Abstract
The effects of activating mutations associated with night blindness on the stoichiometry of rhodopsin interactions with G protein-coupled receptor kinase 1 (GRK1) and arrestin-1 have not been reported. Here we show that the monomeric form of WT rhodopsin and its constitutively active mutants M257Y, G90D, and T94I, reconstituted into HDL particles are effectively phosphorylated by GRK1, as well as two more ubiquitously expressed subtypes, GRK2 and GRK5. All versions of arrestin-1 tested (WT, pre-activated, and constitutively monomeric mutants) bind to monomeric rhodopsin and show the same selectivity for different functional forms of rhodopsin as in native disc membranes. Rhodopsin phosphorylation by GRK1 and GRK2 promotes arrestin-1 binding to a comparable extent, whereas similar phosphorylation by GRK5 is less effective, suggesting that not all phosphorylation sites on rhodopsin are equivalent in promoting arrestin-1 binding. The binding of WT arrestin-1 to phospho-opsin is comparable to the binding to its preferred target, P-Rh*, suggesting that in photoreceptors arrestin-1 only dissociates after opsin regeneration with 11-cis-retinal, which converts phospho-opsin into inactive phospho-rhodopsin that has lower affinity for arrestin-1. Reduced binding of arrestin-1 to the phospho-opsin form of G90D mutant likely contributes to night blindness caused by this mutation in humans.
Keywords: Arrestin, GPCR, phosphorylation, GRK, monomer, nanodiscs
Introduction
Rhodopsin has long served as a prototypical G protein-coupled receptor (GPCR) [1, 2]. Light activation of the receptor leads to the activation of transducin, but the signaling cascade is rapidly turned off through the action of GPCR kinase 1 (GRK1, a.k.a. rhodopsin kinase) and the binding of visual arrestin-11. This process is essential for maintaining high visual sensitivity and for avoiding damage caused by sustained signaling. More than 100 disease-causing mutations in rhodopsin have been identified (http://www.retina-international.org/files/sci-news/rhomut.htm), several of which result in constitutive activity. These variants underlie a range of phenotypes, from relatively mild night blindness to severe retinal degeneration (retinitis pigmentosa) [3]. The effect of these mutations that make rhodopsin constitutively active on receptor desensitization has not been reported. GRKs are directly activated by their interaction with active GPCRs [4]. We have recently demonstrated that monomeric wild type (WT) light-activated rhodopsin (Rh*) incorporated into high-density lipoprotein (HDL) particles is phosphorylated by GRK1 to the level comparable to that of Rh* in native disc membranes [5], and that monomeric phosphorylated light-activated rhodopsin (P-Rh*) effectively binds visual arrestin-1 [5, 6] with physiologically relevant affinity (KD ~3-4 nM) [5] and with the same 1:1 binding stoichiometry previously demonstrated for rhodopsin in native disc membranes in vivo and in vitro [7]. This convenient model membrane system allows us to readily detect differences in the phosphorylation and arrestin-1 binding of rhodopsin variants in different functional states. Differences in GRK1 and arrestin coupling could play a major role in the phenotype of constitutively active disease causing variants.
Here we characterized WT and three mutant rhodopsins with constitutive activity (M257Y, G90D, and T94I), including T94I and G90D found in human patients with stationary night blindness [8]. We also compared the binding of WT, two phosphorylation-independent forms, and constitutively monomeric mutant of arrestin-1 to different functional forms of these rhodopsin variants. We found that in all cases the resulting monomeric phosphorylated receptor effectively binds all forms of arrestin-1. Unexpectedly, we also found that for virtually all arrestin-rhodopsin combinations, phospho-opsin (P-Ops) is the second most preferred arrestin-1 target after P-Rh*. There are three subfamilies of GRKs found in vertebrates [9] (GRK1, GRK2, and GRK5), and each is expected to exhibit unique selectivity among individual receptors and/or to phosphorylate different combinations of Ser/Thr residues in the same receptor. We therefore also tested if these same rhodopsin variants could lead to changes in the amount of phosphorylation of rhodopsin by GRK1, GRK2 and GRK5, which may in turn lead to differential recruitment of arrestin and desensitization. We found that all three GRKs effectively phosphorylate monomeric forms of WT and mutant rhodopsin, but this phosphorylation does not lead to similar levels of arrestin-1 recruitment.
Materials and methods
Materials
[γ-32P]ATP, [14C]leucine, and [3H]leucine were from Perkin-Elmer (Waltham, MA). All restriction and DNA modifying enzymes were from New England Biolabs (Ipswich, MA). Rabbit reticulocyte lysate was from Ambion (Austin, TX), SP6 RNA polymerase was prepared as described [10]. Cell culture reagents and media were from Mediatech (Manassas, VA) or Invitrogen (Carlsbad, CA). Lipids were obtained from Avanti Polar Lipids (Alabaster, AL), β-decyl-maltoside (DM) from Anatrace (Maumee, OH) and Bio-Beads SM-2 Absorbent from Bio-Rad (Hercules, CA). Membrane scaffold protein 1E3D1 (MSP1E3D1) was prepared as described [11]. All other reagents were from Sigma-Aldrich (St. Louis, MO).
Mutagenesis and plasmid construction
For in vitro transcription bovine arrestin-1 was subcloned into pGEM2 (Promega; Madison, WI) with “idealized” 5-UTR [10] between Nco I and Hind III sites, as described [12]. All mutations were introduced in transcription construct by PCR, using the strategy previously described [13]. All constructs were confirmed by dideoxy sequencing.
In vitro transcription and translation were performed as described recently [14-16].
GRK purification
All GRKs were purified from baculovirus-infected High-5 cells. Bovine GRK2 was expressed as the S670A mutant, which eliminates a C-terminal MAP kinase site [17] and contains a C-terminal hexahistidine tag. Bovine GRK1 and GRK5 were expressed as truncated forms ending at residues 535 and 561, respectively, followed by the exogenous amino acids VDHHHHHH to facilitate purification by Ni2+-NTA chromatography. All GRKs were purified essentially as previously described for bovine GRK1535-H6 [18].
Rhodopsin phosphorylation was performed in kinase buffer (KB)(20 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 1 mM DTT, 0.5 mM ATP). Different forms of rhodopsin (1.2-2.8 uM) were phosphorylated by GRK1 (2.6 uM) for 30 min at room temperature (22°C) under the constant illumination (incandescent light). Alternatively, rhodopsin (800 nM) was phosphorylated by GRK2 or GRK5 (240 nM) for 20 min at 22°C. Analytical phosphorylation was performed in 10 μl assays in the presence of [γ-32P]ATP to yield final specific activity of 800-1,200 cpm/pmol. 10 μl aliquots of preparative phosphorylation reactions were supplemented with [γ-32P]ATP to yield the same specific activity. Samples containing [γ-32P]ATP were stopped by the addition of equal volume of SDS sample buffer and subjected to disc-electrophoresis on 4% stacking and 10% running gel. The gels were stained with Coomassie (GelCode Blue, Pierce) to visualize protein bands, destained with water, dried, and exposed to X-ray film. Rhodopsin bands were excised and the radioactivity was quantified by liquid scintillation counting, whereupon the stoichiometry of the phosphorylation was calculated. Aliquotes of phosphorylated opsin were regenerated with 3-fold molar excess of 11-cis-retinal for 2 h at room temperature. Non-regenerated aliquots from the same batch were used as P-opsin.
