Abstract
Block copolymers are well-known to self-assemble into a range of 3-dimensional morphologies. However, due to their nanoscale dimensions, resolving their exact structure can be a challenge. Transmission electron microscopy (TEM) is a powerful technique for achieving this, but for polymeric assemblies chemical fixing/staining techniques are usually required to increase image contrast and protect specimens from electron beam damage. Graphene oxide (GO) is a robust, water-dispersable, and nearly electron transparent membrane: an ideal support for TEM. We show that when using GO supports no stains are required to acquire high contrast TEM images and that the specimens remain stable under the electron beam for long periods, allowing sample analysis by a range of electron microscopy techniques. GO supports are also used for further characterization of assemblies by atomic force microscopy. The simplicity of sample preparation and analysis, as well as the potential for significantly increased contrast background, make GO supports an attractive alternative for the analysis of block copolymer assemblies.
Introduction
The self-assembly of amphiphilic macromolecules in solution is widely studied for the advancement of drug and gene delivery systems,1 nanoreactors,2 in separation science and in nano-electronics.3 With the increasing popularity and commercial availability of controlled radical polymerization initiators/chain transfer agents, the synthesis of amphiphilic macromolecules has become synthetically simple and hence widely accessible.4,5 Furthermore, a wide range of pre-made amphiphilic polymers are commercially available requiring simple dissolution in a suitable solvent for self-assembly to occur. Such amphiphiles have been proposed for a range of nanoscale applications, however to achieve this, the characterization of their size and morphology must be accurately and readily determined.
Although many analysis techniques can be used to infer information about solution-assembled structures, the most accurate methods used to study morphology and size are based on scattering (light, neutron, and X-ray) and microscopy (electron and atomic force) techniques. Scattering techniques are solution based enabling information to be obtained in the ‘natural state’ of the particle; however, limited information is available using standard light scattering (especially for non-spherical particles) and all scattering techniques are indirect, requiring the fitting of data to a model. Neutron and X-ray scattering are particularly powerful; however, they tend not to be readily available and often require detailed and specialized interpretation of the scattering data.
Microscopy techniques complement scattering data; they directly observe individual particles, typically when dried to a substrate or a support. Scanning electron microscopy (SEM) has been used to identify structures,6–8 but its low spatial resolution (∼5 nm) means that it can offer limited information about typical polymer nanostructures prepared by self-assembly. Atomic force microscopy (AFM) has proven useful in identifying the 3D nature of structures when dried to a surface,9–11 but is limited by tip convolution effects and neither SEM nor AFM can resolve internal structure. The highest spatial resolution can be achieved using transmission electron microscopy (TEM), but often the relatively low mass elements which make up the polymers lead to low contrast and hence difficulties in imaging. In TEM, the image contrast is due to scattering of the electrons as they pass through the sample and regions with high atomic number scatter more electrons than low atomic number areas of the same thickness.12,13 Therefore, scattering arises from differences in density and thickness of the specimen. As the sample must be supported under the electron beam, typically a thin amorphous carbon film is utilized. Because the carbon support film is similar in thickness and composition to typical block copolymer assemblies, resolving copolymer structure against the background of a carbon film can be challenging. To overcome this lack of contrast, chemical fixing/staining techniques are often employed. It is well known that these staining techniques can cause artifacts, leading to difficulties in analysis and unreliable results.14 It has also become common practice to screen a series of stains to find the most appropriate for a given system; this, however is a time-consuming and often subjective process.
The development of cryo-TEM15–17 has enabled the imaging of polymeric nanostructures within a vitrified thin film of water, in their ‘natural state’. The low background of the thin vitrified support means that no staining is required and good contrast for polymeric nanostructures can be obtained. As a result the combination of cryo-TEM and neutron scattering experiments has become the gold standard for analysis of self-assembled amphiphiles in solution.18 However, given the limited accessibility and increased time and costs associated with cryo-TEM, dry-state staining TEM is still very prevalent in the literature.9,19–21 One perceived disadvantage of this dry-state method is that drying of the sample on the substrate can cause deformation or complete destruction of the sample, however it has been observed that many polymer based assemblies tend to be quite rigid and therefore retain their shape.
