Abstract
The farnesoid X receptor (FXR) is a bile acid-activated transcription factor belonging to the nuclear receptor superfamily. FXR deficiency in mice results in cholestasis, metabolic disorders, and tumorigenesis in liver and intestine. FXR is known to contribute to pathogenesis by regulating gene transcription; however, changes in the post-transcriptional modification of proteins associated with FXR modulation have not been determined. In the current study, proteomic analysis of the livers of wild-type (WT) and FXR knockout (FXR-KO) mice treated with a FXR synthetic ligand or vehicle was performed. The results identified five proteins as novel FXR targets. Since FXR deficiency in mice leads to liver tumorigenesis, poly (ADP-ribose) polymerase family, member 1 (Parp1) that is important for DNA repair, was validated in the current study by quantitative real-time PCR, and 1- and 2-dimensional gel electrophoresis/western blot. The results showed that Parp1 mRNA levels were not altered by FXR genetic status or by agonist treatment. However, total Parp1 protein levels were increased in FXR-KO mice as early as 3 month old. Interestingly, total Parp1 protein levels were increased in WT mice in an age-dependent manner (from 3 to 18 months), but not in FXR-KO mice. Finally, activation of FXR in WT mice resulted in reduction of phosporylated Parp1 protein in the liver without affecting total Parp1 protein levels. In conclusion, this study reveals that FXR genetic status and agonist treatment affects basal levels and phosphorylation state of Parp1, respectively. These alterations, in turn, may be associated with the hepatobiliary alterations observed in FXR-KO mice and participate in FXR agonist-induced protection in the liver.
Keywords: Farnesoid X receptor, Nuclear receptor, Parp1, Proteomics, Post-translational modification, Mice
Introduction
The farnesoid X receptor (FXR) is a ligand-activated transcription factor belonging to the nuclear receptor superfamily. The endogenous ligands of FXR are bile acids (Forman et al., 1995; Makishima et al., 1999; Parks et al., 1999; Wang et al., 1999). FXR regulates several key hepatic functions including bile acid, cholesterol and lipid homeostasis and glucose metabolism (Guo et al., 2003; Lambert et al., 2003; Ma et al., 2006; Mak et al., 2002; Sinal et al., 2000; Sirvent et al., 2004; Zhang et al., 2006). Moreover, FXR deficiency leads to liver and intestine tumorigenesis in mice (Kim et al., 2007; Maran et al., 2009; Modica et al., 2008; Yang et al., 2007). Animal studies suggest that FXR is a potential target for the prevention and/or treatment of gallstone disease, hepatocellular carcinoma (HCC), diabetes, atherosclerosis, biliary cirrhosis and colon cancer (Zhu et al., 2011).
Most studies of FXR have determined FXR-mediated regulation on gene transcription with very few studies determining the changes of protein levels. However, regulations at post-transcriptional, translational, and post-translational levels are equally important as regulation at the transcriptional level to affect cellular function. Moreover, transcriptional regulation may not always affect protein level due to multiple layers of modification following gene transcription.
Therefore, in the current study, we have determined the effects of FXR modulation at the protein level by proteomic analysis of the livers of wild-type (WT) and FXR-knockout (KO) mice treated with the FXR synthetic ligand (GW4064) or control vehicle. From this analysis, we have identified novel hepatic protein targets associated with FXR modification.
Materials and methods
Animals and treatments
WT and FXR-KO male mice in the C57BL/6J genetic background were used. All mice were housed in a pathogen-free animal facility under standard 12-h light/dark cycle, with free access to food and water ad libitum. The study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institute of Health. Animal protocols and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Kansas Medical Center. For proteomic analysis, 3-month old male C57BL/6J (WT) and FXR-KO mice (n = 3 per group) were gavaged with vehicle (1% methocellulose, 1% Triton X-100 in phosphate-buffered saline [PBS]) or GW4064 (100 mg/kg) twice (first dose at 6 pm and second dose at 8 am next day), with livers harvested 4 h after the second treatment. GW4064 was synthesized by the Chemical Discovery Laboratory at the University of Kansas, Lawrence, KS. In addition, livers of male WT and FXR-KO mice without treatment (n = 3 per group) at 3-, 6-, 12- and 18-month old were used for western blot and at 3-month old were used for Q-PCR analysis.
Proteomic analysis
Protein extraction
Individual livers were homogenized on ice in Urea/Thiourea lysis buffer containing 7 M urea, 2 M Thiourea, 4% CHAPS, 40 mM Tris (pH 8–9), 0.5% Triton X-100, 100 mM DTT 15.4 mg/ml, 0.2% Bio-Lyte ampholytes (Bio-Rad, cat. no. 163-2094, Hercules, CA), and 1 mM PMSF. The homogenates were incubated at room temperature for 30 min and centrifuged at 20,000 ×g for 60 min at 15 °C. The supernatants were collected, and the protein concentration was determined by Dye Reagent Concentrate (Bio-Rad, cat. no. 500-0006, Hercules, CA).
Two-dimensional gel electrophoresis
Forty micrograms of protein and 7.5 µl of standard marker (Bio-Rad, cat. no. 161-0320, Hercules, CA) were solubilized in 300 µl of isoelectric focusing (IEF) rehydration buffer (7 M urea, 2 M thiourea, 2% CHAPS, 100 mM DTT, 0.01% Bromophenol blue, and 0.2% Bio-Lyte ampholytes) to a total volume of 125 µl, which was then rehydrated to 7-cm/PH 3–10 IPG strips (Bio-Rad, cat. no. 161-2000, Hercules, CA). Passive rehydration was performed for 1 h and active rehydration at 50 V for 15 h. Isoelectric focusing was started at 250 V for 30 min and continued at 2000 V for 2 h with a final progression to 4000 V for a total of 20000 V/h. The IEF strips were equilibrated in first 1% DTT-containing and then 2.5% iodoacetamide-containing equilibration buffer (50 mM Tris–HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 0.01% bromophenol blue), each for 15 min. The strips were then placed on the top of a 10%–15% gradient SDS–PAGE gel and ran at 74 V for 30 min followed by 120 V for another 30 min until the bromophenol blue was near the bottom of the gel. The proteins were stained using the Applied Sliver Stain Plus Kit (Bio-Rad cat. no. 161-0449, Hercules, CA). Images were scanned on a GS-800 densitometer (Bio-Rad) and analyzed initially by the PD-Quest software (Bio-Rad, Hercules, CA). Protein spots were matched, and gels were normalized by standard markers in all gels. Protein spots that had at least 2-fold FXR-dependent changes were further analyzed.
