ABSTRACT
Wzx flippases are crucial for bacterial cell surface polysaccharide assembly as they transport undecaprenyl pyrophosphate-linked sugar repeat units from the cytoplasmic to the periplasmic leaflets of the inner membrane (IM) for final assembly. Our recently reported three-dimensional (3D) model structure of Wzx from Pseudomonas aeruginosa PAO1 (WzxPa) displayed a cationic internal vestibule and functionally essential acidic amino acids within transmembrane segment bundles. Herein, we examined the intrinsic transport function of WzxPa following its purification and reconstitution in phospholipid liposomes. WzxPa was capable of mediating anion flux, consistent with its cationic interior. This flux was electrogenic and modified by extraliposomal pH. Mutation of the above-mentioned acidic residues (E61, D269, and D359) reduced proton (H+)-modified anion flux, showing the role of these amino acid side chains in H+-dependent transport. Wzx also mediated acidification of the proteoliposome interior in the presence of an outward anion gradient. These results indicate H+-dependent gating and H+ uptake by WzxPa and allow for the first H+-dependent antiport mechanism to be proposed for lipid-linked oligosaccharide translocation across the bacterial IM.
IMPORTANCE
Many bacterial cell surface polysaccharides that are important for survival and virulence are synthesized at the periplasmic leaflet of the inner membrane (IM) using precursors produced in the cytoplasm. Wzx flippases are responsible for translocation of lipid-linked sugar repeat units across the IM and had been previously suggested to simply facilitate passive substrate diffusion. Through our characterization of purified Wzx in a reconstitution system described herein, we have observed protein-dependent intrinsic transport producing a change in the electrical potential of the system, with H+ identified as the coupling ion. These results provide the first evidence for coupled (i.e., secondary active) transport by these proteins and, in conjunction with structural data, allow for an antiport mechanism to be proposed for the directed transport of lipid-linked sugar substrates across the IM. These findings bring our understanding of lipid-linked polysaccharide transporter proteins more in line with the efflux pumps to which they are evolutionarily related.
Introduction
Pseudomonas aeruginosa is a ubiquitous Gram-negative bacterium and prevalent opportunistic pathogen of compromised individuals with conditions as diverse as cancer, burn wounds, AIDS, and cystic fibrosis (1). As with many infectious agents, its virulence is directly and indirectly modulated by cell surface glycans, such as lipopolysaccharide (LPS), in the outer membrane (OM) (2). LPS is composed of a proximal lipid A membrane anchor, a medial core oligosaccharide (OS) linker, and a distal O-antigen (O-Ag) polymer. Heteropolymeric O-specific (B-band) O-Ag is the predominant surface antigen, the presence of which affects a range of virulence phenotypes, including motility, type III effector secretion, and the modulation of host complement (2).
B-band O-Ag is synthesized via the Wzx/Wzy-dependent assembly pathway (3, 4). Following trisaccharide O-Ag repeat (O-unit) synthesis on the lipid carrier undecaprenyl pyrophosphate (UndPP), O units are flipped from the cytoplasmic to the periplasmic leaflet of the inner membrane (IM) through the cationic lumen of Wzx (5–7) and polymerized by Wzy via a putative catch-and-release mechanism (8–10). O-Ag polymer length is regulated by Wzz1 and Wzz2 (10–12), followed by ligation to lipid A-core oligosaccharide by WaaL (13, 14) to yield a complete LPS molecule, which is exported to the OM via the Lpt pathway (15).
Wzx proteins have been identified in a wide range of Gram-negative and Gram-positive bacteria and in the Archaea (3, 16). These flippases are part of the polysaccharide transporter (PST) family, which is a part of the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Also a part of the MOP superfamily and closely phylogenetically related to PST proteins are the multidrug and toxin extrusion (MATE) family of IM efflux pumps (16), which display ion-dependent antiport activity (17). Data to elucidate the mechanism of Wzx function were lacking until a recent investigation in which our group characterized the functional importance of various amino acids in the context of a novel tertiary structure homology model for Wzx from P. aeruginosa PAO1 (WzxPa) (6), which was based on the X-ray crystal structure of the related MATE protein NorM from Vibrio cholerae O1 El Tor (NorMVc) (18). This has led to the proposition of antiport function for Wzx flippases (3, 6).
Herein, we have examined the intrinsic transport properties of WzxPa (17). Through liposome reconstitution of purified WzxPa, we measured anion flux from proteoliposomes in response to increasing external H+ (but not Na+) concentrations; this result was not observed in several mutant variants with previously demonstrated complementation deficiencies in genetic screens. Fluorometric analysis of reconstituted WzxPa revealed that it also mediated proteoliposome acidification in the presence of an outward anion gradient. Together, these findings demonstrate H+-mediated anion flux via WzxPa that supports a proposed antiport mechanism for O-unit translocation in P. aeruginosa PAO1.
RESULTS
Validation of a liposome reconstitution system for examining gating stimuli.
Examination of ion coupling in MOP superfamily proteins has typically been carried out using whole cells or spheroplasts derived from wild-type and mutant strains (19–23); as such, any confounding effects from other integral IM transport proteins still present in the cellular or spheroplast membrane cannot be discounted. To characterize the mechanism of WzxPa function in the absence of other transport proteins, WzxPa was first overexpressed and purified as a tobacco etch virus (TEV) protease-cleavable fusion to green fluorescent protein (GFP)-His8 (Fig. 1A). GFP fluorescence was exploited to optimize overexpression and detergent solubilization conditions (24); it also facilitated tracking of the protein through in-gel fluorescence detection (Fig. 1B).
FIG 1 .
Purification and liposome reconstitution of Wzx-GFP-His8. Key: ◂, Wzx-GFP-His8 monomer (predicted molecular mass = 74.6 kDa); ◂◂, Wzx-GFP-His8 dimer; ●, protein aggregate; *, cleaved GFP-His8. The identities of all protein bands were confirmed via gel excision, trypsin digestion, and matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry analysis. (A) SDS-PAGE of purified Wzx-GFP-His8. (B) In-gel fluorescence scan of gel in panel A. (C) Proteoliposomes (250 µl) sedimented in an ultracentrifuge, resuspended in 1× sample buffer, resolved via SDS-PAGE, and imaged via in-gel fluorescence scan. (D) Detection of TEV protease-cleaved GFP-His8 from intact and ruptured proteoliposomes for orientation determination. (E) Densitometry analysis of GFP-His8 fluorescence in panel D (n = 4). Results are statistically significant (Student’s t test, P = 0.0008). (F) Flux assay schematic (refer to Materials and Methods and Results for further details). (G) Sample I− flux assay trace. Purified Wzx-GFP-His8 was reconstituted in egg phosphatidylcholine (PC) liposomes (1:400 protein-to-lipid ratio by mass) in the presence of MOPS-KI and then gel filtered to switch the external buffer to MOPS-KGlu containing potential gating stimuli. Valinomycin (20 nM) was added (t = 0 s) to initiate flux. Proteoliposomes were ruptured by addition of 0.3% TX to ensure I− entrapment, as evidenced by the sudden spike in I− detected by the probe upon detergent treatment. The slope before valinomycin addition (cyan line) was subtracted from that after addition (magenta line) to calculate the adjusted rate of I− release (µM/s). The rate of probe saturation (green line) was used as an internal normalization control to compensate for dilution variation in different gel-filtered preparations. Samples of egg PC-alone liposomes were simultaneously prepared and analyzed to provide a baseline reference for I− leakage.