Recent advances in low background TEM specimen supports give the opportunity to develop a simple alternative to staining, with some complementary advantages to cryo-TEM. The thinnest conceivable support is graphene, a hexagonal crystal of carbon atoms which is only one atom thick. Graphene is strong and almost electron transparent; using graphene supports for high contrast imaging of individual atoms has been demonstrated using TEM analysis.22,23 However, monolayer graphene supports are relatively expensive and very few species adsorb to the homogeneous graphene surface. In comparison, heavily oxidised graphene (or graphene oxide) is readily available in large quantities,24 it is water dispersable and is also robust and nearly electron transparent. Graphene oxide (GO) supports are cheap and easy to make;25 a drop of GO solution is placed on a perforated-carbon TEM grid and allowed to dry. The surface functionalities on GO promote the adhesion of many species, and the low background facilitates high-resolution imaging. GO supports have recently been used for imaging of atomically resolved molecules in motion,26 and vitrified biological molecules.27 The strength of the GO, despite being only one atom thick, allows monolayer GO supports to be imaged by AFM,28 and they are equally robust to exposure in the SEM.25
In this report the exploration of GO as a support for analysis of block copolymer assemblies by multiple microscopy techniques is demonstrated through analysis of the three most common types of micellar assemblies (polymersomes, spherical micelles, and cylindrical micelles). The reader is encouraged to think of this report, not as an in-depth study/comparison of the different microscopy techniques herein, but rather as a demonstration of how a wide variety of complementary data can be collected by the non-microscopy experts who regularly rely on these techniques. The data obtained using GO-grids without staining is compared to data obtained using conventional staining techniques and cryo-TEM. A quantitative analysis of the contrast mechanisms is work ongoing by the authors, to be published as a more specialised microscopy paper. Complementary data from AFM and SEM is also presented, including analysis of the same individual polymersomes on a GO substrate using all three techniques. This is not possible using stained samples or cryo-TEM and presents a significant advance in the detailed characterization of polymeric nanostructures. We also highlight how the low background of GO also readily enables other electron microscopy techniques such as tomography and scanning transmission electron microscopy (STEM).
Experimental
All chemicals were used as received from Aldrich, Fluka, or Acros unless otherwise stated. Tert-butyl acrylate and styrene monomers were distilled over CaH2 prior to use and stored at 5 °C. AIBN [azobisisobutyronitrile] was recrystallized twice from methanol and stored in the dark at 5 °C. DDMAT [S-dodecyl-S′-(α′,α′-dimethyl-α″-acetic acid)],29 poly(lactide)40-b-poly(acrylic acid)286 cylindrical micelles (5)30 and solutions of graphene oxide25 were synthesized as previously reported. Copper TEM grids were purchased from Agar, dry-stained images were obtained on 300 mesh formvar/carbon grids, GO imaging was performed on 400 mesh lacy carbon films, with deposited GO films and cryo-TEM images were taken on Quantifoil holey carbon grids with 200 mesh.
GO solutions (0.10–0.15 mg mL−1) were sonicated for 30 s prior to use. Lacey carbon grids (400 Mesh, Cu) (Agar Scientific) were cleaned using air plasma from a glow-discharge system (2 min, 20 mA): this step is not essential but increases the hydrophilicity of the lacy carbon improving the coverage of GO to typically >70%.27 One drop (∼0.08 mL) of the sonicated GO solution was deposited onto each grid and left to air-dry for ∼30 min. Once dry, the grids could be stored for several weeks before sample deposition. A video abstract is also available on how to prepare the GO grids.28 To deposit aqueous micellar assemblies, a small drop (∼2–10 μL) of sample was pipetted onto a pre-prepared GO grid and left to air dry, or blotted away after several minutes depending on sample type and concentration (typically from 0.2–2.0 mg mL−1). Once dry, the grids could be stored for several months and imaged on numerous occasions without significant damage to the GO.
Results and discussion
The formation of polymersomes has attracted particular interest for nanoreactor31–33 and biomedical34,35 applications as the hydrophobic membrane and hydrophilic interior allow for the incorporation and segregation of various reactive/active groups.36,37 Analysis of these structures requires that the membrane be resolved, and often the thickness of this membrane is integral to their application. Light scattering techniques can be used to obtain a radius of gyration/hydrodynamic radius ratio (Rg/Rh), which can indicate whether a structure is hollow or solid. However, this does not prove a membrane structure and can be difficult to obtain if the sample is not stable across changes in concentration.
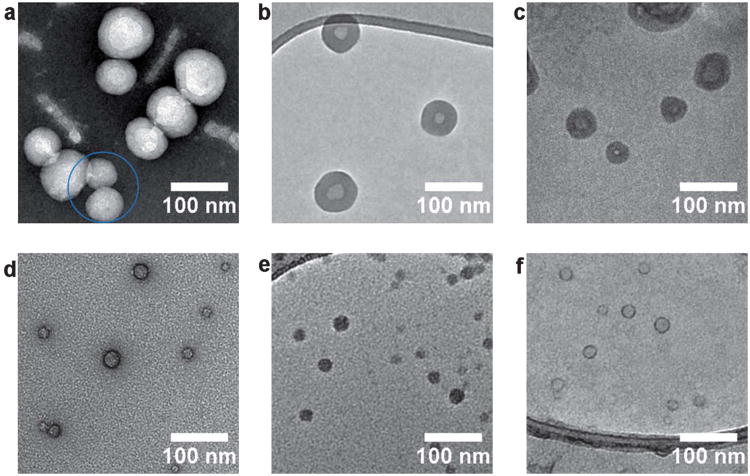
