Abstract
A thorough understanding of the events during mammalian spermatogenesis requires studying specific molecular signatures of individual testicular cell populations as well as their interaction in co-cultures. However, most purification techniques to isolate specific testicular cell populations are time-consuming, require large numbers of animals, and/or are only able to isolate a few cell types. Here we describe a cost-effective and timesaving approach that uses a single protocol to enrich multiple testicular cell populations (Sertoli, Leydig, and several spermatogenic cell populations) from as few as one mouse. Our protocol combines rigorous enzymatic digestion of seminiferous tubules with counter-current centrifugal elutriation, yielding specific testicular cell populations with >80%–95% purity.
Keywords: spermatogenesis, testis, Sertoli cell, Leydig cell, germ cell, spermatocyte, spermatid, elutriation
Spermatogenesis is a continuous process in which spermatogonial stem cells mature in a step-wise manner into specific germ cells before terminally differentiating to form spermatozoa. This dynamic process depends upon Sertoli cells that provide nourishment and structural support to germ cells throughout their development and Leydig cells that synthesize steroidal hormones necessary for germ cell differentiation (1,2). Surprisingly, even after decades of research in the field of male fertility, critical spermatogenic events, including Sertoli cell–germ cell interaction and mechanism of androgen action, remain to be completely understood. A more in-depth understanding of these spermatogenic events would require studying specific molecular signatures of individual testicular cells, as well as their interaction in co-cultures.
Isolation of enriched population of spermatogenic cells as well as Sertoli and Leydig cells from testes is one of the crucial steps to address these important issues. Over the years, various approaches have been used to successfully isolate testicular cells. One approach isolates cells using fluorescence-activated cell sorting (FACS) (3-6). Though potentially useful for specific applications, cells isolated using FACS may not be appropriate for functional studies, as common fluorochromes (e.g., propidium iodide) used for cell sorting are toxic (3). Although supravital dyes, sorting by exclusion of stained cells, and sorting based only on cell size and density may be used to circumvent this problem, there is still concern over the possible harmful effects of the laser beams used in FACS (4-6). Velocity sedimentation separation using STA-PUT chambers is another approach used to isolate spermatogenic cells (7). Although an elegant method to isolate pure germ cell populations, the STA-PUT method is not considered feasible for the isolation of somatic testicular cells (8). Recently, serial digestions using low concentrations of collagenase has also been proposed as a method to isolate Leydig cells from other testicular cell types (9). However, such a method does not take into account the separation of different germ cell populations and Sertoli cells within the seminiferous tubules.
To circumvent these problems, we have coupled a rigorous enzymatic digestion approach with a counter-current centrifugal elutriation approach that uses cellular mass and density to isolate various testicular cell types (10-12). An advantage to our method is that testicular cells are not subjected to considerable physiological alterations during isolation, as they are processed in physiological media/buffers. In addition, our ability to isolate various testicular cell types from one mouse is not only timesaving and cost-effective, but could also be useful when conducting studies in mice in which the desired genotypes are often difficult to obtain. Furthermore, isolation of different cell types from the same set of mice could be quite useful, as it will avoid any variability in cell preparation.
Briefly, our protocol was conducted as follows (summarized in Figure 1). Details of reagents used and solution preparation are available in the Supplementary material. Testes from 6- to 8-week-old mice were collected in Enriched DMEM:F12 (Mediatech, Manassas, VA, USA) and placed on ice. After removal of the testes’ tunica albuginea, seminiferous tubules were dissociated and transferred immediately into 10 mL Enzymatic Solution 1. Tubules were incubated for 15–20 min at 35°C in a shaking water bath at 80 oscillations (osc)/min (12,13), then layered over 40 mL 5% Percoll (Sigma-Aldrich, St. Louis, MO, USA)/95% 1× Hank’s balanced salt solution (HBSS; Invitrogen, Carlsbad, CA, USA) in a 50-mL conical tube and allowed to settle for 20 min. Leydig cells were isolated from the top 35 mL Percoll as described in the Supplementary material.
Figure 1. Protocol work flow and timing.