To create a defined environment, WzxPa was reconstituted in proteoliposomes through a recently described procedure (25) used to characterize the bacterial OM protein AlgE, required for alginate secretion in P. aeruginosa (26). TEV protease digestion of intact and disrupted proteoliposomes followed by fluorescence detection of cleaved GFP-His8 (Fig. 1D) revealed that ~20% of particles were protected from digestion in intact proteoliposomes (Fig. 1E); this suggests that these particles are oriented with the periplasmic face of WzxPa exposed on the surface of the proteoliposomes (facing the bath solution) and the cytoplasmic face present within the interior (see Fig. S1 in the supplemental material).
Based on the positively charged interior conduit identified in our homology model (6), we first tested the ability of liposome-reconstituted WzxPa to mediate anion flux (Fig. 1F). Potassium iodide (K+I−) was trapped inside proteoliposomes during reconstitution, followed by gel filtration to remove the external I− and introduce candidate transport-coupling ions. Flux of I− was monitored over time with an I−-sensitive electrode, first allowing for a baseline leakage rate to be established. We hypothesized that intralumenal positive charge would accumulate if the flux through an “open” WzxPa was anion selective, thereby limiting its electrodiffusion. In this case, addition of the K+ ionophore valinomycin would overcome this limitation, resulting in enhanced flux through an “open” WzxPa. Upon valinomycin addition, a robust increase in the I− flux rate was indeed observed (Fig. 1G), substantiating our claim that WzxPa mediates anion-selective flux. Even after extended time post-valinomycin addition, detergent-lysed proteoliposomes contained an abundance of I−, indicating that intraliposomal I− had not become limiting (Fig. 1G).
WzxPa-mediated I− flux is unaffected by Na+.
Most experimentally characterized MATE proteins have been found to utilize Na+ as the coupling ion during extrusion of charged and hydrophobic toxic compounds from inside the cell via antiport (17); in these investigations, Na+ at 20 mM was shown to elicit optimal MATE protein activity (27–30). To test the role of Na+ in the WzxPa transport function, we added sodium glutamate (Na+Glu−) to the bath containing WzxPa proteoliposomes, while maintaining the conditions in the assay as described previously. However, upon valinomycin addition, no significant I− flux was detected above background levels in the presence of Na+ for WzxPa proteoliposomes (see Fig. S2 in the supplemental material).
H+ titer affects WzxPa-mediated I− flux.
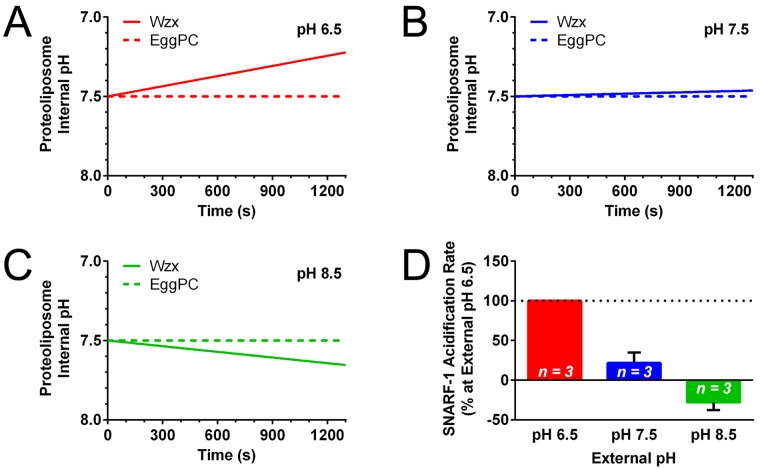
Antiport activity by a subset of MATE proteins has been shown to be coupled to H+ (17). To test the role of protons in WzxPa-mediated anion flux, the external H+ concentration surrounding WzxPa proteoliposomes was altered through gel filtration of proteoliposomes into morpholinepropanesulfonic acid (MOPS)-potassium glutamate (KGlu) buffer at pH 6.5, 7.5, or 8.5 to provide a 10-fold increase, unchanged, or 10-fold decrease in the H+ titer relative to the pH inside the proteoliposomes (pH 7.5), respectively. At external pH 6.5 (Fig. 2A and 2D), a significant I− flux increase was observed (~200%) compared to results for external pH 7.5 (Fig. 2B and 2D). Conversely, when less H+ was present externally (pH 8.5) than in the proteoliposome interior, significantly less I− release was detected (Fig. 2C and 2D). These results indicate that H+ is modifying halide flux through WzxPa. Moreover, the I− flux observed in response to valinomycin addition is consistent with the activity of WzxPa as an electrogenic transporter/channel.
FIG 2 .
Effect of pH on I− flux from Wzx-GFP-His8 proteoliposomes. Proteoliposomes were exchanged via gel filtration into MOPS-KGlu buffer at different pH values: pH 6.5 (A), pH 7.5 (B), or pH 8.5 (C). Valinomycin was added at t = 0. (D) Iodide release rate normalized to that observed for MOPS-KGlu buffer, pH 7.5 (n = 5). Compared against results at pH 7.5, the differences for pH 6.5 and pH 8.5 are both statistically significant (Student’s t test, P = 0.0303 and 0.0098, respectively).
Anionic residue mutants of WzxPa display compromised H+-dependent halide flux.
Based on previous mutagenesis and complementation studies by our group, the single substitutions E61A, D269A, and D359A yielded defective WzxPa variants unable to restore B-band O-Ag biosynthesis in a chromosomal wzx knockout mutant (6). From the tertiary structure homology model of WzxPa (6), these side chain carboxylates were present in α-helical transmembrane segment (TMS) bundles (rather than being positioned to face the interior vestibule). To independently confirm these positions, the FILM3 modeling approach was employed to use peptide fragment assembly and correlated mutation analysis (31) to produce a de novo tertiary structure model of WzxPa in the absence of the manual query template alignment biases potentially imposed during homology modeling (32). Comparison of the homology and FILM3 structures yielded analogous TMS packing and protein folds, with a template modeling (TM) score of 0.67296 calculated via the TM-align algorithm (a TM score of >0.5 indicates the same fold) (see Fig. S3 in the supplemental material) (33).
Equivalent positioning of the above-mentioned three residues in helical bundles was also observed (Fig. 3A). While the previously published WzxPa structural homology model presented the protein in a periplasm-open apo-form state, the FILM3 model displays a more inverted conformation open to the cytoplasm. Since proteins that undergo conformational changes require different sets of contacts to stabilize each state, models built using predicted contacts are likely to adopt an “average” conformation; this is due to the objective function of the program attempting to simultaneously satisfy these different contact sets. This further supports the existence of a cytoplasm-open state for WzxPa. Through positioning analogous to that of residues shown to bind the antiport coupling ion in NorMVc (D36, E255, and D371) and NorM from Neisseria gonorrhoeae (NorMNg; D41, E261, and D377) (34), similar roles were proposed for amino acids E61, D269, and D359 from WzxPa (6). Upon comparison with wild-type WzxPa via the I− flux assay, each mutant displayed a significant reduction in H+-dependent halide flux, confirming the roles of these residues in responsiveness to ion coupling (Fig. 3B and 3C).
FIG 3 .

