SUMMARY
SKN-1/Nrf plays multiple essential roles in development and cellular homeostasis. We demonstrate that SKN-1 executes a specific and appropriate transcriptional response to changes in available nutrients, leading to metabolic adaptation. We isolated gain-of-function (gf) alleles of skn-1, affecting a domain of SKN-1 that binds the transcription factor MXL-3 and the mitochondrial outer membrane protein PGAM-5. These skn-1(gf) mutants perceive a state of starvation even in the presence of plentiful food. The aberrant monitoring of cellular nutritional status leads to an altered survival response in which skn-1(gf) mutants transcriptionally activate genes associated with metabolism, adaptation to starvation, aging, and survival. The triggered starvation response is conserved in mice with constitutively activated Nrf and may contribute to the tumorgenicity associated with activating Nrf mutations in mammalian somatic cells. Our findings delineate an evolutionarily conserved metabolic axis of SKN-1/Nrf, further establishing the complexity of this pathway.
INTRODUCTION
Transcription factors that influence multiple pathways must choose from a variety of targets to ensure an appropriate cellular response. SKN-1/Nrf (NF-E2-related factor) responds to environmental and endogenous stressors including electrophiles, pathogens, and xenobiotics, but plays additional essential roles in regulating development and cellular homeostasis (Bowerman et al., 1992; Wang et al., 2010). In mammals, the Kelch-like ECH-associated protein 1 (Keap1) negatively regulates Nrf by sequestration in the cytoplasm and presentation to the proteasome for degradation (Itoh et al., 1999). An orthologous mechanism of modulating SKN-1 activity is orchestrated by C. elegans WDR-23 (Choe et al., 2009). Mutations in skn-1 and wdr-23 have been shown to influence cell and organism survival in C. elegans (Bishop and Guarente, 2007; Curran and Ruvkun, 2007; Tullet et al., 2008; Wang et al., 2010). SKN-1 can activate and repress the expression of a number of transcripts under basal and stress-activated conditions (An et al., 2005; Inoue et al., 2005; Bishop and Guarente, 2007; Kell et al., 2007; Kahn et al., 2008; Tullet et al., 2008; Oliveira et al., 2009; Li et al., 2011). Although SKN-1 and Nrf have been shown to occupy consensus DNA elements in the promoters of target genes, the mechanism of target selection by SKN-1/Nrf under varying stress conditions is not well understood (Blackwell et al., 1994; Moi et al., 1994; Oliveira et al., 2009) (Figure S1).
The breadth of cellular pathways influenced by SKN-1 requires a sophisticated mechanism for regulating an appropriate transcriptional response and importantly restraining the activation of unnecessary transcripts. In C. elegans SKN-1 is essential for the embryonic development of the digestive system and is required for normal life span and the adaptation to electrophile and pathogen stress in larvae and adult animals (Bowerman et al., 1992; Bowerman et al., 1993; Maduro et al., 2001; An and Blackwell, 2003; Maduro et al., 2007; Lin et al., 2009). Previous studies of skn-1 utilize loss-of-function alleles or activation of SKN-1 by inhibition of the negative regulator wdr-23 (Choe et al., 2009; Hasegawa and Miwa, 2010). SKN-1 is also modulated by the insulin-like/IGF-1 pathway for the promotion of longevity (Tullet et al., 2008).
The ability of an organism to balance energy demands with nutritional supply is paramount for survival. Under conditions of nutrient deprivation organisms induce the expression of starvation response genes to facilitate adaptation to the change in available nutrients and a reliance on energy stores (Lakowski and Hekimi, 1998; Longo, 1999). Here we show the first gain-of-function alleles of skn-1, which alter the expression of genes tied to metabolism, starvation adaptation, growth, and reproduction. These mutations disrupt the association of SKN-1 with the mitochondria outer-membrane protein, PGAM-5, independent of the canonical wdr-23 pathway. SKN-1 is the C. elegans ortholog of mammalian Nrf, and we find that constitutive activation of Nrf induces a similar starvation adaptation response in mice. We conclude that SKN-1/Nrf functions in an evolutionarily conserved pathway to modulate metabolism and adaptation to starvation.
RESULTS
Identification of a Novel SKN-1 Regulatory Domain That Interacts with PGAM-5 and MXL-3
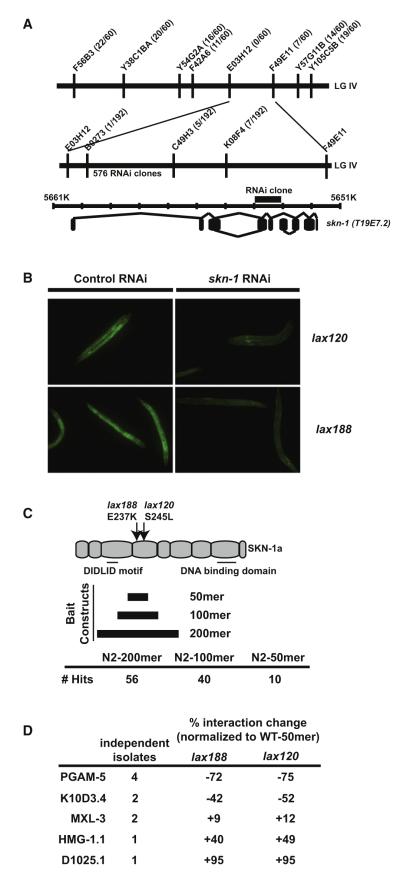
To identify novel regulators of SKN-1 activation, we mutagenized C. elegans harboring the well-established gst-4p::gfp reporter of SKN-1 activity, hypothesizing that it should be feasible to identify genetic mutants that disrupt specialized SKN-1 activation (Figure S2). In support of this idea, we identified several classes of mutants with a range of SKN-1 activity (our unpublished results). One class of mutants that includes alleles lax120 and lax188 was dominant (Figure S2) and mapped to a genetic position near the center of LG IV (Figure 1A). An RNA interference (RNAi) screen of the genes in this genomic region identified a single RNAi clone that targeted T19E7.2/skn-1, which could suppress the dominant gain-of-function phenotype (Figure 1B). Sequencing of the skn-1 locus identified a single missense mutation in lax120 and lax188, changing serine 245 to leucine and glutamic acid 237 to lysine, respectively. The similar phenotypes and close proximity of these mutations suggested the presence of a functional protein domain in this previously uncharacterized region of the SKN-1A/C protein (Figure 1C).
Figure 1. The Dominant SKN-1 Domain Functions Independently of the Canonical WDR-23 Pathway.
(A) SNP mapping with the Hawaiian wild-type strain CB4856 assigned alleles lax120 and lax188 to LGIV between B0273 and C49H3. 576 RNAi clones covering the genes between these SNPs were tested for suppression of the dominant phenotype.
(B) A skn-1/T19E7.2 RNAi clone was the only construct capable of suppression of the dominant phenotype.
(C) lax120 and lax188 encode single missense mutations in the skn-1 coding sequence. Y2H analysis was performed with bait constructs that code for a 200, 100, or 50 amino acid region mutated in the dominant skn-1 alleles.
(D) skn-1(lax120) and skn-1(lax188) mutations alter the interaction of the SKN-1 50 amino acid domain with Y2H targets.
