Abstract
Objective
The purpose of the present study is to discover the extent to which distinct DSM disorders share large, highly recurrent copy number variants (CNVs) as susceptibility factors. We also seek to identify gene mechanisms common to groups of diagnoses and/or specific to a given diagnosis based on associations with CNVs.
Method
Systematic review of 820 PubMed articles on autism spectrum disorder (ASD), intellectual disability (ID), schizophrenia, and epilepsy produced 54 CNVs associated with one or several disorders. Pathway analysis on genes implicated by CNVs in different groupings was conducted.
Results
The majority of CNVs were found in ID with the other disorders somewhat subsumed, yet certain CNVs were associated with isolated or groups of disorders. Based on genes implicated by CNVs, ID encompassed 96.8% of genes in ASD, 92.8% of genes in schizophrenia, and 100.0% of genes in epilepsy. Pathway analysis revealed that synapse processes were enriched in ASD, ID, and schizophrenia. Disease-specific processes were identified in ID (actin cytoskeleton processes), schizophrenia (ubiquitin-related processes), and ASD (synaptic vesicle transport and exocytosis).
Conclusions
Intellectual disability may arise from the broadest range of genetic pathways, and specific subsets of these pathways appear relevant to other disorders or combinations of these disorders. It is clear that statistically significant CNVs across disorders of cognitive development are highly enriched for biological processes related to the synapse. There are also disorder-specific processes that may aid in understanding the distinct presentations and pathophysiology of these disorders.
Keywords: autism, epilepsy, intellectual disability, schizophrenia, copy number variation
Genome-wide association studies (GWAS) have identified a large number of recurrent copy number variants (CNVs) that are associated with disease and are also frequently shared as susceptibility factors by several disorders of cognitive development.1–7 However, the distribution of these CNVs across clinically distinct neurodevelopmental disorders, such as autism spectrum disorders (ASD), intellectual disability (ID), schizophrenia, or epilepsy, has not been systematically studied. For example, are there CNVs that are common to all disorders? Are there CNVs that are specific only to a subset of these disorders? Are there CNVs, either deletions or duplications, that are specific to only one disorder? Detailed and systematic examination of the distribution of disease-associated CNVs across these disorders will aid in the nosology of psychiatric disorders and will also provide insight into shared and distinct biological processes underlying the DSM diagnoses.
Overlapping symptoms are found across DSM categories of neurodevelopmental disorders. In particular, ID, ASD, and schizophrenia are all considered together here as disorders of cognitive development, as all conditions share central symptoms such as cognitive impairment and all have neurodevelopmental causes in a majority of cases.4 Epilepsy is similarly considered among this group of clinical conditions given high rates of co-occurrence and many known shared etiologies with the other diagnoses.8 Likewise, based on a large number of studies, specific CNVs have been identified across DSM categories including ID, ASD, schizophrenia, and epilepsy. For example, 16p11.2 CNVs are significantly associated with disease in ASD (deletions9–12 and duplications9–11), schizophrenia (duplications only13–17), and ID (deletions18–21 and duplications19–21). 16p11.2 CNVs have also been found in patients with epilepsy8. In addition, 1q21.1 CNVs have been confirmed in case-control studies as significantly associated with ID (deletions and duplications19, 20, 22) and schizophrenia (deletions14, 17, 23–25 and duplications14). By a similar logic, attention-deficit/hyperactivity disorder (ADHD) may be grouped with the above conditions as a disorder of cognitive development. In one ADHD GWAS study, large rare CNVs were highest among ADHD patients with co-morbid ID and identified the locus of 16p13.11 as significantly enriched.26 However, given the relatively low number of large, genome-wide studies in CNVs associated with ADHD, we have excluded ADHD from the current analysis.
Studies of CNVs also promise to identify potential susceptibility mechanisms associated with specific neurodevelopmental disorders. For example, post-synaptic mechanisms have been implicated in ASD as a result of genome-wide CNV studies.27 Specific post-synaptic pathways include SHANK328, 29 and neuronal cell-adhesion (NLGN3 and NLGN4X30). Some of these loci may be shared by other conditions, in particular ID, but also schizophrenia31 and epilepsy32 in other cases. The present study attempts to clarify the distribution of disease-associated CNVs across four disorders of cognitive development, namely ID, ASD, schizophrenia, and epilepsy.
First we identify the specific CNVs (considering deletions and duplications separately) that are associated with ASD, ID, schizophrenia, and epilepsy. We then examine the groups of CNVs that emerge as common to all disorders, specific to subsets of disorders, or unique to a given single disorder. Next we apply gene pathway analysis techniques to specific subsets of genes within CNVs associated with isolated disorders or given combinations of disorders. We first analyze groups of CNVs based on their association with one given DSM category. We then examine genetic pathways associated with CNVs from specific combinations of disorders (i.e. CNVs found in two or more DSM disorders) in order to uncover potential shared genetic mechanisms. In addition, we examine subgroups of CNVs that are associated with relatively isolated conditions (for example, ID without autism, schizophrenia, or epilepsy) and schizophrenia (without ID, ASD, or epilepsy), with the hope of uncovering mechanisms that may be relatively specific to a given disorder. Our study provides an important and unique approach to the study of CNVs in neurodevelopmental disorders and begins to uncover some of the shared as well as distinct pathways that are at the root causes of the DSM diagnoses.
METHOD
Identification of highly recurrent CNVs
A systematic review of CNVs was pursued based on the outline in Figure S1A, available online. CNVs associated with neurodevelopmental disorders were searched in PubMed through June 2012. Specific searches were employed for ASD (“copy number” and “autism”), ID (“copy number”, “intellectual disability”, and “mental retardation”), schizophrenia (“copy number” and “schizophrenia”), and epilepsy (“copy number” and “epilepsy”). Searches were limited to English language, humans, and publications after 2005. CNVs were considered highly recurrent and associated with disease if they were identified in a PubMed publication with the following features: (a) genetically tested in a large disease cohort (N>400); (b) included a comparison control sample; (c) statistically compared the CNV frequencies in cases and controls; (d) and the CNV was significantly enriched in the case sample. CNV significance was based on criteria established by the given publication. In all cases, this represented p<0.05, but the majority of studies used much stricter genome-wide criteria for significance. All CNV coordinates are reported in NCBI build 36, hg18. In general, we were able to consider deletions and duplications as separate CNVs.
Classification of CNV to specific disorders and determination of CNV co-ordinates/genes
Highly recurrent CNVs were assigned to one or more disorder based on the following approach (Figure S1B, available online). CNVs were assigned to the phenotype for which they met rigorous case-control criteria based on criteria described above. This was “strict” criteria for association. Further, significant CNVs were assigned to additional disorder categories based on “broad” criteria if a study participant with a primary disorder is reported to have symptoms meeting criteria for another disorder. An example of broad criteria is if a child with a recurrent CNV from an ASD study is described as having co-occurring ID, then the given CNV was coded for both ASD and ID disorders.