Flux phenotypes for functionally defective Wzx mutants. (A) Positions of E61, D269, and D359 within the Wzx homology model (blue) (21) and FILM3-generated model (gray). (B) WT and mutant Wzx-GFP-His8 proteoliposomes in MOPS-KGlu, pH 7.5. (C) Iodide release rate normalized to that observed for the WT. Compared against results for the WT, differences for the E61A, D269A, and D359A mutants were statistically significant (Student’s t test, P < 0.0001). Differences between the mutants were not statistically significant (E61A versus D269A, P = 0.3959; E61A versus D359A, P = 0.9924; D269A versus D359A, P = 0.4150).
WzxPa mediates lumenal acidification of liposomes.
Proteoliposomes were reconstituted in the presence of the membrane-impermeant pH-sensitive dye 5 (and 6-)-carboxy-SNARF-1 (SNARF-1). Changes in pH result in a ratiometric shift in fluorescence intensities between the two emission peaks of the dye; higher acidity or alkalinity results in increased intensity of the 580-nm or 640-nm peak, respectively (35). To examine the utility of using SNARF-1 for monitoring internal changes in the proteoliposomal proton concentration, SNARF-1-loaded proteoliposomes were gel filtered into buffers at pH 6.5, 7.5, or 8.5 as previously described, followed by treatment with the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP) to artificially promote H+ transport across the membrane. Dual-wavelength fluorescence emission was monitored over time, with the 580-nm/640-nm ratio determined. Lumen acidification (i.e., faster 580-nm intensity increases) would manifest as a linear regression line with a positive slope; the steeper the slope, the faster the rate of lumen acidification. At external pH 6.5, CCCP addition resulted in significant 580-nm peak intensity increases, while at external pH 7.5, no significant deviation from 0 was detected. At external pH 8.5, when H+ was higher inside than outside the proteoliposomes, the 640-nm peak intensity increased faster, as H+ was removed from the proteoliposome lumen (see Fig. S4 in the supplemental material).
Next, we assessed changes in proteoliposomal proton flux mediated by WzxPa. Valinomycin was added to SNARF-1-loaded proteoliposomes (to enable continuous I− flux), and changes in SNARF-1 fluorescence were monitored (see Fig. S5 in the supplemental material). With an inward-directed H+ gradient (at external pH 6.5), a considerable increase in lumen acidity was observed (Fig. 4A and 4D). Upon reversal of the H+ gradient (at external pH 8.5), a net decrease in lumen acidity was observed; this is consistent with the second population of reconstituted proteins (which have their periplasmic faces exposed on the proteoliposome interior) removing H+ from within the proteoliposome interior (Fig. 4C and 4D). Given the requirement of valinomycin in the I− flux experiments to initiate halide flux in an electrogenic manner (Fig. 2), this would posit that H+ should be transported by WzxPa even in the absence of an H+ gradient. Consequently, in the presence of an internal and external pH of 7.5, proteoliposome lumen acidification was still detected (Fig. 4B and 4D) (albeit at a lower rate than observed for external pH 6.5), indicating WzxPa-mediated H+ uptake.
FIG 4 .
Fluorescence-based detection of H+ shifts inside proteoliposomes. The membrane-impermeant pH-sensitive dye carboxy-SNARF-1 was incorporated inside proteoliposomes during reconstitution, followed by gel filtration to alter the external buffer pH to 6.5 (A), 7.5 (B), or 8.5 (C). Displayed is a representative linear regression of the internal proteoliposome pH (calculated from the 580-nm/640-nm fluorescence emission ratio) over time after valinomycin addition and mixing. Net acidification of the proteoliposome interior would be due to H+ transit from the outside of the liposome, resulting in the intensity of the 580-nm peak increasing over time, yielding a linear regression fit with a positive slope. Conversely, loss of H+ from the proteoliposome interior would cause net alkalinization, resulting in the intensity of the 580-nm peak increasing over time, yielding a linear regression fit with a negative slope. (D) Rate of carboxy-SNARF-1 acidification normalized to external pH 6.5 (n = 3). All rate differences were statistically significant (P ≤ 0.05) as calculated using the Student t test: pH 6.5 versus pH 7.5, P = 0.0009; pH 6.5 versus pH 8.5, P < 0.0001; pH 7.5 versus pH 8.5, P = 0.0390.
DISCUSSION
The PST and MATE protein families are ubiquitous in bacteria and are the two most closely related groups in the MOP exporter superfamily (16). However, while MATE proteins (e.g., NorM) have been well studied, PST proteins (e.g., Wzx) have remained poorly characterized. NorM and related proteins utilize antiport mechanisms in which two solutes are transported in opposite directions across the IM (36). While the majority of MATE proteins characterized have been shown to use Na+ as the coupling ion, a subset have demonstrated H+-dependent antiport (17), including NorM (i.e., PmpM) from P. aeruginosa PAO1 (NorMPa) (20), PfMATE from the archaeon Pyrococcus furiosus (23), and eukaryotic hMATE1/2 from Homo sapiens (37, 38).
Recent X-ray crystallographic studies and molecular dynamics (MD) simulations of NorMNg and NorMVc, respectively, have revealed considerable internal TMS rearrangement upon coupling-ion binding, even in the absence of bound substrate, yielding a water-accessible pathway through the protein (18, 39, 40). Proton transport uncoupled from substrate transport has also been demonstrated for PfMATE (23). Given the structural and evolutionary analogies between these proteins and WzxPa (3, 6, 16), we tested the ability of known MATE coupling ions to induce such changes in the latter that would allow flux of a signal anion from the interior of proteoliposomes. Use of a defined and established reconstitution system (25, 26) allowed us to examine these characteristics in isolation and avoid any effects of other proteins that could influence the flux phenotype (3, 22, 23). WzxPa activity was found to respond to H+ (but not Na+), mirroring the phenotype observed for NorMPa and PfMATE (20, 23). Moreover, WzxPa was shown to mediate H+ uptake even in the absence of a directed H+ gradient, further supporting its capacity to catalyze movement of H+. The need for valinomycin treatment to register anion flux points to the electrogenic nature of the flippase with a proton: anion stoichiometry that is not 1:1. In addition, the observation that H+ uptake still occurs with symmetric pH gradients but an outward anion gradient provides further evidence for an antiport mechanism (36).
The 3D structural context of the E61, D269, and D359 WzxPa flippase functional groups suggested functional roles similar to those of the NorMVc residues D36, E255, and D371 (34, 41). Consistent with those in NorMVc (40), the functional defects of these WzxPa residues corresponded to compromised coupling-ion responsiveness. The only other Wzx protein characterized via site-directed mutagenesis is that from Escherichia coli O157:H7 (WzxEc), for which the anionic residues D85 and D326 were shown to be functionally important (42). Through the use of in silico and in vitro techniques (43), a topological map of WzxEc was also proposed (44). Comparison of the WzxEc topology map and WzxPa homology model (6) indicated putative positional equivalence of these WzxEc residues to E61 and D269 from WzxPa (3). While H+ is the coupling ion for WzxPa and NorMPa, NorM (YdhE) from E. coli utilizes Na+ to drive antiport activity (45); as such, if WzxEc were to display Na+-dependent gating, it might point to the conserved use of a particular coupling ion between related transport proteins in a given species.
Residues R59, Y60, F139, R146, and K272 from WzxPa are also functionally important. Putative substrate interaction roles for these residues have been highlighted, as follows: (i) the WzxPa O-unit trisaccharide substrate has a net negative charge (2), (ii) aromatic side chains are often found in sugar- and carbohydrate-binding sites in proteins (46, 47), and (iii) most are in direct contact with the internal flippase cavity (6), similar to the case of substrate-binding D41, S61, F265, Q284, S288, D355, and D356 from NorMNg (39).