To determine if this region of the SKN-1 polypeptide facilitates an interaction with specific proteins, we performed a yeast-two-hybrid (Y2H) screen using a bait construct consisting of a 200 amino acid region centered on the lax120 and lax188 mutations (Figure 1C). We identified 56 prey constructs from this initial screen and sequenced the plasmids to identify the potential protein interactors of this SKN-1 domain. To enhance the specificity of the interaction domain we retested the 56 plasmids in a directed Y2H assay utilizing a refined bait construct containing 100 amino acids of this new SKN-1 domain. This second tier of screening eliminated 16 constructs that failed to retest. We performed a final test of the remaining 40 prey plasmids with a further refined bait construct that contained only 50 amino acids of the SKN-1 domain and that did not contain any of the upstream DIDLID sequence (Walker et al., 2000). Bioinformatic analysis of this new 50 amino acid domain reveals a casein kinase 2 domain, which is disrupted in lax188, multiple potential phosphorylation sites, and an enrichment for charged residues (Figure S2).
Ten plasmids passed all three tiers of screening. Four of the ten plasmids contained the pgam-5 locus, which codes for a mitochondrial outer membrane protein. Two plasmids contained the sequence for the nutrient-responsive transcription factor mxl-3; another two plasmids coded for the conserved serine proteinase inhibitor K10D3.4; one plasmid coded for the high-mobility group protein, HMG-1.1; and the last plasmid coded for a protein with unknown function, D1025.1. The identification of multiple independent isolates of these genes combined with the fact that these plasmids facilitated the strongest interaction with all skn-1 bait constructs suggests that PGAM-5, MXL-3, and K10D3.4 are bona fide SKN-1 interactors (Figure S2). The ability of SKN-1A and SKN-1C to bind MXL-3 and PGAM-5 was confirmed by in vitro coimmunoprecipitation studies utilizing an engineered carboxyl-terminal HA epitope tag in SKN-1 (Figure S2). These results indicate a direct biochemical interaction of MXL-3 and PGAM-5 with SKN-1.
The dominant mutations in skn-1 should alter the association of the SKN-1 protein with these new interactors. We tested this directly by repeating the Y2H assay with a 50 amino bait construct encoding the lax188(E237K) or lax120(S245L) mutation in combination with the isolated prey plasmids. Yeast harboring both plasmids were screened for any change in the bait-prey interaction, as measured by X-α-Gal activity and resistance to the antibiotic aureobasidin A. The lax188 and lax120 mutations decreased the interaction of the SKN-1 domain with PGAM-5 by 72% and 75%, respectively (Figure 1D). Interestingly, both mutations resulted in a modest increase in the interaction with MXL-3, HMG-1.1, and D1025.1. These findings suggest that this SKN-1 domain interacts with PGAM-5, presumably at the mitochondria, and with MXL-3, which may help SKN-1 to specify transcriptional targets.
PGAM-5 is a predicted phosphoglycerate mutase that uses alternate catalytic activity as a protein serine/threonine phosphatase (Takeda et al., 2009). The interaction with PGAM-5 at the mitochondria would optimize the position of SKN-1 to function as a sensor of mitochondrial function, stress, energy output, or cellular metabolic status. The serine 245 mutation in lax120 combined with the fact that PGAM-5 can function as a serine/threonine phosphatase is intriguing and may clue us in to the biochemical function of the SKN-1/PGAM-5 interaction. Based on these data, the lax120 and lax188 mutations destabilize the association of SKN-1 with PGAM-5 or decrease the affinity of SKN-1 as a biochemical substrate for PGAM-5 catalytic activity (Figure S1).
To identify the domain in the interacting proteins that facilitate binding to SKN-1, we created new Y2H prey constructs coding for truncated versions of the binding proteins and tested their ability to interact with the SKN-1 domain (Figure S2). The N-terminal domain of MXL-3 was capable of binding to the SKN-1 50-, 100-, and 200-mer constructs. The N terminus of MXL-3 is also highly charged, which may facilitate the interaction and binding to the equally highly charged 50-mer domain of SKN-1 through electrostatic interactions. None of the truncated versions of PGAM-5 revealed a positive yeast-2-hybrid interaction with SKN-1, potentially due to misfolding of the prey fusion protein.
Identification of Mitochondria Associated SKN-1
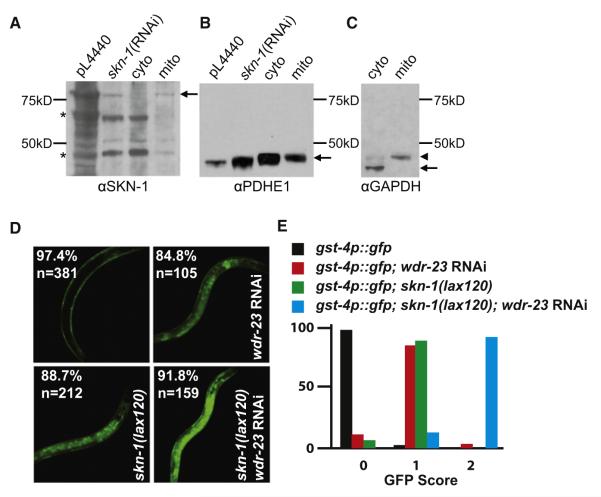
A mitochondrial pool of SKN-1 has not been previously identified. To determine if endogenous SKN-1 was associated with this organelle, we isolated an enriched mitochondrial fraction from whole worm lysates by differential centrifugation. We identified a SKN-1-specific band with a predicted molecular weight of the SKN-1A isoform that was enriched in the mitochondria subcellular fraction along with the mitochondrial marker PDHE1 (Brys et al., 2010; Greiss et al., 2008; Li et al., 2009) (Figures 2A and S3). While the strength and the function of SKN-1 on the mitochondrial surface are unclear, the steady-state presence of SKN-1 on isolated mitochondria would suggest that the association is relatively stable. It is notable that the skn-1(lax120) and skn-1(lax188) gain-of-function (gf) mutants display a Sma (small) body-size phenotype (Figure S2), a trait shared with many mitochondria mutants with reduced ATP production (Lee et al., 2003). Taken together, these observations suggest that a pool of SKN-1 (hereafter referred to as mito-SKN-1) is present at the mitochondria, potentially facilitated through an interaction with the mitochondrial outer membrane protein PGAM-5.
Figure 2. Identification of a Unique Mitochondria-Associated SKN-1 Activation Pathway.
(A) SKN-1 is enriched in a mitochondria fraction of whole C. elegans. The pL4440 and skn-1 RNAi controls are from whole-worm extracts. In the left panel, asterisks denote bands that are not clearly decreased by skn-1 RNAi, in contrast to 85 kD SKN-1 itself.
(B) Partial purification was achieved by spinning down the mito pellet (mito), as indicated by the presence of remaining mitochondrial marker, PDHE1 (Greiss et al., 2008; Li et al., 2011; Brys et al., 2010), in the cytosolic fraction (cyto), but the mitochondria fraction lacks the cytoplasmic marker GAPDH.
(C) Arrowhead indicates remaining PDHE1 that was not removed by stripping.
(D) Synergistic activation of the SKN-1 transcriptional reporter (gst-4p::gfp) by the canonical wdr-23 pathway and the dominant skn-1 mutations. Percentage represents fraction of total animals, n, with phenotype.
(E) GFP score of indicated genotypes based on fluorescence intensity.
