We might define intergenic OMIC units as contiguous regions of open chromatin defined in functional experiments, contiguous regions of high species conservation. Or we might include some of the genes from the “I” expression arrays for those regions that are “known” to play regulatory role for the genes (e.g. eQTL regions for the gene).
2.3. Correlated Meta-Analysis of p-values
Suppose we have conducted N different OMIC scans on M common OMIC units of inference. Let PN×M=(pij) for i=1,…,N and j=1,…,M be the N×M matrix of p-values for the N scans of across the M OMICs units For each i, j let Z = Φ-1(1-P) denote the element-wise monotonic “complement probit” transformation of p-values to z-scores, where Φ(z) denotes the cumulative distribution function of the unit normal, N(0,1), i.e
For each row “j,” corresponding to a particular OMIC unit of inference, we look at the N×1submatrix formed by taking only the jth -row of PN×M , and denote this vector by P(j)N×1. It is this set of p-values that we wish to meta-analyze to test the combined effect of the jth OMIC unit across all N scans.
We apply a theorem from multivariate normal (MVN) distribution theory, whose proof is found in Anderson (2003):
Theorem 1
If Zk×1 is a MVN random variable, Zk×1 ∼ N[μk×1,Σk×k], and D1×k is any vector of constants, then the linear combination D1×k Zk×1 ∼ N[D1×k μk×1, D1×k Σk×k D′k×1]. In particular, when k=N, D1×N = 11×N (i.e. the 1×N vector of all “1”s) and μN×1 = 0N×1 (i.e. the N×1 vector of all “0”s), then out meta-analysis Z-value is given by
| (1) |
We then convert back from the z-score scale back to the p-value scale using the monotonic inverse transformation to the one above, to get the meta-analysis p-value: Pmeta = 1 - Φ(Zmeta) . Note that if the jth OMIC unit does not have a significance test result for one or more of the N scans, then the corresponding entries in P(j)N×1 will be missing (this will happen if data are low quality for that OMIC unit in one or more scans or if OMIC unit harmonization is not complete across scans for whatever reason). In such cases, we may use the basic property of the MVN distribution that every sub-dimensional space is also MVN distributed. Specifically, if Nj < N is the number of non-missing p-values for the jth OMIC unit, and we denote by P(j)*Nj×1 the Nj×1 submatrix of P(j)N×1 with all missing p-value rows deleted, then Theorem (1) still applies to k=Nj, the corresponding sub-dimensional components of D(j)*1×Nj, μ(j)*Nj×1 and Σ(j)*Nj×Nj being the non-missing submatricies of D1×N, μN×1 and ΣN×n , respectively.
We illustrate the application of this theorem to the meta-analysis of OMIC scans with two extreme mathematical examples.
Example 1: k independent OMIC scans
For each common OMIC unit, j=1, 2, …, M we denote the “k” p-values at that jth OMIC unit by the k×1 vector p(j)k×1 = (p(j)1, p(j)2. …, p(j)k)′. We transform these elementwise to z-scores via the complement probit transformation given in Equation (1), above, so that Z(j)k×1 = (Z(j)1, Z(j)2. …, Z(j)k)′. Then ∀ j=1, 2, …, M we have Z(j)k×1 ∼ N(0k×1, ,Σk×k = Ik×k], where Ik×k is the k×k identity matrix, so that
Example 2: k completely correlated (equal) scans
Zk×1 = (Z1, Z2. …, Zk)′. Then Zk×1 ∼ N(0, ,Σk×k = 1k×k], where 1k×k is the k×k matrix, with all elements equals to “1.”. Because each Zi∼N[0,1], then for each i, Zi=Z1 (i.e., they are actually all equal), so that
which is the same result we get from applying Theorem 1.
These two examples represent the extreme cases and make sense. In Example 1, when all scans are really independent, the Σk×k matrix is the identity, and our correlated meta-analysis method is the same as the traditional meta-method. In the other extreme, in Example 2, when all scans are completely correlated, the Σk×k matrix becomes the Ik×k matrix, and there is really just one scan, so the meta-analysis should recognize this and just return the original scan, which our correlated meta-method does, with no inflation of type I error.