Rhodopsin expression and purification
Bovine rhodopsin containing the desired mutations was cloned into the pACMVtetO vector for tetracycline-inducible expression in mammalian cells. The vector was stably transfected into HEK293S-GnTI− cells with restricted and homogenous N-glycosylation [19]. Cells were expanded as adhesion cultures in DMEM/F12 medium (Invitrogen) supplemented with FBS (10%), PenStrep (Gibco), Geneticin-G418 (200 μg/ml) and blasticidin (5 μg/ml). Cells were harvested and further expanded as suspension cultures in 2l Erlenmeyer shaker flasks or a 10l wave bioreactor (GE Healthcare) containing PEM medium (Invitrogen) supplemented with FBS (5%) and PenStrep (Gibco). Protein expression was induced by addition of 2 μg/ml tetracycline and 5 mM sodium butyrate. Cells were harvested after 72 h and stored at −80°C. For purification of recombinant rhodopsin cell pellets were solubilized for 1 h at 4°C with PBS buffer containing protease inhibitors (complete protease inhibitor cocktail tablets, Roche) and 1.25% DM (β-decyl-maltoside). Opsin was bound to 1D4 antibody [20] coupled to CNBr-activated Sepharose 4B resin (GE Healthcare) and washed with PBS pH7.0, 0.125% DM. 11-cis retinal (50 μM) was added and the resin incubated overnight at 4°C to reconstitute ground-state rhodopsin. All steps involving retinal were performed under dim red light. The resin was washed with PBS pH 7.0, 0.125% DM followed by 10 mM HEPES pH 7.0, 0.125% DM. The purified protein was eluted in the same buffer supplemented with an elution peptide resembling the C-terminus of rhodopsin (TETSQVAPA, 80 μM) and concentrated using 50 kDa cutoff Vivaspin concentrators.
Rhodopsin reconstitution into HDL particles
HDL particles were self-assembled essentially as described [21]. Briefly, palmitoyloleoylphosphatidylcholine (POPC) and palmitoyloleoylphosphatidylglycerol (POPG) were mixed at a 3:2 molar ratio and dried under nitrogen. Residual chloroform was further evaporated in a Savant SpeedVac for 3 h at RT. Lipids were solubilized to 24 mM in buffer A (20 mM HEPES, 100 mM NaCl and 1 mM EDTA, pH 8.0) containing 2.4 % DM by sonication and freeze/thaw cycles until the solution was transparent at 4 °C. Under dim red light, 8 mM lipids, 0.8 % DM, 100 μM MSP1E3D1 and a maximum of 10 μM purified mutant or WT rhodopsin were gently mixed with one volume of equilibrated Bio-Beads and rocked at 4 °C overnight. HDL particles containing a single rhodopsin molecule were purified by size exclusion chromatography in buffer A and incorporated rhodopsin was quantified by absorbance at 500 nm (40,600 M−1cm−1).
Direct binding assay
Translated radiolabeled arrestin-1 (2 nM; 100 fmol/assay) was incubated with 0.3 μg of indicated functional form of rhodopsin in 50 μl of 50 mM Tris-HCl, pH 7.4, 100 mM potassium acetate, 1 mM EDTA, 1 mM DTT for 5 min at 30°C under room light (Rh*, P-Rh*, and P-Ops) or in the dark (Rh and P-Rh). Samples were cooled on ice, and bound arrestin-1 eluting with rhodopsin-containing HDL particles was separated from free at 4°C by gel-filtration on 2-ml column of Sephadex G-100 (under dim red light in case of dark forms Rh and P-Rh) and quantified by liquid scintillation counting. Non-specific “binding” was determined in samples containing empty HDL particles and subtracted [5].
Results
GRK1, GRK2, and GRK5 effectively phosphorylate monomeric WT rhodopsin and its constitutively active mutants
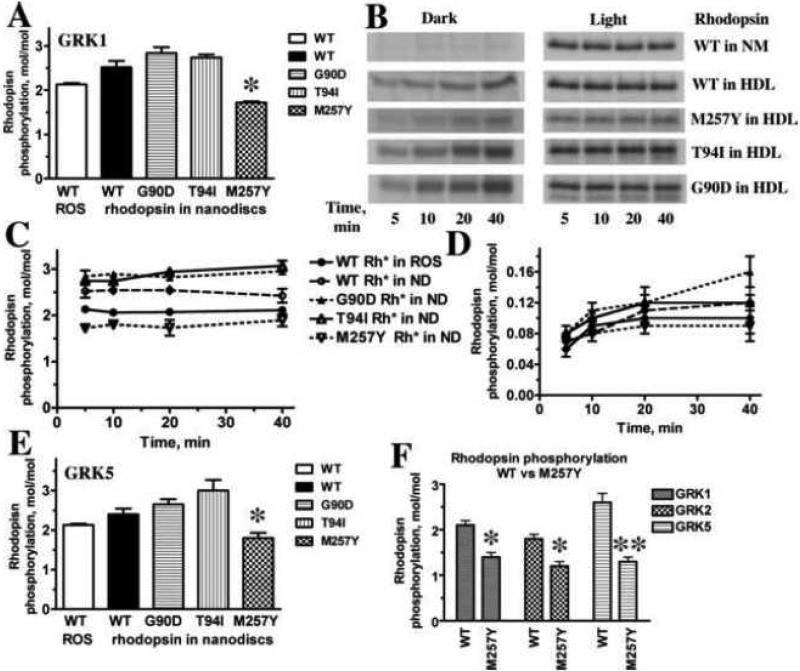
Rh* in rods is rapidly inactivated by a two-step mechanism: phosphorylation followed by arrestin-1 binding [22]. High-affinity arrestin-1 binding has previously been shown to require both rhodopsin activation and the presence of at least three receptor-attached phosphates [23]. The absence of GRK1 in vivo, which ablates rhodopsin phosphorylation, completely prevents arrestin-1 binding that is necessary for rapid Rh* inactivation [24]. We previously showed that GRK1 phosphorylates monomeric WT Rh* in HDL particles at least as efficiently as Rh* in native disc membranes [5]. Here we tested whether monomeric constitutively active rhodopsin mutants in HDL particles can be phosphorylated by GRK1 (Fig. 1). For these experiments we selected M257Y, one of the best characterized constitutively active forms of rhodopsin [25, 26], as well as G90D [27, 28] and T94I [29, 30], both of which are associated with night blindness in humans. The structures of M257Y [25] and G90D [31] are available, whereas that of T94I was modeled using the structure of closely related G90D [31]. Because many pre-activated rhodopsin mutants are less stable than WT, these forms were expressed on the background of a mutant variant carrying double mutation N2C/D282C. This mutant forms a stabilizing disulfide bond between the N-terminus and loop E3, permitting opsin purification in detergent solutions due to increased thermal stability [32, 33]. This extra disulfide bridge does not significantly affect rhodopsin structure [32] or its ability to activate transducin [17, 34], and has been instrumental for the structure determination of several constitutively active mutants [25, 35, 36]. As additional controls, we used purified bovine rhodopsin in native disc membranes and expressed and purified WT rhodopsin in HDL particles that also carried the N2C/D282C mutation. All forms of rhodopsin were phosphorylated by GRK1 to levels comparable to that of WT rhodopsin in native discs, suggesting that these disease-causing mutants interact with and activate GRK1 similar to WT rhodopsin. Interestingly, both G90D and T94I showed slightly higher levels of GRK1 phosphorylation, whereas the phosphorylation of M257Y was consistently lower (Fig. 1A). The phosphorylation of WT rhodopsin in vivo and in native membranes in vitro is strictly dependent on its light activation [37-39]. Therefore, we compared the kinetics of WT rhodopsin phosphorylation with GRK1 in the dark and light in native membranes and HDL particles (Fig. 1B-D). The process was equally light-dependent in both cases: dark phosphorylation was below 5% of the level achieved upon light activation. Because rhodopsin mutants associated with night blindness were reported to exist in an active-like conformation even in the dark [21, 28, 30, 40], we performed similar experiments with these forms of rhodopsin, all of which were reconstituted with 11-cis-retinal (Fig. 1B,D). As with WT rhodopsin, the phosphorylation was completed within the first 5 minutes, and the level did not appreciably change up to 40 min of incubation. The mutants were phosphorylated to a far lesser extent in the dark, although phosphate incorporation appreciably increased with time from 5 to 40 min (Fig. 1B,D). Only G90D demonstrated light-independent phosphate incorporation that was slightly greater than 5% of the level in the light (Fig. 1D). Thus, none of the mutants in the dark assumed conformations necessary for efficient GRK1 phosphorylation. These data suggest that their light-independent signaling in the dark [21] cannot be quenched by normal GRK1 phosphorylation and subsequent arrestin binding, which likely contributes to the observed constitutive rod desensitization in patients carrying T94I or G90D mutations and in mouse models of this disorder [41].
Fig. 1. Phosphorylation of WT rhodopsin and constitutively active mutants by GRK1, GRK2, and GRK5.
A. WT rhodopsin in native disc membranes (WT ROS) or indicated forms of purified rhodopsin reconstituted in monomeric form into HDL particles (7.8 pmol) was phosphorylated by purified GRK1 in the presence of [γ-32P]ATP, subjected to SDS-PAGE, and incorporated radioactivity was quantified, as described in Methods. *, p<0.05, as compared to WT rhodopsin in nanodiscs. B. Representative autoradiograms of indicated forms of rhodopsin in native disc membranes (NM) or HDL particles phosphorylated by GRK1 in the dark or in room light for indicated times. C,D. Calculated stoichiometry of phosphorylation of indicated forms of rhodopsin by GRK1 in the light (C) or dark (D). E. Calculated stoichiometry of phosphorylation of indicated forms of rhodopsin by GRK1. *, p<0.05, as compared to WT, G90D, and T94I rhodopsin in nanodiscs. F. Purified reconstituted into HDL particles WT rhodopsin or M257Y mutant (7.8 pmol) were phosphorylated by indicated purified GRKs in the presence of [γ-32P]ATP, subjected to SDS-PAGE, and incorporated radioactivity was quantified, as described in Methods. *, p<0.05; **, p<0.01, as compared to WT rhodopsin. Means ± SD of three independent experiments performed in duplicate are shown in panels A, C, D, E, F.
GRK1 is almost exclusively expressed in photoreceptors [42], whereas both GRK2 [43] and GRK5 [44] are ubiquitous and represent the most abundant forms in most tissues, including the brain [45-48], which expresses the greatest variety of GPCRs. GRK2 and GRK5 represent two subfamilies of GRK distinct from that of GRK1 [49]. Both of these GRKs phosphorylate Rh* in native discs fairly efficiently [44, 50]. Therefore, to test whether these non-visual GRKs are activated by a monomer of active receptor, we compared the phosphorylation of WT and mutant Rh* in HDL particles by GRKs 1, 2, and 5 (Fig. 1B). All three GRKs yielded very similar levels of Rh* phosphorylation. Interestingly, M257Y was phosphorylated by all three GRKs less efficiently than other forms of rhodopsin (Fig. 1E,F). These data suggest that M257Y is somewhat deficient in its ability to bind and/or activate GRKs, and that the mechanism of activation of all GRKs is similar enough to yield a virtually identical response to subtle structural differences induced by the M257Y substitution [25].
GPCR phosphorylation by GRKs facilitates arrestin binding [51]. Therefore, next we tested whether the phosphorylation of WT rhodopsin by representatives of three GRK subfamilies differentially promotes arrestin-1 binding (Fig. 2A). To this end, we used a direct binding assay with radiolabeled arrestin-1 produced in cell-free translation [52, 53]. Because HDL particles are much smaller than native disc membranes, we used gel-filtration on Sephadex G100, instead of Sepharose 2B in our standard assay [52, 53], to separate arrestin-1 bound to rhodopsin-containing HDL particles and free arrestin-1 [5, 31]. We measured arrestin-1 interaction with inactive (dark) P-Rh, which is likely driven solely by the binding to rhodopsin-attached phosphates, and to P-Rh*, which reflects the ability of simultaneous activation and phosphorylation to facilitate the recruitment of arrestin-1 [12, 14, 54, 55]. The binding to inactive P-Rh generated by all GRKs was essentially the same, whereas light-activated rhodopsin phosphorylated by GRK5 demonstrated lower arrestin-1 binding (Fig. 2A). This difference suggests that the phosphates attached by this kinase interact with the phosphate-binding arrestin residues normally, but appear to be less efficient in cooperating with the active rhodopsin conformation to induce the conformational change in arrestin-1 necessary for high-affinity binding [56]. Conceivably, GRK5 phosphorylates different sites in the rhodopsin C-terminus, which are less conducive to arrestin-1 activation.
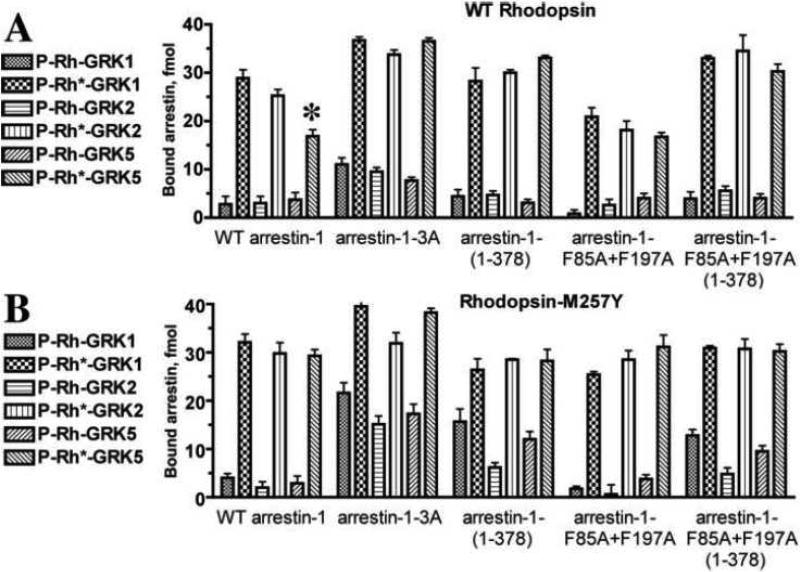
Fig. 2. The binding of arrestin-1 mutants to P-Rh and P-Rh* forms of WT and constitutively active M257Y rhodopsin does not depend on GRK subtype.