Regulation of SKN-1(gf) Is Independent of the WDR-23 and GSK-3 Pathways
Our data indicate that the mitochondria-associated pool of SKN-1 could be modulated in a WDR-23-independent manner. WDR-23 is a molecularly well-defined regulator of SKN-1 activity, but we did not pull out any wdr-23 prey constructs from our Y2H screen using the new SKN-1 protein domain. In addition, we directly tested a wdr-23 prey vector with the SKN-1 50-mer-bait construct and could not detect a measureable interaction (Figure S2). To further confirm our hypothesis that the 50-mer-bait can be regulated independently of WDR-23 we measured the activation of the gst-4p::gfp reporter in the skn-1(lax120) and skn-1(lax188) backgrounds with and without wdr-23 (Figure 2D). In support of two independent mechanisms of SKN-1 regulation we observed a synergistic activation of the SKN-1 target gene gst-4 when the skn-1(gf) alleles were combined with wdr-23 inactivation (Figure 2E).
WDR-23 modulates the activity and abundance of SKN-1 by sequestering SKN-1 and delivering it to the proteasome for degradation. As verification of our model we measured an increase in the expression of wdr-23 in both mutant backgrounds (Table 1). The activation of wdr-23 was also dependent on skn-1, suggesting a molecular feedback loop of this response pathway and identifying wdr-23 as the major mechanism of terminating SKN-1 activity regardless of the mode of activation.
Table 1.
Activation of Mito-SKN-1 Regulates a Unique Transcriptional Response
|
skn-1(lax120)
|
skn-1(lax188)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| fold changeb | std devc | t-testd | fold changeb | std devc | t-testd | |||
| Stress | Glutathione | gcs-1 | 4.48 | 1.00 | 0.001 | 4.00 | 0.95 | <0.001 |
| gst-4 | 21.31 | 1.06 | <0.001 | 22.16 | 1.01 | <0.001 | ||
| Detox | ugt-11 a | 8.75 | 0.96 | 0.001 | 4.61 | 1.05 | <0.001 | |
| Aging/Dauer | Aging | wdr-23 a | 2.81 | 1.01 | 0.001 | 2.85 | 0.98 | <0.001 |
| dod-17 | 5.00 | 0.94 | 0.002 | 4.38 | 1.01 | <0.001 | ||
| dod-24 | 7.69 | 1.15 | 0.005 | 7.49 | 1.20 | <0.001 | ||
| Dauer | dod-6 | 2.60 | 0.92 | NS | 2.33 | 1.22 | 0.004 | |
| C32H11.4 | 18.85 | 1.08 | <0.001 | 14.68 | 1.15 | <0.001 | ||
| Unknown | dao-4 | −9.92 | 1.21 | 0.001 | −2.37 | 1.09 | 0.001 | |
| R05G9R.1 | −8.38 | 2.05 | 0.018 | −4.49 | 1.11 | <0.001 | ||
| zk180.6 a | −7.66 | 2.49 | 0.031 | −3.78 | 1.09 | <0.001 | ||
| Metabolism | Amino Acid | tyr-5 a | −42.71 | 2.38 | 0.009 | −48.67 | 1.01 | <0.001 |
| aat-5 | 1.77 | 0.87 | NS | 1.51 | 1.12 | 0.004 | ||
| atgp-2 | 1.88 | 0.85 | 0.045 | 1.81 | 1.07 | 0.002 | ||
| aat-2 | 2.31 | 1.02 | <0.001 | 3.21 | 1.00 | <0.001 | ||
| K07E1.1 | 4.97 | 1.08 | 0.002 | 2.83 | 0.98 | <0.001 | ||
| Lipid | F09C8.1 | −14.06 | 1.89 | 0.009 | −9.03 | 0.82 | <0.001 | |
| Y4C6B.6 | −3.23 | 0.95 | 0.010 | −3.20 | 0.96 | <0.001 | ||
| acdh-6 a | −2.93 | 1.18 | 0.010 | −2.89 | 1.00 | <0.001 | ||
| lips-3 a | −2.37 | 0.96 | 0.010 | −2.96 | 1.06 | 0.011 | ||
| lips-10 | 2.74 | 1.14 | 0.007 | 1.90 | 1.13 | <0.001 | ||
| zk742.4 | 9.36 | 1.08 | 0.001 | 7.80 | 1.09 | <0.001 | ||
| Quinone | R08E5.3 a | −6.67 | 2.67 | 0.039 | −4.67 | 0.94 | <0.001 | |
| sqrd-1 | −6.18 | 2.40 | 0.040 | −3.59 | 1.15 | <0.001 | ||
| Unknown | Y42G9A.3 a | 3.77 | 1.08 | 0.004 | 3.42 | 1.07 | <0.001 | |
| Reproduction | Germline | bcc-1 | 2.32 | 0.96 | 0.001 | 2.35 | 0.95 | <0.001 |
| glh-1 | 2.42 | 0.80 | 0.007 | 2.24 | 1.14 | <0.001 | ||
| Piwi/Paz | ppw-2 | 2.75 | 0.65 | NS | 2.38 | 1.21 | 0.001 | |
| Signaling | Insulin | ins-35 a | 3.20 | 1.00 | 0.001 | 3.64 | 1.01 | <0.001 |
| Kinase | kin-15 | 3.61 | 0.90 | 0.003 | 1.52 | 0.91 | 0.002 | |
| kin-16 a | 4.04 | 1.05 | 0.003 | 2.15 | 1.00 | <0.001 | ||
| Transcription | mxl-3 a | −1.03 | 0.92 | NS | −1.47 | 0.87 | 0.026 | |
| skn-1 | 1.12 | 0.89 | NS | 1.03 | 1.05 | NS | ||
| Transporter | T21C9.3 a | 3.67 | 1.03 | 0.001 | 2.60 | 0.83 | 0.001 | |
Bold type indicates directionality of gene expression changes indicative of starvation.
Mean fold change relative to wild type animals.
Standard error of the mean.
Significance from t-test, NS, not significant.
GSK-3 has been shown to inhibit the localization of SKN-1 to the nucleus and subsequently repress the activation of SKN-1 targets (An et al., 2005). To determine if the skn-1(gf) mutants are still responsive to GSK-3 regulation, we treated skn-1(lax120) and skn-1(lax188) mutant animals with gsk-3 RNAi. The animals fed the gsk-3 RNAi clone had higher expression of the SKN-1 target gst-4 than animals fed control RNAi clones (Figure S3). These data indicate that the SKN-1 gain-of-function mutants are still responsive to the gsk-3 pathway suggesting the mitochondrial pool of SKN-1 can be activated by mechanisms independent of both wdr-23 and gsk-3.