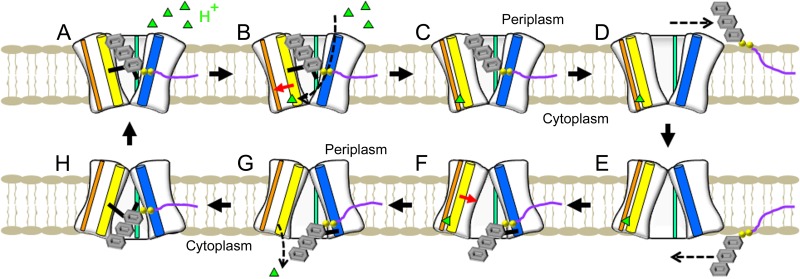
Taking into consideration knowledge derived from the latest data in the Wzx and NorM literature, including X-ray crystallography (23, 18, 39), mutagenesis (6, 23, 39, 41), modeling (6), biochemical studies (4), MD simulations (40), and phylogenetic analysis (16), as well as the proteoliposomal I− and H+ flux results obtained in this study, we have adapted the MATE efflux model described by Lu et al. (39) to propose the following model for the mechanism of O-unit flipping across the IM. In a periplasm-facing O-unit-bound WzxPa (Fig. 5A; see also Fig. S6B in the supplemental material), H+ is bound by the D269–D359 (TMS8–TMS11) dyad via recruitment by E61 (TMS2) (Fig. 5B; see also Fig. S6B), causing K272 (TMS8) and TMS11 to shift away from TMS1 and toward TMS10 (Fig. 5C; see also Fig. S6C). This allows the flipped O unit to disengage F139 (TMS4) and laterally diffuse into the outer leaflet of the IM via the exit portal formed by TMS1 and TMS9; this opening also facilitates transit of the Und moiety between the IM leaflets (Fig. 5D; see also Fig. S6D). The H+-bound O-unit-free WzxPa reverts to a cytoplasm-facing inverted conformation (Fig. 5E; see also Fig. S6E) in which another UndPP-linked O unit enters the protein from the TMS1–TMS9 entryway and is loaded via stereospecific interaction with R59 and Y60 (TMS2) (Fig. 5F; see also Fig. S6F). Binding of the new O unit induces a reset in TMS8 and TMS11 positioning (Fig. 5G), thus weakening the D269–D359 dyad H+ binding and resulting in H+ release into the cytoplasm (Fig. 5H; see also Fig. S6G). WzxPa then relaxes to a periplasm-facing state, resulting in transbilayer flipping of the O unit across the IM, restarting the cycle (Fig. 5A; see also Fig. S6H).
FIG 5 .
Proposed mechanism of Wzx function. (A) O-unit-bound Wzx lacking a coupling ion. (B to D) Binding of H+ (green triangle) induces movement of TMS8 and TMS11 away from the vestibule (B), disengaging retention interactions (C) and allowing for lateral diffusion of the flipped UndPP-linked O unit to the outer leaflet of the IM (D). (E and F) Wzx transitions to an inverted H+-bound structure open to the cytoplasm (E), allowing for loading of a new UndPP-linked O unit from the cytoplasmic leaflet of the IM (F). (G) O-unit binding triggers movement of TMS8 and TMS11 toward the vestibule, weakening dyad binding of H+ and resulting in H+ release to the cytoplasm. (H) Inverted O-unit-bound Wzx, lacking a coupling ion, transitions back to a periplasm-facing conformation, resulting in flipping of the UndPP-linked O unit between IM leaflets. Cylinder legend: blue, TMS1 and TMS2 combined; cyan, TMS4; yellow, TMS8 and TMS11 combined; orange, TMS10.
In conclusion, these findings provide the basis for interpreting the structure and function of Wzx flippases, a widespread yet poorly understood class of proteins.
MATERIALS AND METHODS
DNA manipulations.
Site-directed mutagenesis, transformation, and plasmid recovery were carried out as previously described (6, 9) on the pWaldo-wzx-GFP plasmid encoding Wzx-GFP-His8 (4) to obtain plasmids encoding E61A, D269A, and D359A mutant constructs using previously designed oligonucleotide primers (6). A tobacco etch virus (TEV) protease cleavage site is present between the terminus of Wzx and the start of GFP-His8 (24).
Overexpression and purification of Wzx-GFP-His8.
E. coli BL21(DE3) was transformed with the wild-type (WT) and mutant pWaldo-wzx-GFP constructs via heat shock. Transformants were grown on lysogeny broth (LB) agar and in LB broth supplemented with kanamycin (50 µg/ml). Broth cultures were inoculated to an initial optical density at 600 nm (OD600) of 0.01, and grown with shaking (220 rpm, 37°C) until an OD600 of 0.25 to 0.35 was reached. Cultures were shifted to 30°C for 30 min and then induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Roche) for 5 to 7 h (30°C, 220 rpm) (24).
Cell pellets were resuspended in 25 mM MOPS and 75 mM potassium iodide (pH 7.5) (MOPS-KI) buffer, with 1× Complete protease inhibitor lacking EDTA (Roche) and lysed via two passages (20,000 lb/in2) through an Emulsiflex C3 high-pressure homogenizer (Avestin). Unlysed cells and crude debris were sedimented at 12,000 × g (30 min, JA25.50 rotor) in a Beckman superspeed centrifuge, followed by sedimentation at 120,000 × g (1.25 h, Ti70 rotor) in a Beckman ultracentrifuge to obtain the membrane pellet. This was resuspended in MOPS-KI with 1× Complete lacking EDTA, 0.2% lauryldimethylamine-N-oxide (LDAO) detergent, and 20 mM imidazole via three passages in a Dounce homogenizer. Samples were sedimented in an ultracentrifuge to obtain the solubilized membrane fraction.
Solubilized membrane samples were mixed with Talon affinity resin (Clontech) at a ratio of 5:1, incubated on a Nutator platform at 4°C (2 h), and then packed in a disposable purification column at 4°C (Bio-Rad). Samples were washed and eluted with increasing concentrations of imidazole in MOPS-KI buffer containing 0.1% LDAO. Elution fractions were pooled and concentrated in a 30-kDa cutoff Vivaspin column (Sartorius) in a Sorval centrifuge (3,000 × g). MOPS-KI (0.1% LDAO, no imidazole) was added and spun through the sample thrice. Protein concentrations of purified samples were measured using the detergent-compatible bicinchonic acid assay kit (Thermo) in a 96-well plate using a BMG FluoStar Optima microplate reader. Purified proteins and proteoliposomes were heated in Laemmli buffer (37°C, 30 min) and resolved via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) along with ECL Plex fluorescent Rainbow marker (GE Healthcare). In-gel fluorescence scans were obtained with a Typhoon 9410 imager and ImageQuant TL software (GE Healthcare). The blue 2 (488), green (532), and red (633) excitation lasers and the 526SP, 580BP, and 670BP emission filters were used to detect GFP, Cy3, and Cy5 fluorescence, respectively.
Liposome reconstitution of WT and mutant Wzx-GFP-His8 constructs.