Protein identification by MALDI-TOF MS analysis
The protein spots on Coomassie blue stained two-dimensional gel were excised and subjected to in-gel tryptic digestion by sequence grade modified trypsin (Promega, Madison, WI). After in-gel digestion, the peptides were mixed with 1 µl α-cyano-4-hydroxycinnanmic acid. Peptides samples were subject to peptide mass fingerprinting (PMF) using matrix associated laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS). In general, peptide digest from each 2D spot was pressure loaded on a C18 reverse phase nano-column (75 µm ID fused silica packed in-house with 9 cm of 100 Å, 5µ, Magic C18 particles, Michrom Bioresources). The column was mounted on the electrospray stage of a FT IC mass spectrometer (LTQ FT, Thermo Finnigan) and the peptides were separated on-line with an Eksigent 2D HPLC with a 0–90% acetonitrile gradient in 120 min at a flow rate of 0.25 µl/min. An electrospray voltage of 1.9 kV was used with the ion transfer temperature set to 200 °C. The mass spectrometer was controlled by the Xcalibur software to perform continuous mass scan analysis on the FT followed by MS/MS scans on the ion trap of the six most intense ions with a dynamic exclusion of two repeat scans of the same ion, 30 s repeat duration and 90 s exclusion duration. Normalized collision energy for MS/MS was set to 35%. For data analysis, all MS/MS scans were searched by the Sequest algorithm included in Bioworks 3.2 (Thermo Finnigan) using the human International Protein Index database (IPI human, v3.17). The results of the search were filtered using the following set of criteria for high confidence: minimum cross-correlation score of (Xcorr) of 1.9, 2.5 and 3.5 for 1, 2 and 3 charges ions, respectively, in addition to a delta correlation score (Δcorr) greater than 0.1, a probability of a random match lower than 0.001 and a minimum of 2 different peptides per protein.
RNA isolation and quantitative real-time PCR (Q-PCR)
Total RNA was isolated from frozen livers using TRIzol reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). The concentration of total RNA was determined by spectrophotometry, and RNA integrity was confirmed by MOPS gel electrophoresis. The mRNA expression levels of Fxr, Shp, Ostβ, and Parp1 were quantified using SYBR green chemistry (Fermentas, Glen Burnie, MD) with a standard protocol in an ABI Prism 7900 Detection system (Applied Biosystems, Foster City, CA). The mRNA results were normalized to Gapdh mRNA levels.
Western blot
Livers were homogenized in radioimmunoprecipitation assay lysis buffer (50 mM Tris–HCl, PH 7.4, 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA, 1% PMSF, and 1% protease inhibitor cocktail [Thermo Scientific, Rockford, IL]). Protein concentrations were determined by the BCA protein assay kit (Pierce, Rockford, IL). The samples were pooled from each group, and 50 µg of protein was electrophoresed on 10% SDS–polyacrylamide gels. The protein was transferred to a PVDF membrane and blocked with 5% nonfat milk for 2 h at room temperature. The blots were incubated with rabbit anti-Parp1 (Cell signaling, Danvers, MA) antibody at 4 °C overnight and then incubated with a goat anti-rabbit HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at room temperature for 2 h. Bands were visualized with ECL western blotting detection reagents (GE Healthcare Bio-Sciences, Piscataway, NJ). Gapdh (Millipore, Billerica, MA) was used as the loading control. Densitometric quantification of immunoblots was performed using ImageJ 1.43 software (NIH, Bethesda, MD).
Two-dimensional gel electrophoresis western blot
Two-dimensional gel electrophoresis was performed using 100 µg of pooled protein samples extracted from livers of WT mice treated with vehicle or GW4064 as described in the methods. Two gels were run for each group with one gel used for Coomassie blue staining and the other gel for western blot analysis. The procedures and antibodies for western blot were described above, and ImageJ 1.43 software was used for the densitometric quantification of immunoblots. The gel stained with Coomassie blue was used to identify spots detected in the western blot gel.
Immunoprecipitation and protein phosphorylation analysis
For immunoprecipitation assays, aliquots of total liver protein (in lysis buffer containing both protease and phosphorase inhibitors) were pre-cleared for 2 h at 4 °C using protein G agarose slurry (Millipore). The mixtures were then centrifuged at 8,000× g for 5 min at 4 °C with the cleared supernatants retained. Anti-Parp1 antibodies (Pierce) were added to the supernatants and incubated overnight at 4 °C with rotation. The Parp1/antibody complexes were captured by the addition of protein G sepharose slurry and incubating for 3 h at 4 °C with rotation. Following washes with lysis buffer, immunoprecipitates were boiled in SDS loading buffer (10% β-mercaptoethanol, 125 mM Tris–HCl (pH 6.8), 2% SDS, 10% glycerol, 0.005% bromophenol blue) for 10 min before subjecting to western blot analysis as described above. The phosphoprotein antibody sampler pack (Invitrogen) was used to determine the phosphorylation of serine (S), threonine (T) and tyrosine (Y) sites in Parp1 protein.
Statistical analysis
Data are presented as mean ± SD. The differences between two groups were analyzed by Student's t-tests after determining samples were normally distributed. The difference among multiple groups was analyzed by one-way ANOVA followed by Duncan's test. The results were considered statistically significant with p < 0.05.