SKN-1(gf) Induces the Expression of Metabolism and Starvation Response Genes
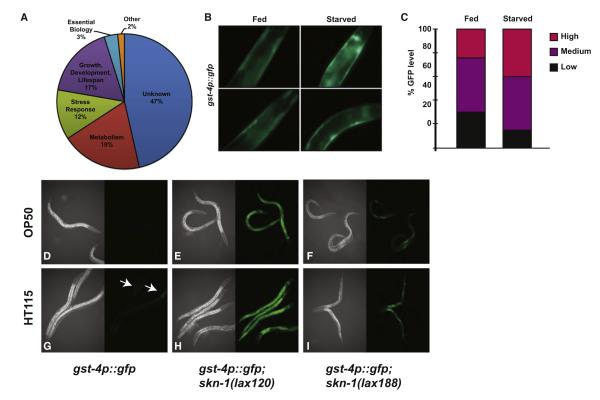
Our model predicts that the activation of mito-SKN-1 would induce a specific transcriptional response in C. elegans. We examined the transcriptional profile of synchronous skn-1(lax120) and skn-1(lax188) animals compared to age-matched wild-type controls (Figure 3A; Tables 1 and S1). The functional GO terms most highly represented in the mito-SKN-1 activation mutants were genes tied to metabolism and starvation response pathways followed by genes involved in growth, development, and life span. Two metabolism-related genes upregulated in the skn-1 gain-of-function mutants were previously identified as being upregulated by SKN-1. Y42G9A.3 is induced by SKN-1 under normal conditions while zk742.4 is induced by SKN-1 in response to exposure to arsenic and t-BOOH. Of particular interest are metabolism and starvation adaption genes not previously reported to be SKN-1 responsive following exposure to environmental and electrophile stress conditions. These metabolism genes include tyr-5/Tyrosinase, F09C8.1/Phospholipase, sqrd-1/mitochondrial oxidoreductase, Y4C6B.6/glucosylceramidase, acdh-6/acyl CoA dehydrogenase, lips-3/lipase-related, aat-5/amino acid transporter catalytic subunit, atgp-2/amino acid transporter glycoprotein subunit, aat-2/ amino acid transporter catalytic subunit, and lips-10/lipase related. In addition, the aging/dauer-related genes zk180.6/uncharacterized protein, R08E5.3/methyltrasnferase, and wdr-23/SKN-1 negative regulator, which previously had not been identified as SKN-1 targets, are differentially expressed under starvation conditions. The expression profile of these genes is altered in the same direction as in wild-type worms subjected to starvation (Kang and Avery, 2009). Taken together, these results indicate that the worms harboring the skn-1 gain-of-function alleles inappropriately perceive a state of starvation. We further confirmed the specificity of the gain-of-function alleles and WDR-23 pathways by comparing the abundance of mito-SKN-1 target mRNAs in the activated SKN-1(gf) strains with and without wdr-23 regulation. We identified a synergistic change in gene expression of mito-SKN-1(gf)-dependent transcripts when we depleted wdr-23 by RNAi (Table S2). Additivity was observed for transcripts induced and repressed by mito-SKN-1 activation indicating the existence of two parallel pathways that can activate SKN-1.
Figure 3. SKN-1 Is Activated by Starvation and Changes in Available Nutrients.
(A) GO-terms of genes identified as dysregulated by transcriptional analysis of the lax120 and lax188 mutants.
(B) SKN-1 activity sensor worms were grown on food or under starvation conditions and then imaged for GFP expression indicative of SKN-1 activation of the gst-4p::gfp reporter. (C) Quantification of GFP levels observed as described in (B).
(D–I) Indicated strains were fed the E. coli B strain OP50.
(D)–(F) or the E.coli K12 strain HT115 (G)–(I) and imaged for GFP expression as described in (B).
The altered expression of metabolism and nutrient-mobilization genes is notable as skn-1 loss-of-function mutants have been shown to suppress the longevity phenotype associated with dietary restriction (Bishop and Guarente, 2007). The genes that changed most significantly and the direction of change are reminiscent of a sensed state of starvation or severe dietary restriction (Szewczyk et al., 2006; Baugh et al., 2009). Although the transcriptional analysis of the mito-SKN-1 mutants is representative of the entire organism and not tissue specific, we observed the strongest mito-SKN-1 activation in the hypodermal and intestinal tissues (Figure 1B), which collectively comprise a significant portion of the total worm body. To test if starvation was capable of inducing a SKN-1 transcriptional response we compared the expression of the gst-4p::gfp reporter of SKN-1 activity in the presence and absence of food. Consistent with the activation observed in skn-1(gf) mutants we detected GFP expression in the hypodermal tissues when animals were removed from food (Figures 3B and 3C). The intestine and hypodermis are also the major sites of lipid storage in C. elegans, which may explain the observed changes to the metabolism and starvation response transcripts.
Recent work has shown that the bacterial diet itself can have a strong impact on the physiology of the worm, and in some cases even reverse or suppress genetic phenotypes (Soukas et al., 2009; Maier et al., 2010). The HT115 RNAi strain is an E. coli K12 background whereas the standard C. elegans food source, OP50, is of the E.coli B genotype. The genetic differences between the K12 and B genotypes can alter the nutritional composition of the food source as well as the “taste” of the food to the worm as measured by occupancy on the lawn of bacteria (Watts and Browse, 2002; Shtonda and Avery, 2006; Soukas et al., 2009; Watts, 2009). Worms on a HT115 diet spend less time on the bacterial lawn and as such experience oscillating periods of caloric restriction. To test if the E.coli B and K12 diets influence SKN-1 activity we grew the gst-4p::gfp sensor strain on both strains. On the HT115 diet we observed GFP expression within intestinal cells not normally observed when the animals are fed the OP50 diet (Figures 3D–3I).
Since diet can have a strong impact on physiology, we examined the expression of mito-SKN-1 transcripts on animals fed OP50/E.coli B and compared to animals raised on a HT115/E.coli K12 food source. We identified mito-SKN-1 transcripts that were differentially expressed when fed an E.coli K12 diet. Notably, we identified a switch from transcriptional repression on OP50 to activation on HT115 for genes involved in aging and metabolic pathways (Table S2). These changes were most obvious for the aging/dauer-related genes dao-4/novel protein, ZK180.6/uncharacterized protein, and R05G9R.1/mucin-2-like, as well as the metabolism/signaling-related genes mxl-3/Max-like transcription factor, acdh-6/acyl CoA dehydrogenase, and lips-3/lipase related (Table S2).
The physical interaction of SKN-1 with MXL-3 may help specify the transcription of metabolism and starvation genes in the skn-1(gf) background. To test this we combined skn-1(gf) alleles with a loss-of-function allele of mxl-3(ok1947) and examined the transcript levels of the genes altered in the gain-of-function background (Figure 1, Table S3). The genes tied to metabolism and aging/dauer regulatory pathways were the most affected by the absence of mxl-3. The mxl-3 null allele partially suppressed the increased expression of acdh-6/acyl CoA dehydrogenase, lips-10/lipase-related, R01E6.5/Filaggrin-2-like, dao-4/novel protein, dod-24/downstream of DAF-16, dod-17/downstream of DAF-16, ugt-11/UDP-Glucuronosyl Transferase, C32H11.4/hydrolase, and ins-35/insulin peptide genes. The mxl-3 deletion also reversed the decreased expression of F09C8.1/Phospholipase and R08E5.3/methyltransferase in the skn-1(gf) background. Intriguingly, the skn-1;mxl-3 double mutants synergized the increased expression of wdr-23, indicating a concerted effort to suppress the gain-of-function activity of SKN-1. The dysregulated activation of SKN-1 results in a transcriptional profile indicative of a perceived state of starvation. The change in expression of metabolism genes in the SKN-1(gf) mutants and the dependence of mxl-3 introduces a new regulatory axis for organismal nutrient mobilization to maintain energy homeostasis.
A Perceived State of Starvation Alters Metabolic Adaptation Capacity in skn-1(gf) Mutants
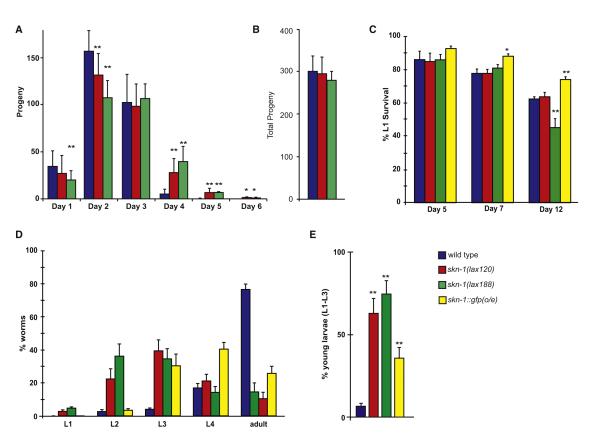
The presence of specific and independent mechanisms of SKN-1 activation predicts that there would be a unique physiological role for these pathways. To identify a specific function for mito-SKN-1 we characterized the mito-SKN-1(gf) mutants for reproduction and life-span phenotypes and challenged their ability to adapt to physiologic stress conditions. The skn-1 signaling pathway has functions within multiple longevity regulatory cascades; however, it was not clear if the gain-of-function mutants would have an aging phenotype based on the altered expression of both pro- and anti-aging genes (Figure 3A, Table 1). We assessed the effect of the SKN-1 activating mutations on adult lifespan (Figures 4A and S4). Neither allele of skn-1 had a significant effect on the rate of aging compared to wild-type animals.