Egg phosphatidylcholine (PC) (200 µg) in chloroform (Avanti) was dried in a glass vial under argon gas. Dried egg PC was resuspended in MOPS-KI (0.1% LDAO) via vortex mixing (10 s) twice and sonication in a chilled bath (5 s) thrice. Protein sample volume was added to the resuspended lipid at a protein-to-lipid ratio of 1:400 by weight such that the final mixture volume remained 1 ml. Protein-lipid mixtures were extruded 15 times through an Eclipse 25 G × 1½ in. needle (BD Biosciences) and incubated on ice for 1 h. Protein-lipid mixtures were passed down a detergent-removal spin column (Thermo), which had been prepared using MOPS-KI lacking detergent according to the manufacturer’s instructions. All reconstitutions were performed in pH 7.5 buffer unless otherwise specified, resulting in an internal proteoliposome pH of 7.5. For fluorimetry analysis of intraliposomal pH changes, the pH-sensitive dye (35) 5 (and 6-)-carboxy-seminaphthorhodafluor 1 (SNARF-1) (100 µM; Life Technologies) was added to the protein-lipid mixture immediately before detergent removal. To ensure maximal membrane and protein integrity, liposome samples were always freshly prepared and used immediately for gel filtration and downstream analyses; they were never frozen and thawed.
Determination of Wzx-GFP-His8 orientation in liposomal membrane.
Proteoliposomes (100 µl) were aliquoted in triplicate, with the first left untreated. Thirty units of TEV protease (Invitrogen) were added to the second and third aliquots, with the latter sample lysed with Triton X-100 (TX) (0.5%; Sigma). All three samples were left shaking in the dark at room temperature (9 h). Insoluble components were sedimented in a tabletop Beckman ultracentrifuge (MLA130 rotor, 120,000 × g, 30 min, 4°C). The supernatant containing cleaved GFP-His8 was mixed with 2× SDS-PAGE sample buffer and heated at 37°C (30 min). Samples (40 µl) were resolved on a 12% SDS-PAGE gel and imaged using a Typhoon scanner as described above. Densitometry of GFP-His8 fluorescence was performed using ImageJ software. Results were normalized to the intensity observed in the presence of TEV protease and TX. All statistical analyses were carried out using GraphPad Prism 6 software.
Proteoliposome-based I− and H+ flux assays for Wzx-GFP-His8.
Iodide efflux from proteoliposomes was measured as previously described for the eukaryotic cystic fibrosis transmembrane conductance regulator (25) and the prokaryotic outer-membrane beta-barrel AlgE required for alginate secretion (26). In brief, I− was removed from the extraliposomal buffer via triplicate gel filtration passage through columns packed with preswelled Sephadex G-50 resin (GE Healthcare); this resin was saturated in I−-free buffer containing 20 mM MOPS and 75 mM potassium glutamate (MOPS-KGlu) external solution. Sephadex G-50 resin was preswelled with MOPS-KGlu buffer at different pH levels (adjusted with concentrated potassium hydroxide to pH 6.5, 7.5, or 8.5) or supplemented with 20 mM sodium glutamate (pH 7.5) to test the effects on I− flux of altered extraliposomal pH or the presence of extraliposomal sodium, respectively. The potassium-selective ionophore valinomycin was added (10 nM) to equilibrate changes in membrane potential generated by I− leakage through Wzx-GFP-His8. The protonophore carbonylcyanide m-chlorophenylhydrazone (CCCP) was added (20 µM) to shuttle H+ across the liposomal membrane. For all pH values, I− flux readings were obtained within the linear range of the I− electrode (see Fig. S7 in the supplemental material).
Iodide efflux measurements were carried out as previously described (25, 48, 49). External I− concentrations were continuously monitored with an I−-selective electrode (Lazar Research Laboratories) connected to a Digidata 1320A data acquisition system running Clampex 8 software (Axon Instruments). Fluorimetry analyses were carried out in a quartz cuvette (Hellma) using a QuantaMaster 80 fluorimeter (Photon Technology International). SNARF-1 was excited at 514 nm, with emission from the acid-sensitive and base-sensitive peaks monitored at 580 nm and 640 nm, respectively, with real-time correction. Valinomycin (10 nM) or CCCP (20 µM) was added (t = −300 s) to the gel-filtered proteoliposomes, and after a final mix (t = 0 s) the run was monitored for an extended period to observe shifts in the relative intensities of the 580-nm and 640-nm emission peaks. At each time point, the 580-nm/640-nm fluorescence emission ratio was determined and used to calculate the internal proteoliposome pH. These data were fit to a linear regression line of the ratio plotted over time.
De novo structure modeling.
A WzxPa multiple sequence alignment (MSA) was generated using the software program jackhmmer (50). Three search iterations were performed with an E value inclusion threshold of 10−6 against the UNIREF100 data bank (51), with duplicate rows and columns containing gaps in the target sequence removed, resulting in a final MSA of 22,241 aligned sequences. The program PSICOV (52) was then used to generate a list of predicted contacts from the MSA along with precision estimates for each contact. Secondary structure and TMS predictions generated using the PSIPRED (53) and MEMSAT-SVM (54) programs were combined using a simple consensus scoring scheme ensuring that the predicted topology was enforced. Using the list of predicted contacts where estimated precision was >0.5 and the consensus secondary structure as inputs, 200 models were generated using the FILM3 de novo structure prediction method (31), 100 with z coordinate constraints derived from topology prediction and 100 without. The lowest-energy model was then identified using the standard FILM3 objective function, and the 100 lowest-energy models were fitted to it by rigid body superposition. The mean pairwise TM score (55) was calculated for all models in the resulting ensemble, producing a value of 0.25, which indicates good homogeneity among the candidate structures. In development of FILM3, this value showed a strong correlation with the observed TM score of the final model (Pearson’s r = 0.77), allowing the TM score of the Wzx model to be predicted using linear regression as 0.59, therefore indicating a model with high probability of a correct fold. The FILM3 combinatorial refinement protocol was applied to the ensemble, but the resulting model displayed an implausible topology. We therefore selected the lowest-energy structure as the final model, which we refined using the software program MODELLER (24) to produce reasonable loop and side chain conformations.