2.4. Estimating Σk×kfrom the tetrachoric correlations amongst the OMIC scans themselves
Unlike the two simple examples above, in practice with real data, we will not know the values of the Σk×k matrix. However, if entire OMIC scans are available to us, we can exploit this fact to estimate Σk×k , by making using of the following assumption:
Assumption 1
In any OMIC scan, by far (in fact, by several orders of magnitude) most of the statistical tests will be under the NULL hypothesis --.e. the OMIC units of inference are actually statistically independent of the phenotype being scanned.
With this assumption, we can use the observed correlations between OMIC scans to obtain our estimate of Σk×k., and then apply Theorem 1 to conduct our correlated meta-analysis.
Note that this is a biologically motivated assumption, which stems from our understanding of the OMIC architecture of phenotypes and traits, i.e. that for any fixed trait, most of the genome, exome, proteome, etc. is neutral. There may be (hopefully is) some “contamination” of the alternative hypotheses somewhere within the OMIC scan (otherwise, our scans are fruitless). But OMIC analysis experience tells us (as does population genetic theory), that the number of true OMIC signals should be far outnumbered by the number of noise OMIC units of inference. Nonetheless, if we simply estimate Σk×k across scans between corresponding OMIC units there are two problems. The first is that the p-value scale is uniformly distributed under the null hypothesis and we are assuming that we are dealing with a MVN distribution in Theorem 1. But this can be easily handled by making use of the complement probit transformation discussed above. The second, bigger problem is that the contamination of the OMIC units that are under the alternative should not so easily be dismissed as trivial. Yes, Assumption 1 tells us that they are small in frequency, but it says nothing about their impact on the estimate of Σk×k. Correlated, highly significant true signal results may be small in number but highly influential on the estimate of Σk×k. Worse, we do not want to over-estimate Σk×k. because we are downweighting the results of our meta-analysis by its magnitude. Σk×k is supposed to be estimating the correlation between OMIC scans only for those OMIC units under the H0, because we want to avoid accumulating evidence across highly correlated scans that are only due to correlations of type I errors (due to overlapping subjects, relatedness, etc.). But we do NOT want to down weight evidence at OMIC units that are under the alternative hypothesis. In fact, we want the meta-p-values to be as significant as possible here. But we do not know which OMIC units are under the null and which are under the H1 (or we would not be doing the meta-analysis in the first place). We can minimize the impact of this contamination of the alternative hypothesis by using the tetrachoric correlation instead of the Pearson correlation in Σk×k. To do this, we first truncate all of the individual scan Z scores into two categories (Z < 0) vs (Z > 0). Then, at each common OMIC unit, if there are M scans, we form the M dimensional 2×2×…×2 table of scores across all scans, from which we estimate the tetachoric correlation matrix Σk×k. The tetrachoric correlation is less sensitive to contamination from the alternative hypothesis, because it lumps them with all moderately significant and even non-significant findings at the P<0.5 level. Thus, it provides some protection for over correction of the correlation amongst OMIC scans.
3. Results
3.1. Simulations
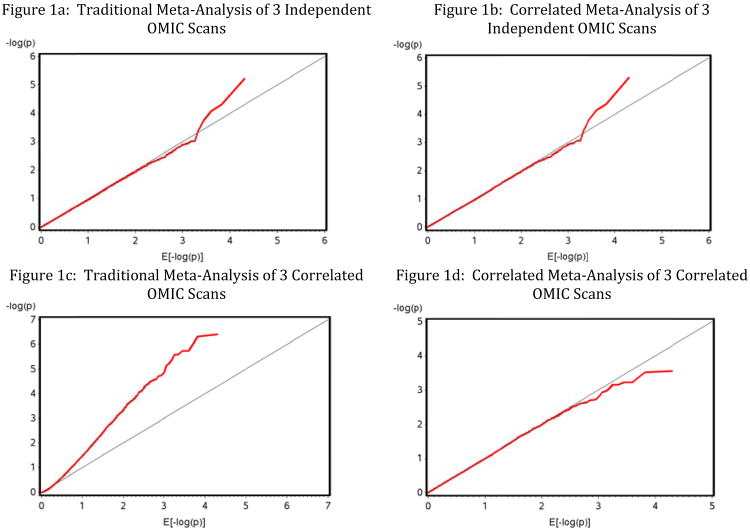
To validate the correlated meta-analysis method, we performed a series of simulation experiments. We generated 100 simulation replications of 3 OMIC scans on 10,000 OMIC units each, with a known correlational structure. For each replication, we conducted both the traditional meta-analysis (which assumes the 3 scans are independent) as well as our correlated meta-analysis procedure, which estimates the tetrachoric correlations between scans of the truncated p-values across the OMIC units of inference, and then uses that correlation matrix to correct the meta-analysis inference, as described above. The results of our simulations are shown in Table 1, where we catalog the distribution of estimates across the 100 simulation experiments, and Figure 1, where we show the Quantile-Quantile (Q-Q) plot for a typical (the first) replication's meta-analysis of the 3 scans.