Since the seminal work of Eisenberg and co-workers38,39–42 polyacrylic acid-b-polystyrene has become an archetypal amphiphilic block copolymer for self-assembly into higher ordered structures. Using a range of block ratios and assembly conditions it has been shown that these block copolymers can form spherical micelles, cylindrical micelles, polymersomes, multilamellar structures, and compound micelles.43 In this work, PAA11-b-PS250 was synthesized by the reversible addition fragmentation chain transfer (RAFT) polymerization and assembled using the solvent switch method (see Supporting Information for details). It should be noted that in addition to polymersomes this sample also contained some multilamellar structures, in agreement with previous reports,40 which were not used for analysis (images are included in the supporting information). Fig. 1a–c shows TEM images of the self-assembled structures obtained from the amphiphile PAA11-b-PS250 using a formvar carbon grid stained with uranyl acetate (UA), a GO TEM grid (with no staining), and cryo-TEM analysis (a–c respectively). With the GO grid sample preparation used (see experimental section) typically >50% of the TEM grid is covered by GO, much of the coverage is monolayer with other areas being a few layers thick (diffraction patterns from single and a few layered GO are shown in the ESI†). The GO and GO TEM grids can be stored in air for many months with no sign of deterioration and are extremely stable under the electron beam. Fig. 1a–c show the membrane structure which is indicative of polymersome formation. The contrast in the stained image (Fig. 1a) is due to increased electron scattering where the stain is present, and in this case the stain used here has not been absorbed by the polymersomes, thus the polymersomes appear bright. However, the stained image also shows artifacts such as rod-like features, which are not found when analyzing the same sample by cryo-TEM or on a GO support (a common feature of using a negative staining technique). Furthermore, it is known that different stains can give different contrasts and different artifacts, for example with phosphotungstic acid (PTA) staining there is a complete absence of a membrane structure (see ESI,† Fig. S1). In addition the fine structure of the polymersomes is obscured by the stain: looking at the particles in the blue circle of Fig. 1a (UA stained), in some cases the bilayer is difficult to distinguish. By comparison the polymersome structure is clearly and unambiguously resolved in Fig. 1b without the application of a stain through the utilization of GO as a low background support for imaging. The contrast in this GO image is mainly due to electrons scattered by the polymersomes, and hence reflects their actual structure. This distinction between image contrast in stained and unstained samples is reinforced, in Fig. 1 (d–f) by analysis of a self-assembled PAA100-b-PS46 amphiphile which is known to form spherical micelles.44 When a UA stain is applied (Fig. 1d) it selectively binds to the acrylic acid corona so that a ring-like feature is noted, perhaps suggesting a membrane like-structure. Imaging the same sample unstained on a GO support (Fig. 1e) shows spherical micelles; the true nature of the self-assembled structures. Fig. 1f shows that applying the UA stain to the GO grid gives a similar artificial ring-like contrast indicating that if stains are required, e.g. to identify the location of a particular polymer block within a particle, they can be used on GO grids.45 A comparison of the polymersome images taken by cryo-TEM (Fig. 1c) and on GO supports (Fig. 1b) indicates that the background is significantly lower on the GO than on the vitrified water support (cryo-TEM), permitting clearer resolution of the particle structure and simplifying software detection of particle statistics. Combined with the inherent difficulties in performing cryo-TEM, this makes statistical analysis of a large number of particles considerably easier using GO as a substrate. Average membrane thicknesses for the polymersomes were measured using each imaging technique. The data obtained for GO (21 ± 2 nm) and UA staining (21 ± 2 nm) were comparable, however, slightly lower membrane thickness (17 ± 3 nm) were obtained by cryo-TEM analysis. This could indicate a small distortion of the membrane in the dry state TEM compared to the cryo-TEM analysis, and highlights that although the GO is an excellent low background support for analysis, drying effects cannot be ignored.
Fig. 1.
Representative TEM images of the PAA11-b-PS250 polymersomes using a carbon formvar grid and (a) UA staining, (b) imaged on a GO grid and (c) imaged using cryo-TEM analysis and PAA100-b-PS46 spherical micelles imaged using a carbon formvar grid and (d) UA staining, (e) imaged a GO grid and (f) imaged on a GO grid with UA staining.
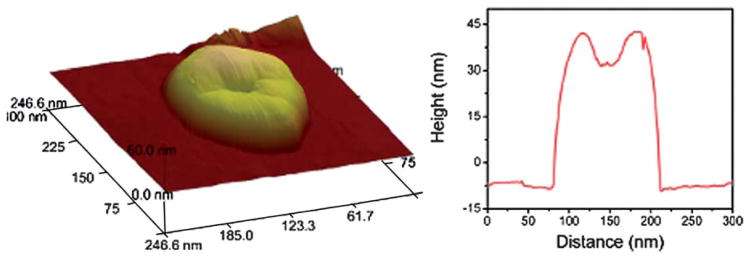
Imaging by cryo-TEM, using positive staining or using the GO-grids are complementary techniques, however, a significant advantage of the GO-grids is that it allows for the direct comparison by TEM and AFM on the same support. This is important as conventional TEM (dry-state or cryo) gives images that are 2D projections of 3D objects from which it is impossible to infer the overall 3D morphology; however AFM analysis can provide information on the 3D structure of the materials. Fig. 2 shows an AFM image of a large polymersome (∼125 nm in diameter); in this particle the centre appears collapsed, as expected when drying a large hollow structure to a substrate. Typically, when TEM/AFM are used in combination to study polymer assemblies, individual samples are prepared for each technique on different substrates and therefore drying effects are likely to be different in each case. Performing both analysis techniques on the same sample on the same substrate allows more accurate correlation of the results and hence more robust analysis of polymeric nanostructures. 3D imaging can also be performed using electron tomography (discussed below); however, the collection of data for many particles will be much more time consuming and require more complex treatment of the collected images.