SUPPLEMENTAL MATERIAL
Schematic of Wzx-GFP-His8 orientation determination assay. Proteoliposomes containing Wzx-GFP-His8 are digested with TEV protease in the presence and absence of Triton X-100 detergent (TX100). In the absence of detergent-mediated proteoliposome permeabilization, only the GFP-His8 exposed on the outside should be amenable to protease cleavage. In the presence of detergent, all GFP-His8, regardless of its original inside/outside position, should be amenable to protease cleavage; cleavage from this treatment is set as the total fluorescence to which other values are normalized. Comparison of fluorescence readings from the intact and detergent-ruptured sample sets allows for the proportional determination of Wzx–GFP-His8 proteins in a given proteoliposome preparation that are oriented with their N and C termini facing the inside of the proteoliposome. Download
Effect of Na+ on I− flux. Valinomycin was added at t = 0. (A) Wzx-GFP-His8 proteoliposomes in standard Na+-free MOPS-KGlu buffer. (B) Wzx-GFP-His8 proteoliposomes in MOPS-KGlu buffer supplemented with 20 mM Na+. (C) Iodide release rate normalized to that observed for standard MOPS-KGlu buffer (n = 4). The difference between the presence and absence of Na+ was not statistically significant (Student’s t test, P = 0.6750). Download
Comparison of WzxPa structural models. The WzxPa structural homology model was aligned with the de novo FILM3-generated model (based on correlated mutations of aligned protein sequences). Alignment via TM-align yielded a TM score of 0.67296 (a TM score > 0.5 indicates that the structures share the same fold). (A) Top (periplasmic) view. (B) Side view. TMS have been labeled 1 to 12. Download
Fluorescence-based detection of H+ shifts inside proteoliposomes in response to CCCP. The pH-sensitive dye carboxy-SNARF-1 was incorporated inside proteoliposomes during reconstitution, followed by gel filtration to alter the external buffer pH to pH 6.5, 7.5, or 8.5. (A) Displayed is a linear regression of the change in fluorescence emission intensity at 580 nm relative to that at 640 nm (post-CCCP addition [t = −300 s] and -mixing [t = 0 s]). Net acidification of the proteoliposome interior would be due to H+ transit from the outside of the liposome, resulting in the intensity of the 580-nm peak increasing over time, yielding a linear regression fit with a positive slope. Conversely, loss of H+ from the proteoliposome interior would cause net alkalinization, resulting in the intensity of the 640-nm peak increasing over time, yielding a linear regression fit with a negative slope. Raw fluorescence emission data were used to calculate linear regression lines for pH 6.5 (B), pH 7.5 (C), and pH 8.5 (D). Legend: red, fluorescence emission at 580 nm; blue, fluorescence emission at 640 nm; green, 580-nm/640-nm ratio of fluorescence emission at each time point used for calculation of linear regression lines in panel A. Download
Raw data for fluorescence-based detection of H+ shifts inside proteoliposomes in response to valinomycin (Fig. 4). The pH-sensitive dye carboxy-SNARF-1 was incorporated inside proteoliposomes during reconstitution, followed by gel filtration to alter the external buffer pH to pH 6.5, 7.5, or 8.5. A linear regression of the change in fluorescence emission intensity at 580 nm relative to that at 640 nm (post-valinomycin addition [t = −300 s] and -mixing [t = 0 s]) was calculated. Net acidification of the proteoliposome interior would be due to H+ transit from the outside of the liposome, resulting in the intensity of the 580-nm peak increasing over time, yielding a linear regression fit with a positive slope. Conversely, loss of H+ from the proteoliposome interior would cause net alkalinization, resulting in the intensity of the 640-nm peak increasing over time, yielding a linear regression fit with a negative slope. Legend: red, fluorescence emission at 580 nm; blue, fluorescence emission at 640 nm; green, 580-nm/640-nm ratio of fluorescence emission at each time point. Download
Putative TMS rearrangements in the proposed Wzx flippase mechanism. Amino acids of functional importance are displayed as spheres. TMS domains proposed to undergo positional shifts are depicted as tubes. Structures have been colored with a gradient, from blue (N terminus) to red (C terminus). Certain TMS have been rendered semitransparent to not occlude the view of internal rearrangement events. Entry/exit routes of particular molecules are displayed as hatched arrows. (A) Periplasmic views of WzxPa when open to the periplasm (structural homology model) and cytoplasm (FILM3-generated structure; current investigation). TMS have been labeled 1 to 12. (B and C) Periplasm-facing, O-unit-bound conformation (B) that recruits H+ (black diamond) via E61 (TMS2) to be bound by the D269–D359 dyad, causing TMS8 (containing K272) and TMS11 to shift toward TMS10 (C). (D) The flipped UndPP-linked O unit can disengage from F139 and laterally diffuse out of the exit portal in the periplasmic leaflet of the IM formed by TMS1 and TMS9. (E and F) Loss of the O unit (in the H+-bound state) results in conformational inversion to a cytoplasm-facing state (E) in which an UndPP-linked O unit at the cytoplasmic leaflet of the IM can enter the flippase via the opening formed by TMS1 and TMS9 and be loaded onto residues R59 and Y60 (TMS2) (F). (G) Binding of the new O unit induces positional reset of TMS8 and TMS11, weakening the D269–D359 dyad and resulting in H+ release into the cytoplasm. (H) Loss of H+ drives conformational relaxation of the O-unit-bound flippase back to a periplasm-facing state, resulting in O-unit translocation across the IM. Download
Iodide electrode responses at different pH values. Standard curves were obtained for 2-fold dilutions of I− (in MOPS-KGlu buffer) at pH 6.5, 7.5, and 8.5, from 128 µM down to 250 nM. The pH of each solution was adjusted with concentrated KOH. All millivolt readings were recorded after 500 s to ensure proper equilibration. Values used to calculate the probe calibration are displayed as triangles. Download
ACKNOWLEDGMENTS
We thank the members of the Bear lab for technical assistance and helpful discussions and Nicholas Nickerson for critical reading and feedback on the manuscript.
Work in the laboratories of J.S.L. and C.E.B. has been supported by operating grants from Cystic Fibrosis Canada (CFC) and the Canadian Institutes of Health Research (CIHR) (grant MOP-14687 to J.S.L. and grant MOP-97954 to C.E.B.). S.T.I. is the recipient of a CIHR Frederick Banting and Charles Best Canada Graduate Scholarship doctoral award, a CIHR Michael Smith Foreign Study award, a CFC doctoral studentship, and an Ontario Graduate Scholarship in Science and Technology. P.D.W.E. is supported by postdoctoral fellowships from CFC and CIHR. T.N. is supported by a Special Training Fellowship in Biomedical Informatics from the United Kingdom Medical Research Council. J.S.L. holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology.
Footnotes
Citation Islam ST, Eckford PDW, Jones ML, Nugent T, Bear CE, Vogel C, Lam JS. 2013. Proton-dependent gating and proton uptake by Wzx support O-antigen-subunit antiport across the bacterial inner membrane. mBio 4(5):e00678-13. doi:10.1128/mBio.00678-13.