Table 1. Distributions (Mean, Min, Max) across 100 simultation replications of parameter estimates from Traditional vs. Correlated meta-analyses of 3 OMIC scans.
| Parameter | Expected Value | Parameter Estimates using Traditional Meta | Parameter Estimates using Correlated Meta | ||||
|---|---|---|---|---|---|---|---|
| Mean | Min | Max | Mean | Min | Max | ||
| 3 Independent OMIC Scans | |||||||
| ρ(Z1,Z2) | 0 | [0] | [0] | [0] | -0.00082 | -0.04044 | 0.04886 |
| ρ(Z1,Z3) | 0 | [0] | [0] | [0] | 0.00121 | 0.04380 | 0.03524 |
| ρ(Z2,Z3) | 0 | [0] | [0] | [0] | 0.00066 | -0.04143 | 0.03101 |
| μ(ZMeta) | 0 | 0.00016 | -0.02125 | 0.02762 | 0.00160 | -0.02127 | 0.02761 |
| σ(ZMeta) | 1 | 1.00001 | 0.98363 | 1.01877 | 0.99977 | 0.97254 | 1.02056 |
| 3 Correlated OMIC Scans | |||||||
| ρ(Z1,Z2) | 0.5 | [0] | [0] | [0] | 0.50153 | 0.47154 | 0.52387 |
| ρ(Z1,Z3) | 0.2 | [0] | [0] | [0] | 0.20110 | 0.16289 | 0.23495 |
| ρ(Z2,Z3) | 0.9 | [0] | [0] | [0] | 0.89994 | 0.88774 | 0.91027 |
| μ(ZMeta) | 0 | 0.00015 | -0.02820 | 0.04023 | 0.00011 | -0.01961 | 0.02798 |
| σ(ZMeta) | 1 | 1.43792 | 1.40103 | 1.46045 | 0.99983 | 0.98170 | 1.01774 |
Figure 1. Q-Q plots under the H0 for replication 1 in a simulation of a meta-analysis of 3 OMIC scans.
As can be seen in Table 1, when the 3 scans are actually independent (top half of the table), our correlated meta-method correctly senses this and accurately estimates that the 3 tetrachoric correlations between the scans (ρ12, ρ13 and ρ23) are all nearly zero for all 100 replications (as evidenced by the fact that the mean, min and max are all roughly equal to one another and to the expected value of zero). Not shown in the table, we also compared the tetrachoric estimates of the correlations between scans, to the Pearson correlations on the continuous (non-truncated) transformed p-values, which should be more correct under the null. The average pairwise difference between the tetrachoric and pearson correlation estimates between the 3 scans in Table 1 across all simulation replications were -0.001, 0.0004, and -0.0003 for ρ(Z1,Z2), ρ(Z1,Z3), ρ(Z2,Z3), respectively in the top half of the table (independent scans), and 0.001, -d0.001, −0.002 for the bottom half of the table (dependent scans). More importantly, no tetrachoric correlation estimate differed from its corresponding Pearson correlation estimate by more than 0.037 in any simulation replication under any condition. Thus, in all cases (not just on average), the tetrachoric correlation provides accurate estimates of the correlation between scans under the null, but unlike the Pearson correlations, it reduces the impact of of correlations between true positives (for which we do NOT want to “correct away,” instead we want such evidence to accumulate in favor of the alternative).
This results in little bias as well as little loss in power in our correlated meta-test as compared to the traditional meta-analysis (which fixes the 3 between study correlations to zero). Thus, both the tradtional and the correlated meta analysis produce meta-Z scores that are nearly normal and have mean nearly zero and variance nearly one, as expected. The Q-Q plots in Figures 1a and 1b (for the 3 independent scans), verify that there is no inflation of type I error in this case, but more importantly from the correlated meta-analysis view, there is no over conservatism. The p-values are just as true estimating the 3 correlations to be nearly zero as they are assuming them to be zero, so there is no harm in applying the correlated meta-analysis procedure even when the 3 scans really are independent. The correlated meta will tell us what it thinks are the correlations, and will correct for them, regardless of their magnitude.