Fig. 2.
AFM image and corresponding line profile of a PAA11-b-PS250 polymersome on the same GO grid that was used for TEM analysis.
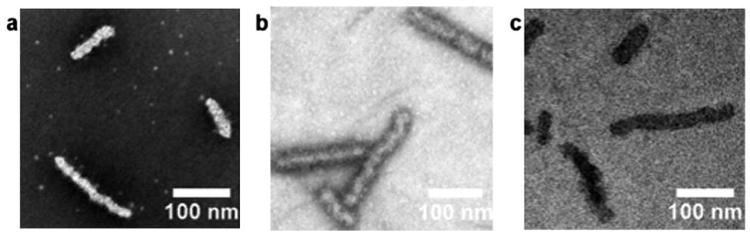
Non-spherical morphologies are also of great interest10,46,47 and as researchers begin to exploit phenomena beyond simple solvophobic effects, their synthesis is becoming more accessible. Microscopy analysis is even more important for these structures as most light scattering models break down for non-spherical aggregates, making these anisotropic structures more difficult to analyze using readily available techniques. As previously reported,30 the crystallization behaviour of poly-(L-lactic acid) (PLLA) can be exploited in order to drive the assembly of block copolymers into cylindrical morphologies. Previous studies focused on measuring the growth of the cylinders by TEM using conventional staining techniques with confirmation of the natural state morphology by cryo-TEM. Fig. 3 shows a comparison of representative TEM images of the same sample (PLLA40-b-PAA286, 4) obtained using (a) a formvar carbon grid and PTA staining, (b) a GO TEM-grid,(c) and cryo-TEM. All three images show similar cylindrical structures; however, the lower background of the GO support (in b) reveals a core-shell contrast that is not visible using the other techniques. This increase in contrast is proposed to allow for more detailed analysis of the exact core-shell nature of the nanostructures and may potentially shed light on the growth mechanism of their formation.
Fig. 3.
Representative TEM images of the PLLA40-b-PAA298 cylinders using (a) a carbon formvar grid with PTA staining, (b) a GO grid and (c) cryo-TEM.
Statistical analysis of the size of the cylinders, extracted from the TEM images, is given in Table 1. The diameters calculated from the images taken using different stains are inconsistent, and do not agree with cryo-TEM analysis. Images taken on GO supports give diameters consistent with cryo-TEM, and AFM analysis of the same sample suggests that there is little if any distortion after drying, as might be expected for cylinders with crystalline PLLA cores. The length distributions follow a similar trend.
Table 1. Summary of the length and width measurements obtained using different TEM techniques for sample (4).
| Technique | Lav (TEM)/nma | Dav (TEM)/nma | Hav (AFM)/nmb |
|---|---|---|---|
| AM staining | 127 (±7) | 26 (±6) | n/a |
| PTA staining | 111 (±8) | 26 (±6) | n/a |
| UA staining | 113 (±7) | 42 (±7) | n/a |
| GO substrate | 106 (±6) | 33 (±5) | 30 (±4) |
| Cryo | 103 (±9) | 32 (±4) | n/a |
100 particles counted for dry state imaging and 43 counted in cryo.
67 particles counted.
The hydrophilic nature of GO makes it readily applicable to aqueous based assemblies; however, the analysis of an organic solvent based sample has also been achieved by our group.48 In addition, should it be required, hydrophobicity can be dialed into the GO substrate by either sonication in water/acetone mixtures49 or simply by heating the GO TEM grid on a hot plate prior to use. The latter can be seen in the gradual increase of contact angle (from about ∼12° to ∼90°) with increasing heating times for a series of GO thin films (see ESI†).
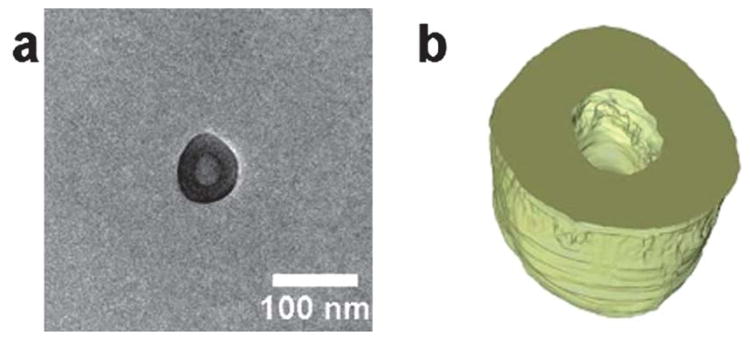
Resolving the full 3D morphology of polymeric assemblies, including the inner structure, can be achieved through electron tomography (ET) analysis. Significant advances have been made in cryo-electron tomography (cryo-ET), but despite the excellent work by Sommerdijk and co-workers50,51 cryo-ET is not yet widely used to study solution-assembled polymeric nanostructures. This is due in part to the relatively high background of the vitreous support, and as a result of support beam damage during the prolonged exposure required for cryo-ET. However, the extremely low background and stability of the GO support make electron tomography and higher resolution images more accessible. Fig. 4a shows the bright tilt TEM image of the PAA11-b-PS250 polymersomes, acquired at 0° tilt and Fig. 4b shows the surface-rendered tomographic reconstruction of the polymersome. We propose that imaging and tomography analysis on GO for more advanced structures, e.g. multi-compartmental or Janus micelles, could prove invaluable and in the authors' view, the availability of high contrast dry-state tomography should encourage the wider use of this technique. While specimen beam damage is obviously sample specific, it should be noted that the ET was performed at 200 kv and, data were collected over a period of >1 h, during which time no deterioration of the sample was found.