Three CNV categories were defined: A, B, and C (Figure S2, available online). Category A consists of stereotyped, recurrent CNVs whereby multiple cases of a particular CNV encompassed the same region. Category A CNVs are widely known to be generated in stereotyped fashion due to flanking segmental duplications and non-allelic homologous recombination (such as 16p11.2 deletions/duplications). Category B consists of CNVs that may be variable but hit a singular, common gene (such as NRXN1 deletions/duplications). Category C consists of non-stereotyped, recurrent CNVs that overlap with a common region, which contains many genes (for example, 1p36 deletions/duplications). To determine CNV category, we reviewed Decipher CNV cases (when possible) and entered CNV coordinates into the UCSC Genome Browser database searching for “segmental dups” (http://www.genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=263029387&c=chr2&g=genomicSuperDups). If a CNV was positive for flanking segmental duplications, then it provided further evidence for Category A inclusion.
We used a systematic approach to determine CNV intervals. For a given locus within a given disorder, if multiple intervals were determined, we chose intervals that were inclusive of all such CNVs, ie the largest interval covering all CNVs. If a CNV was found in two or more disorders, for the gene pathway analysis examining overlap across those disorders, we used the minimum interval, in the intersection of all relevant intervals. Genes were included in the pathway analysis if they were within or intersected by the genetic intervals determined as described above.
Mean CNV sizes for all disorders were compared with Wilcoxon rank test using STATA33 11.0. Fisher’s exact test in R was used to compare both the number of deletions and duplications, as well as CNV type across disorders.
Visualization of gene groupings by Venn diagrams
Gene groupings were visualized via VennMaster software.34 List of genes in all categories were imported as text files to the program.
Gene pathway analysis and test subgroups
Gene groups were analyzed by hypergeometric statistical methods using the Database for Annotation, Visualization and Integrated Discovery v6.7 software.35 DAVID parameters used the recommended Gene Ontology categories (GOTERM_BP_FAT, GOTERM_CC_FAT, and GOTERM_MF_FAT) with “high” classification stringency to ensure a strong association between genes. An additional pathway analysis investigated if there were differences in gene ontology depending on CNV type. Three CNV categories were defined: A, B, and C (Figure S2, available online).
RESULTS
Distribution of CNVs Across Disorders
Based on Pubmed searches and review of references, a total of 820 unique articles were identified: 223 ASD, 373 ID, 164 schizophrenia, and 60 epilepsy articles. After full review of all articles, 37 articles met stringent search criteria: 8 ASD9–12, 27, 36–38, 9 ID18–20, 22, 39–43, 15 schizophrenia13–17, 23–25, 37, 44–49, and 5 epilepsy8, 50–53 (Table S1, available online). We categorized CNVs to disorders based “strict” criteria and “broad” criteria (see Method and Figure S1, available online). From these articles, 79 CNVs (counting deletions and duplications separately) met strict criteria: 39 were assigned to ID (26 deletions, 13 duplications), 14 to ASD (8 deletions, 6 duplications), 23 to schizophrenia (13 deletions, 10 duplications), and 3 to epilepsy (3 deletions, no duplications) In summary, 54 unique loci (now combining deletions and duplications) encompassing 1,416 unique genes, were determined to be highly recurrent and associated with disease (Table 1). Ten out of the 54 CNVs did not contain any genes and were not included in the pathway analysis. The assignment of CNVs to the broad criteria are also shown in Table 1.
Table 1.
List of Highly Recurrent, Eligible Copy Number Variants (CNVs) From Genome-Wide Association Studies (GWAS) Studies.
| CNV | Group | Disorder (Strict Criteria) | Disorder (Broad Criteria) | Coordinates | Size (kb) | Number of Genes (Strict Criteria) | Coordinate Source |
|---|---|---|---|---|---|---|---|
| 1p33 (AGBL4) | B | ASDDel | IDDel, ASDDel | 49,685,647–49,770,826 | 85 | 1 | Pinto et al. 201036 |
| 1p36 | C | IDB | IDB, ASDDel, EpilepsyDel | 0–10,000,000 | 10,000 | 222 | Cooper et al. 201119 |
| 1q21.1 | A | IDB, SchizophreniaB | IDB, ASDB, SchizophreniaB, EpilepsyB | 144,600,000–146,300,000 | 1,700 | 29 | Levinson et al. 201114, Mefford et al. 200822, Cooper et al. 201119, Kaminsky et al. 201120, ISC 200823, Stefansson et al. 200824, Kirov et al. 200925, Grozeva et al. 201217 |
| 2p16.3 (NRXN1, DKFZp313P2036) | B | SchizophreniaDel | IDDel, SchizophreniaDel, EpilepsyDel | 49,900,000–51,500,000 | 1,600 | 2 | Rujescu et al. 200944, Levinson et al. 201114 |
| 2p21 (PRKCE, SRBD1, UNQ6975) | C | IDDup | IDDup | 45,200,000–45,900,000 | 700 | 3 | Cooper et al. 201119 |
| 2p25.3(MYT1L, PXDN) | C | SchizophreniaDup | SchizophreniaDup | 1,618,945–1,857,129 | 238 | 2 | Lee et al. 201245 |
| 2q13 | A | IDB | IDB, ASDB, EpilepsyDel | 110,180,000–110,340,000 111,050,000–112,950,000 |
160 1,900 |
36 | Cooper et al. 201119 |
| 2q37 | C | IDDel | IDDel, ASDDel, EpilepsyDel | 239,370,000–242,120,000 | 2,750 | 42 | Cooper et al. 201119 |
| 3p26.2(SUMF1) | B | ASDDel, SchizophreniaDel | ASDDel, SchizophreniaDel | 4,199,731–4,236,304 4,063,809–4,074,877 |
37 11 |
1 | Glessner et al. 200927, Glessner et al. 201013 |
| 3p26.3 (CNTN4) | B | ASDDup | ASDDup | 2,548,148–2,548,531 | 0.4 | 1 | Glessner et al. 200927 |
| 3q26.31(NLGN1) | B | ASDDup | ASDDup | 174,754,378–174,771,975 | 18 | 1 | Glessneret al. 200927 |
| 3q29 | A | IDDel, SchizophreniaB | IDDel, SchizophreniaB, EpilepsyDel | 197,240,451–198,829,062 | 1,589 | 36 | Kaminsky et al. 201120, Levinson et al. 201114, Vacic et al. 201115, Mulle et al. 201046, Grozeva et al. 201217 |
| 4p16.3 (Wolf-Hirschhorn Syndrome) (SCARNA22, WHSC1, WHSC2) | C | IDDel | IDDel, EpilepsyDel | 1,840,000–1,980,000 | 140 | 3 | Cooper et al. 201119 |
| 4q21.21-q21.22 | C | IDDel | IDDel, ASDDel, EpilepsyDel | 81,950,000–83,350,000 | 1,400 | 4 | Cooper et al. 201119 |
| 5q35.2-q35.3 (Sotos Syndrome) | C | IDDel | IDDel, EpilepsyDel | 175,650,000–176,990,000 | 1,340 | 45 | Cooper et al. 201119, Kaminsky et al. 201120, |
| 6q26 (PARK2) | B | ASDDel, SchizophreniaDup | ASDDel, SchizophreniaDup | 162,584,576–162,587,001 162,835,583–162,997,592 |