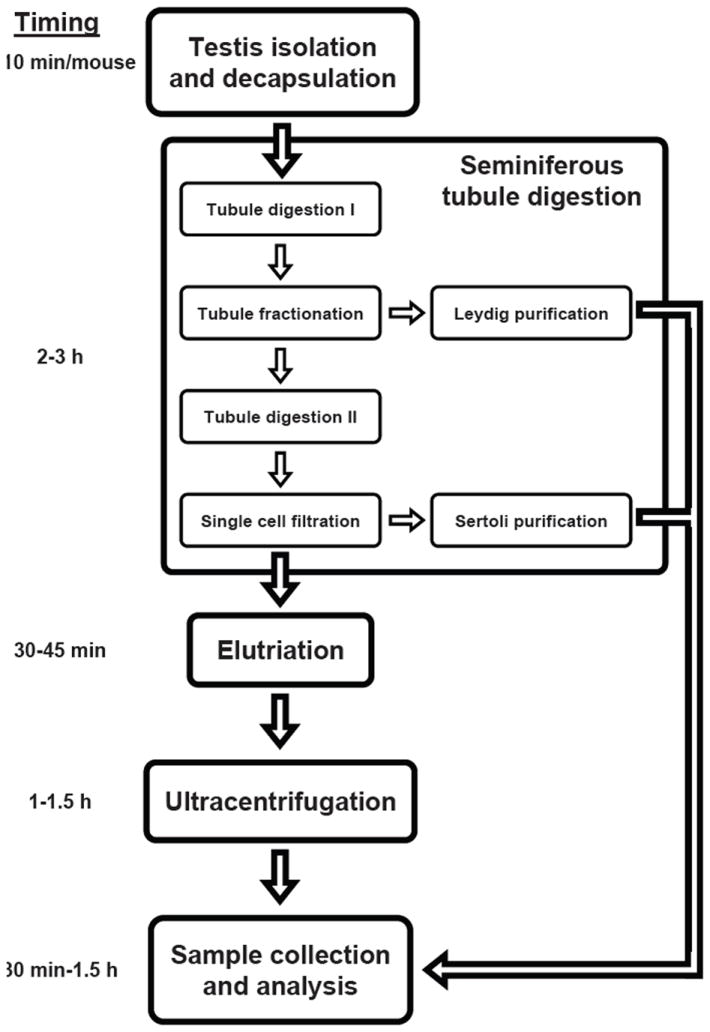
Schematic summarizing the major steps and timing of the protocol. Details of the method describing the protocol can be found in the text and the Supplementary material.
The bottom 5 mL Percoll were transferred to a fresh 50-mL conical tube containing 10 mL Enzymatic Solution 2. Tubules were incubated for 20 min at 35°C and 80 osc/min. After incubation, 3 mL charcoal-stripped FBS (Thermo Fisher Scientific, Waltham, MA, USA) were immediately added to halt the digestion. The digested product was filtered through a 70-μm cell strainer and again through a 40-μm cell strainer. Sertoli cells were isolated from cells retained by the filters as described in the Supplementary material.
The flow-through, composed of a single-cell suspension of spermatogenic cells, was centrifuged at 500× g for 10 min at 4°C. The pellet was resuspended in 10 mL Enzymatic Solution 3 and placed on ice. To fractionate germ cell populations, centrifugal elutriation was performed on an elutriator (Avanti J-26 XPI Centrifuge, JE-5.0 Elutriator Rotor, 5-mL Elutriation Chamber; Beckman Coulter, Brea, CA, USA). Details for preparing the elutriator and collecting cell fractions are described in the Supplementary material.
All fractions were immediately centrifuged at 500× g at 4°C for 10 min. Pellets from round spermatid fractions were resuspended in 7 mL 25% Percoll in 1× HBSS (vol/vol). Pellets from pachytene spermatocyte fractions were resuspended in 7 mL 21% Percoll in 1× HBSS (vol/vol). The round spermatid fraction was layered over the 28%–45% Percoll gradients, and the pachytene spermatocyte fraction was layered over the 26%–38% Percoll gradients. Percoll gradient preparation is described in the Supplementary material. Gradients were centrifuged at 10,000 rpm for 15 min without the brake. A 3-mL fraction was collected from the top of each gradient, followed by three 5-mL fractions from the top of each gradient. Each fraction was diluted to 20 mL with 1× Dulbecco’s phosphate-buffered saline (DPBS; Invitrogen), centrifuged at 500× g at 4°C for 10 min. Percoll was removed, and smears for each fraction were air-dried on slides.
Cells from each fraction were counted on the hemocytometer to determine the number of cells obtained (Supplementary Table S1). To determine the purity of the cell populations, slides with germ cell populations were stained with the Periodic Acid-Schiff (PAS) kit (Sigma-Aldrich) according to the manufacturer’s instructions, and slides with Sertoli or Leydig cell populations were stained by immunofluorescence (Figure 2 and Supplementary Figure S1) as described in the Supplementary material. After determining fractions that have the highest number of desired cells for each population (>80%– 95%), total RNA was extracted from half of the cells using Qiazol (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The total amounts of RNA obtained were determined by OD260 measurements (Supplementary Table S1), and quantitative RT-PCR (qRT-PCR) analyses were conducted as described in the Supplementary material (Table 1).
Figure 2. PAS and immunofluorescence staining of enriched testicular fractions.