Results
Novel FXR targets identified in mouse livers by proteomic analysis
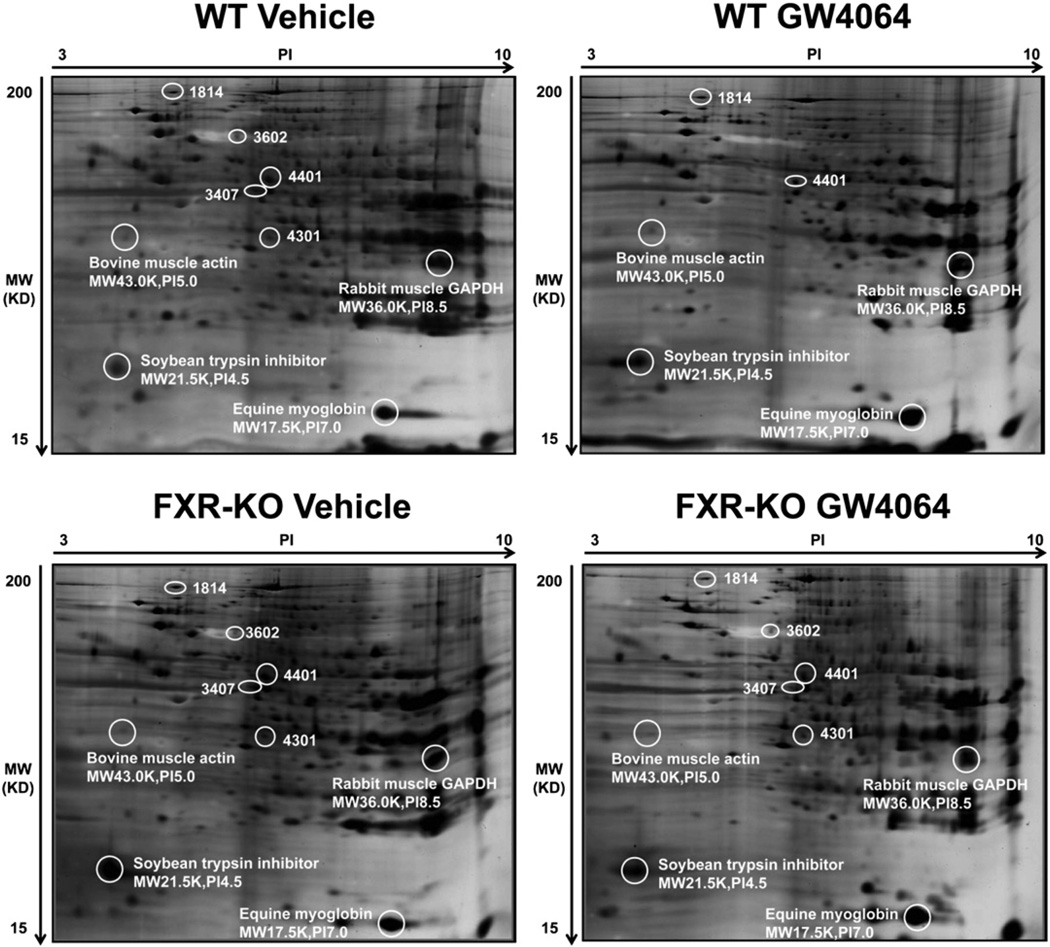
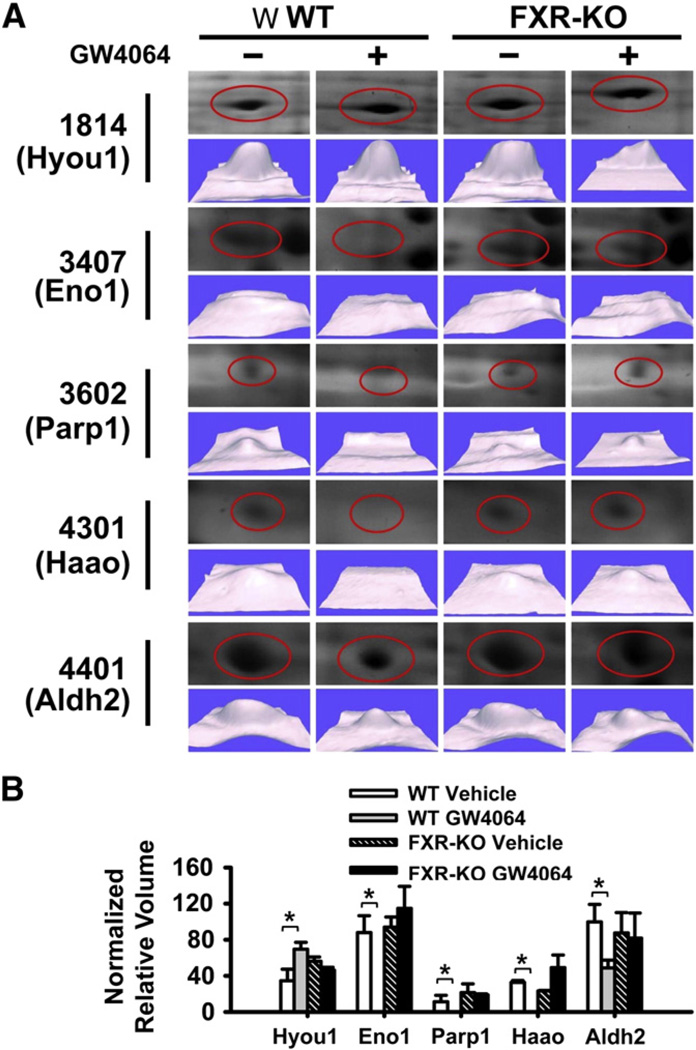
In the 2D-DIGE assay, no spot was identified with more than 2-fold difference in density between WT and FXR-KO mice. However, in WT mice treated with GW4064, 5 spots were identified with more than a 2-fold difference (Table 1, Figs. 1 and 2). These five spots were identified as the following proteins: hypoxia up-regulated 1 (Hyou1), enolase1 (Eno1), poly (ADP-ribose) polymerase family, member 1 (Parp1), 3-hydroxyanthranilate 3,4-dioxygenase (Haao), and mitochondrial aldehyde dehydrogenase 2 (Aldh2) (Table 1). Specifically, GW4064 increased the relative protein amount of Hyou1 and decreased the relative protein volume of the other four proteins (Fig. 2B).
Table 1.
Liver proteins changed by FXR modulation in mice by 2D-DIGE assay.
| Spot | Protein name | Gene symbol |
Protein score |
Sequence coverage (%) |
|---|---|---|---|---|
| 1814 | Hypoxia up-regulated 1 | hyou1 | 73 | 99.642 |
| 3407 | Enolase1 | eno1 | 128 | 100 |
| 3602 | Poly(ADP-ribose) polymerase Family, member 1 | parp1 | 77 | 99.844 |
| 4301 | 3-Hydroxyanthranilate 3,4-dioxygenase | haao | 108 | 100 |
| 4401 | Mitochondrial aldehyde dehydrogenase 2 | aldh2 | 116 | 100 |
Fig. 1.
Novel FXR target proteins identified in mouse livers by proteomic analysis. Total protein was isolated from four groups of mouse livers and was used for 2D-DIGE assay. Forty micrograms of protein and 7.5 µl of standard marker were separated on 7-cm/PH 3–10 IPG strips followed by separation on 10–15% gradient SDS–PAGE gels. Four standard markers, including bovine muscle actin, rabbit muscle GAPDH, soybean trypsin inhibitor and equine myoglobin, were labeled with their protein names, molecular weights and pI values on the gels. The gels were stained by silver staining.