Figure 4. Activated Mito-SKN-1 Promotes a Perceived State of Starvation.
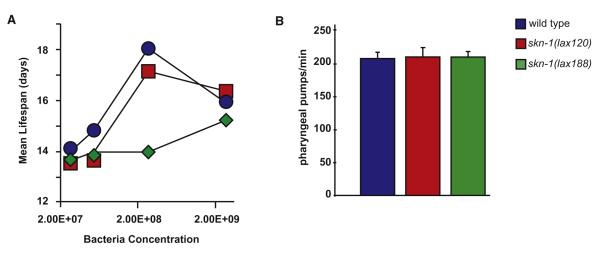
(A) Bacterial dilution (bDR) life span; the mean life span of adult worms grown on solid media with four concentration of bacteria at 25°C. (B) Feeding rates as assessed by pharyngeal pumping assays for indicated strains. Error bars represent average +SD.
Food availability also has a strong influence on life span. As such, we tested the ability of our mutants to respond to dietary restriction (Figures 4A and S4). We examined the life span of both skn-1(lax120) and skn-1(lax188) mutants on a range of bacteria concentrations. skn-1(lax188) failed to display a lifespan extension phenotype with bacterial dilution (bDR), while skn-1(lax120) mutant animals showed an attenuated response. These results are consistent with the diminished L1 survival phenotype (Figure S5) in the skn-1(lax188) mutant and further support the idea that these animals perceive a reduction in food supply under ad libitum conditions leading to a more extreme starvation response under bDR growth conditions. An alternative mechanism of caloric restriction in C. elegans results from genetic mutations that decrease pharyngeal pumping and thus reduce the ingestion of E. coli (Lakowski and Hekimi, 1998). As such, we measured pharyngeal pumping, feeding rates, and bacterial clearance in the skn-1(gf) mutants and found no statistically significant difference from wild-type (Figures 4B and S4, Table S4).
skn-1 is essential for development as skn-1(z-m-) embryos are not viable (Bowerman et al., 1992). However, the lax120 and lax188 mutations both facilitate an extended reproductive period in hermaphrodites by approximately 2 days (Figure 5A). Intriguingly, this observed suppression of reproductive senescence does not come at the expense of reduced brood size as worms with either mutation yield a similar number of viable progeny as compared to wild-type animals (Figure 5B). We did however notice a low penetrance (less than 10%) bag of worms (Bag) phenotype, where progeny hatch within the parent at higher temperatures (unpublished observation) (Lakowski and Hekimi, 1998). C. elegans require a demanding supply of nutrients once they reach reproductive maturity. If removed from food, hermaphrodites cease egg laying, allowing progeny to hatch in utero leading to a Bag phenotype (Dong et al., 2000). The low penetrance Bag phenotype in the mito-SKN-1(gf) mutants is consistent with the idea that these animals experience a perceived state of starvation.
Figure 5. Activated SKN-1 Promotes Extended Reproduction and Developmental Delay in Response to Food.
(A) Nonmated reproductive span of worms fed OP50 bacteria at 20°C.
(B) Total progeny from (A).
(C) L1 diapause survival was measured by movement and staining with bromophenol blue following 5, 7, and 12 days in the absence of food.
(D) Growth and development of animals in L1 diapause following feeding.
(E) Percentage of animals arrested early in development after recovery from starvation. (* < 0.05, ** < 0.01, *** < 0.001). Error bars represent SEM.
The transcriptional changes in the skn-1(gf) mutants are reminiscent of an animal coping with nutrient deprivation conditions. C. elegans can survive extended periods of starvation under two stages of diapause— the first is during larval stage 1 (L1) and the next as dauers, an alternative third larval stage (L3). Since the skn-1(gf) worms demonstrated transcriptional changes associated with a metabolic profile similar to a response to starvation, we directly tested the ability of these mutants to survive without food for extended periods of time. We performed L1 survival assays as previously reported (Baugh et al., 2009) and assessed the ability of skn-1(lax120), skn-1(lax188), and skn-1-overexpressing skn-1b/c::gfp worms to withstand starvation after 5, 7, and 12 days at room temperature (~21°C) (Figures 5C and S5, Table S5). At 5 days, there is no clear difference in the survival of the skn-1(gf) worms as compared to wild-type or skn-1b/c::gfp worms. However, with increasing “age,” the skn-1b/c::gfp worms start to exhibit significantly increased survival over wild-type worms. Intriguingly, the stronger skn-1(gf) allele, skn-1(lax188), has a modest increase at day 7, but by day 12 has a dramatically decreased survival. In contrast, the skn-1(lax120) mutants were somewhat comparable to wild-type worms in this assay. Our results suggest that given that skn-1(lax188) animals are already experiencing a starvation-like response, the prolonged absence of food is detrimental to their long-term survival.
Worms are constantly sensing their environment for nutritional cues, and when cultured on regular growth media, they crawl toward the bacterial lawn to feed. Indeed, L1-arrested animals typically recover from their diapause and resume reproductive growth upon transfer to regular growth media (Figure S5, dark blue bars). We assessed the ability of the “starving” skn-1(gf) mutants to recover after 8 days of starvation on regular media (Figures 5D and S5; Table S5). As shown in Figure 5E, after 2 days of incubation at 20 degrees, the majority of wild-type worms (>80%) grow into gravid adults. In contrast, the skn-1(gf) worms have a much slower growth profile. In particular, a small fraction of the skn-1(gf) worms are still arrested as L1s, with the rest of the population demonstrating asynchronous growth but primarily comprising of L2 and L3 stage animals. Despite this obvious lag in growth, these worms eventually do develop into fertile adults (data not shown). The skn-1 transgenic worms show a markedly reduced growth rate but are still able to form gravid adults (~30% adults) after 2 days. We conclude that even in the presence of food, these skn-1(gf) animals behave as if they are undergoing a constant state of starvation. In support of this, we show the upregulation of several metabolic transcripts, inability to survive extended starvation, and a slow recovery time despite plentiful levels of food. Additionally, our data also indicate that the gf alleles do not behave similarly to the skn-1 overexpressing strain, suggesting that the increased abundance of SKN-1 does not necessarily correlate with increased function/activity. Thus under stressful survival conditions, such as when food resources are limited, SKN-1 activation may provide a protective role in ensuring that the animal can cope with extended periods of starvation.