2 162 |
1 | Glessner et al. 200927, Vacic et al. 201115 |
| 7q11.22-q11.23 (Williams-Beuren Syndrome) | A | IDB, ASDDup | IDB, ASDDup, EpilepsyB | 72,380,000–73780449 | 1,400 | 32 | Cooper et al. 201119, Kaminsky et al. 201120, Sanders et al. 20119 |
| 7q36.3 (BC041429, BC042556, VIPR2) | B | SchizophreniaDup | SchizophreniaDup | 158,448,321–158,630,000 | 182 | 3 | Vacic et al. 201115, Levinson et al. 201114 |
| 8p23.1 | C | IDDel | IDDel | 8,156,705–11,803,128 | 3,646 | 37 | Kaminsky et al. 201120 |
| 9q34.3 (9q subteleometric deletion syndrome) | C | IDDel | IDDel, ASDDel, EpilepsyDel | 136,950,000–140,200,000 | 3,250 | 132 | Cooper et al. 201119 |
| 10q11.21 (RET) | B | SchizophreniaDel | SchizophreniaDel | 42,932,615–42,934,354 | 2 | 1 | Glessner et al. 201013 |
| 10q23.2(GRID1, KIAA1220) | B | IDDel, ASDDel | IDDel, ASDDel | 87,941,666–87,949,029 | 7 | 2 | Glessner et al. 200927, Cooper et al. 201119 |
| 11q25(GLB1L3) | B | SchizophreniaDel | SchizophreniaDel | 133,650,000–133,690,000 | 40 | 1 | Levinson et al. 201114 |
| 15q11.2 | A | IDDel, SchizophreniaDel, EpilepsyDel | IDDel, ASDDel, Schizophrenia Del, EpilepsyDel | 20,306,549–20,691,555 | 385 | 6 | Mefford et al. 200941, Cooper et al. 201119, Stefansson et al. 200824, Zhao et al. 201247, Kirov et al. 200925, Grozeva et al. 201217, de Kovel et al. 201050, Mefford et al. 20108 |
| 15q11-q13 (Prader-Willi/Angelman Syndrome) | A | IDB, ASDDup, SchizophreniaDup | IDB, ASDDup, SchizophreniaDup, EpilepsyB | 21,309,483–26,208,861 | 4,899 | 104 | Kaminsky et al. 201120, Cooper et al. 201119, Glessner et al. 200927, Ingason et al. 2011a48 |
| 15q13.3 | A | IDB, SchizophreniaDel, EpilepsyDel | IDB, ASDB, SchizophreniaDel, EpilepsyB | 28,000,000–31,000,000 | 3,000 | 78 | ISC 200823, Sharp et al. 200840, Cooper et al. 201119, Kaminsky et al. 201120, Levinson et al. 201114, Stefansson et al. 200824, Vacic et al. 201115, Kirov et al. 200925, Grozeva et al. 201217, de Kovel et al. 201050, Dibbens et al. 200951, Mefford et al. 20108, Helbig et al. 200952 |
| 16p11.2 | A | IDB, ASDB, SchizophreniaDup | IDB, ASDB, SchizophreniaDup, EpilepsyB | 29,474,810–30,110,000 | 635 | 46 | Cooper et al. 201119, Sanders et al. 20119, Bachmann-Gagescu et al. 201018, Rosenfeld et al. 201042, Kaminsky et al. 201120, Weiss et al. 200810, Kumar et al. 200812, Marshall et al. 200811, McCarthy et al. 200916, Glessner et al. 201013, Levinson et al. 201114, Vacic et al. 201115, Grozeva et al. 201217 |
| 16p11.2-p12.2 | A | IDB | IDB, ASDDup, EpilepsyB | 28,680,000–29,020,000 | 340 | 15 | Cooper et al. 201119 |
| 16p12.1 | A | IDDel | IDDel, ASDDel, EpilepsyDel | 21,850,000–22,374,785 | 525 | 13 | Cooper et al. 201119, Girirajan et al. 201039 |
| 16p13.11 | A | IDB, SchizophreniaDup, EpilepsyDel | IDB, SchizophreniaDup, EpilepsyDel | 14,700,000–16,770,000 | 2,070 | 27 | Hannes et al. 200943, Mefford et al. 200941, Cooper et al. 201119, Kaminsky et al. 201120, Ingason et al. 2011b49, de Kovel et al. 201050, Heinzen et al. 201053, Meffford et al. 20108 |
| 16p13.2 (AK057657, C16orf72) | B | SchizophreniaDup | SchizophreniaDup | 9,090,000–9,120,000 | 30 | 2 | Levinson et al. 201114 |
| 16q22.1 | C | SchizophreniaDel | SchizophreniaDel | 68,743,639–68,770,545 | 27 | 4 | Glessner et al. 201013 |
| 17p12 | C | SchizophreniaDel | SchizophreniaDel | 14,048,304–15,357,533 | 1,309 | 9 | Kirov et al. 200925 |
| 17p12-p11.2 (Potocki-Lupski/Smith-Magenis Syndromes) | A | IDB | IDB, ASDDup, EpilepsyB | 16,650,000–20,420,000 | 3,770 | 113 | Cooper et al. 201119, Kaminsky et al. 201120 |
| 17p13.3-p13.2 (Miller-Dieker Syndrome) | C | IDDel | IDDel, ASDDel, EpilepsyDel | 500,000–1,300,000 | 800 | 13 | Cooper et al. 201119 |
| 17q12 | A | IDB, ASDDel, SchizophreniaDel | IDB, ASDB, SchizophreniaDel, EpilepsyB | 31,893,783–33,277,865 | 1,384 | 20 | Moreno-De-Luca et al. 201037, Cooper et al. 201119, Kaminsky et al. 201120 |
| 17q21.3 | A | IDDel | IDDel, ASDDel, EpilepsyDel | 41,060,000–41,650,183 | 590 | 10 | Cooper et al. 201119, Kaminsky et al. 201120 |
| 18q12.3 (LOC284260) | B | SchizophreniaDel | SchizophreniaDel | 38,310,567–38,311,765 | 1 | 1 | Glessner et al. 201013, |
| 18q21.31(KIAA0439, NEDD4L, NEDL3) | B | SchizophreniaDup | SchizophreniaDup | 53,860,000–54,220,000 | 360 | 3 | Levinson et al. 201114 |
| 19q13.33(CLEC11A, SHANK1, SYT3) | C | ASDDel | ASDDel | 55,808,307–55,935,995 | 128 | 3 | Sato et al. 201238 |
| 22q11 (Velocardiofacial/DiGeorge Syndrome) | A | IDB, ASDDup, SchizophreniaDel | IDB, ASDB, SchizophreniaDel, EpilepsyB | 17,400,000–18,676,130 | 1,276 | 40 | Cooper et al. 201119, Kaminsky et al. 201120, Pinto et al. 201036, ISC 200823, Levinson et al. 201114, Vacic et al. 201115, Kirov et al. 200925, Glessner et al. 201013, Mulle et al. 201046, Grozeva et al. 201217 |
| 22q11.2 | A | IDDel | IDDel, ASDDel, EpilepsyDel | 20,240,000–21,980,000 | 1,740 | 31 | Cooper et al. 201119 |
| 22q13 (Phelan-Mcdermid Syndrome) (SHANK3) | B | IDDel | IDDel, ASDDel, EpilepsyDel | 49,460,000–49,520,000 | 60 | 1 | Cooper et al. 201119 |
| Xp22.1 (DDX53, PTCHD1) | C | ASDDel | IDDel, ASDDel, EpilepsyDel | 22,829,183–23,214,712 23,116,188–23,280,628 |
386 164 |
2 | Pinto et al. 201036 |
| Highly Recurrent CNV Regions with No Genes | |||||||
| CNV | Disorder | Coordinates | Coordinate Source | ||||
| 1q25.2 | ASDDup | 174,500,555–174,543,675 | 43 | 0 | Glessner et al. 200927 | ||
| 2p16.3 | ASDDel | 51,120,644–51,147,600 | 27 | 0 | Glessner et al. 200927 | ||
| 2p24.3 | ASDDup | 13,119,667–13,165,898 | 46 | 0 | Glessner et al. 200927 | ||
| 3p26.3 | ASDDel | 1,915,190–1,915,922 | 0.7 | 0 | Glessner et al. 200927 | ||
| 3q26.1 | SchizophreniaDel | 165,610,000–165,660,000 | 50 | 0 | Levinson et al. 201114 | ||
| 4p16.1 | SchizophreniaDel | 9,881,886–9,884,092 | 2 | 0 | Glessner et al. 201013 | ||
| 4q31.21 | ASDDup | 144,847,402–144,854,579 | 7 | 0 | Glessner et al. 200927 | ||
| 7q36.3 | SchizophreniaDup | 158,731,401–158,810,016 | 79 | 0 | Vacic et al. 201115 | ||
| 9q34.3 | SchizophreniaDel | 140,145,139–140,152,969 | 8 | 0 | Glessner et al. 201013 | ||
| 22q11 | ASDDup | 19,351,264–19,358,946 | 8 | 0 | Glessner et al. 200927 |
Note: Fifty-four loci, including 10 non-genic CNVs are listed. For each CNV, deletions (del), duplications (dup), and both (B) are noted for significant disorder(s). All coordinates in NCBI 36/hg 18 build. Studies in bold indicate that coordinates from these studies reported in table.