A. Translated radiolabeled WT arrestin-1 (WT) and indicated mutants (100 fmol) were incubated with 0.3 μg of WT rhodopsin phosphorylated by indicated GRKs in the dark (P-Rh) or room light (P-Rh*) in 50 μl at 37°C for 5 min. The samples were cooled on ice, and bound arrestins were separated from free by gel-filtration on 2-ml Sephadex G-100 columns, as described in methods. Bound arrestins eluted with rhodopsin-containing HDL particles were quantified by scintillation counting. *, p<0.05, as compared to WT rhodopsin phosphorylated with GRK1 and GRK2. B. The same experiment described in A was performed with M257Y rhodopsin. Means ± SD of two independent experiments performed in duplicate are shown in both panels.
Triple alanine substitution of the hydrophobic residues Phe375, Val376, and Phe377 (3A mutation) forcibly detaches the arrestin-1 C-tail from the body of the molecule, thereby bypassing the need for receptor-attached phosphates and allowing arrestin-1 to bind efficiently to unphosphorylated Rh* [57]. Deletion of the arrestin-1 C-tail in the 1-378 mutant (Tr) has the same functional consequences as the 3A mutation: it promotes the binding to non-preferred functional forms of rhodopsin, dark P-Rh and Rh* [57, 58]. To test whether phospho-rhodopsin generated by GRK5 does not activate arrestin-1 as well as that generated by GRK1 and GRK2, we measured the binding of both pre-activated arrestin-1 mutants to phospho-rhodopsin produced by all three GRKs. The binding of 3A and Tr arrestin-1 to inactive P-Rh and P-Rh* phosphorylated by the three different GRKs was indistinguishable (Fig. 2A). Thus, the effects observed are not specific for a particular arrestin-1 mutant. The results suggest that arrestin-1 activation promoted by GRK5 phosphorylation of WT rhodopsin was indeed less efficient than by GRK1 and GRK2 phosphorylation. However, we observed no differences between these three GRKs in case of the M257Y mutant (Fig. 2B).
Bovine, mouse, and human arrestin-1 robustly self-associates at physiological concentrations found in photoreceptors [7, 59, 60], forming dimers and tetramers [61-64]. The elucidation of the structure of a physiologically relevant solution tetramer [63], which is distinct from that found in crystals [65, 66], enabled the construction of constitutively monomeric arrestin-1- F85A,F197A by alanine substitution of Phe85 and Phe197 [63, 64]. The binding of this mutant to rhodopsin was previously characterized in native disc membranes [64, 67]. Therefore, we tested whether the inability of arrestin-1 to self-associate affects its binding to monomeric rhodopsin in HDL particles. We found that this variant showed somewhat reduced binding to all forms of rhodopsin, but the relative effect of phosphates attached by GRKs 1, 2, and 5 was essentially the same as with WT arrestin-1 (Fig. 2A). It was recently demonstrated in mouse arrestin-1 that mutations disabling self-association can be combined with activating mutations that enhance its binding to all forms of rhodopsin [67]. Because this study was only performed with rhodopsin in native disc membranes, we tested the arrestin-1-(1-378)(F85A,F197A) combination mutant and found that truncation similarly enhances the binding of constitutively monomeric arrestin-1 to rhodopsin phosphorylated by all three GRKs (Fig. 2A). The same effects of arrestin-1 mutations on its binding to rhodopsin monomer in HDL particles and in native disc membranes shows that these interactions are mechanistically indistinguishable, suggesting that in native membrane arrestin-1 also binds a single molecule of P-Rh*. These data do not exclude the possibility that arrestin-1 can engage (albeit with much lower affinity) a second molecule of opsin in native membrane that is virtually packed with rhodopsins [68, 69], because we only tested HDL particles containing a single rhodopsin molecule.
Next we performed the same set of experiments with the constitutively active rhodopsin mutant M257Y, which was extensively characterized functionally [26] and structurally [25]. Phosphorylation by GRKs 1, 2, and 5 similarly promoted arrestin binding to M257Y rhodopsin, despite an overall lower level of M257Y phosphorylation by all three kinases (Fig. 1F). The difference between the effects of phosphorylation by GRK5, as compared to GRKs 1 and 2, apparent in case of WT rhodopsin, completely disappeared in the case of M257Y (Fig. 2B). Two additional differences were revealed. First, the absolute binding levels of WT arrestin-1 and its constitutively monomeric F85A,F197A mutant to P-Rh* form of the M257Y was somewhat higher than to WT rhodopsin (Fig. 2A,B). Interestingly, the binding to M257Y-P-Rh* of the enhanced 3A and Tr mutants, as well as the monomeric form of the latter, was essentially the same as to WT P-Rh* (Fig. 2A,B). These results suggest that pre-activation of either rhodopsin or arrestin-1 yields maximum possible enhancement of the interaction, i.e., mutational activation of both is not additive. In addition, the binding of pre-activated arrestin-1 mutants to dark phosphorylated M257Y was higher than to the same form of WT rhodopsin (Fig. 2A,B). Thus, in case of arrestin-1 binding to a non-preferred form of rhodopsin, the effects of pre-activation of rhodopsin and arrestin-1 appear to be near-additive.