Constitutive Activation of Mammalian Nrf Induces a Starvation Response
Cellular adaptation to starvation is a conserved stress response (Hardie, 2011). In light of the evolutionary conservation of the SKN-1-like cap-n-collar transcription factors, we were curious if Nrf activation could also mediate a starvation-like response in mammals. To determine if Nrf activation leads to a perceived state of starvation, we measured the transcriptional profile of tissue from an established Keap1−/− knockout mouse line (Ohta et al., 2008) (Tables 2 and S6). Despite the fact that this tissue would display dysregulation of transcripts from all Keap1-Nrf dependent pathways, we were curious if we could detect any alteration of starvation-response genes. Genes tied to metabolism GO terms were significantly enriched in the Keap1−/− tissues, similar to that found in C. elegans (Table S7). In support of a SKN-1/Nrf function in maintaining metabolic homeostasis, we discovered genes associated with starvation responses were significantly upregulated in the Keap1 knockout mice as compared to wild-type litter mates (Table 2). We confirmed the transcriptional changes identified in our microarray data by additional qRT-PCR analysis. A hallmark of a state of starvation is the activation of the energy sensors AMP-activated protein kinase and adenylate kinase (Hardie, 2011). The catalytic domain of AMPK, Prkaa2 as well as Ak7, was upregulated in the lung tissue of the Keap1 knockout mice by 2.07- and 3.04-fold, respectively. Importantly, the levels of cAMP-dependent protein kinase subunits, which have the opposite effects on energy homeostasis, remained unchanged. Of additional interest were the upregulation of genes involved in mitochondrial and peroxisomal fatty acid oxidation, lipases, and lipid transporters (Table 2). These genes have been shown to change in the same direction in mouse liver during periods of starvation (Bauer et al., 2004). Remarkably, the activated Nrf lung tissue has a more than 300-fold increase in Ces1g. Overexpression of mouse esterase-x/carboxylesterase 1 (Ces1g) has been shown to promote beta-oxidation (Ko et al., 2009). Two C. elegans Ces1g-like orthologs coded in an operon T28C12.4 and T28C12.5 were both significantly upregulated in the skn-1(lax120) and skn-1 (lax188) mutant backgrounds (Table S1). These results suggest that the constitutively activated Nrf in the lung tissue of Keap1 knockout mice induces a perceived state of starvation, similar to that observed in the intestine and hypodermis of C. elegans mito-SKN-1(gf) mutants. As such, in addition to the canonical electrophile stress responses, the SKN-1/Nrf system also functions in an evolutionarily conserved pathway to modulate cellular metabolism.
Table 2.
Nrf Activation in Mouse Lung Tissue Triggers a Conserved Starvation Response
| Fold changea | Std Devb | ||
|---|---|---|---|
| Detox | |||
|
| |||
| Aldh2 | 3.19 | 0.41 | Ethanol oxidation |
| Adh7 | 24.64 | 3.17 | |
| Aldh1a1 | 6.49 | 0.83 | |
| Aldh3a1 | 61.98 | 7.97 | |
| Gss | 3.16 | 0.41 | Glutathione |
| Gclm | 5.69 | 0.73 | |
| Gclc | 13.94 | 1.79 | |
| Gsr | 4.60 | 0.59 | Redox |
| Gpx2 | 31.98 | 4.11 | |
| Gsta3 | 21.33 | 2.74 | Transferase |
| Gstm1 | 25.81 | 3.32 | |
|
| |||
| Metabolism | |||
|
| |||
| Gls | 1.52 | 0.20 | Amino Acid |
| Degradation | |||
| Ces1h | 12.85 | 1.65 | Fatty Acid Oxidation |
| Ces1g | 334.91 | 43.04 | |
| Acss3 | 2.80 | 0.36 | |
| Slc27a2 | 1.54 | 0.20 | |
| Acot1 | 1.64 | 0.21 | |
| Acaa1 | 1.70 | 0.22 | |
| Acad10 | 1.76 | 0.23 | |
| Pte2a/Acot3 | 2.68 | 0.34 | |
| Acsm3 | 3.70 | 0.48 | |
| Acsm1 | 4.10 | 0.53 | |
| Hmgcs2 | 6.19 | 0.79 | |
| Acox2 | 9.69 | 1.25 | |
| Awat1 | 14.06 | 1.81 | |
| Pnpla7 | 1.99 | 0.26 | Lipase |
| Pnpla2 | 2.75 | 0.35 | |
| Pla2g7 | 5.13 | 0.66 | |
| Liph | 6.41 | 0.82 | |
| Taldo1 | 2.50 | 0.32 | Pentose Phosphate |
| Pgd | 4.84 | 0.62 | |
| Ak7 | 3.04 | 0.39 | Sensor |
| Akd1 | 3.67 | 0.47 | |
| Prkaca | 1.00 | 0.13 | |
| Prkacb | 1.06 | 0.14 | |
| Prkab1 | 1.09 | 0.14 | |
| Akap14 | 1.88 | 0.24 | |
| Prkaa2 | 2.07 | 0.27 | |
| Irs2 | 1.93 | 0.25 | Signaling |
| Adora3 | 2.30 | 0.30 | |
| Igf2bp1 | 2.42 | 0.31 | |
| Adora1 | 3.43 | 0.44 | |
| Apoe | 2.67 | 0.34 | Transport |
| Apod | 5.27 | 0.68 | |
|
| |||
| Signaling | |||
|
| |||
| Aurkb | −2.87 | 0.04 | Growth |
| Keap1 | −4142.71 | 0.00 | Nfe2l2 |
| Nfe2l2 | 1.16 | 0.15 | |
| Map3k6 | 4.19 | 0.54 | Stress |
| Cebpd | 7.11 | 0.91 | |
| Hif3a | 10.82 | 1.39 | |
| Mef2c | −2.85 | 0.05 | Transcription Factors |
| Mafb | 1.80 | 0.23 | |
| Mafg | 2.34 | 0.30 | |
| Junb | 2.60 | 0.33 | |
| Uchl1 | 2.24 | 0.29 | Proteasome |
qRT-PCR was performed on cDNA templates generated from wild type and Keap1 knockout littermates.
Average fold change compared to wild type animals.
Standard deviation of fold change.
DISCUSSION
Transcription factors like SKN-1/Nrf, which are involved in multiple aspects of cell biology, must be capable of initiating an appropriate transcriptional response when activated under a specific cellular condition. This specification can be accomplished by posttranslational modification, changing the association with transcriptional activators and repressors, sequestration away from the nucleus, and/or degradation of the transcription factor (Wang et al., 2006; Wolff and Dillin, 2006; Villeneuve et al., 2010). The SKN-1/Nrf family of transcription factors is a well-established regulator of the cellular response to electrophiles and pathogens. By characterizing the first skn-1(gf) alleles, we have identified a novel function of SKN-1 in the regulation of metabolism and adaptation to starvation. This new function is mediated through a pool of SKN-1 that seems to be sequestered at the mitochondrial presumably at the outer membrane. The idea of a distinct organelle population of SKN-1 that is adapted to respond to specific changes in the cellular environment is a new and exciting way of thinking about the mechanism an organism utilizes for directing an appropriate transcriptional response.
Despite the functional conservation between Nrf and SKN-1, a clear KEAP1 ortholog in C. elegans has not been identified. Our results suggest that despite little sequence identity between KEAP1 and WDR-23, these two proteins function in an evolutionarily conserved manner to negatively regulate SKN-1 activity and in the formation of a mitochondrial SKN-1 complex. Although our data support the model where SKN-1 can interact with PGAM-5 independently of WDR-23, the interaction of Nrf with PGAM5 is dependent on KEAP1. These results are intriguing from an evolutionary standpoint and imply that the mechanism of recruiting SKN-1/Nrf evolved twice, clearly demonstrating the importance of the mitochondrial localized SKN-1 population. WDR-23, however, is also capable of binding PGAM-5 and MXL-3, suggesting aspects of coordinated control of these two pathways (Figure S1). Despite this mechanistic difference, the increased activity of mitochondrial SKN-1 in the gain-of-function mutants or activated Nrf in the KEAP1 knockout mutant results in a perceived state of starvation and consequently the activation of metabolism and starvation-response pathways.