Fig. 2.
A detailed analysis of 5 proteins with at least 2-fold FXR-dependent changes in 2D-DIGE analysis of livers of mice following FXR activation. (A) 2D-DIGE analysis of the 5 proteins that were different between livers of WT and FXR-KO mice treated with vehicle or GW4064. The gels were stained with silver staining and were identified by MALDI-TOF MS analysis. (B) Normalized relative volume of each protein in mouse livers. n = 3 mice per group. An asterisk indicates P<0.05 between vehicle and GW4064 treated groups.
Modulating FXR function did not affect Parp1 mRNA levels
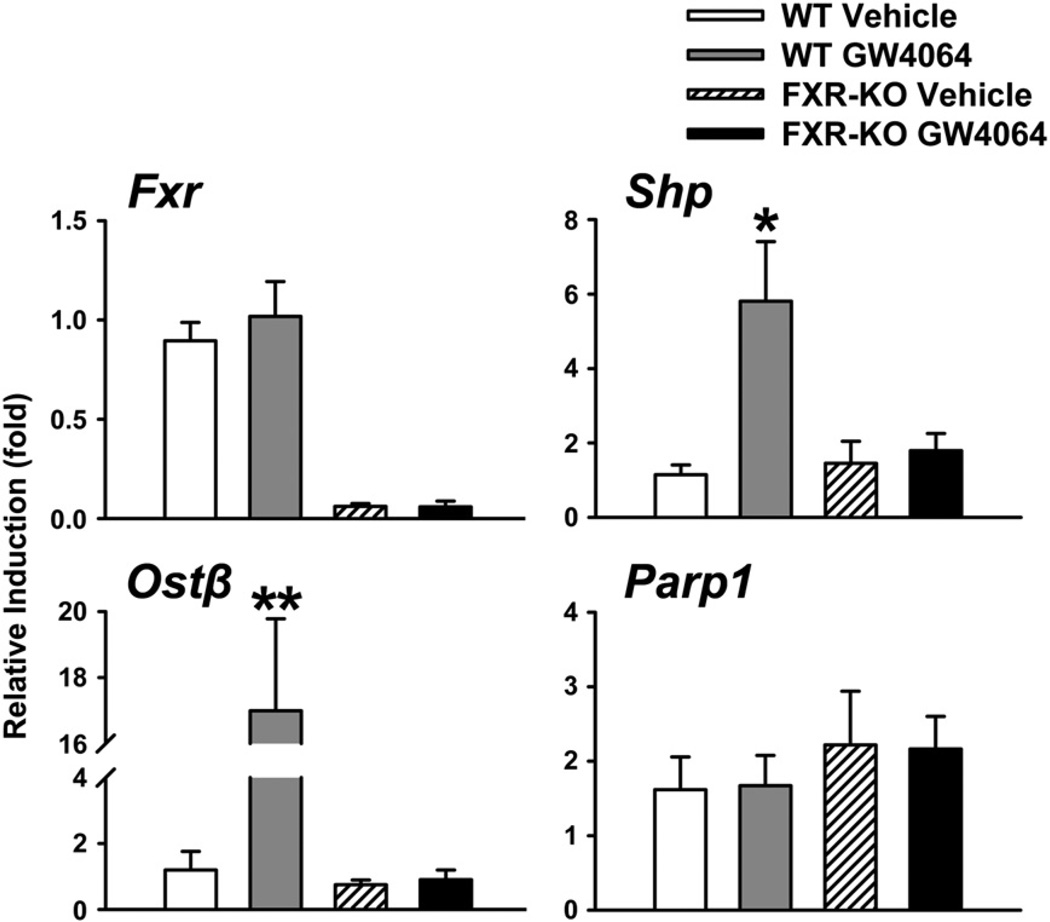
Studies have shown that deficiency of FXR leads to spontaneous HCC development in mice (Kim et al., 2007; Yang et al., 2007), but the underlying mechanism is not clear. In the current study, activation of FXR in WT mice by GW4064 treatment significantly reduced Parp1 protein levels in the liver in a FXR-dependent manner (Fig. 2). Since Parp1 plays critical roles in DNA repair and carcinogenesis (Krishnakumar and Kraus, 2010; Min et al., 2010), we decided to further focus on determining the effect of FXR modulation on Parp1 expression.
Shown in Fig. 3, FXR activation in WT mice was first confirmed by induction of classical FXR target genes, including small heterodimer partner (Shp) and organic solute transporter β (Ostβ) (Li et al., 2010; Teodoro et al., 2011). The mRNA levels of Parp1 did not change in WT mice treated with GW4064 (Fig. 3). Parp1 mRNA levels were slightly increased in FXR-KO mice compared to those in WT mice, although this change was not statistically significant (Fig. 3).
Fig. 3.
The mRNA levels of Fxr, Shp, Ostβ, and Parp1 with FXR modulation in the liver. FXR was activated in the livers of WT mice by GW4064 treatment. FXR-KO mice and vehicle treatment were served as controls. Shp and Ostβ are classical FXR target genes and their expression levels served as positive controls to indicate FXR activation. An asterisk indicates P<0.05 between vehicle and ligand treatment groups. Double asterisks mean P<0.01 between vehicle and ligand treatment groups.