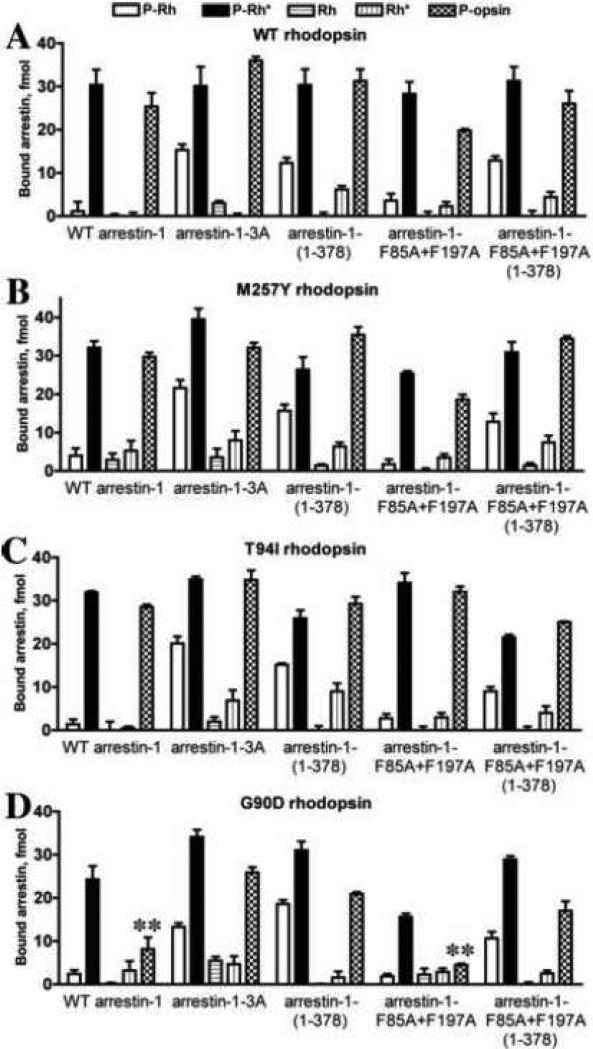
To fully characterize the effects of rhodopsin mutations on its interactions with arrestin-1, we tested the binding of five physiologically relevant functional forms of each mutant: 1) inactive P-Rh (generated by high-gain phosphorylation [70, 71]); 2) the preferred arrestin target P-Rh*; 3) inactive unphosphorylated (Rh), which constitutes the bulk of rhodopsin in dim light where rods function; 4) light-activated unphosphorylated (Rh*), which is directly generated by light; and 5) P-Ops, the immediate product of P-Rh* decay (Fig. 3). Surprisingly, the binding of WT arrestin-1 to P-Ops was almost as high as to P-Rh*, being many fold higher than to all other forms of rhodopsin (Fig. 3A). The constitutively monomeric F85A+F197A arrestin-1 mutant showed the same pattern (Fig. 3A). Importantly, every pre-activated arrestin-1 mutant tested also demonstrated similarly high binding to P-Ops and to P-Rh* (Fig. 3A). Both 3A mutation and the deletion of the C-tail significantly increased arrestin-1 binding to dark P-Rh, as was previously shown in experiments with rhodopsin in native discs [57, 58]. Unexpectedly, we found that neither mutation increases arrestin-1 binding to unphosphorylated Rh* to the extent observed with rhodopsin in native discs in vitro [16, 57, 58] or in vivo [72]. This result suggests that either arrestin-1 binding to Rh* is more demanding in terms of a particular membrane environment (large lipid bilayer surface of native disc, as compared to small area of bilayer surrounded by membrane scaffold protein in HDL particle), or it actually involves additional interactions with neighboring rhodopsin molecules, as was recently suggested [68, 69].
Fig. 3. The binding of arrestin-1 mutants to five functional forms of WT rhodopsin and three constitutively active variants.
A. Translated radiolabeled WT arrestin-1 (WT) and indicated mutants (100 fmol) were incubated with 0.3 μg of WT rhodopsin (unphosphorylated, Rh and Rh*) or phosphorylated by GRK1 (P-Rh, P-Rh*, and P-opsin) in the dark (Rh and P-Rh) or room light (Rh*, P-Rh*, and phospho-opsin) in 50 μl at 37°C for 5 min. The samples were cooled on ice, and bound arrestins were separated from free by gel-filtration on 2-ml Sephadex G-100 columns, as described in methods. Bound arrestins eluted with rhodopsin-containing HDL particles were quantified by scintillation counting. B, C, D. The same experiment described in A was performed with M257Y, T94I, or G90D rhodopsin mutants, respectively. Means ± SD of two independent experiments performed in duplicate are shown in all panels. **, p<0.01, as compared to the binding of the same arrestin-1 (WT or F85A+F197A constitutively monomeric mutant) to p-Ops form of WT rhodopsin.
The interactions of different forms of arrestin-1 with the constitutively active rhodopsin M257Y mutant largely follow the same pattern: very high binding to P-Ops, comparable to that to P-Rh*, and increased binding of all pre-activated arrestin-1 mutants to dark P-Rh (Fig. 3A). Interestingly, the binding of all forms of arrestin to M257Y-Rh* was consistently higher than to the same form of WT rhodopsin (Fig. 3A,B). Rhodopsin with T94I mutation that causes night blindness in humans [29] performed essentially like M257Y (Fig. 3B,C), which has comparable light-independent activity. However, the binding of both WT arrestin-1 and its oligomerization-deficient F85A+F197A mutant to G90D rhodopsin, which causes very similar disease phenotype in human patients [27], showed a striking reduction of the interaction with P-Ops (Fig. 3D). Arrestin-1 pre-activation by 3A mutation or C-tail deletion significantly increased the binding to G90D P-Ops, although not to the level observed with M257Y or T94I mutants (Fig. 3B-D). Also, the binding of enhanced arrestins to the Rh* form of G90D mutant is closer to that observed with WT rhodopsin than to the other two constitutively active rhodopsin mutants, M257Y and T94I (Fig. 3). This finding is in agreement with previous report [73] and suggests that pre-activated mutant rhodopsins demonstrating similar ability to activate transducin, can have different propensity to bind arrestin-1.
Discussion
Recently, GRK1 was shown to effectively phosphorylate monomeric light-activated rhodopsin in HDL particles [5], and normal arrestin-1 binding to the P-Rh* monomer has been independently demonstrated by two groups [5, 6]. However, the ability of other GRKs to phosphorylate monomeric GPCRs has not been reported. Therefore, here we determined the ability of two ubiquitously expressed non-visual GRKs, GRK2 and GRK5 [43, 44], to phosphorylate monomeric rhodopsin reconstituted into HDL particles. Our data show that both phosphorylate the monomeric form of WT rhodopsin, its constitutively active M257Y mutant, and two pre-activated mutants associated with night blindness in humans to a similar level observed with the rhodopsin kinase, GRK1 (Fig. 1). The phosphorylation of WT rhodopsin and the M257Y mutant by GRK2 enhances arrestin-1 binding essentially to the same extent as their phosphorylation by GRK1, whereas the phosphorylation by GRK5 is less efficient in this regard (Fig. 2). When the pre-activated forms of arrestin-1, which are less dependent on rhodopsin phosphorylation [57], are used, this difference is no longer apparent (Fig. 2). Yet GRK5 attaches a higher average number of phosphates per rhodopsin molecule (Fig. 1B,C,D). Thus, either it preferentially places phosphates in positions that are not conducive to arrestin-1 activation, or it phosphorylates a smaller fraction of the total rhodopsin population to the level required for high-affinity arrestin-1 binding, which is three or more phosphates per molecule [14]. Considering that the average number of phosphates transferred to rhodopsin by all three GRKs is comparable (Fig. 1), this result is the first indication that not all rhodopsin phosphorylation sites may be functionally equivalent. This issue requires further in-depth investigation using rhodopsin mutants lacking specific sites. Interestingly, even though the phosphorylation of the M257Y variant of rhodopsin by all GRKs is lower than that of WT rhodopsin (Fig. 1A,B), arrestin-1 binding to both dark and light-activated forms of M257Y mutant is the same or even higher (Fig. 2A,B). Thus, either predisposition of M257Y to assume the active conformation overcomes the relative paucity of attached phosphates, or a higher fraction of M257Y acquires the minimum number of phosphates necessary for arrestin-1 activation.