ASD = autism spectrum disorder; ID = intellectual disability.
Strict Criteria for CNV Assignment
The distribution of CNVs conferring susceptibility across the four disorders was characterized using Venn diagrams for CNV groupings and gene groupings. These data for strict criteria are shown in Figure 1 and Table S2, available online. ID had the largest grouping of CNVs out of all DSM disorders (27 of 54, 50% of CNVs) and these CNVs were the most gene rich (27 of 44, 61.4% of genic CNVs which encompassed 1,198 of 1,416 or 84.6% of total genes). As a consequence, one striking result of this study is that, based on gene distributions, all disorders were largely subsumed by the ID gene group even for the strict criteria (Figure 1A). This pattern was supported also when gene groupings were separated into those implicated by deletions or duplications (Figure 1B and 1C). With the distribution of genes across disorders using strict criteria, ID was associated to an even greater degree with 96.8% (244 of 252) of genes in ASD, 92.8% (386 of 416) of genes in schizophrenia, and 100% (111 of 111) of genes in epilepsy. Broad criteria again demonstrated high association with ID: 99.3% (933 of 940) of genes in ASD, 91.5% (303 of 331) of genes in schizophrenia, and 100% (1,008 of 1,008) of genes in epilepsy.
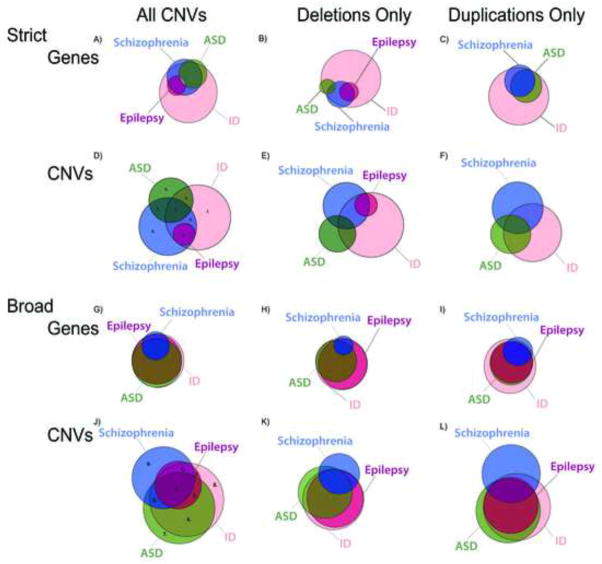
Figure 1.
Distribution of copy number variants (CNVs) and genes for autism spectrum disorder (ASD), intellectual disability (ID), schizophrenia, and epilepsy. Note: Figure 1A shows the distribution of genes contained within CNVs using strict criteria among all 4 disorders, as well as separates strict gene distribution by deletions (1B) and duplications (1C). Figure 1D shows the distribution of CNVs using strict criteria and is also separated into deletions (1E) and duplications (1F). Figure 1G shows the distribution of genes contained within CNVs for broad criteria, as well as deletions (1H) and duplications (1I). Broad CNV distribution is shown in Figure 1J and also categorized into deletions (1K) and duplications (1L). For Figure 1D, #1–8 represent strict CNV distributions that were analyzed in the pathway analysis (results in Table S4, available online), while in Figure 1J, #1–9 represent broad CNV distributions (results in Table S5, available online).
Visualization of the data with regard to the distribution of CNVs across disorders for strict criteria using CNV groupings also demonstrated a high degree of overlap between all disorders and ID, yet less so than when visualized by gene groupings. ID was associated with 46.2% (6 of 13) of CNVs associated with autism, 43.0% (9 of 21) of CNVs in schizophrenia, and 100% (3 of 3) of CNVs in epilepsy. Almost half of ASD CNVs (6 of 13, 46.2%) and all epilepsy CNVs (3 of 3, 100%) were also found in schizophrenia. There was no overlap among significant ASD and epilepsy CNVs using strict criteria. The distinction between the extent of overlap across disorders based on gene-content as compared to based on CNVs appears to reflect the genic content of CNVs implicated in each disorder under strict criteria. Specifically, the average number of genes per CNV for each disorder was as follows: ID—49 genes; ASD—28 genes; schizophrenia—32 genes; and epilepsy—47 genes. Notably, ID CNVs were significantly larger than ASD CNVs (p=.009) (Figure 2). Also CNVs were categorized as either deletion or duplication. As shown in Figure 2A, ID demonstrated a majority of deletions, 67% as compared 33% duplications, yet by contrast schizophrenia exhibited 57% deletions and 43% duplications. However, there were no statistically significant differences between deletions and duplications across DSM disorders.
Figure 2.