REFERENCES
- 1. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 2. Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D. 2011. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Islam ST, Lam JS. 2013. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ. Microbiol. 15:1001–1015 [DOI] [PubMed] [Google Scholar]
- 4. Islam ST, Taylor VL, Qi M, Lam JS. 2010. Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. mBio 1(3):e00189-10. 10.1128/mBio.00189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burrows LL, Lam JS. 1999. Effect of wzx (rfbX) mutations on A-band and B-band lipopolysaccharide biosynthesis in Pseudomonas aeruginosa O5. J. Bacteriol. 181:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Islam ST, Fieldhouse RJ, Anderson EM, Taylor VL, Keates RA, Ford RC, Lam JS. 2012. A cationic lumen in the Wzx flippase mediates anionic O-antigen subunit translocation in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 84:1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu D, Cole RA, Reeves PR. 1996. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J. Bacteriol. 178:2102–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Kievit TR, Dasgupta T, Schweizer H, Lam JS. 1995. Molecular cloning and characterization of the rfc gene of Pseudomonas aeruginosa (serotype O5). Mol. Microbiol. 16:565–574 [DOI] [PubMed] [Google Scholar]
- 9. Islam ST, Gold AC, Taylor VL, Anderson EM, Ford RC, Lam JS. 2011. Dual conserved periplasmic loops possess essential charge characteristics that support a catch-and-release mechanism of O-antigen polymerization by Wzy in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 286:20600–20605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woodward R, Yi W, Li L, Zhao G, Eguchi H, Sridhar PR, Guo H, Song JK, Motari E, Cai L, Kelleher P, Liu X, Han W, Zhang W, Ding Y, Li M, Wang PG. 2010. In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat. Chem. Biol. 6:418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burrows LL, Chow D, Lam JS. 1997. Pseudomonas aeruginosa B-band O-antigen chain length is modulated by Wzz (Ro1). J. Bacteriol. 179:1482–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Daniels C, Griffiths C, Cowles B, Lam JS. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4:883–897 [DOI] [PubMed] [Google Scholar]
- 13. Abeyrathne PD, Daniels C, Poon KK, Matewish MJ, Lam JS. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187:3002–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abeyrathne PD, Lam JS. 2007. WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core. Mol. Microbiol. 65:1345–1359 [DOI] [PubMed] [Google Scholar]
- 15. Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier MH., Jr. 2003. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 270:799–813 [DOI] [PubMed] [Google Scholar]
- 17. Kuroda T, Tsuchiya T. 2009. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794:763–768 [DOI] [PubMed] [Google Scholar]
- 18. He X, Szewczyk P, Karyakin A, Evin M, Hong WX, Zhang Q, Chang G. 2010. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467:991–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Begum A, Rahman MM, Ogawa W, Mizushima T, Kuroda T, Tsuchiya T. 2005. Gene cloning and characterization of four MATE family multidrug efflux pumps from Vibrio cholerae non-O1. Microbiol. Immunol. 49:949–957 [DOI] [PubMed] [Google Scholar]
- 20. He GX, Kuroda T, Mima T, Morita Y, Mizushima T, Tsuchiya T. 2004. An H+-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J. Bacteriol. 186:262–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huda MN, Chen J, Morita Y, Kuroda T, Mizushima T, Tsuchiya T. 2003. Gene cloning and characterization of VcrM, a Na+-coupled multidrug efflux pump, from Vibrio cholerae non-O1. Microbiol. Immunol. 47:419–427 [DOI] [PubMed] [Google Scholar]
- 22. Rick PD, Barr K, Sankaran K, Kajimura J, Rush JS, Waechter CJ. 2003. Evidence that the wzxE gene of Escherichia coli K-12 encodes a protein involved in the transbilayer movement of a trisaccharide-lipid intermediate in the assembly of enterobacterial common antigen. J. Biol. Chem. 278:16534–16542 [DOI] [PubMed] [Google Scholar]
- 23. Tanaka Y, Hipolito CJ, Maturana AD, Ito K, Kuroda T, Higuchi T, Katoh T, Kato HE, Hattori M, Kumazaki K, Tsukazaki T, Ishitani R, Suga H, Nureki O. 2013. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496:247–251 [DOI] [PubMed] [Google Scholar]
- 24. Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. 2006. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 3:303–313 [DOI] [PubMed] [Google Scholar]
- 25. Eckford PD, Li C, Ramjeesingh M, Bear CE. 2012. Cystic fibrosis transmembrane conductance regulator (CFTR) potentiator VX-770 (Ivacaftor) opens the defective channel gate of mutant CFTR in a phosphorylation-dependent but ATP-independent manner. J. Biol. Chem. 287:36639–36649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Whitney JC, Hay ID, Li C, Eckford PD, Robinson H, Amaya MF, Wood LF, Ohman DE, Bear CE, Rehm BH, Howell LP. 2011. Structural basis for alginate secretion across the bacterial outer membrane. Proc. Natl. Acad. Sci. U. S. A. 108:13083–13088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J, Morita Y, Huda MN, Kuroda T, Mizushima T, Tsuchiya T. 2002. VmrA, a member of a novel class of Na+-coupled multidrug efflux pumps from Vibrio parahaemolyticus. J. Bacteriol. 184:572–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huda MN, Morita Y, Kuroda T, Mizushima T, Tsuchiya T. 2001. Na+-driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a non-halophilic bacterium. FEMS Microbiol. Lett. 203:235–239 [DOI] [PubMed] [Google Scholar]
- 29. Long F, Rouquette-Loughlin C, Shafer WM, Yu EW. 2008. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli. Antimicrob. Agents Chemother. 52:3052–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morita Y, Kataoka A, Shiota S, Mizushima T, Tsuchiya T. 2000. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J. Bacteriol. 182:6694–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nugent T, Jones DT. 2012. Accurate de novo structure prediction of large transmembrane protein domains using fragment-assembly and correlated mutation analysis. Proc. Natl. Acad. Sci. U. S. A. 109:E1540–E1547. 10.1073/pnas.1120036109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ravna AW, Sylte I. 2012. Homology modeling of transporter proteins (carriers and ion channels), p 281–299 In Orry AJW, Abagyan R, Homology modeling: methods and Protocols, vol 857 Humana Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 33. Zhang Y, Skolnick J. 2005. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33:2302–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Veen HW. 2010. Structural biology: last of the multidrug transporters. Nature 467:926–927 [DOI] [PubMed] [Google Scholar]
- 35. Whitaker JE, Haugland RP, Prendergast FG. 1991. Spectral and photophysical studies of benzo[c]xanthene dyes: dual emission pH sensors. Anal. Biochem. 194:330–344 [DOI] [PubMed] [Google Scholar]
- 36. Forrest LR, Krämer R, Ziegler C. 2011. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta 1807:167–188 [DOI] [PubMed] [Google Scholar]
- 37. Masuda S, Terada T, Yonezawa A, Tanihara Y, Kishimoto K, Katsura T, Ogawa O, Inui KJ. 2006. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J. Am. Soc. Nephrol. 17:2127–2135 [DOI] [PubMed] [Google Scholar]
- 38. Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y. 2005. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc. Natl. Acad. Sci. U. S. A. 102:17923–17928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu M, Symersky J, Radchenko M, Koide A, Guo Y, Nie R, Koide S. 2013. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. U. S. A. 110:2099–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vanni S, Campomanes P, Marcia M, Rothlisberger U. 2012. Ion binding and internal hydration in the multidrug resistance secondary active transporter NorM investigated by molecular dynamics simulations. Biochemistry 51:1281–1287 [DOI] [PubMed] [Google Scholar]