For the 3 correlated OMIC scans (bottom half of Table 1), we generated the scans pairwise correlated at 0.5, 0.2 and 0.9, respectively, and then applied traditional as well as our correlated meta-analysis method. As can be seen, our correlated meta-method accruately estimated the 3 correlations using the tetrachoric approach, across all 100 replications. More importantly, our correlated meta-analysis method correctly produces Z-scores with the proper 0,1 first and second moments. Whereas the traditional meta-analysis, yields a meta-Z-score with a badly inflated variance (Standard Error is approximately 1.4). This results in the traditional meta being overly liberal, since it calculates P-values assuming it's meta-Z score has a variance of 1, instead of 1.4. The inflation is readily apparent in the Q-Q plot of Figure 1c, compared to 1d.
3.2. Example Pleiotropy Scanning in the NHLBI Family Heart Study
Finally, we demonstrate the utility of our correlated meta-analysis procedure on a real data example, from a GWAS on the NHLBI Family Heart Study. In Table 2, we show the results of combining two GWASes from the same study on two highly correlated traits to look for pleiotropy. Note that this is an extreme example of overlapping subjects (ALL 2,767 of them overlap), so the Lin and Sullivan method would not be of much help here, but if we do the traditional meta-analysis, we are in real danger of having type I errors accumulate in this situation. For traits Body Mass Index (BMI) and Waist Circumference (WC), the tetrachoric correlation between the scans of these two highly correlated variables is estimated to be 0.70. Ignoring this correlation with the traditional meta-analysis, results in meta-P∼10-10 for the SNPS listed which are far above the GW threshold for significance. However, the correlated meta-analysis method finds a more moderate level of evidence at P∼10-6, suggesting that the traditional analysis is very much inflating the evidence. On the other hand, for HOMA (a measure of insulin resistance based upon insulin/glucose levels) and WC, the estimated tetrachoric correlation is 0.14. Here the traditional and correlated meta-analyses are in better agreement that indeed, there appears to be genome-wide significant pleiotropy at this locus for these two traits.
Table 2. Correlated vs. Traditional Meta-Analysis of GWAS SNPs to assess Pleiotropy of related traits in the NHLBI Family Heart Study.
| Table 2a: SNPS in the NEGR1 gene (chrom 1) for pleiotropy to BMI (Body Mass Index) and Waist Circumference (WC) | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | P-value BMI | Beta BMI | SE (beta) BMI | P-value WC | Beta WC | SE (beta) WC | P-value Traditional Meta | P-value Correlated Meta |
| rs577674 | 9.52E-06 | 0.58 | 0.13 | 8.38E-06 | 0.58 | 0.13 | 6.52E-10 | 1.59E-06 |
| rs522451 | 9.96E-06 | -0.59 | 0.13 | 9.90E-06 | -0.59 | 0.13 | 8.01E-10 | 1.80E-06 |
| rs473019 | 1.00E-05 | -0.59 | 0.13 | 9.99E-06 | -0.59 | 0.13 | 8.11E-10 | 1.81E-06 |
| rs580626 | 1.00E-05 | -0.59 | 0.13 | 1.00E-05 | -0.59 | 0.13 | 8.16E-10 | 1.82E-06 |
| Table 2b: SNPs in the ABCF2 gene (chrom 7) to assess Pleiotropy for pleiotropy to HOMA (a measure of Insulin Resistance) and Waist Circumference (WC) | ||||||||
| SNP | P-value HOMA | Beta HOMA | SE (beta) HOMA | P-value WC | Beta WC | SE (beta) WC | P-value Traditional Meta | P-value Correlated Meta |
| rs12538823 | 8.96E-06 | 0.24 | 0.05 | 1.77E-04 | 0.18 | 0.05 | 1.36E-08 | 9.75E-08 |
| rs7786151 | 9.84E-06 | -0.24 | 0.05 | 1.92E-04 | -0.18 | 0.05 | 1.61E-08 | 1.13E-07 |
| rs12113924 | 4.15E-06 | -0.25 | 0.05 | 2.28E-04 | -0.18 | 0.05 | 8.97E-09 | 6.77E-08 |
| rs3800794 | 4.04E-06 | -0.25 | 0.05 | 2.29E-04 | -0.18 | 0.05 | 8.84E-09 | 6.68E-08 |
4. Discussion
Our correlated meta-analysis method provides a simple, robust approach to integrate information across multiple OMIC scans so as to avoid inflation of type I error due to hidden dependencies. Our method makes few statistical assumptions. It first estimates empirically the degree of non-independence between the OMIC scans, and then uses this estimate to corrects the meta-inference. The method does not require any additional knowledge of numbers of overlapping subjects, nor any preliminary forensic analyses, and provides the correct type I error when scans are correlated, regardless of the number and character of its source causes, as part of the meta-analysis itself. It is applicable for combining OMIC scans on quantitative, qualitative or any combinations of phenotypes. It can even be used to scan for evidence of pleiotropic effects when subjects are completely overlapping. When OMIC scans actually are independent, it estimates this correctly, and becomes the same as the traditional meta-analysis test. Thus, the method can be safely used in any situation, when there is any doubt that there may be violations of study independence assumptions.