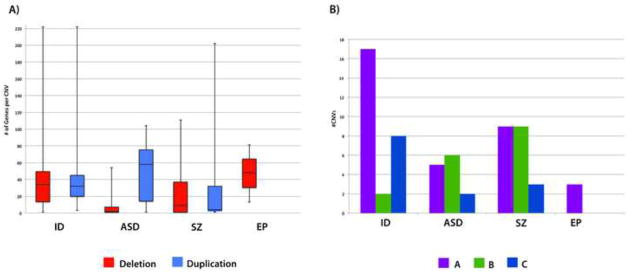
Figure 2A shows the number of genes per copy number variant (CNV) for all significant CNVs in autism spectrum disorder (ASD), intellectual disability (ID), schizophrenia (SZ), and epilepsy (EP) and is separated into deletions (red) and duplications (blue). Note: The median number of genes per CNV is shown in the center of each box and the whiskers indicate the range. Figure 2B shows the number of CNV categories (e.g. A, B, and C) for all 4 disorders. ID had a greater number of category A CNVs in both ASD (p=.004) and epilepsy (p<.001). Also, ID had a significantly greater number of category C CNVs for ASD (p=.04) and epilepsy (p=.002).
Despite the high degree of overlap that was apparent, disorders showed distinct profiles of type of CNV (Figure 2B). We categorized CNVs as category A if they showed highly stereotyped intervals likely based on non-allelic homologous recombination due to flanking segmental duplications, category B if the CNV involved single genes, or category C if the CNVs affected multiple genes and occurred in a given locus but with highly variable intervals (See Method and Figure S2, available online). There were significantly more category A CNVs in ID (n=17) than in either ASD (n=5, p=.004) and epilepsy (n=3, p<.001). Similar findings were noted for category C with ID (n=8) encompassing significantly more than ASD (n=2, p=.04) or epilepsy (n=0, p=.002). There were significantly more category B CNVs in schizophrenia (n=9) than in either ID (n=2, p=.03) and epilepsy (n=0, p<.001). In general, the patterns seemed to favor more category A and C in ID, and more category B (single gene CNVs) in autism and schizophrenia.
Disorder-specific CNVs and Broad Criteria for Assignment of Disorders
In addition to the high degree of sharing, each disorder (with the exception of epilepsy) has a set of genes implicated by CNVs that were disorder-specific under strict criteria: ID (16 CNVs, 719 genes; 59.3% of ID CNVs), ASD (5 CNVs, 8 genes, 38.5% of CNVs), and schizophrenia (10 CNVs, 28 genes, 47.6%) had a subset of genes from CNVs that were disorder-specific.
Schizophrenia was the disorder with the least amount of overlap with ID. Using strict criteria there were a total of ten CNVs that were specific to schizophrenia (Table 1). In general, these CNVs were smaller on average at approximately 379 kb and contained fewer genes, 2.8 genes on average. The majority of these schizophrenia-only CNVs were Category B (7 of 10).
Using the broad criteria, the extent of disorder-specific genes was substantially reduced. Using broad criteria, disorder-specific CNVs and genes were reduced in number: ID (2 CNVs, 40 genes; 6.7% of ID CNVs), ASD (3 CNVs, 5 genes, 10.7%), and schizophrenia (9 CNVs, 26 genes, 42.9%). Of note, given the high degree of overlap across disorders using broad criteria, schizophrenia was the disorder that stood out in greatest distinction from ID and other conditions.
PATHWAY ANALYSIS
Our systematic groupings of CNVs provided opportunities to investigate gene pathways that may be unique or shared across DSM diagnoses or within sub-groups of these disorders. Pathway analysis identified significant biological, cellular, and molecular gene processes, some of which were shared across disorders and others were unique to given sub-groupings. We first examined enriched gene pathways in gene groups associated with each of the four DSM diagnoses separately under strict criteria (e.g. ASD, ID, schizophrenia) (Figure 3A–C and Table S3, available online). Epilepsy was excluded because of the small number of CNVs under strict criteria, only three in total. The majority of significant findings across disorders pinpointed processes at the synapse. For example, synapse processes were significant for ASD (p<.001), ID (p=.007), and schizophrenia (p=.003). In addition, synaptic transmission was significant in ASD (p<.001) and schizophrenia (p<.001). Post-synaptic membrane was highly enriched in ASD (p=.003) and ID (p=.003). Extracellular ligand-gated ion channel activity was significant in ASD (p=.002), ID (p=.02), and schizophrenia (p=.007). Also, glycerophospholipid metabolic process was noted in ID (p=.02) and schizophrenia (p=.002).
Figure 3.
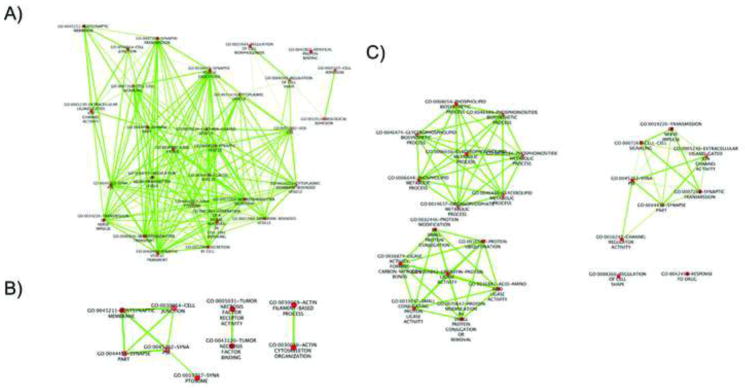
Enriched gene networks of significant pathway analysis results for autism spectrum disorder (ASD) (3A), intellectual disability (ID) (3B), and schizophrenia (3C). Note: ASD processes shown include vesicle and synaptic processes. Synaptic, necrosis factor, and actin filament-based processes are enriched in ID. Significant schizophrenia networks consist of phospholipid, ubiquitin, and synaptic processes.
Analysis of gene groupings based on DSM disorders also identified pathways that were specific to individual disorders. Significant ASD processes included exocytosis (p=.002), vesicle (p=.009), synaptic vesicle transport (p=.002), and regulation of neurotransmitter levels (p<.001). Notable ID processes identified actin cytoskeleton organization (p<.001), actin filament-based process (p<.001), tumor necrosis factor binding (p<.001), and regulation of acute inflammatory response (p=.02). Schizophrenia pathways were related to protein ubiquitination (p<.001) and ubiquitin-protein ligase activity (p<.001).
We then examined “co-morbid” groupings, for example, CNVs that are shared by two disorders such as ASD and ID, or CNVs found in 3 diagnostic groups (Table S4, available online). CNVs that only overlapped among ID-ASD CNVs were enriched for cell junction (p=.01), calcium-independent cell–cell adhesion (p=.03), and protein heterodimerization activity (p=.03). Significant CNVs that only overlapped with ID-schizophrenia identified endoplasmic reticulum membrane (p<.001) and glycerophospholipid biosynthetic process (p=.003). Significant results from “ID-ASD-schizophrenia” grouping included GABA receptor activity (p=.003), synaptic transmission (p=.006), and vesicle (p=.006). Interesting gene mechanisms emerged from the relatively “pure” groupings (Table S4, available online). For example, isolated ID processes included tumor necrosis factor binding (p<.001), positive regulation of apoptosis (p=.02), and actin cytoskeleton organization (p=.03). Channel regulator activity (p=.03) was the only significant process for isolated schizophrenia. Pathway analysis of gene groupings determined by broad criteria replicated many of the findings described above (Table S5, available online).