This HDL-based assay system facilitates the analysis of the effects of constitutively active rhodopsin mutants on phosphorylation by GRKs and arrestin-1 binding. Rhodopsin mutations G90D and T94I cause night blindness in humans [27, 29]. Significant light-independent activity of these mutants [28, 30, 40] is believed to be the cause of the disease phenotype, because these forms of rhodopsin even in darkness produce an “equivalent light” [27, 40] sufficient to fully desensitize rods. The ability of monomeric WT rhodopsin to couple to the visual G protein transducin [21, 74, 75], serve as a substrate for GRK1 [5], and bind WT arrestin-1 with high affinity and 1:1 stoichiometry [5, 6] has been clearly established. Phosphorylation and arrestin-1 binding of mutant rhodopsins in cell membranes was also demonstrated [73]. Mutation-induced changes in the mode of interaction of rhodopsin with any of these three partners would have important implications for the understanding of the molecular basis of the disease phenotype and for possible therapeutic approaches. Our data show that monomeric forms of all constitutively active rhodopsin mutants, including G90D and T94I that cause night blindness, are effectively phosphorylated by GRK1 and show the ability to bind arrestin-1, although the G90D mutant does so less efficiently than WT rhodopsin (Figs. 1,3). A recent crystal structure of G90D opsin shows that the introduced Asp forms a salt bridge with K296 [36], which likely restricts the conformational dynamics of TM6/TM7 that in turn modulate arrestin-1 binding [76]. The WT-like arrestin binding to T94I opsin is consistent with this interpretation, as the uncharged Ile would not be able to form similar ionic interactions between TM2 and TM7.
Collectively our data demonstrate that all of the variants of rhodopsin that we tested are equally good substrates for three different GRKs representing the three vertebrate GRK subfamilies [9](Figs. 1,2) and all efficiently bind arrestin-1 (Figs. 2,3). Moreover, mutations rendering arrestin-1 pre-activated or constitutively monomeric do not appreciably affect its binding to monomeric P-Rh*. Thus, the disease-causing T94I mutation in rhodopsin does not appear to affect its interactions with GRKs and arrestin-1, whereas G90D reduces arrestin-1 binding, particularly to P-Ops. Because G90D and T94I mutations produce similar disease phenotypes, these results suggest that alterations in the inactivation mechanism are not the only cause of the pathology observed in human patients.
Our study yielded three unexpected findings. First, we observed very high binding of WT arrestin-1 to WT P-Ops (Fig. 3). Direct binding data are consistent with recent report that P-Rh* and P-Ops induce similar dramatic changes in the NMR spectrum of labeled arrestin-1, distinct from those induced by Rh* or dark P-Rh [77]. This finding suggests that in rod photoreceptors arrestin-1 likely remains bound after P-Rh* decays to P-Ops. Thus, arrestin-1 is likely released later than was previously believed [53] when opsin is regenerated with 11-cis retinal, which converts the receptor to inactive P-Rh that has low affinity for arrestin-1 (Fig. 3). This sequence of events makes sense biologically. Single photon sensitivity of rods [78] requires extremely low noise. The suppression of the non-zero ability of opsin to activate transducin [79, 80] by arrestin-1 that remains bound until opsin is regenerated into completely silent dark P-Rh by covalent attachment of inverse agonist 11-cis-retinal would be an important factor in noise reduction. Our finding is consistent with the reports that arrestin-1 plays a role in rhodopsin regeneration [81-83] and identifies a probable molecular mechanism of this phenomenon.
Second, we found that the P-Ops form of G90D is dramatically different from WT rhodopsin and other mutants in this regard: it demonstrates much lower propensity to bind arrestin-1 (Fig. 3). Since G90D mutant binds 11-cis-retinal less effectively than WT rhodopsin, a larger proportion of the mutant likely exists in the form of opsin in vivo. Thus, mutation-caused defect in arrestin-1 binding is likely to significantly increase overall noise in photoreceptors expressing the G90D mutant, which could contribute to rod desensitization and the night blindness phenotype. However, the P-Ops form of the T94I mutant with similar phenotypic manifestations binds arrestin-1 as robustly as WT rhodopsin, indicating that premature release of P-Ops is not the only disease-causing mechanism.
Third, we found that enhanced arrestin-1 mutants that avidly bind unphosphorylated Rh* in native disc membranes [57, 58, 67, 84] demonstrate much less avid binding to monomeric Rh* (Fig. 3). It was previously shown that the complex of constitutively active arrestin-1 mutants with Rh* is almost as salt-resistant as the complex of WT arrestin-1 with P-Rh* [12, 54], in sharp contrast to the WT arrestin-1-Rh* complex [53]. This led to the idea that the interactions of pre-activated arrestins with Rh* are mechanistically similar to WT arrestin-P-Rh* binding [85, 86]. Our data suggest that this is not necessarily the case. Naturally, in therapeutically relevant situations unphosphorylated rhodopsin will be in native disc membranes of the patient. Nonetheless, better understanding of the distinct mechanisms of the interactions of enhanced arrestin-1 mutants with Rh* would help design mutant forms of arrestin-1 that can yield more efficient compensation for defects in rhodopsin phosphorylation.
Many human disorders outside of the visual system, ranging from cartilage diseases to several forms of cancer, are associated with excessive signaling by different GPCRs [87]. It was recently demonstrated that certain mutations on the receptor-binding surface of inherently promiscuous non-visual arrestins can dramatically restrict their receptor preference [88]. This opens the possibility of specific targeting of individual overactive GPCRs by existing enhanced forms of non-visual arrestins [89, 90] to rebalance the signaling [85, 86]. Moreover, in some arrestin-receptor combinations GPCR phosphorylation does not appear to play as decisive a role as in arrestin-1 binding to rhodopsin [13]. Therefore, our results suggest that further functional and structural characterization of arrestin complexes with unphosphorylated receptors is necessary.
Conclusions
Constitutively active rhodopsin mutants, including G90D and T94I that cause night blindness in humans, reconstituted in monomeric form into HDL particles, are effectively phosphorylated by GRK1, GRK2, and GRK5.
In all cases phosphorylation is light dependent, suggesting that constitutive activity of these mutants is biased towards transducin and does not make them good substrates for GRKs in the dark.
Phospho-opsin forms of WT and mutant rhodopsin is the second most preferred target of arrestin proteins after light-activated phosphorylated rhodopsin.
Phospho-opsin form of G90D mutant demonstrates significantly lower arrestin binding. Since G90D is impaired in retinal binding, its opsin form is abundant. Due to reduced arrestin affinity it likely contributes to the disease phenotype via increased background signaling in patients.
Highlights.
Constitutively active rhodopsin mutants associated with night blindness are effectively phosphorylated by GRK1, GRK2, and GRK5 in the light, but not in darkness.
WT and pre-activated forms of arrestin-1 effectively bind these mutants reconstituted in monomeric form into nanodiscs.