What facilitates the interaction of SKN-1 with the mitochondria? Our data suggest that the SKN-1 gain-of-function mutants have a reduced interaction with PGAM-5. If PGAM-5 is the anchor for SKN-1 to the mitochondria then perhaps this reduced interaction promotes activation. An alternative hypothesis is that SKN-1 is anchored to the mitochondrial outermembrane through a predicted transmembrane domain in the N terminus of the SKN-1A isoform. Future studies will be directed to uncover the mechanisms that coordinate SKN-1 association with the mitochondria and subsequent activation/dissociation.
The aberrant regulation of the SKN-1 gain-of-function mutants is also independent of the glycogen synthase kinase pathway, GSK-3 (An et al., 2005). SKN-1 has been shown to be regulated by GSK-3 but the phosphorylation event occurs on serine residues in the 300–400 amino acid region of SKN-1. lax120 has a mutation in serine 245 in the SKN-1A protein. This serine residue however is a poor GSK-3 target as it lacks the required priming serine residue for GSK-3 function (An et al., 2005).
We have identified new SKN-1-interacting proteins that refine the appropriate SKN-1 transcriptional response. The bZip transcription factor MXL-3 partners with SKN-1 for the specification of the response to starvation. The new highly charged 50 amino acid SKN-1 domain we have identified as responsible for binding to MXL-3 also facilitates an interaction with mitochondrial phosphatase PGAM-5 and serine proteinase inhibitor K10D3.4. The expression of pgam-5 and mxl-3 is clearly expressed in the worm nervous and pharyngeal tissues (Figure S2). The behavioral response to limited nutrients and starvation is controlled by a well-established neuroendocrine circuit (Avery et al., 1993; Baumeister et al., 2006; Bishop and Guarente, 2007). Our findings incorporate mito-SKN-1, PGAM-5, and MXL-3 into that behavioral circuit. We have tested the mechanistic role of mxl-3 on the expression of transcriptional targets altered in the skn-1 gain-of-function mutants. In support of the metabolic specificity of this pathway, we find that the expression of metabolism transcripts altered by SKN-1 are also influenced by MXL-3. Recent examination of SKN-1 function under normal conditions identifies different but overlapping sets of genes that are also regulated in response to different stresses (Oliveira et al., 2009). MXL-3 may provide a paradigm for how SKN-1 could interact combinatorially with other transcription factors to regulate different subsets of genes under conditions of variable availability of nutrients.
Although both gain-of-function alleles lead to a perceived state of starvation and similar transcriptional profiles, there are molecular and physiological differences between the two alleles. We identified allele-specific changes in the effect of the E. coli K12 diet and of wdr-23 inactivation for some mito-SKN-1-dependent transcripts (signaling: ins-35, kin-15, and kin-16; metabolism: ZK742.4, lips-10, and aat-2; aging/dauer: ZK180.6 and R05G9R.1) (Table S2). These allele-specific changes are of particular interest based on the observation that the skn-1(lax120) mutant animals remain partially responsive to dietary restriction whereas skn-1(lax188) mutants are not. These results further specify unique properties of the different mito-SKN-1(gf) mutants and substantiate the idea that mito-SKN-1, perhaps in concert with MXL-3, influence metabolism and the organismal response to changes in available nutrients.
Activating Nrf mutations have also been identified in numerous somatic tumors, especially lung cell carcinomas (Ohta et al., 2008; Shibata et al., 2008). These tumors tend to be extremely resistant to both chemical and radiation-based therapies. It has been shown that the resistance phenotype is partially dependent on the activation of Nrf-dependent transcripts (Singh et al., 2008). Hyperproliferative tumor cells can rapidly exhaust available energy supplies forcing the necessity to adapt to a nutritionally limiting environment (Jang et al., 2011). The lung is not canonically thought of as a metabolic tissue as it does not have a major contribution to the maintaining energy homeostasis of the organism. The lung is, however, exceptionally very active metabolically, fabricating lipids in the form of surfactants rich in phospholipids and sterols. The activation of mitochondrial lipid oxidation could provide a mechanism to ensure adequate ATP levels for continued cancer growth prior to new vasculature provided by angiogenesis. Nrf has been identified as a potential target for cancer treatment to suppress cellular stress adaption mechanisms. Our results suggest that an understanding of the different specific mechanisms of Nrf activation is likely to be important for elucidating its effects on cell survival and carcinogenesis (Mitsuishi et al., 2012). Further characterization of the role of mito-SKN-1/Nrf will provide insights into the physiologic roles for the mitochondrial-specific population of this essential biological regulator and may lead to novel strategies for combatting cancers resistant to traditional therapies.
EXPERIMENTAL PROCEDURES
Standard Growth Conditions
C. elegans were raised on standard 6 cm nematode growth media plates supplemented with streptomycin and seeded with Escherichia coli OP50. For RNAi experiments NGM plates containing 5 mM IPTG and 100 ug ml−1 carbencillin were seeded with overnight cultures of double-stranded RNAi-expressing HT115 bacteria. Plates were allowed to induce overnight followed by transfer of age-synchronous populations of C. elegans.
Strains Used in This Study
Strains used in this study were N2 Bristol (wild-type), CB4856 (Hw), CL2166 [gst-4p::gfp]III, SPC167[skn-1(lax120)IV;gst-4p::gfp], SPC168[skn-1(lax188) IV;gst-4p::gfp], SPC207[skn-1(lax120)IV], and SPC227[skn-1(lax188)IV].
Statistics
Statistical analyses were performed with JMP 8 software.
Isolation of Dominant SKN-1 Activation Mutants
A C. elegans strain harboring the SKN-1 transcriptional reporter gst-4p::gfp was mutagenized via Ethyl methanesulfonate (EMS). F1 generation worms with increased GFP expression, indicating SKN-1 activation, were isolated for further characterization. F2 generation animals were singled and F3 progeny were analyzed for 100% penetrance of the activated GFP phenotype. These homozygous animals were then mated back to the unmutagenized parental strain. Successful matings that yielded 100% F1 progeny expressing the activated GFP phenotype were confirmed as dominant mutants.
Yeast-2-Hybrid
Bait and prey plasmids were generated by cloning 50-mer, 100-mer, and 200-mer oligonucleotides of the skn-1(lax120) and skn-1(lax188) mutations into pLexA and a C. elegans cDNA library into pACT2.2 (Addgene), respectively. Interaction was tested on synthetic complete agar that lacked leucine and tryptophan and was supplemented with X-α-gal. Interactors were identified by transforming Y2HGold (Clontech) with bait (pLexA-SKN-1) and the cDNA prey library. Positive clones were grown in the absence of tryptophan and sequenced. Sequenced clones were then retested individually.
Microarrays
RNA was processed using the Affymetrix GeneChip 3′ IVT express Kit. Mouse samples were hybridized to Affymetrix MouseGene 1.0 ST array. Five knockout mice and three wild-type littermates were used for these studies. C. elegans samples were hybridized to Affymetrix GeneChip C. elegans Genome Arrays. Three biologically unique wild-type samples were compared to three biological replicates of skn-1(lax120) and four biological replicates of skn-1(lax188) were utilized for this study.