Finally, we conducted pathway analysis based on the category of the CNV (ie type A, B, or C). This showed distinct mechanisms for each CNV categories A, B, and C (Table S6, available online). Of note, category B (single gene) findings highlight synaptic processes, such as regulation of synaptic transmission (p=.001) and postsynaptic membrane (p=.02). Interesting, category A (recurrent intervals) identified Ras protein signal transduction as enriched (p=.03).
DISCUSSION
Classic studies heralded the importance of CNVs in neuropsychiatric disorders.54–58 Explosive progress in this area resulted from microarray methods that permitted genome-wide discovery of CNVs in large population samples.59–61 While evidence for association of large, highly recurrent CNVs with disease represents among the strongest findings in psychiatric genetics, this result has to contend with two challenges: 1) each CNV generally contains numerous genes; and 2) there is a high degree of loci sharing across disorders. The present study seeks to address these challenges through a novel and highly systematic approach to establishing the distribution of CNVs (deletions and duplications separately) as susceptibility factors across common disorders of cognitive development. The most prominent finding of our study was the high extent to which all disorders are subsumed by ID. Even under strict criteria, 96% of all genes identified by CNVs were associated with ID and greater than 90% of genes for the other disorders were also found in ID. In addition to this contribution, our study has elucidated mechanisms that are enriched in a given disorder or may be shared by disorders.
There are very few previous studies that have taken this sort of approach to studying CNVs in DSM diagnoses. Crespi et al.62 examined 7 highly recurrent CNVs (all included in the present analysis) implicated in ASD and schizophrenia to determine the genetic relationship between the two psychiatric disorders. The degree of overlap supported either a (1) diametric hypothesis whereby ASD and schizophrenia are on diametric ends of the diagnostic spectrum or (2) an overlapping hypothesis whereby the two disorders share some overlapping genetic risk. An alternative “subsumed” hypothesis, where ASD is a sub-category of schizophrenia was ruled out. With regard to autism and schizophrenia, our results also largely rule out the subsumed hypothesis, and are most in agreement with an overlapping hypothesis. However, with regard to ID and the remaining disorders, a subsumed hypothesis is most consistent. This is particularly so for autism and epilepsy. While schizophrenia stood out the most from ID, under broad criteria schizophrenia’s relationship with ID would be most consistent with a subsumed model as well. However, the schizophrenia specific CNVs should be interpreted with caution as some have been found thus far only in a single study. The interpretation of these findings suggests that ID may represent a disorder with the broadest possibilities of genetic mechanisms and susceptibility pathways, and the other disorders appear to have more restricted susceptibilities.
The results presented here also strongly implicate gene networks that regulate synaptic function, which have been identified in previous studies, and this was found here across all disorders. A recent gene-set enrichment analysis of de novo CNVs in schizophrenia found synaptic processes to be highly enriched.63 Specifically postsynaptic gene networks, including the NMDAR complex, were significant. A pathway analysis of rare de novo CNVs in ASD was recently conducted using a network-based analysis of genetic associations (NETBAG).64 Enriched gene networks identified processes related to actin network dynamics and reorganization, synaptogenesis, axonogenesis, cell–cell adhesion, small GTPase signaling, and neurite development. One GWAS ASD study conducted a pathway analysis of rare CNVs (deletions only as they were significant over controls) and identified cell proliferation, cell projection, cell motility, GTPase/Ras signaling, and kinase activity/regulation.36 The same study analyzed ASD/ID genes and found enriched processes related to microtubule cytoskeleton, glycosylation, and central nervous system (CNS) development/adhesion. Our results support these findings and demonstrate that cytoskeleton organization may be more associated with the ID phenotype.
While there were notable overlapping gene networks, the pathway analysis also identified more distinct disorder-specific processes. For example, processes related to actin cytoskeleton organization and tumor necrosis factor were preferentially enriched in ID. Ubiquitin processes were highly associated with schizophrenia. Category B CNVs have the ability to pinpoint individual genes and taken as a group, these genes were enriched for processes related to synapse/postsynaptic density and enriched in autism and schizophrenia relative to ID. Caution would be warranted in interpreting these studies of Catergory B CNVs if the studies reviewed had chosen these as candidate genes based on function; however, each of the Category B genes found in this study has been identified at some point in genome-wide studies. Category C also identified synaptic processes, however, not to the degree of Category B. Notably, Category A (those CNVs with stereotyped intervals) identified the Ras signaling pathway as an enriched gene class. Indeed, the genes involved in a number of well-known monogenic ID syndromes, such as Tuberous sclerosis, the diverse syndromes resulting from PTEN mutations and others, are well known to interact with the Ras pathway, and this pathway has been suggested previously to play a role at least in the pathophysiology of autism.65
The strength of the current study is that we have provided a novel analysis of CNV distribution across disorders which is critical to understanding the nosology and pathophysiology of neuropsychiatric disorders, especially given the high degree of comorbidity. There are several limitations with the current analysis. The broad CNV overlap data are likely not complete as not all studies detail secondary medical/psychiatric diagnoses and often such diagnoses are difficult to make in the setting of a primary disorder. While these data may be incomplete, the broad CNV distribution analysis does have a high degree of overlap already. In addition, as described, we found fewer studies in the area of epilepsy so these data in particular may be incomplete. Also, we were not able to substantially compare the frequency of CNVs across disorders. This is hampered in particular because of potentially different ascertainment biases for each disorder. An example of a difference in ascertainment methods is that ID studies include data from clinical referrals, whereas CNV data are acquired for schizophrenia generally only in the research setting where it may be difficult to ascertain the most severely affected patients. These biases need to be considered carefully and in theory could have contributed to a broadening of the ID diagnostic group; however, we suspect that these potential biases are unlikely to impact the major findings of our study that emphasize the vast degree of loci sharing across these disorders. In follow-up studies to this one when sample sizes get even larger, there may be a dimension to this sort of analysis whereby the frequency of a given CNV in a disorder is considered as opposed to simply the categorical association alone which was the subject of the current analysis. Finally, the high degree of overlap across disorders is clear in this study for these most common among the rare, large CNVs; however, it is possible that as sample sizes reach hundreds-of-thousands, we may observe very rare CNVs (frequency approximately 1/10,000 or less), and subsequently the distribution of these very rare CNVs across disorders will need to be studied.