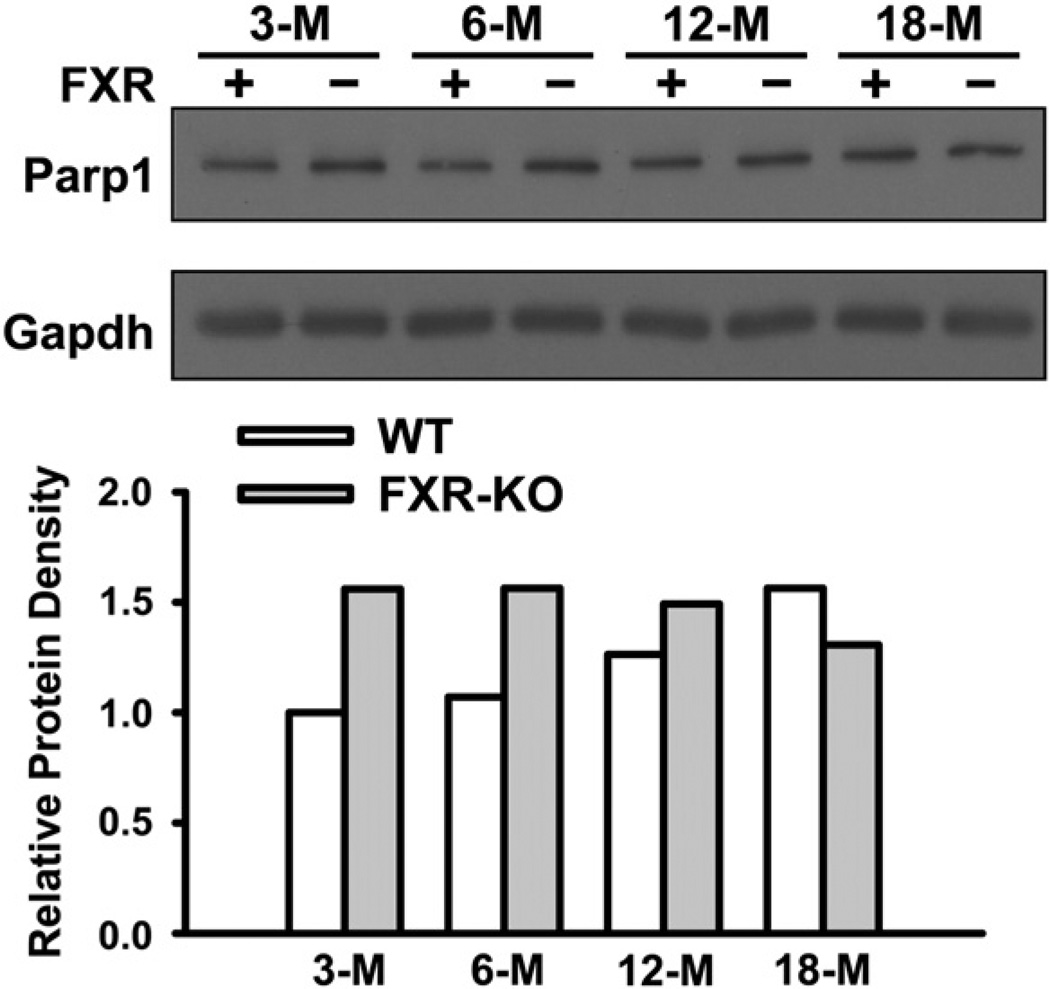
Induction of Parp1 total protein levels by FXR deficiency
To further determine the changes in total Parp1 protein levels in WT and FXR-KO mice, Parp1 protein was measured in livers of 3-, 6-,12-, and 18-month old WT and FXR-KO mice. There was an age-dependent increase in the total hepatic Parp1 protein in WT mice (Fig. 4). Furthermore, Parp1 protein levels appeared to be markedly increased in young FXR-KO mice compared to age-matched WT mice. However, in contrast to WT mice, aging did not affect Parp1 protein levels in FXR-KO mice (Fig. 4). Therefore, WT and FXR-KO mice tended to have similar Parp1 protein levels by 18 months of age.
Fig. 4.
Induction of Parp1 total protein levels in livers of FXR-KO mice. Western blot was performed with pooled liver protein samples (n = 3 per group) from 3-, 6-, 12-, and 18-month old WT (FXR +) and FXR-KO (FXR −) mice. Fifty µg of protein were electrophoresed on 10% SDS–polyacrylamide gels. Gapdh was used as a loading control. The density of bands was normalized to Gapdh and quantified by ImageJ software.
Activation of FXR reduced a modified form of Parp1 protein, but not the total Parp1 protein
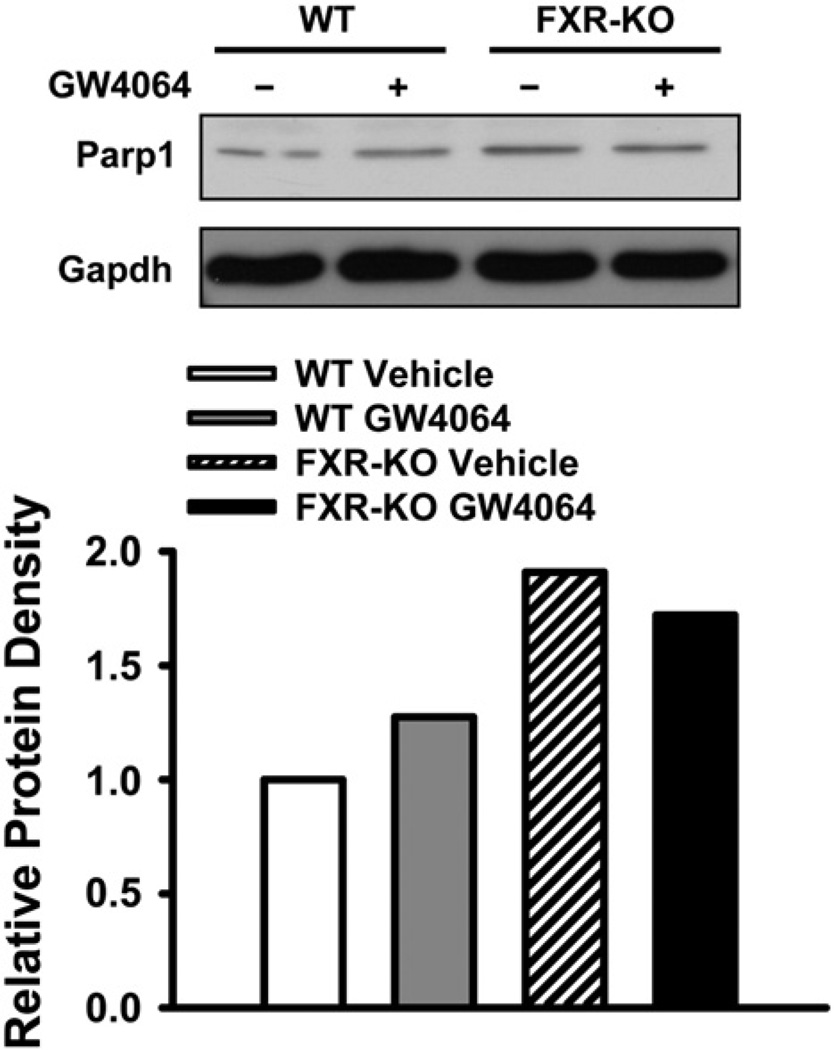
We have shown that deficiency of FXR increased total Parp1 protein levels in the livers of mice (Fig. 4). Therefore, the effect of FXR activation on Parp1 protein levels was determined in livers of 3-month old WT and FXR-KO mice treated with vehicle or GW4064. The result showed that activation of FXR by GW4064 did not result in significant changes of total Parp1 protein levels (Fig. 5).