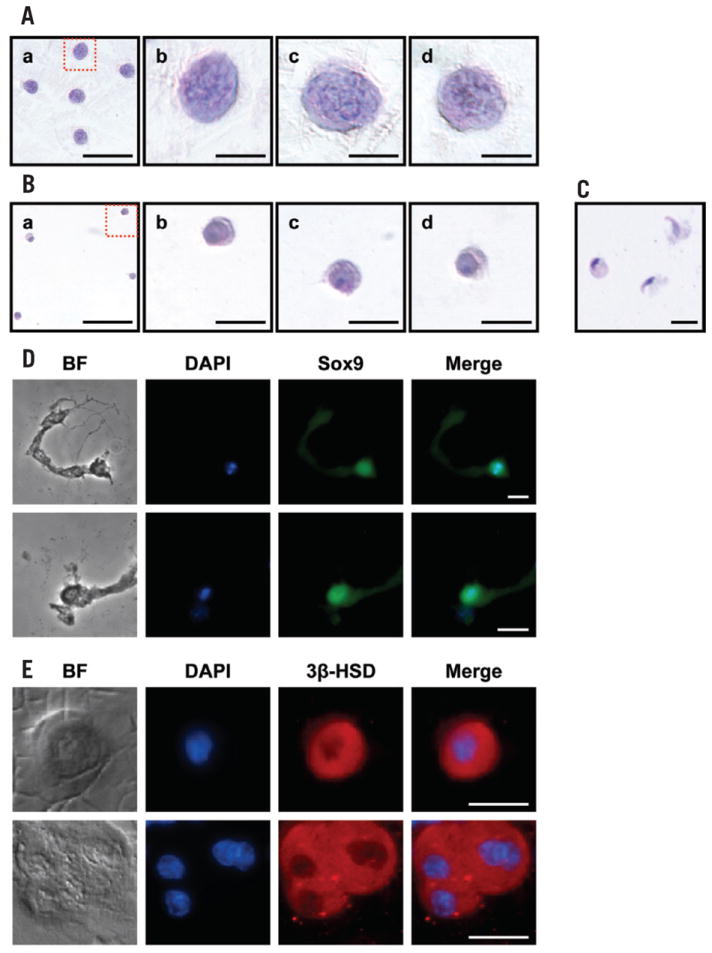
(A–C) Smears of enriched germ cell fractions were PAS-stained according to the manufacturer’s instructions. (A) Pachytene spermatocytes; area in dotted red box (panel a) is magnified in panel b. Irregularly shaped masses of chromatin (darker regions in the nucleus), characteristic of pachytene spermatocytes, are clearly visible in panels b–d. Scale bars, 50 μm (panel a) and 10 μm (panels b–d). (B) Round spermatids; area in dotted red box (panel a) is magnified in panel b. Scale bars, 50 μm (panel a) and 10 μm (panels b–d). (C) Elongating spermatids. Scale bar, 10 μm. (D and E) Smears of enriched testicular somatic cell fractions were subjected to immunofluorescence staining as described in the Supplementary material. (D) Sertoli cells were incubated with anti- Sox9 (a Sertoli cell-specific nuclear protein) primary antibody followed by Alexa Fluor 488 goat anti-rabbit IgG secondary antibody. Scale bars, 10 μm. (E) Leydig cells were incubated with anti-3β-HSD (a Leydig cellspecific protein) primary antibody followed by Alexa Fluor 594 donkey anti-goat IgG secondary antibody. Scale bars, 10 μm. Sertoli and Leydig cell smears were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; blue) for nuclear staining. Photomicrographs were taken on the Nikon Eclipse TE2000-U (Nikon Instruments, Melville, NY, USA). Figures shown are representative of six independent experiments.
Table 1.
Expression of markers genes in purified cell populations
| Marker | Germ cell | Somatic cell | Primary | |||
|---|---|---|---|---|---|---|
| Gene | ES | RS | PS | Leydig | Sertoli | Source |
| Sycp3 | 0.15 ± 0.04 | 0.54 ± 0.25 | 7.08 ± 2.35 | Undet. | 0.36 ± 0.18 | PS |
| Acrv1 | 0.54 ± 0.05 | 10.6 ± 3.2 | 0.58 ± 0.18 | Undet. | 0.31 ± 0.18 | RS |
| Dbil5 | 4.58 ± 0.12 | 1.72 ± 0.54 | 0.75 ± 0.25 | Undet. | 0.20 ± 0.10 | ES/RS |
| Cyp11a1 | 0.002 | Undet. | 0.10 | 0.97 | Undet. | Leydig |
| Rhox5 | 0.63 ± 0.22 | 0.63 ± 0.37 | 0.46 ± 0.21 | Undet. | 4.55 ± 0.75 | Sertoli |
Values represent qRT-PCR analyses of RNA from purified mouse testicular cell populations with Sertoli, Leydig, and germ cell-specific markers, normalized to Rps2 RNA using the comparative threshold cycle (ΔΔCT) method. The values shown are representative of six independent experiments, relative to those for total testes RNA. Primer sequences are available in Supplementary Table S2. PS, pachytene spermatocyte; RS, round spermatid; ES, elongating spermatid; Undet., undetected, value was below limit of detection for qRT-PCR analyses (CT > 38).