Genome-wide approaches to study CNVs are currently being applied to other psychiatric disorders such as ADHD and bipolar disorder. Further research is necessary to conclusively determine whether large, recurrent CNVs that confer susceptibility to these psychiatric disorders overlap, subsume, or are independent of the current models. Early results have shown mixed findings in terms of genetic burden of CNVs in the few studies emerging for these disorders. GWAS ADHD results did not find an increase in rare CNVs66 yet an increase was noted in large, rare CNVs.26 This finding has now been replicated in a very convincing CNV study in a large cohort of twins with attentional problems.67 Specific CNVs identified in ADHD include 15q13.368 and 16p13.1126 (especially significant in comorbid ADHD and ID) and these CNVs have both been found in ID, ASD, schizophrenia, and epilepsy. Bipolar disorder GWAS studies have been mixed in terms of CNV burden.69–71 McCarthy et al.16 conducted a meta-analysis and found 16p11.2 (specifically duplications) highly enriched in bipolar disorder, which has also been identified in ID, ASD, schizophrenia, and epilepsy. However, additional studies have not identified any significant specific CNVs.69, 71 Overall, our studies strongly suggest that, in general, there is a little evidence that single CNVs confer susceptibility to individual disorders. Instead it seems as if individual CNVs large confer susceptibility to a broad array of disorders. These results lead to the question: As CNVs confer susceptibility to neuropsychiatric disorders (a fact that is now unequivocal), how is disorder specificity established? Is it stochastic, dependent on interaction with other genetic and/or environmental factors? Indeed, one recent study has demonstrated that approximately 10% of people with ID and a single large CNV may have a second large CNV.72 This study suggests at least two possible hypotheses with regard to genetics. First, that disorder specificity may not be encoded in individual CNVs but in combination of CNVs; or alternatively, second, that disorder specificity may be coded in the overall burden measures with distinct thresholds for different disorders. Of course, these hypotheses will await deep genotyping in large population cohorts with quality phenotypic assessments. Further, we also should point out, that while in general there is a low level of specificity, there are some exceptions. For example, there are some CNVs that generally lead to specific syndromes, for example, 7q11.2 deletions are almost always recognized in William syndrome and rarely recognized in other disorders.73 Also, in our data, schizophrenia may standout as the disorder with the least amount of overlap with ID. For example, 16p11.2 deletions and duplications are associated with ASD9–12 and ID18–20, 42, but only 16p11.2 duplications are associated with schizophrenia.13–16, 71
Through discovery of point mutations, novel genome-wide sequencing approaches will pinpoint individual genes in disorders of cognitive development. These new approaches provide further precision regarding susceptibility mechanisms as compared to CNV studies. However, early sequencing studies already suggest that the heterogeneity of individual molecular causes will remain broad.74, 75 Early studies have already implicated genetic loci shared by multiple disorders. For example, de novo SCN1A have been discovered in autism.76, 77 SCN1A mutations had been previously well-known for their role in epilepsy with and without intellectual disability.78–80 Further sequencing studies and examination of smaller CNVs may refine the rules regarding overlapping susceptibility. Nonetheless, large, highly recurrent CNVs will remain as important causes of developmental disorders, and indeed a follow-up study to this one may need to examine combinations of large CNVs or genetic factors across these disorders.
Supplementary Material
Acknowledgments
This research received support from a Career Award in Medical Science from the Burroughs Wellcome Fund (E.M.M.) and from the National Institute of Mental Health (NIMH) grant 1K23MH080954-05 (E.M.M.). Dr. Gamsiz is the first Brown University Alpert Medical School Translational Neuroscience Postdoctoral Fellow jointly sponsored by the Lifespan Research Institute, the Lifespan Division of Psychiatry, the Brown Institute for Brain Science, and the Norman Prince Neurosciences Institute. Funding for the project was provided by the Wellcome Trust.
Footnotes
Disclosure: Drs. Gamsiz and Morrow, Mr. Pescosolido, and Mr. Nagpal report no biomedical financial interests or potential conflicts of interest.
Supplemental material cited in this article is available online.
This study makes use of data generated by the DECIPHER Consortium. A full list of centers that contributed to the generation of the data is available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Mr. Matthew F. Pescosolido, Institute for Brain Science at Brown University. Developmental Disorders Genetics Research Program, Emma Pendleton Bradley Hospital, and the Warren Alpert School of Medicine at Brown University
Dr. Ece D. Gamsiz, Institute for Brain Science at Brown University. Developmental Disorders Genetics Research Program, Emma Pendleton Bradley Hospital, and the Warren Alpert School of Medicine at Brown University
Mr. Shailender Nagpal, Institute for Brain Science at Brown University
Dr. Eric M. Morrow, Institute for Brain Science at Brown University. Developmental Disorders Genetics Research Program, Emma Pendleton Bradley Hospital, and the Warren Alpert School of Medicine at Brown University
References
- 1.Guilmatre A, Dubourg C, Mosca AL, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Archives of General Psychiatry Sep. 2009;66(9):947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta AR, State MW. Recent advances in the genetics of autism. Biological Psychiatry. 2007;61(4):429–437. doi: 10.1016/j.biopsych.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Kooy RF. Distinct disorders affecting the brain share common genetic origins. F1000 biology reports. 2010;2:11. doi: 10.3410/B2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrow EM. Genomic copy number variation in disorders of cognitive development. J Am Acad Child Adolesc Psychiatry. 2010;49(11):1091–1104. doi: 10.1016/j.jaac.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends in Genetics. 2009;25(12):528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sisodiya SM, Mefford HC. Genetic contribution to common epilepsies. Current Opinion in Neurology. 2011;24(2):140–145. doi: 10.1097/WCO.0b013e328344062f. [DOI] [PubMed] [Google Scholar]
- 7.State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68(2):254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genetics May. 2010;6(5):e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders SJ, Hus V, Luo R, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11. 23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. The New England journal of medicine. 2008 Feb 14;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 11.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet Feb. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008 Feb 15;17(4):628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 13.Glessner JT, Reilly MP, Kim CE, et al. Strong synaptic transmission impact by copy number variations in schizophrenia. Proceedings of the National Academy of Sciences. 2010 Jun 8;107(23):10584–10589. doi: 10.1073/pnas.1000274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levinson DF, Duan J, Oh S, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. American Journal of Psychiatry. 2011;168(3):302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vacic V, McCarthy S, Malhotra D, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011 Mar 24;471(7339):499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarthy SE, Makarov V, Kirov G, et al. Microduplications of 16p11. 2 are associated with schizophrenia. Nature genetics. 2009;41(11):1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grozeva D, Conrad DF, Barnes CP, et al. Independent estimation of the frequency of rare CNVs in the UK population confirms their role in schizophrenia. Schizophrenia research Mar. 2012;135(1–3):1–7. doi: 10.1016/j.schres.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann-Gagescu R, Mefford HC, Cowan C, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genetics in medicine: official journal of the American College of Medical Genetics. 2010;12(10):641–647. doi: 10.1097/GIM.0b013e3181ef4286. [DOI] [PubMed] [Google Scholar]