Fig. 5.
Activation of FXR does not affect total Parp1 protein levels. Pooled liver protein samples (50 µg/well) from 3-month old WT and FXR-KO mice treated with vehicle or GW4064 (n = 3 per group) were used for western blot after separation on 10% SDS–polyacrylamide gels. Gapdh was used as a loading control. The density of bands was normalized to Gapdh and quantified by ImageJ software.
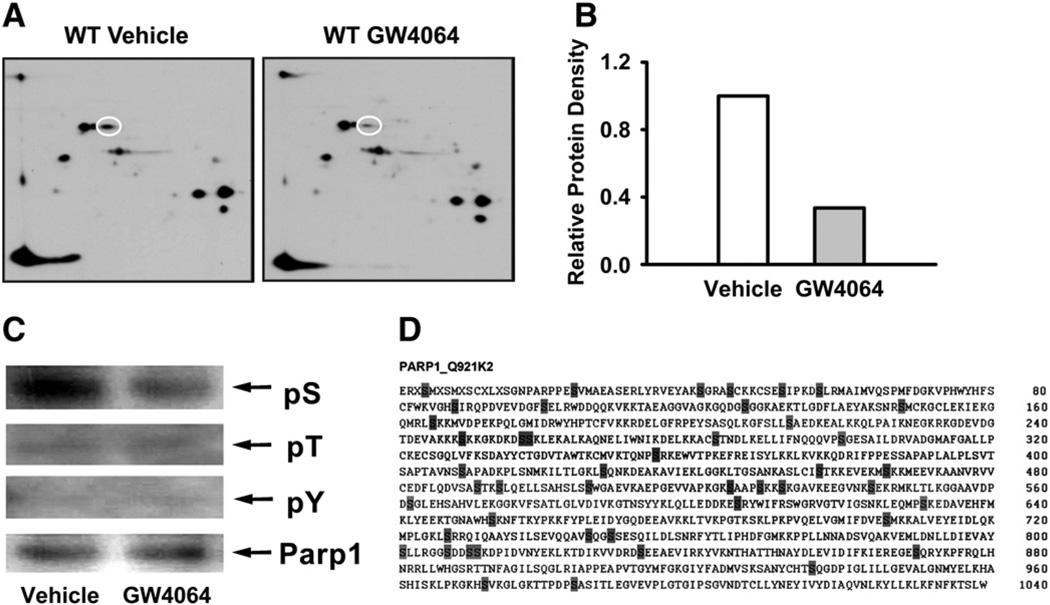
We then carefully examined the proteomic results obtained from Fig. 1 and found that the absence of detectable Parp1 in the WT mice treated with GW4064 was a form of Parp1 with a pI value of 6.7. The theoretical pI value of Parp1 by calculation is approximately 9 without consideration of protein modifications (Fig. 1). These results indicate that FXR activation reduces a modified form of Parp1 rather than total Parp1 protein, indicating that FXR modulation may alter the post-translational modification of Parp1 protein. To further test this hypothesis, we performed a two-dimensional gel electrophoresis followed by western blot analysis to determine the pI and amount of Parp1 following GW4064 administration. The result showed that a modified form of Parp1 was indeed reduced by FXR activation (Figs. 6A and B).
Fig. 6.
FXR activation reduced serine-phosphorylated Parp1 protein. (A) WT mice were treated with vehicle or GW4064 (n = 3 per group), and the liver proteins were isolated and pooled for each group. A total of 100 µg of protein and 7.5 µl of standard marker were separated on 7-cm/PH 3–10 IPG strips, followed by 10–15% gradient SDS–PAGE gels and western blot. (B) The density of spots was normalized and quantified by ImageJ software. (C) Phosphorylated Parp1 at sites of serine, threonine, and tyrosine were determined by immunoprecipitation. (D) Predicted serine phosphorylation sites in Parp1. The amino acid sequence of mouse Parp1 was analyzed using the NetPhos 2.0 software program with the corresponding probability scores >0.50 marked.
Parp1 is subject to several post-translational modifications, including SUMOylation, acetylation, and phosphorylation (Pic et al., 2011). Phosphorylation is one of the most important post-translational modifications that shift the pI value by several pH units. Indeed, there were more than 2 pI-units difference between the theoretical pI and the experimental pI of the Parp1 protein (Fig. 1), which suggests that altered phosphorylation may be a major modification of Parp1 in this study. To determine whether phosphorylation of Parp1 at specific phosphorylation sites are altered by FXR activation, antibodies against phospho-serine, -threonine or -tyrosine were used on samples immunoprecipitated with Parp1. Within the Parp1 protein, phosphorylation was associated primarily on serine site(s), while it was low on theronine and tyrosine sites. Furthermore, activation of FXR with GW4046 decreased the levels of phosphoserine in immunoprecipitated Parp1 protein in WT mice (Fig. 6C). There are 89 serines within Parp1 (almost 9% amino acid composition). To shift the pI value from 9.0 to 6.7, theoretically, 22 serines would need to be phosphorylated. Forty six of the 89 serines within Parp1 were predicted to be potential phosporylation sites (Blom et al., 2004) (Fig. 6D). The specific site of serine phosphorylation in Parp1 after FXR activation could be determined in future studies.
Discussion
We report a novel study of altered protein expression in mouse livers following FXR activation. Novel protein targets, including Parp1, were identified using a proteomics approach. FXR deficiency resulted in increased total Parp1 protein levels without affecting the mRNA levels of Parp1. Moreover, activation of FXR by the FXR agonist, GW4064, reduced the levels of serine-phosphorylated Parp1 protein without affecting Parp1 expression at the mRNA or total protein level.
In the current study, 5 proteins (Hyou1, Enol1, Parp1, Haao and Aldh2) were altered at least 2-fold in a FXR-dependent manner by proteomic analysis. Hyou1 is involved in cytoprotection in hypoxia-induced cellular perturbation (Kitao et al., 2004), Parp1 participates in the regulation of various cellular processes such as differentiation, proliferation and tumor transformation, and also plays a role in molecular events involved in the recovery from DNA damage (Ji and Tulin, 2010). Haao is an enzyme that catalyzes the formation of quinolonate from tryptophan via the biosynthetic pathway (Zhang et al., 2005). Eno1 and Aldh2 are both associated with metabolism of glucose, tryptophan and alcohol (Crabb et al., 2004; Gerlt et al., 2005). Among these 5 proteins, only Hyou1 was identified as heat shock protein 8, which belongs to the heat shock 70 kDa protein family, in a previous FXR-proteomics study (Gardmo et al., 2011).