Phospho-opsin is the second most preferred arrestin-1 target after light-activated phosphorylated rhodopsin.
Phospho-opsin form of G90D mutant is impaired in arrestin-1 binding, which likely contributes to the disease phenotype.
Acknowledgements
The authors thank Dr. T. H. Bayburt for reconstituting native bovine rhodopsin into HDL particles. Supported in part by NIH grants EY011500, GM077561, GM081756 (VVG), HL086865, HL071818, MH089378 (JJGT), GM033775 (SGS), AHA grant N014938 (KTH), Swiss National Science Foundation (SNSF) grant 31003A_132815 (JS and GFXS) and 31003A_141235 (JS).
Abbreviations
- GPCR
G protein-coupled receptor
- GRK
G protein-coupled receptor kinase
- WT
wild type
- Rh
dark unphosphorylated rhodopsin
- Rh*
light-activated unphosphorylated rhodopsin
- P-Rh
dark phosphorylated rhodopsin
- P-Rh*
light-activated phosphorylated rhodopsin
- P-Ops
phospho-opsin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributors SAV, MKO, AS, VP, KTH, and AG performed experiments and contributed to manuscript writing; SGS, JJGT, GFXS, JS, and VVG designed the study, coordinated experiments, and wrote the manuscript. All authors have approved the final article.
We use systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48 kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin; for reasons that are not clear its gene is called “arrestin 3” in HUGO database).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Hofmann KP, Scheerer P, Hildebrand PW, Choe H-W, Park JH, Heck M, Ernst OP. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Deupi X, Standfuss J, Schertler G. Biochem Soc Trans. 2012;40:383–388. doi: 10.1042/BST20120001. [DOI] [PubMed] [Google Scholar]
- 3.Lem J, Fain GL. Trends Mol Med. 2004;10:150–157. doi: 10.1016/j.molmed.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K, Buczylko J, Kaplan MW, Polans AS, Crabb JW. J Biol Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 5.Bayburt TH, Vishnivetskiy SA, McLean M, Morizumi T, Huang C-c, Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukamoto H, Sinha A, Dewitt M, Farrens DL. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Proc Nat Acad Sci USA. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. Proc Natl Acad Sci USA. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurevich EV, Tesmer JJ, Mushegian A, Gurevich VV. Pharmacol Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurevich VV. Methods Enzymol. 1996;275:382–397. doi: 10.1016/s0076-6879(96)75023-1. [DOI] [PubMed] [Google Scholar]
- 11.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurevich VV, Benovic JL. J Biol Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 13.Gimenez LE, Kook S, Vishnivetskiy SA, Ahmed MR, Gurevich EV, Gurevich VV. J Biol Chem. 2012;287:9028–9040. doi: 10.1074/jbc.M111.311803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV. J Biol Chem. 2007;282:32075–32083. doi: 10.1074/jbc.M706057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, Gurevich VV. J Biol Chem. 2011;286:24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vishnivetskiy SA, Chen Q, Palazzo MC, Brooks EK, Altenbach C, Iverson TM, Hubbell WL, Gurevich VV. J Biol Chem. 2013;288:3394–3405. doi: 10.1074/jbc.M112.445437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitcher JA, Tesmer JJ, Freeman JL, Capel WD, Stone WC, Lefkowitz RJ. J Biol Chem. 1999;274:34531–34534. doi: 10.1074/jbc.274.49.34531. [DOI] [PubMed] [Google Scholar]
- 18.Huang CC, Yoshino-Koh K, Tesmer JJ. J Biol Chem. 2009;284:17206–17215. doi: 10.1074/jbc.M809544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Proc Nat Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molday RS, MacKenzie D. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 21.Whorton MR, Jastrzebska B, Park PSH, Fotiadis D, Engel A, Palczewski K, Sunahara RK. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, Gurevich EV. Prog Retin Eye Res. 2011;30:405–430. doi: 10.1016/j.preteyeres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurevich VV, Gurevich EV. Trends Pharmacol Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Chen CK, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Proc Nat Acad Sci USA. 1999;96:3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, Schertler G, Standfuss J. Proc Natl Acad Sci U S A. 2012;109:119–124. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han M, Smith SO, Sakmar TP. Biochemistry. 1998;37:8253–8261. doi: 10.1021/bi980147r. [DOI] [PubMed] [Google Scholar]
- 27.Sieving PA, Richards JE, Naarendorp F, Bingham EL, Scott K, Alpern M. Proc Natl Acad Sci U S A. 1995;92:880–884. doi: 10.1073/pnas.92.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao VR, Cohen GB, Oprian DD. Nature. 1994;367:639–642. doi: 10.1038/367639a0. [DOI] [PubMed] [Google Scholar]
- 29.al-Jandal N, Farrar GJ, Kiang AS, Humphries MM, Bannon N, Findlay JBC, Humphries P, Kenna PF. Hum Mutat. 1999;13:75–81. doi: 10.1002/(SICI)1098-1004(1999)13:1<75::AID-HUMU9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Gross AK, Rao VR, Oprian DD. Biochemistry. 2003;42:2009–2015. doi: 10.1021/bi020613j. [DOI] [PubMed] [Google Scholar]
- 31.Singhal A, Ostermaier MK, Vishnivetskiy SA, Panneels V, Homan KT, Tesmer JJ, Veprintsev D, Deupi X, Gurevich VV, Schertler GF, Standfuss J. EMBO Rep. 2013 doi: 10.1038/embor.2013.44. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GFX. J Mol Biol. 2007;372:1179–1188. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie G, Gross AK, Oprian DD. Biochemistry. 2003;42:1995–2001. doi: 10.1021/bi020611z. [DOI] [PubMed] [Google Scholar]
- 34.Standfuss J, Zaitseva E, Mahalingam M, Vogel R. J Mol Biol. 2008;380:145–157. doi: 10.1016/j.jmb.2008.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fung JJ, Deupi X, Pardo L, Yao XJ, Velez-Ruiz GA, Devree BT, Sunahara RK, Kobilka BK. EMBO J. 2009;28:3315–3328. doi: 10.1038/emboj.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kühn H, Dreyer WJ. FEBS Lett. 1972;20:1–6. doi: 10.1016/0014-5793(72)80002-4. [DOI] [PubMed] [Google Scholar]
- 38.Kühn H. Nature. 1974;250:588–590. doi: 10.1038/250588a0. [DOI] [PubMed] [Google Scholar]
- 39.McDowell JH, Kühn H. Biochemistry. 1977;16:4054–4060. doi: 10.1021/bi00637a018. [DOI] [PubMed] [Google Scholar]
- 40.Jin S, Cornwall MC, Oprian DD. Nat Neurosci. 2003;6:731–735. doi: 10.1038/nn1070. [DOI] [PubMed] [Google Scholar]
- 41.Dizhoor AM, Woodruff ML, Olshevskaya EV, Cilluffo MC, Cornwall MC, Sieving PA, Fain GL. J Neurosci. 2008;28:11662–11672. doi: 10.1523/JNEUROSCI.4006-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lorenz W, Inglese J, Palczewski K, Onorato JJ, Caron MG, Lefkowitz RJ. Proc Natl Acad Sci U S A. 1991;88:8715–8719. doi: 10.1073/pnas.88.19.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benovic JL, DeBlasi A, Stone WC, Caron MG, Lefkowitz RJ. Science. 1989;246:235–240. doi: 10.1126/science.2552582. [DOI] [PubMed] [Google Scholar]