Acknowledgments
This work is partially supported by grants AG023746, HL088215, DK075681 and DK089256 from the USA National Institutes of Health
Contributor Information
Michael A Province, Email: mprovince@wustl.edu, Division of Statistical Genomics, Washington University School of Medicine, Box 8506, 4444 Forest Park, Blvd, St. Louis, MO, 63105, USA.
Ingrid B Borecki, Email: iborecki@wustl.edu, Division of Statistical Genomics, Washington University School of Medicine, Box 8506, 4444 Forest Park, Blvd, St. Louis, MO, 63105, USA.
References
- Anderson TW. An Introduction to Multivariate Statistical Analysis. Wiley; NY: 2003. ISBN-13:978-0471360919. [Google Scholar]
- Province MA. The significance of not finding a gene. Am J Hum Genet. 2001 Sep;69(3):660–3. doi: 10.1086/323316. Epub 2001 Jul 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Sullivan PF. Meta-analysis of genome-wide association studies with overlapping subjects. Am J Hum Genet. 2009 Dec;85(6):862, 72. doi: 10.1016/j.ajhg.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley RD, Abrams KR, Sutton AJ, Lambert PC, Thompson JR. Bivariate random-effects meta-analysis and the estimation of between-study correlation. BMC Med Res Methodol. 2007 Jan 12;7:3. doi: 10.1186/1471-2288-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conneely KN, Boehnke M. Meta-analysis of genetic association studies and adjustment for multiple testing of correlated SNPs and traits. Genet Epidemiol. 2010 Nov;34(7):739–46. doi: 10.1002/gepi.20538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YH, Zillikens MC, Wilson SG, Farber CR, Demissie S, Soranzo N, Bianchi EN, Grundberg E, Liang L, Richards JB, Estrada K, Zhou Y, van Nas A, Moffatt MF, Zhai G, Hofman A, van Meurs JB, Pols HA, Price RI, Nilsson O, Pastinen T, Cupples LA, Lusis AJ, Schadt EE, Ferrari S, Uitterlinden AG, Rivadeneira F, Spector TD, Karasik D, Kiel DP. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 2010 Jun 10;6(6):e1000977. doi: 10.1371/journal.pgen.1000977. PubMed PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Rosenberg PS, Goedert JJ, O'Brien TR. International Meta-analysis of HIV Host Genetics. Commentary: meta-analysis of individual participants' data in genetic epidemiology. Am J Epidemiol. 2002 Aug 1;156(3):204–10. doi: 10.1093/aje/kwf031. [DOI] [PubMed] [Google Scholar]
- Turchin MC, Hirschhorn JN. Gencrypt: one-way cryptographic hashes to detect overlapping individuals across samples. Bioinformatics. 2012 Mar 15;28(6):886–8. doi: 10.1093/bioinformatics/bts045. Epub 2012 Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung J. A note on combining dependent tests of significance. Biometrical Journal. 1999;41(7):849–855. [Google Scholar]
- Province MA. Meta-Analyses of Correlated Genomic Scans. Genetic Epidemiology. 2005;29:137. [Google Scholar]
- Moutselos K, Maglogiannis I, Chatziioannou A. Delineation and interpretation of gene networks towards their effect in cellular physiology- a reverse engineering approach for the identification of critical molecular players, through the use of ontologies. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:6709–12. doi: 10.1109/IEMBS.2010.5626249. [DOI] [PubMed] [Google Scholar]