These four novel proteins found in the current study were not reported previously by Gardmo et al. (2011) and this may be due to the following reasons: (1) the current study used a FXR synthetic ligand, GW4064 rather than INT-747. INT-747 is a derivative of chenodeoxycholic acid (CDCA) and is potent in activating FXR (Pellicciari et al., 2002). GW4064 is a selective and potent activator of FXR with an EC50 of 90 nM (Maloney et al., 2000). These two agonists may have different potencies in activating FXR in the liver, which may lead to differential protein expression profile; (2) silver and Coomassie blue staining were used for the detection of proteins in the 2D-DIGE assay in our study, which differs from the cyanine dyes used in the other study (Gardmo et al., 2011).
Our results clearly showed that the hepatic total Parp1 protein levels were consistently induced by FXR deficiency or aging, but not both. Moreover, the mRNA levels of Parp1 were not affected by FXR modulation. In addition, the total Parp1 protein levels were not altered by FXR activation in WT mice, but it was in the KO. These results led us to hypothesize that FXR affects Parp1 post-transcriptional modification. This was confirmed by differences in the observed versus theoretical pI and the selective down-regulation of the phosphoserine form of Parp1. Many proteins exist in several charged isoforms that can be resolved on the 2D-DIGE assay but appear as a single band on western blot. In addition, the immunoblotting results could be affected significantly by the specificities and sensitivities of the antibodies used (Li et al., 2011).
Parp1 is one of the most abundant nuclear proteins involved in DNA repair and intracellular transport (Abd Elmageed et al., 2012; Schreiber et al., 2006). Parp1 inhibitors have been used as chemo- and radio-sensitizers to enhance and/or prolong the antitumor effects of several anticancer therapies (Sandhu et al., 2010; Yap et al., 2011). Our previous studies have shown that FXR-KO mice develop spontaneous HCC, and deficiency of FXR enhances intestinal tumorigenesis (Kim et al., 2007; Maran et al., 2009; Modica et al., 2008; Yang et al., 2007). Our results showed that Parp1 was induced in the liver by FXR deficiency and FXR-KO mice have increased bile acids. It is likely that bile acids accumulate to such a toxic level in livers of FXR-KO mice that cell and DNA damage occur. This damage leads to activation of Parp1, which in turn helps to repair damaged DNA or trigger apoptosis. However, Parp1 protein levels were not further induced in FXR-KO mouse livers with increased bile acids (data not shown), indicating either saturation of Parp1 induction by bile acids or bile acids are not the only mechanism to induce Parp1 expression in FXR-KO mice. Moreover, Parp1 phosphorylation was reduced in livers of WT mice treated with GW4064, further indicating that FXR modulation affect Parp1 protein in a variety of manners. FXR is a transcription factor and thus is not likely to be directly involved in post-translational modifications of proteins. In addition, no report has shown direct transcriptional regulation of genes encoding kinases and/or phosphatases by FXR. However, it cannot be excluded of a possible involvement of an intermediate FXR-dependent protein(s) as a potential modulator of Parp1. Furthermore, FXR is known to regulate homeostasis of bile acids and other lipids, which are signaling molecules that are known to directly or indirectly activate the intracellular signaling pathways (reviewed by Zhu et al., 2011). Therefore, it is possible that FXR alters protein modification via interactive cellular networks. Future studies are warranted to determine the underlying mechanism.
Although studies have found that mice with Parp1 deletion are susceptible to HCC development, the function of Parp1 in HCC is yet not well known (Min et al., 2010). More recently, studies showed that Parp1 binds to the HBV core promoter (HBVCP) and increases the replication efficiency of HBV, which potentially contributes to the development of HCC (Ko and Ren, 2011). Moreover, other reports have shown that the protein amplified in liver cancer 1 (Alc1) is recruited to the nucleus and activated via interaction with Parp1 (Ahel et al., 2009; Gottschalk et al., 2009). Alc1 is a chromatin remodeling enzyme and has been shown to be over-expressed in more than 50% of human HCC samples (Ma et al., 2008; Wong et al., 2003). Over-expression of Alc1 exhibits induced colony formation in vitro and increased tumorigenesis rate in vivo (Ma et al., 2008). These results suggest that Alc1 functions as an oncogene to enhance HCC development. Moreover, IPA analysis of the Parp1 network in HCC shows potential protein candidates that may be associated with Parp1 during liver carcinogenesis (Supplemental Fig. 1). These results also predict other potential biological functions of Parp1 in HCC development. Thus, activation of Parp1 may be related to the activation of oncogenes, such as Alc1, which may contribute to liver carcinogenesis in FXR-KO mice. In contrast, inhibition of Parp1 by FXR activation may prevent HCC development. In the future, the role of Parp1 in HCC development especially with FXR deficiency warrants further study.
In summary, the present study identified 5 novel hepatic FXR targets at the protein level in livers of mice by proteomics analysis. Among these new targets, FXR deficiency or aging increased total Parp1 protein levels, whereas FXR activation reduced serine-phosphorylated but not total Parp1 protein.
Supplementary Material
Acknowledgments
We appreciated Dr. Antonio Artigues for his excellent technical support in proteomics analysis.
Financial support
This study was supported by the National Institutes of Health (DK081343 and DK090036, R03-HD062734, and ES005022). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Abbreviations
- 2D-DIGE
two-dimensional gel electrophoresis
- Alc1
amplified in liver cancer 1
- Aldh2
mitochondrial aldehyde dehydrogenase 2
- CDCA
chenodeoxycholic acid
- Eno1
enolase1
- FXR
farnesoid X receptor
- FXR-KO
FXR-knockout
- HBVCP
HBV core promoter
- HCC
hepatocellular carcinoma
- Hyou1
hypoxia up-regulated 1
- Haao
3-hydroxyanthranilate 3,4-dioxygenase
- IPA
ingenuity pathway analysis
- Q-PCR
quantitative real-time PCR
- Parp1
poly (ADP-ribose) polymerase family, member 1
- pI
isoelectric point
- OSTβ
organic solute transporter β
- SHP
small heterodimer partner
- WT
wild-type
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.taap.2012.11.012.
Competing interests
The authors have declared that no competing interests exist.
Contributor Information
Fei Li, Email: xw_lifei@yahoo.com.cn.
Grace L. Guo, Email: guo@eohsi.rutgers.edu.