L1 Arrest, Survival, and Recovery Assays
Following hypochlorite treatment, eggs were seeded into 24 well plates (4 wells per strain) with S-Basal + Cholesterol. For scoring the viability, one drop from each well was spotted onto a coverslip. Movement was used as a parameter to score viability. Bromophenol blue staining was used to confirm dead worms. Animals were scored 5, 7, and 12 days. At day 8 a cohort of animals were placed on OP50 for starvation recovery assays. Animals were scored for developmental progress once wild-type animals had reached reproductive maturity.
Life-Span Analysis
Life-span experiments were performed as previously described in (Curran and Ruvkun, 2007). The strains were egg prepped to generate synchronous populations, and 20–30 L4 animals were placed on NGM plates seeded with OP50 and containing 50 μg ml−1 5-fluoro-2′-deoxyuridine (FUdR) to prevent progeny hatching. The worms were kept at 20°C and scored every two days for survival by gentle prodding with platinum wire. Dietary restriction lifespan experiments were performed at 25°C on plates containing ampicillin to inhibit bacterial growth. Worms were transferred daily to fresh plates with the same bacterial concentration.
Additional methods can be found in the Supplemental Information.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chandra Tucker, Duke University for the C. elegans Y2H library (Developed by Maureen Barr’s laboratory). We are grateful to Daniel Wai, Children’s Hospital Los Angeles for analysis of Affymetrix microarray data. We also thank Drs. Caleb Finch, Alexander Soukas, and Shanshan Pang for critically reading the manuscript and all members of the Curran laboratory for technical support and fruitful discussions. S.P.C. conceived and designed the study. T.K.B. and M.Y. collaborated with SPC to plan and organize experiments. J.P., J.Y.L., S.D.N., T.N.N., K.G.C., S.R.S., T.S., and S.P.C. conducted the experiments. Some of the strains used in the study were obtained from the Caenorhabditis Genetics Center (CGC), which is supported by the National Institutes of Health—NCRR. Research in this study was supported by NIH R01GM62891 (T.K.B.) and R00AG032308 (S.P.C.). S.P.C. is an Ellison Medical Foundation Junior Scholar in Aging.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes five figures, seven tables, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2012.09.007.
REFERENCES
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Vranas K, Lucke M, Inoue H, Hisamoto N, Matsumoto K, Blackwell TK. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc. Natl. Acad. Sci. USA. 2005;102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Bargmann CI, Horvitz HR. The Caenorhabditis elegans unc-31 gene affects multiple nervous system-controlled functions. Genetics. 1993;134:455–464. doi: 10.1093/genetics/134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Hamm AC, Bonaus M, Jacob A, Jaekel J, Schorle H, Pankratz MJ, Katzenberger JD. Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol. Genomics. 2004;17:230–244. doi: 10.1152/physiolgenomics.00203.2003. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- Baumeister R, Schaffitzel E, Hertweck M. Endocrine signaling in Caenorhabditis elegans controls stress response and longevity. J. Endocrinol. 2006;190:191–202. doi: 10.1677/joe.1.06856. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Draper BW, Mello CC, Priess JR. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74:443–452. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early. C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Brys K, Castelein N, Matthijssens F, Vanfleteren JR, Braeckman BP. Disruption of insulin signalling preserves bioenergetic competence of mitochondria in ageing Caenorhabditis elegans. BMC Biol. 2010;8:91. doi: 10.1186/1741-7007-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell. Biol. 2009;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong MQ, Chase D, Patikoglou GA, Koelle MR. Multiple RGS proteins alter neural G protein signaling to allow. C. elegans to rapidly change behavior when fed. Genes Dev. 2000;14:2003–2014. [PMC free article] [PubMed] [Google Scholar]
- Greiss S, Hall J, Ahmed S, Gartner A. C. elegans SIR-2.1 trans-location is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev. 2008;22:2831–2842. doi: 10.1101/gad.482608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. Sensing of energy and nutrients by AMP-activated protein kinase. Am. J. Clin. Nutr. 2011;93:891S–6. doi: 10.3945/ajcn.110.001925. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Miwa J. Genetic and cellular characterization of Caenorhabditis elegans mutants abnormal in the regulation of many phase II enzymes. PLoS ONE. 2010;5:e11194. doi: 10.1371/journal.pone.0011194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, Matsumoto K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang T, Calaoagan JM, Kwon E, Samuelsson S, Recht L, Laderoute KR. 5′-AMP-activated protein kinase activity is elevated early during primary brain tumor development in the rat. Int. J. Cancer. 2011;128:2230–2239. doi: 10.1002/ijc.25558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- Kang C, Avery L. Systemic regulation of starvation response in Caenorhabditis elegans. Genes Dev. 2009;23:12–17. doi: 10.1101/gad.1723409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell A, Ventura N, Kahn N, Johnson TE. Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Radic. Biol. Med. 2007;43:1560–1566. doi: 10.1016/j.freeradbiomed.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko KW, Erickson B, Lehner R. Es-x/Ces1 prevents triacylglycerol accumulation in McArdle-RH7777 hepatocytes. Biochim. Biophys. Acta. 2009;1791:1133–1143. doi: 10.1016/j.bbalip.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Li J, Cai T, Wu P, Cui Z, Chen X, Hou J, Xie Z, Xue P, Shi L, Liu P, et al. Proteomic analysis of mitochondria from Caenorhabditis elegans. Proteomics. 2009;9:4539–4553. doi: 10.1002/pmic.200900101. [DOI] [PubMed] [Google Scholar]
- Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KT, Broitman-Maduro G, Hung WW, Cervantes S, Maduro MF. Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans. Dev. Biol. 2009;325:296–306. doi: 10.1016/j.ydbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD. Mutations in signal transduction proteins increase stress resistance and longevity in yeast, nematodes, fruit flies, and mammalian neuronal cells. Neurobiol. Aging. 1999;20:479–486. doi: 10.1016/s0197-4580(99)00089-5. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Mengarelli I, Rothman JH. Maternal deployment of the embryonic SKN-1—>MED-1,2 cell specification pathway in C. elegans. Dev. Biol. 2007;301:590–601. doi: 10.1016/j.ydbio.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell. 2001;7:475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Maier W, Adilov B, Regenass M, Alcedo J. A neuromedin U receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol. 2010;8:e1000376. doi: 10.1371/journal.pbio.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, Suzuki T, Kobayashi A, Yokota J, Sakiyama T, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda BB, Avery L. Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk NJ, Udranszky IA, Kozak E, Sunga J, Kim SK, Jacobson LA, Conley CA. Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J. Exp. Biol. 2006;209:4129–4139. doi: 10.1242/jeb.02492. [DOI] [PubMed] [Google Scholar]
- Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S, Noguchi T, Kinoshita H, Sekine Y, Iemura S, et al. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc. Natl. Acad. Sci. USA. 2009;106:12301–12305. doi: 10.1073/pnas.0901823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve NF, Lau A, Zhang DD. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin-ring ubiquitin ligases. Antioxid. Redox Signal. 2010;13:1699–1712. doi: 10.1089/ars.2010.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, See R, Batchelder C, Kophengnavong T, Gronniger JT, Shi Y, Blackwell TK. A conserved transcription motif suggesting functional parallels between Caenorhabditis elegans SKN-1 and Cap‘n’Collar-related basic leucine zipper proteins. J. Biol. Chem. 2000;275:22166–22171. doi: 10.1074/jbc.M001746200. [DOI] [PubMed] [Google Scholar]
- Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, Blackwell TK. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6:6. doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Watts JL. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol. Metab. 2009;20:58–65. doi: 10.1016/j.tem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Browse J. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2002;99:5854–5859. doi: 10.1073/pnas.092064799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Exp. Gerontol. 2006;41:894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.