Fig. 4.
(a) 2D Bright field TEM image of a polymersome 3, (b) cross section of the 3D reconstruction of the polymersome.
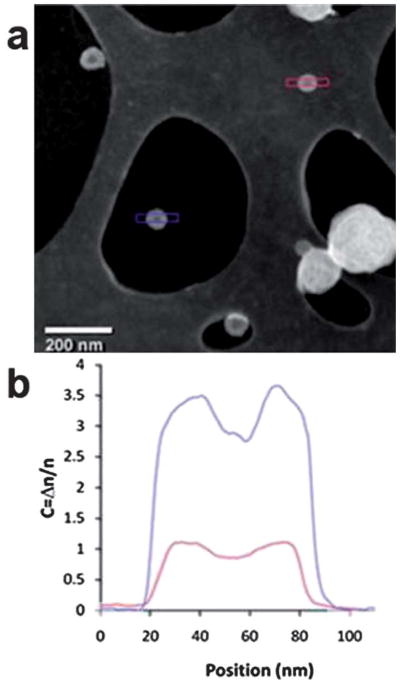
Imaging polymeric particles without staining and in the dry state allows a wider range of analytical TEM techniques, such as those regularly used in hard material analysis, to be applied. For example, features observed in the images recorded from nanoscale polymer assemblies taken using conventional bright-field TEM imaging result from elastic scattering in the sample and interference effects due to the coherence of the electron beam. These phase contrast effects are highly sensitive to focus, even to the extent that the contrast can be reversed with focus, hence imaging is mostly done at a slight under focus. An alternative is to take images in the high angle annular dark field (HAADF) mode of STEM, which collects high-angle elastically scattered electrons. HAADF-STEM is an incoherent imaging technique where the image contrast depends on the atomic number of the scattering atoms, density of the sample, and its thickness (so called “mass-thickness” contrast). Because polymers consist mainly of carbon, contrast of polymeric assemblies in this mode is dominated by changes in density and thickness, making interpretation of HAADF-STEM images more straightforward than conventional TEM images, as no phase contrast is involved. Fig. 5a shows a HAADF image of the PAA11-b-PS250 polymersomes, on the GO substrate which was previously used for bright field imaging. Using this method the membrane structure can be clearly resolved; however what is more significant is that the signal-to-noise ratio is significantly greater for the polymersome on the GO support than for the polymersome on lacy carbon. The line profiles in Fig. 5b demonstrate this increase in contrast by at least a factor of 3, and the corresponding improvement in the signal-to noise ratio. Quantitative analysis of these images could be used to test models for the polymersome structure; thus, the increased contrast on GO would be a significant advantage. This demonstration of HAADF-STEM imaging of block copolymer assemblies on GO indicates several exciting future avenues of research. Combining HAADF-STEM and tomography, as is conventionally done for electron tomography of inorganic structures, should allow less ambiguous and higher resolution analysis of polymeric nanostructures. Furthermore, the inelastic electron/soft material interactions are relatively strong;52 thus chemical analysis can be performed in STEM through measurement of the inelastic scattering. Therefore STEM of polymeric materials on GO gives the opportunity not only for readily interpretable images but also simultaneous collection of electron energy loss and elemental dispersive X-ray spectra at high spatial resolutions.
Fig. 5.
(a) HAADF image containing polymersomes on GO (blue) and on the lacy carbon support (red) and (b) the plot profile along two polymersomes on different supports.