References
- Abd Elmageed ZY, Naura AS, Errami Y, Zerfaoui M. The poly(ADP-ribose) polymerases (PARPs): new roles in intracellular transport. Cell. Signal. 2012;24:1–8. doi: 10.1016/j.cellsig.2011.07.019. [DOI] [PubMed] [Google Scholar]
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Crabb DW, Matsumoto M, Chang D, You M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc. Nutr. Soc. 2004;63:49–63. doi: 10.1079/pns2003327. [DOI] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- Gardmo C, Tamburro A, Modica S, Moschetta A. Proteomics for the discovery of nuclear bile acid receptor FXR targets. Biochim. Biophys. Acta. 2011;1812:836–841. doi: 10.1016/j.bbadis.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch. Biochem. Biophys. 2005;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl) ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB, Jr, Kliewer SA, Gonzalez FJ, Sinal CJ. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J. Biol. Chem. 2003;278:45062–45071. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. The roles of PARP1 in gene control and cell differentiation. Curr. Opin. Genet. Dev. 2010;20:512–518. doi: 10.1016/j.gde.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao Y, Hashimoto K, Matsuyama T, Iso H, Tamatani T, Hori O, Stern DM, Kano M, Ozawa K, Ogawa S. ORP150/HSP12A regulates Purkinje cell survival: a role for endoplasmic reticulum stress in cerebellar development. J. Neurosci. 2004;24:1486–1496. doi: 10.1523/JNEUROSCI.4029-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HL, Ren EC. Novel poly (ADP-ribose) polymerase 1 binding motif in hepatitis B virus core promoter impairs DNA damage repair. Hepatology. 2011;54:1190–1198. doi: 10.1002/hep.24502. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert G, Amar MJ, Guo G, Brewer HB, Jr, Gonzalez FJ, Sinal CJ. The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J. Biol. Chem. 2003;278:2563–2570. doi: 10.1074/jbc.M209525200. [DOI] [PubMed] [Google Scholar]
- Li G, Thomas AM, Hart SN, Zhong X, Wu D, Guo GL. Farnesoid X receptor activation mediates head-to-tail chromatin looping in the Nr0b2 gene encoding small heterodimer partner. Mol. Endocrinol. 2010;24:1404–1412. doi: 10.1210/me.2010-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Glinskii OV, Zhou J, Wilson LS, Barnes S, Anthony DC, Glinsky VV. Identification and analysis of signaling networks potentially involved in breast carcinoma metastasis to the brain. PLoS One. 2011;6:e21977. doi: 10.1371/journal.pone.0021977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma NF, Hu L, Fung JM, Xie D, Zheng BJ, Chen L, Tang DJ, Fu L, Wu Z, Chen M, Fang Y, Guan XY. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology. 2008;47:503–510. doi: 10.1002/hep.22072. [DOI] [PubMed] [Google Scholar]
- Mak PA, Kast-Woelbern HR, Anisfeld AM, Edwards PA. Identification of PLTP as an LXR target gene and apoE as an FXR target gene reveals overlapping targets for the two nuclear receptors. J. Lipid Res. 2002;43:2037–2041. doi: 10.1194/jlr.c200014-jlr200. [DOI] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Maloney PR, Parks DJ, Haffner CD, Fivush AM, Chandra G, Plunket KD, Creech KL, Moore LB, Wilson JG, Lewis MC, Jones SA, Willson TM. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 2000;43:2971–2974. doi: 10.1021/jm0002127. [DOI] [PubMed] [Google Scholar]
- Maran RR, Thomas A, Roth M, Sheng Z, Esterly N, Pinson D, Gao X, Zhang Y, Ganapathy V, Gonzalez FJ, Guo GL. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharmacol. Exp. Ther. 2009;328:469–477. doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min W, Cortes U, Herceg Z, Tong WM, Wang ZQ. Deletion of the nuclear isoform of poly(ADP-ribose) glycohydrolase (PARG) reveals its function in DNA repair, genomic stability and tumorigenesis. Carcinogenesis. 2010;31:2058–2065. doi: 10.1093/carcin/bgq205. [DOI] [PubMed] [Google Scholar]
- Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008;68:9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, Morelli A, Parks DJ, Willson TM. 6alpha-Ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 2002;45:3569–3572. doi: 10.1021/jm025529g. [DOI] [PubMed] [Google Scholar]
- Pic E, Gagne JP, Poirier GG. Mass spectrometry-based functional proteomics of poly(ADP-ribose) polymerase-1. Expert Rev. Proteomics. 2011;8:759–774. doi: 10.1586/epr.11.63. [DOI] [PubMed] [Google Scholar]
- Sandhu SK, Yap TA, de Bono JS. Poly(ADP-ribose) polymerase inhibitors in cancer treatment: a clinical perspective. Eur. J. Cancer. 2010;46:9–20. doi: 10.1016/j.ejca.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Sirvent A, Claudel T, Martin G, Brozek J, Kosykh V, Darteil R, Hum DW, Fruchart JC, Staels B. The farnesoid X receptor induces very low density lipoprotein receptor gene expression. FEBS Lett. 2004;566:173–177. doi: 10.1016/j.febslet.2004.04.026. [DOI] [PubMed] [Google Scholar]
- Teodoro JS, Rolo AP, Palmeira CM. Hepatic FXR: key regulator of whole-body energy metabolism. Trends Endocrinol. Metab. 2011;22:458–466. doi: 10.1016/j.tem.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- Wong N, Chan A, Lee SW, Lam E, To KF, Lai PB, Li XN, Liew CT, Johnson PJ. Positional mapping for amplified DNA sequences on 1q21–q22 in hepatocellular carcinoma indicates candidate genes over-expression. J. Hepatol. 2003;38:298–306. doi: 10.1016/s0168-8278(02)00412-9. [DOI] [PubMed] [Google Scholar]
- Yang F, Huang X, Yi T, Yen Y, Moore DD, Huang W. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-ribose) polymerase (PARP) inhibitors: exploiting a synthetic lethal strategy in the clinic. CA Cancer J. Clin. 2011;61:31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Colabroy KL, Begley TP, Ealick SE. Structural studies on 3-hydroxyanthranilate-3,4-dioxygenase: the catalytic mechanism of a complex oxidation involved in NAD biosynthesis. Biochemistry. 2005;44:7632–7643. doi: 10.1021/bi047353l. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li F, Guo GL. Tissue-specific function of farnesoid X receptor in liver and intestine. Pharmacol. Res. 2011;63:259–265. doi: 10.1016/j.phrs.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.