- 44.Kunapuli P, Benovic JL. Proc Natl Acad Sci U S A. 1993;90:5588–5592. doi: 10.1073/pnas.90.12.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bychkov ER, Ahmed MR, Gurevich VV, Benovic JL, Gurevich EV. Neurobiol Dis. 2011;44:248–258. doi: 10.1016/j.nbd.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed MR, Gurevich VV, Dalby KN, Benovic JL, Gurevich EV. J Pharmacol Exp Ther. 2008;325:276–283. doi: 10.1124/jpet.107.131987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmed MR, Bychkov E, Gurevich VV, Benovic JL, Gurevich EV. J Neurochem. 2008;104:1622–1636. doi: 10.1111/j.1471-4159.2007.05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bychkov ER, Gurevich VV, Joyce JN, Benovic JL, Gurevich EV. Neurobiol Aging. 2008;29:379–396. doi: 10.1016/j.neurobiolaging.2006.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mushegian A, Gurevich VV, Gurevich EV. PLoS One. 2012;7:e33806. doi: 10.1371/journal.pone.0033806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benovic JL, Mayor FJ, Somers RL, Caron MG, Lefkowitz RJ. Nature. 1986;321:869–872. doi: 10.1038/321869a0. [DOI] [PubMed] [Google Scholar]
- 51.Gurevich VV, Gurevich EV. Pharmacol Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gurevich VV, Benovic JL. J Biol Chem. 1992;267:21919–21923. [PubMed] [Google Scholar]
- 53.Gurevich VV, Benovic JL. J Biol Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 54.Gurevich VV, Benovic JL. Mol Pharmacol. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- 55.Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. J Biol Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 56.Kim M, Vishnivetskiy SA, Van Eps N, Alexander NS, Cleghorn WM, Zhan X, Hanson SM, Morizumi T, Ernst OP, Meiler J, Gurevich VV, Hubbell WL. Proc Nat Acad Sci USA. 2012;109:18407–18412. doi: 10.1073/pnas.1216304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gurevich VV. J Biol Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 58.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez M-G, Gurevich VV. J. Biol. Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 59.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. J Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, Gurevich VV. Neuroscience. 2011;174:37–49. doi: 10.1016/j.neuroscience.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schubert C, Hirsch JA, Gurevich VV, Engelman DM, Sigler PB, Fleming KG. J Biol Chem. 1999;274:21186–21190. doi: 10.1074/jbc.274.30.21186. [DOI] [PubMed] [Google Scholar]
- 62.Hanson SM, Van Eps N, Francis DJ, Altenbach C, Vishnivetskiy SA, Arshavsky VY, Klug CS, Hubbell WL, Gurevich VV. EMBO J. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanson SM, Dawson ES, Francis DJ, Van Eps N, Klug CS, Hubbell WL, Meiler J, Gurevich VV. Structure. 2008;16:924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim M, Hanson SM, Vishnivetskiy SA, Song X, Cleghorn WM, Hubbell WL, Gurevich VV. Biochemistry. 2011;50:2235–2242. doi: 10.1021/bi1018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 66.Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 67.Vishnivetskiy SA, Chen Q, Palazzo MC, Brooks EK, Altenbach C, Iverson TI, Hubbell WL, Gurevich VV. J Biol Chem. 2013;288:3394–3405. doi: 10.1074/jbc.M112.445437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sommer ME, Hofmann KP, Heck M. J Biol Chem. 2011;286:7359–7369. doi: 10.1074/jbc.M110.204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sommer ME, Hofmann KP, Heck M. Nat Commun. 2012;3:995. doi: 10.1038/ncomms2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Binder BM, Biernbaum MS, Bownds MD. J Biol Chem. 1990;265:15333–15340. [PubMed] [Google Scholar]
- 71.Binder BM, O'Connor TM, Bownds MD, Arshavsky VY. J Biol Chem. 1996;271:19826–19830. doi: 10.1074/jbc.271.33.19826. [DOI] [PubMed] [Google Scholar]
- 72.Song X, Vishnivetskiy SA, Gross OP, Emelianoff K, Mendez A, Chen J, Gurevich EV, Burns ME, Gurevich VV. Curr Biol. 2009;19:700–705. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rim J, Oprian DD. Biochemistry. 1995;34:11938–11945. doi: 10.1021/bi00037a035. [DOI] [PubMed] [Google Scholar]
- 74.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Proc Natl Acad Sci U S A. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. J Biol Chem. 2007;282:4875–4881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 76.Banerjee S, Huber T, Sakmar TP. J Mol Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 77.Zhuang T, Chen Q, Cho M-K, Vishnivetskiy SA, Iverson TI, Gurevich VV, Sanders CR. Proc Nat Acad Sci USA. 2013;110:942–947. doi: 10.1073/pnas.1215176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baylor DA, Lamb TD, Yau KW. J Physiol. 1979;288:613–634. [PMC free article] [PubMed] [Google Scholar]
- 79.Okada D, Nakai T, Ikai A. Photochem Photobiol. 1989;49:197–203. doi: 10.1111/j.1751-1097.1989.tb04096.x. [DOI] [PubMed] [Google Scholar]
- 80.Cohen GB, Yang T, Robinson PR, Oprian DD. Biochemistry. 1993;32:6111–6115. doi: 10.1021/bi00074a024. [DOI] [PubMed] [Google Scholar]
- 81.Sommer ME, Smith WC, Farrens DL. J Biol Chem. 2005;280:6861–6871. doi: 10.1074/jbc.M411341200. [DOI] [PubMed] [Google Scholar]
- 82.Sommer ME, Farrens DL. Vision Res. 2006;46:4532–4546. doi: 10.1016/j.visres.2006.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hofmann KP, Pulvermüller A, Buczyłko J, Van Hooser P, Palczewski K. J Biol Chem. 1992;267:15701–15706. [PubMed] [Google Scholar]
- 84.Vishnivetskiy SA, Baameur F, Findley KR, Gurevich VV. J Biol Chem. 2013;288:11741–11750. doi: 10.1074/jbc.M113.450031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gurevich VV, Gurevich EV. Expert Rev Mol Med. 2010;12:e13. doi: 10.1017/S1462399410001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gurevich VV, Gurevich EV. Cell Signal. 2012;24:899–1908. doi: 10.1016/j.cellsig.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schöneberg T, Schulz A, Biebermann H, Hermsdorf T, Römpler H, Sangkuhl K. Pharmacol Ther. 2004;104:173–206. doi: 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 88.Gimenez LE, Vishnivetskiy SA, Baameur F, Gurevich VV. J Biol Chem. 2012;287:29495–29505. doi: 10.1074/jbc.M112.366674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. J Biol Chem. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 90.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. J Biol Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]