- 19.Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nature genetics Sep. 2011;43(9):838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaminsky EB, Kaul V, Paschall J, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genetics in medicine: official journal of the American College of Medical Genetics Sep. 2011;13(9):777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenfeld JA, Coppinger J, Bejjani BA, et al. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J Neurodev Disord Mar. 2010;2(1):26–38. doi: 10.1007/s11689-009-9037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. The New England journal of medicine. 2008 Oct 16;359(16):1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008 Sep 11;455(7210):232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirov G, Grozeva D, Norton N, et al. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009 Apr 15;18(8):1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams NM, Zaharieva I, Martin A, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376(9750):1401–1408. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009 May 28;459(7246):569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nature Genetics Jan. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. American Journal of Human Genetics Dec. 2007;81(6):1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamain S, Quach H, Betancur C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nature Genetics. 2003;34(1):27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gauthier J, Champagne N, Lafrenière RG, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proceedings of the National Academy of Sciences. 2010;107(17):7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CP, Lin SP, Chern SR, et al. A de novo 7.9 Mb deletion in 22q13. 23 qter in a boy with autistic features, epilepsy, developmental delay, atopic dermatitis and abnormal immunological findings. European Journal of Medical Genetics. 2010;53(5):329–332. doi: 10.1016/j.ejmg.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 33.StataCorp. Stata Statistical Software: Release 11. College Station TSL; 2009. [Google Scholar]
- 34.Kestler HA, Muller A, Gress TM, Buchholz M. Generalized Venn diagrams: a new method of visualizing complex genetic set relations. Bioinformatics. 2005 Apr 15;21(8):1592–1595. doi: 10.1093/bioinformatics/bti169. [DOI] [PubMed] [Google Scholar]
- 35.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Pinto D, Pagnamenta AT, Klei L, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010 Jul 15;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno-De-Luca D, Mulle JG, Kaminsky EB, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010 Nov 12;87(5):618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato D, Lionel AC, Leblond CS, et al. SHANK1 Deletions in Males with Autism Spectrum Disorder. Am J Hum Genet. 2012 May 4;90(5):879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Girirajan S, Rosenfeld JA, Cooper GM, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nature genetics Mar. 2010;42(3):203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp AJ, Mefford HC, Li K, et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nature genetics Mar. 2008;40(3):322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mefford HC, Cooper GM, Zerr T, et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res Sep. 2009;19(9):1579–1585. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenfeld JA, Ballif BC, Torchia BS, et al. Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genetics in Medicine. 2010;12(11):694. doi: 10.1097/GIM.0b013e3181f0c5f3. [DOI] [PubMed] [Google Scholar]
- 43.Hannes FD, Sharp AJ, Mefford HC, et al. Recurrent reciprocal deletions and duplications of 16p13.11: the deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. Journal of medical genetics Apr. 2009;46(4):223–232. doi: 10.1136/jmg.2007.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rujescu D, Ingason A, Cichon S, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009 Mar 1;18(5):988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee Y, Mattai A, Long R, Rapoport JL, Gogtay N, Addington AM. Microduplications disrupting the MYT1L gene (2p25.3) are associated with schizophrenia. Psychiatric genetics Aug. 2012;22(4):206–209. doi: 10.1097/YPG.0b013e328353ae3d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulle JG, Dodd AF, McGrath JA, et al. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet. 2010 Aug 13;87(2):229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Q, Li T, Zhao X, et al. Rare CNVs and Tag SNPs at 15q11.2 Are Associated With Schizophrenia in the Han Chinese Population [published online February 8, 2012] Schizophrenia bulletin. 2012 doi: 10.1093/schbul/sbr197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingason A, Kirov G, Giegling I, et al. Maternally derived microduplications at 15q11-q13: implication of imprinted genes in psychotic illness. American Journal of Psychiatry. 2011;168(4):408–417. doi: 10.1176/appi.ajp.2010.09111660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingason A, Rujescu D, Cichon S, et al. Copy number variations of chromosome 16p13.1 region associated with schizophrenia. Mol Psychiatry. 2011;16(1):17–25. doi: 10.1038/mp.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Kovel CGF, Trucks H, Helbig I, et al. Recurrent microdeletions at 15q11. 2 and 16p13. 11 predispose to idiopathic generalized epilepsies. Brain. 2010;133(1):23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dibbens LM, Mullen S, Helbig I, et al. Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance. Human Molecular Genetics. 2009 Oct 1;18(19):3626–3631. doi: 10.1093/hmg/ddp311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helbig I, Mefford HC, Sharp AJ, et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nature genetics Feb. 2009;41(2):160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinzen EL, Radtke RA, Urban TJ, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. American Journal of Human Genetics. 2010 May 14;86(5):707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu B, Roos JL, Levy S, Van Rensburg E, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nature genetics. 2008;40(7):880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 55.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455(7215):919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 56.Ledbetter DH, Riccardi VM, Airhart SD, Strobel RJ, Keenan BS, Crawford JD. Deletions of chromosome 15 as a cause of the Prader–Willi syndrome. New England Journal of Medicine. 1981;304(6):325–329. doi: 10.1056/NEJM198102053040604. [DOI] [PubMed] [Google Scholar]
- 57.Yan WL, Jacobsen LK, Krasnewich DM, et al. Chromosome 22q11. 2 interstitial deletions among childhood-onset schizophrenics and “multidimensionally impaired”. American Journal of Medical Genetics. 1998;81(1):41–43. [PubMed] [Google Scholar]
- 58.Karayiorgou M, Morris MA, Morrow B, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proceedings of the National Academy of Sciences. 1995;92(17):7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sebat J, Lakshmi B, Malhotra D, et al. Strong association of de novo copy number mutations with autism. Science. 2007 Apr 20;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sebat J. Major changes in our DNA lead to major changes in our thinking. Nature genetics. 2007;39:S3–S5. doi: 10.1038/ng2095. [DOI] [PubMed] [Google Scholar]
- 61.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 62.Crespi B, Stead P, Elliot M. Comparative genomics of autism and schizophrenia. Proceedings of the National Academy of Sciences. 2010 Jan 26;107 (Suppl 1):1736–1741. doi: 10.1073/pnas.0906080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirov G, Pocklington A, Holmans P, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Molecular Psychiatry. 2011;17(2):142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70(5):898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008 Oct 31;135(3):401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 66.Elia J, Gai X, Xie H, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Molecular Psychiatry. 2009;15(6):637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ehli EA, Abdellaoui A, Hu Y, et al. De novo and inherited CNVs in MZ twin pairs selected for discordance and concordance on Attention Problems. European journal of human genetics: EJHG Oct. 2012;20(10):1037–1043. doi: 10.1038/ejhg.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams NM, Franke B, Mick E, et al. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13. 3. American Journal of Psychiatry. 2012;169(2):195–204. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malhotra D, McCarthy S, Michaelson JJ, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72(6):951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang D, Cheng L, Qian Y, et al. Singleton deletions throughout the genome increase risk of bipolar disorder. Molecular Psychiatry. 2008;14(4):376–380. doi: 10.1038/mp.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grozeva D, Kirov G, Ivanov D, et al. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Archives of General Psychiatry. 2010 Apr 1;67(4):318–327. doi: 10.1001/archgenpsychiatry.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Girirajan S, Rosenfeld JA, Coe BP, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. The New England journal of medicine. 2012 Oct 4;367(14):1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pober BR. Williams-Beuren syndrome. The New England journal of medicine. 2010 Jan 21;362(3):239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 74.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012 May 10;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neale BM, Kou Y, Liu L, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012 May 10;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012 May 10;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Roak BJ, Deriziotis P, Lee C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature Genetics. 2011;43(6):585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Catarino CB, Liu JYW, Liagkouras I, et al. Dravet syndrome as epileptic encephalopathy: evidence from long-term course and neuropathology. Brain. 2011;134(10):2982–3010. doi: 10.1093/brain/awr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dravet C. The core Dravet syndrome phenotype. Epilepsia. 2011;52:3–9. doi: 10.1111/j.1528-1167.2011.02994.x. [DOI] [PubMed] [Google Scholar]
- 80.Marini C, Scheffer IE, Nabbout R, et al. The genetics of Dravet syndrome. Epilepsia. 2011;52:24–29. doi: 10.1111/j.1528-1167.2011.02997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.