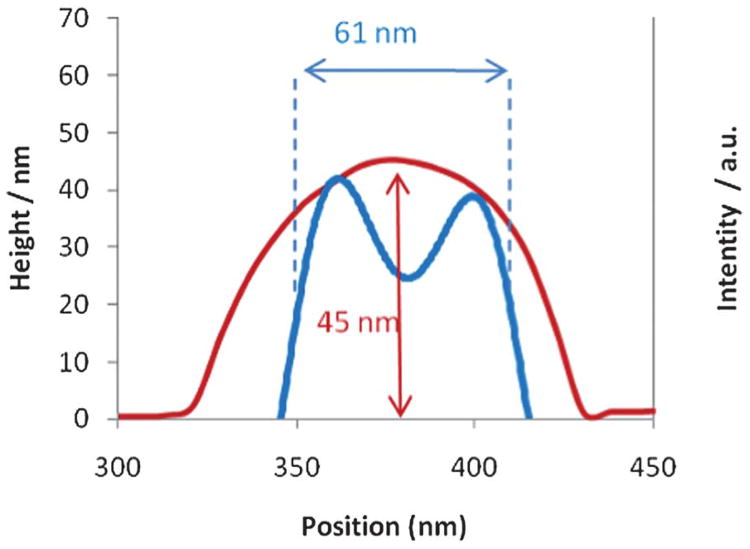
The TEM and AFM results for both the polymersomes and the cylinders (Fig. 1, 2 and 3) were obtained using the same GO grids but from different, random areas of the grid. Using the GO grids, it is not only possible to compare the average results from these techniques but by using a ‘finder’ TEM grid exactly the same area can be examined by multiple techniques (TEM, AFM and SEM in this case) on this single substrate. Fig. 6 shows TEM, AFM, and SEM images of the polymersomes, discussed previously, prepared on a GO support. To the authors' knowledge this is the first time the same individual self-assembled polymeric structures have been imaged using multiple techniques. Imaging the exact same area of the grid further backs up the minimal damage caused when analyzing these samples. The GO causes no charging in the SEM and while some contamination was observed in the SEM, the particles could easily be imaged without the need for coatings which are often used on soft materials. The corresponding plot profiles from AFM and TEM (Fig. 7) show the roughly spherical nature of the particle on the surface (AFM) and its hollow internal structure (TEM). The measured height (45 nm by AFM) and width (61 nm by TEM) show that it is not perfectly spherical, suggesting that drying may have distorted its structure. It should be noted that the apparent width of the polymersome as measured by AFM is increased due to tip convolution effects. Interestingly the reverse side of the GO support can also be imaged using AFM analysis (see Supporting Information) which shows small local distortions in the GO membrane caused by adhesion of the polymersomes. The complementary information on the 3D structure of the polymersome on GO has potential ramifications as it is routine in high-resolution TEM to compare image simulations with experimental observations to attain the maximum structural information. We propose that the enhanced 3D information extracted from the AFM images on GO would help in the construction of an accurate model against which to test the observed TEM images.
Fig. 6.
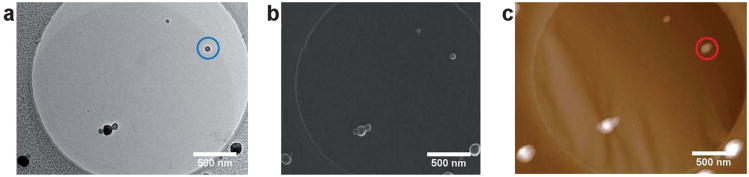
(a) Bright field TEM, (b) SEM, and (c) AFM images of exactly the same area of a GO grid.
Fig. 7.
The plot profiles corresponding to the polymersome in Fig. 6 (circled in blue and red from the TEM and AFM images respectively). The TEM plot profile has been inverted for clarity.
Previous reports25–28 have demonstrated the advantages of using GO as a substrate for TEM, such as the cheap, simple and robust preparation of both the GO and subsequent GO TEM grids which give largely monolayer thickness over the majority of the grid. The good coverage of an almost electron transparent substrate gives ideal imaging conditions for low contrast specimens making analysis far simpler than using staining methods or cryo-TEM. While GO is undoubtedly an excellent substrate for the analysis of soft materials by TEM, its compatibility with other techniques (AFM, SEM and STEM) allows and encourages the collection of data from multiple sources, both minimising misinterpretation and greatly improving image analysis.
Conclusions
The use of GO as a TEM substrate potentially negates the need to use stains for soft materials, reducing image artifacts and subjective interpretations, and simplifying TEM sample preparation. Although distortions due to drying will still be present, by combining TEM data with height information from AFM these drying effects can be measured and better understood. With only one GO TEM-grid needed for analysis by TEM, SEM, and AFM, it is possible to analyze a sample quickly and consistently. This method of analysis is much more accessible than cryo-TEM, provides better contrast, and allows a wider range of advanced microscopy techniques to be used. GO supports do not replace the need for complementary techniques that allow the native structures in solution to be assessed, such as cryo-TEM and scattering techniques (e.g. X-ray and neutron diffraction) but they do offer a readily accessible alternative for routine analysis. In conclusion, the simple use of graphene oxide as a support material for the characterisation of block copolymer assemblies allows high contrast images to be acquired without the use of stains and opens the way for increased resolution, structural determination and chemical analysis.
Supplementary Material
Acknowledgments
The authors thank the University of Warwick and the EPSRC for funding. A.M.S. thanks the Science City Research Alliance and the HEFCE Strategic Development Fund for funding support. T.P.S and T.H.E thank the NIH-NCRR COBRE (#P20RR017716) and NIST, U.S. Department of Commerce (#70NANB7H6178) for financial support. The statements herein do not reflect the views of NIH or NIST. Some items of equipment that were used in this research were funded by Birmingham Science City, with support from Advantage West Midlands and part funded by the European Regional Development Fund. We thank Wellcome Trust grant reference: 055663/Z/98/Z for cryo-instrument use in the electron microscopy facility at Warwick. We acknowledge the University of Delaware Keck Microscopy Facility for use of their TEM and Vitrobot, and the Center for Molecular and Engineering Thermodynamics for use of their DLS and SLS.