- 41. Otsuka M, Yasuda M, Morita Y, Otsuka C, Tsuchiya T, Omote H, Moriyama Y. 2005. Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J. Bacteriol. 187:1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marolda CL, Li B, Lung M, Yang M, Hanuszkiewicz A, Rosales AR, Valvano MA. 2010. Membrane topology and identification of critical amino acid residues in the Wzx O-antigen translocase from Escherichia coli. J. Bacteriol. 192:6160–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Islam ST, Lam JS. 2013. Topological mapping methods for α-helical bacterial membrane proteins–an update and a guide. MicrobiologyOpen 2:350–364. 10.1002/mbo3.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marolda CL, Li B, Lung M, Yang M, Hanuszkiewicz A, Rosales AR, Valvano MA. 2011. Membrane topology and identification of critical amino acid residues in the Wzx O-antigen translocase from Escherichia coli. J. Bacteriol. 193:1291–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu XJ, Su XZ, Morita Y, Kuroda T, Mizushima T, Tsuchiya T. 2003. Molecular cloning and characterization of the HmrM multidrug efflux pump from Haemophilus influenzae Rd. Microbiol. Immunol. 47:937–943 [DOI] [PubMed] [Google Scholar]
- 46. Elumalai P, Rajasekaran M, Liu HL, Chen C. 2010. Investigation of cation–π interactions in sugar-binding proteins. Protoplasma 247:13–24 [DOI] [PubMed] [Google Scholar]
- 47. Malik A, Ahmad S. 2007. Sequence and structural features of carbohydrate binding in proteins and assessment of predictability using a neural network. BMC Struct. Biol. 7:1. 10.1186/1472-6807-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alkhouri B, Denning RA, Kim Chiaw P, Eckford PD, Yu W, Li C, Bogojeski JJ, Bear CE, Viirre RD. 2011. Synthesis and properties of molecular probes for the rescue site on mutant cystic fibrosis transmembrane conductance regulator. J. Med. Chem. 54:8693–8701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pasyk S, Li C, Ramjeesingh M, Bear CE. 2009. Direct interaction of a small-molecule modulator with G551D-CFTR, a cystic fibrosis-causing mutation associated with severe disease. Biochem. J. 418:185–190 [DOI] [PubMed] [Google Scholar]
- 50. Eddy SR. 2011. Accelerated profile HMM searches. PLOS Comput. Biol. 7:e1002195 http://dx.doi.org/10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Magrane M, UniProt 2011. UniProt Knowledgebase: a hub of integrated protein data. Database 2011:bar009 http://dx.doi.org/10.1093/database/bar009 [DOI] [PMC free article] [PubMed]
- 52. Jones DT, Buchan DW, Cozzetto D, Pontil M. 2012. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics 28:184–190 [DOI] [PubMed] [Google Scholar]
- 53. Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. 2010. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 38:W563–W568. 10.1093/nar/gkq427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nugent T, Jones DT. 2012. Detecting pore-lining regions in transmembrane protein sequences. BMC Bioinformatics 13:169. 10.1186/1471-2105-13-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu J, Zhang Y. 2010. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic of Wzx-GFP-His8 orientation determination assay. Proteoliposomes containing Wzx-GFP-His8 are digested with TEV protease in the presence and absence of Triton X-100 detergent (TX100). In the absence of detergent-mediated proteoliposome permeabilization, only the GFP-His8 exposed on the outside should be amenable to protease cleavage. In the presence of detergent, all GFP-His8, regardless of its original inside/outside position, should be amenable to protease cleavage; cleavage from this treatment is set as the total fluorescence to which other values are normalized. Comparison of fluorescence readings from the intact and detergent-ruptured sample sets allows for the proportional determination of Wzx–GFP-His8 proteins in a given proteoliposome preparation that are oriented with their N and C termini facing the inside of the proteoliposome. Download
Effect of Na+ on I− flux. Valinomycin was added at t = 0. (A) Wzx-GFP-His8 proteoliposomes in standard Na+-free MOPS-KGlu buffer. (B) Wzx-GFP-His8 proteoliposomes in MOPS-KGlu buffer supplemented with 20 mM Na+. (C) Iodide release rate normalized to that observed for standard MOPS-KGlu buffer (n = 4). The difference between the presence and absence of Na+ was not statistically significant (Student’s t test, P = 0.6750). Download
Comparison of WzxPa structural models. The WzxPa structural homology model was aligned with the de novo FILM3-generated model (based on correlated mutations of aligned protein sequences). Alignment via TM-align yielded a TM score of 0.67296 (a TM score > 0.5 indicates that the structures share the same fold). (A) Top (periplasmic) view. (B) Side view. TMS have been labeled 1 to 12. Download
Fluorescence-based detection of H+ shifts inside proteoliposomes in response to CCCP. The pH-sensitive dye carboxy-SNARF-1 was incorporated inside proteoliposomes during reconstitution, followed by gel filtration to alter the external buffer pH to pH 6.5, 7.5, or 8.5. (A) Displayed is a linear regression of the change in fluorescence emission intensity at 580 nm relative to that at 640 nm (post-CCCP addition [t = −300 s] and -mixing [t = 0 s]). Net acidification of the proteoliposome interior would be due to H+ transit from the outside of the liposome, resulting in the intensity of the 580-nm peak increasing over time, yielding a linear regression fit with a positive slope. Conversely, loss of H+ from the proteoliposome interior would cause net alkalinization, resulting in the intensity of the 640-nm peak increasing over time, yielding a linear regression fit with a negative slope. Raw fluorescence emission data were used to calculate linear regression lines for pH 6.5 (B), pH 7.5 (C), and pH 8.5 (D). Legend: red, fluorescence emission at 580 nm; blue, fluorescence emission at 640 nm; green, 580-nm/640-nm ratio of fluorescence emission at each time point used for calculation of linear regression lines in panel A. Download
Raw data for fluorescence-based detection of H+ shifts inside proteoliposomes in response to valinomycin (Fig. 4). The pH-sensitive dye carboxy-SNARF-1 was incorporated inside proteoliposomes during reconstitution, followed by gel filtration to alter the external buffer pH to pH 6.5, 7.5, or 8.5. A linear regression of the change in fluorescence emission intensity at 580 nm relative to that at 640 nm (post-valinomycin addition [t = −300 s] and -mixing [t = 0 s]) was calculated. Net acidification of the proteoliposome interior would be due to H+ transit from the outside of the liposome, resulting in the intensity of the 580-nm peak increasing over time, yielding a linear regression fit with a positive slope. Conversely, loss of H+ from the proteoliposome interior would cause net alkalinization, resulting in the intensity of the 640-nm peak increasing over time, yielding a linear regression fit with a negative slope. Legend: red, fluorescence emission at 580 nm; blue, fluorescence emission at 640 nm; green, 580-nm/640-nm ratio of fluorescence emission at each time point. Download
Putative TMS rearrangements in the proposed Wzx flippase mechanism. Amino acids of functional importance are displayed as spheres. TMS domains proposed to undergo positional shifts are depicted as tubes. Structures have been colored with a gradient, from blue (N terminus) to red (C terminus). Certain TMS have been rendered semitransparent to not occlude the view of internal rearrangement events. Entry/exit routes of particular molecules are displayed as hatched arrows. (A) Periplasmic views of WzxPa when open to the periplasm (structural homology model) and cytoplasm (FILM3-generated structure; current investigation). TMS have been labeled 1 to 12. (B and C) Periplasm-facing, O-unit-bound conformation (B) that recruits H+ (black diamond) via E61 (TMS2) to be bound by the D269–D359 dyad, causing TMS8 (containing K272) and TMS11 to shift toward TMS10 (C). (D) The flipped UndPP-linked O unit can disengage from F139 and laterally diffuse out of the exit portal in the periplasmic leaflet of the IM formed by TMS1 and TMS9. (E and F) Loss of the O unit (in the H+-bound state) results in conformational inversion to a cytoplasm-facing state (E) in which an UndPP-linked O unit at the cytoplasmic leaflet of the IM can enter the flippase via the opening formed by TMS1 and TMS9 and be loaded onto residues R59 and Y60 (TMS2) (F). (G) Binding of the new O unit induces positional reset of TMS8 and TMS11, weakening the D269–D359 dyad and resulting in H+ release into the cytoplasm. (H) Loss of H+ drives conformational relaxation of the O-unit-bound flippase back to a periplasm-facing state, resulting in O-unit translocation across the IM. Download
Iodide electrode responses at different pH values. Standard curves were obtained for 2-fold dilutions of I− (in MOPS-KGlu buffer) at pH 6.5, 7.5, and 8.5, from 128 µM down to 250 nM. The pH of each solution was adjusted with concentrated KOH. All millivolt readings were recorded after 500 s to ensure proper equilibration. Values used to calculate the probe calibration are displayed as triangles. Download