In conclusion, we describe a cost-effective and timesaving protocol that is able to isolate multiple somatic and spermatogenic cell populations with high purity from the testis. We have successfully used the purified cells for various downstream assays including genotyping to detect deletion of haploid spermatidand Sertoli cell-specific genes, gene- and miRNA-expression analyses by qRT-PCR or microarray, immunofluorescence staining, and Western blot analyses.
Supplementary Material
Acknowledgments
We would like to thank Dr. Marvin Meistrich and Dr. Ming Zhao for their invaluable instructions. This research was supported by National Institute of Child Health and Development (NICHD) HD057118, National Institutes of Health (NIH). This paper is subject to the NIH Public Access Policy.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary material for this article is available at www.BioTechniques.com/article/113763
References
- 1.O’Shaughnessy PJ, Morris ID, Huhtaniemi I, Baker PJ, Abel MH. Role of androgen and gonadotrophins in the development and function of the Sertoli cells and Leydig cells: data from mutant and genetically modified mice. Mol Cell Endocrinol. 2009;306:2–8. doi: 10.1016/j.mce.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Cheng CY, Mruk DD. A local autocrine axis in the testes that regulates spermatogenesis. Nat Rev Endocrinol. 2010;6:380–395. doi: 10.1038/nrendo.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mays-Hoopes LL, Bolen J, Riggs AD, Singer-Sam J. Preparation of spermatogonia, spermatocytes, and round spermatids for analysis of gene expression using fluorescence-activated cell sorting. Biol Reprod. 1995;53:1003–1011. doi: 10.1095/biolreprod53.5.1003. [DOI] [PubMed] [Google Scholar]
- 4.Lassalle B, Ziyyat A, Testart J, Finaz C, Lefevre A. Flow cytometric method to isolate round spermatids from mouse testis. Hum Reprod. 1999;14:388–394. doi: 10.1093/humrep/14.2.388. [DOI] [PubMed] [Google Scholar]
- 5.Cowden RR, Curtis SK. Acridine orange as a supravital fluorochrome indicating varying degrees of chromatin condensation. Histochemistry. 1974;40:305–310. doi: 10.1007/BF00495037. [DOI] [PubMed] [Google Scholar]
- 6.Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci USA. 2003;100:6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han SY, Zhou L, Upadhyaya A, Lee SH, Parker KL, DeJong J. TFIIAalpha/beta-like factor is encoded by a germ cell-specific gene whose expression is up-regulated with other general transcription factors during spermatogenesis in the mouse. Biol Reprod. 2001;64:507–517. doi: 10.1095/biolreprod64.2.507. [DOI] [PubMed] [Google Scholar]
- 8.Bellve AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Zhong L, Zhu Y, Liu G. Research on the isolation of mouse Leydig cells using differential digestion with a low concentration of collagenase. J Reprod Dev. 2011;57:433–436. doi: 10.1262/jrd.10-123n. [DOI] [PubMed] [Google Scholar]
- 10.Meistrich ML. Separation of spermatogenic cells and nuclei from rodent testes. Methods Cell Biol. 1977;15:15–54. doi: 10.1016/s0091-679x(08)60207-1. [DOI] [PubMed] [Google Scholar]
- 11.Meistrich ML, Longtin J, Brock WA, Grimes SR, Jr, Mace ML. Purification of rat spermatogenic cells and preliminary biochemical analysis of these cells. Biol Reprod. 1981;25:1065–1077. doi: 10.1095/biolreprod25.5.1065. [DOI] [PubMed] [Google Scholar]
- 12.Zhao M, Rohozinski J, Sharma M, Ju J, Braun RE, Bishop CE, Meistrich ML. Utp14b: a unique retrogene within a gene that has acquired multiple promoters and a specific function in spermatogenesis. Dev Biol. 2007;304:848–859. doi: 10.1016/j.ydbio.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anway MD, Folmer J, Wright WW, Zirkin BR. Isolation of sertoli cells from adult rat testes: an approach to ex vivo studies of Sertoli cell function. Biol Reprod. 2003;68:996–1002. doi: 10.1095/biolreprod.102.008045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