Footnotes
Electronic Supplementary Information (ESI) available: Further images, and analysis are included. See DOI: 10.1039/c2sm07040e/
Contributor Information
Neil R. Wilson, Email: Neil.Wilson@warwick.ac.uk.
Rachel K. O'Reilly, Email: r.k.o-reilly@warwick.ac.uk.
Notes and references
- 1.Blanazs A, Armes SP, Ryan AJ. Macromol Rapid Commun. 2009;30:267–277. doi: 10.1002/marc.200800713. [DOI] [PubMed] [Google Scholar]
- 2.Kim KT, Meeuwissen SA, Nolte RJM, van Hest JCM. Nanoscale. 2010;2:844–858. doi: 10.1039/b9nr00409b. [DOI] [PubMed] [Google Scholar]
- 3.Lazzari GLM, Lecommandoux S. Block Copolymers in Nanoscience. Wiley-VCH; 2006. [Google Scholar]
- 4.Read ES, Armes SP. Chem Commun. 2007:3021–3035. doi: 10.1039/b701217a. [DOI] [PubMed] [Google Scholar]
- 5.O'Reilly RK, Hawker CJ, Wooley KL. Chem Soc Rev. 2006;35:1068–1083. doi: 10.1039/b514858h. [DOI] [PubMed] [Google Scholar]
- 6.Xu JT, Tao L, Boyer C, Lowe AB, Davis TP. Macromolecules. 2011;44:299–312. [Google Scholar]
- 7.Yang YQ, Zheng LS, Guo XD, Qian Y, Zhang LJ. Biomacromolecules. 2011;12:116–122. doi: 10.1021/bm101058w. [DOI] [PubMed] [Google Scholar]
- 8.Hossain MD, Tran LTB, Park JM, Lim KT. J Polym Sci Part A Polym Chem. 2010;48:4958–4964. [Google Scholar]
- 9.Li Z, Ma J, Lee NS, Wooley KL. J Am Chem Soc. 2011;133:1228–1231. doi: 10.1021/ja109191z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Presa Soto A, Gilroy JB, Winnik MA, Manners I. Angew Chem Int Ed. 2010;49:8220–8223. doi: 10.1002/anie.201003066. [DOI] [PubMed] [Google Scholar]
- 11.Hamasaka G, Muto T, Uozumi Y. Angew Chem Int Ed. 2011;50:4876–4878. doi: 10.1002/anie.201100827. [DOI] [PubMed] [Google Scholar]
- 12.Williams DB, Carter CB. Transmission Electron Microscopy-A textbook for Materials Science. New York: Plenum Press; 1996. [Google Scholar]
- 13.Loos J, Sourty E, Lu KB, de With G, van Bavel S. Macromolecules. 2009;42:2581–2586. [Google Scholar]
- 14.Talmon Y. J Colloid Interface Sci. 1983;93:366–382. [Google Scholar]
- 15.Zhong S, Pochan DJ. Polym Rev. 2010;50:287–320. [Google Scholar]
- 16.Friedrich H, Frederik PM, de With G, Sommerdijk NAJM. Angew Chem Int Ed. 2010;49:7850–7858. doi: 10.1002/anie.201001493. [DOI] [PubMed] [Google Scholar]
- 17.Cui H, Hodgdon TK, Kaler EW, Abezgauz L, Danino D, Lubovsky M, Talmon Y, Pochan DJ. Soft Matter. 2007;3:945–955. doi: 10.1039/b704194b. [DOI] [PubMed] [Google Scholar]
- 18.Mortensen K, Talmon Y. Macromolecules. 1995;28:8829–8834. [Google Scholar]
- 19.Smith AE, Xu X, Kirkland-York SE, Savin DA, McCormick CL. Macromolecules. 2010;43:1210–1217. [Google Scholar]
- 20.Yan Q, Yuan J, Cai Z, Xin Y, Kang Y, Yin Y. J Am Chem Soc. 2010;132:9268–9270. doi: 10.1021/ja1027502. [DOI] [PubMed] [Google Scholar]
- 21.Yan Q, Zhou R, Fu C, Zhang H, Yin Y, Yuan J. Angew Chem Int Ed. 2011;50:4923–4927. doi: 10.1002/anie.201100708. [DOI] [PubMed] [Google Scholar]
- 22.Meyer JC, Girit CO, Crommie MF, Zettl A. Nature. 2008;454:319–322. doi: 10.1038/nature07094. [DOI] [PubMed] [Google Scholar]
- 23.Cretu O, Krasheninnikov AV, Rodríguez-Manzo JA, Sun L, Nieminen RM, Banhart F. Phys Rev Lett. 2010;105:196102. doi: 10.1103/PhysRevLett.105.196102. [DOI] [PubMed] [Google Scholar]
- 24.Dreyer DR, Park S, Bielawski CW, Ruoff RS. Chem Soc Rev. 2010;39:228–240. doi: 10.1039/b917103g. [DOI] [PubMed] [Google Scholar]
- 25.Wilson NR, Pandey PA, Beanland R, Young RJ, Kinloch IA, Gong L, Liu Z, Suenaga K, Rourke JP, York SJ, Sloan J. ACS Nano. 2009;3:2547–2556. doi: 10.1021/nn900694t. [DOI] [PubMed] [Google Scholar]
- 26.Sloan J, Liu Z, Suenaga K, Wilson NR, Pandey PA, Perkins LM, Rourke JP, Shannon IJ. Nano Lett. 2010;10:4600–4606. doi: 10.1021/nl1026452. [DOI] [PubMed] [Google Scholar]
- 27.Pantelic RS, Meyer JC, Kaiser U, Baumeister W, Plitzko JM. J Struct Biol. 2010;170:152–156. doi: 10.1016/j.jsb.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 28.Wilson NR, Pandey PA, Beanland R, Rourke JP, Lupo U, Rowlands G, Roemer RA. New J Phys. 2010;12:125010. [Google Scholar]
- 29.Skey J, O'Reilly RK. Chem Commun. 2008:4183–4185. doi: 10.1039/b804260h. [DOI] [PubMed] [Google Scholar]
- 30.Petzetakis N, Dove AP, O'Reilly RK. Chem Sci. 2011;2:955–960. [Google Scholar]
- 31.Vriezema DM, Garcia PML, Sancho Oltra N, Hatzakis Ns, Kuiper Sm, Nolte RJM, Rowan AE, van Hest JCM. Angew Chem Int Ed. 2007;46:7378–7382. doi: 10.1002/anie.200701125. [DOI] [PubMed] [Google Scholar]
- 32.Kim KT, Cornelissen JJLM, Nolte RJM, Hest JCMv. Adv Mater. 2009;21:2787–2791. [Google Scholar]
- 33.van Dongen Stijn FM, Nallani M, Cornelissen Jeroen JLM, Nolte Roeland JM, van Hest Jan CM. Chem –Eur J. 2009;15:1107–1114. doi: 10.1002/chem.200802114. [DOI] [PubMed] [Google Scholar]
- 34.Photos PJ, Bacakova L, Discher B, Bates FS, Discher DE. J Controlled Release. 2003;90:323–334. doi: 10.1016/s0168-3659(03)00201-3. [DOI] [PubMed] [Google Scholar]
- 35.Jain JP, Kumar N. Biomacromolecules. 2010;11:1027–1035. doi: 10.1021/bm1000026. [DOI] [PubMed] [Google Scholar]
- 36.Du JZ, O'Reilly RK. Soft Matter. 2009;5:3544–3561. [Google Scholar]
- 37.Onaca O, Enea R, Hughes DW, Meier W. Macromol Biosci. 2009;9:129–139. doi: 10.1002/mabi.200800248. [DOI] [PubMed] [Google Scholar]
- 38.Cameron NS, Corbierre MK, Eisenberg A. Can J Chem. 1999;77:1311–1326. [Google Scholar]
- 39.Zhang LF, Eisenberg A. Science. 1995;268:1728–1731. doi: 10.1126/science.268.5218.1728. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Eisenberg A. Macromolecules. 1996;29:8805–8815. [Google Scholar]
- 41.Zhang LF, Eisenberg A. J Am Chem Soc. 1996;118:3168–3181. [Google Scholar]
- 42.Zhang L, Eisenberg A. Macromolecules. 1999;32:2239–2249. [Google Scholar]
- 43.Zhang L, Eisenberg A. Polym Adv Technol. 1998;9:677–699. [Google Scholar]
- 44.Terreau O, Bartels C, Eisenberg A. Langmuir. 2003;20:637–645. doi: 10.1021/la035557h. [DOI] [PubMed] [Google Scholar]
- 45.Moughton AO, Stubenrauch K, O'Reilly RK. Soft Matter. 2009;5:2361–2370. [Google Scholar]
- 46.Li ZB, Chen ZY, Cui HG, Hales K, Wooley KL, Pochan DJ. Langmuir. 2007;23:4689–4694. doi: 10.1021/la063292b. [DOI] [PubMed] [Google Scholar]
- 47.Gadt T, Ieong NS, Cambridge G, Winnik MA, Manners I. Nat Mater. 2009;8:144–150. doi: 10.1038/nmat2356. [DOI] [PubMed] [Google Scholar]
- 48.Williams PE, Moughton AO, Patterson JP, Khodabakhsh S, O'Reilly RK. Polym Chem. 2011;2:720–729. [Google Scholar]
- 49.Rafiee J, Rafiee MA, Yu ZZ, Koratkar N. Adv Mater. 2010;22:2151–2154. doi: 10.1002/adma.200903696. [DOI] [PubMed] [Google Scholar]
- 50.Nudelman F, de With G, Sommerdijk NAJM. Soft Matter. 2011;7:17–24. [Google Scholar]
- 51.Frederik Pm, Sommerdijk N. Curr Opin Colloid Interface Sci. 2005;10:245–249. [Google Scholar]
- 52.Libera MR, Egerton RF. Polym Rev. 2010;50:321–339. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.