Abstract
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the United States, and, even though 5–15% of the total CRC cases can be attributed to individual genetic predisposition, environmental factors could be considered major factors in susceptibility to CRC. Lifestyle factors increasing the risks of CRC include elevated body mass index, obesity, and reduced physical activity. Additionally, a number of dietary elements have been associated with higher or lower incidence of CRC. In this context, it has been suggested that diets high in fruit and low in meat might have a protective effect, reducing the incidence of colorectal adenomas by modulating the composition of the normal nonpathogenic commensal microbiota. In addition, it has been demonstrated that changes in abundance of taxonomic groups have a profound impact on the gastrointestinal physiology, and an increasing number of studies are proposing that the microbiota mediates the generation of dietary factors triggering colon cancer. High-throughput sequencing and molecular taxonomic technologies are rapidly filling the knowledge gaps left by conventional microbiology techniques to obtain a comprehensive catalog of the human intestinal microbiota and their associated metabolic repertoire. The information provided by these studies will be essential to identify agents capable of modulating the massive amount of gut bacteria in safe noninvasive manners to prevent CRC. Probiotics, defined as “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” (219), are capable of transient modulation of the microbiota, and their beneficial effects include reinforcement of the natural defense mechanisms and protection against gastrointestinal disorders. Probiotics have been successfully used to manage infant diarrhea, food allergies, and inflammatory bowel disease; hence, the purpose of this review was to examine probiotic metabolic activities that may have an effect on the prevention of CRC by scavenging toxic compounds or preventing their generation in situ. Additionally, a brief consideration is given to safety evaluation and production methods in the context of probiotics efficacy.
Keywords: lactic acid bacteria
according to the centers for disease control and prevention (CDC), colorectal cancer (CRC) is the third most commonly diagnosed cancer in the United States. In 2005, more than 141,000 Americans (72,007 men and 69,398 women) were diagnosed with CRC. Men and women had significantly different incidence rates. Black men had the highest rates (64.9/100,000), white men were second (55.4/100,000), followed by Hispanic men (46.8), Asian/Pacific Islander men (40.6), and American Indian/Alaska Native men (34.3). Among women, black women were the most likely to be diagnosed with colorectal cancer in 2005 (49.4/100,000) followed by white women (40.8), Hispanic women (33.9), Asian/Pacific Islander women (32.2), and American Indian/Alaska Native (24.5) (51). In the European Union, CRC is the second most common cancer and second major cause of death in men and women (35).
The present review summarizes the advances in the characterization of the human gastrointestinal (GI) microbiota, how the microbiome might contribute to the development of CRC and, on the basis of this information, how “tailored probiotics” might be used to modulate specific bacterial populations of the GI tract to restore an equilibrated microbiota and intestinal microenvironment, which can contribute to CRC prevention.
Colorectal Cancer: Genetics, Diet, and Lifestyle
Five to fifteen percent of all CRC cases can be attributed to the following hereditary CRC syndromes: Lynch syndrome (also hereditary nonpolyposis CRC or HNPCC), familial adenomatous polyposis (FAP), and MUTYH-associated polyposis (MAP) (50). Lynch syndrome is an autosomal-dominant disease characterized by early-onset CRC and the presence of multiple neoplasms affecting the colon, rectum, and other organs (263). HNPCC is associated with germline mutations in the mismatch repair (MMR genes), mainly MSH2, MLH1, MSH6, and PMS2 (65, 94). FAP is characterized by the early onset of hundreds to thousands of adenomas throughout the large bowel, attributable to a germline mutation in the adenomatous polyposis coli (APC) gene, located on chromosome 5q21. APC is a tumor suppressor gene, first localized in 1987 and cloned in 1991 following mutation analyses in unrelated families with FAP (reviewed in Ref. 71). MAP is an autosomal recessive disorder characterized by adenomatous polyps of the colorectum and a very high risk of colorectal cancer (187).
CRC is thought to develop over many years in a multistep process, known as the “adenoma-carcinoma sequence” (273). Early studies showed the involvement of APC in FAP (113). Subsequent studies revealed that mutations in APC are also found in 63% of sporadic adenomas and up to 80% of sporadic colorectal cancers (146, 179, 208). APC is a multidomain protein that contains binding sites for numerous proteins, including microtubules, the Wnt/Wg pathway components β-catenin and axin, the cytoskeletal regulators EB1 and IQGAP1, and the Rac guanine-nucleotide-exchange factor Asef1 (11). APC acts as a “gatekeeper gene”, maintaining low levels of β-catenin in the absence of a Wnt signal, thus preventing excessive cell proliferation. On the other hand, HNPCC has been used to identify an alternative pathogenesis mechanism that involves “caretaker” MMR genes responsible for recognizing and repairing single-base and larger-strand slippage mismatches in DNA replication (65).
Although there is a strong genetic component in the development of colorectal adenomas or CRCs, it is generally accepted that environmental factors including diet and lifestyle have a major impact on risk. Lifestyle aspects related to increased risk of CRC include elevated body mass index (BMI), obesity, and low physical activity (128, 165). A large number of dietary compounds have been associated with either increased or decreased risk of colon cancer [see Fig. 1, based on reviews or papers by Marshall (165); Butt and Sultan (43); Kim and Kwon (133); Berquin et al. (26); Pool-Zobel and Sauer (207); and Johnson and Lund (128)]. Broadly, elevated risk of CRC has been associated with high consumption of red and processed meat, refined grains, sweets, and alcohol, and to a low consumption of fruits and vegetables (19, 215).
Fig. 1.
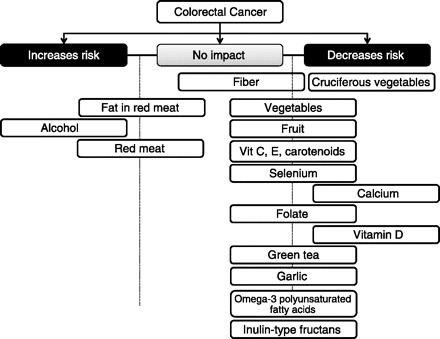
Dietary compounds associated with either increased or decreased risk of colorectal cancer [Based on reviews or papers by Marshall (165), Butt and Sultan (43), Kim and Kwon (133), Berquin et al. (26), Pool-Zobel and Sauer (207), and Johnson and Lund (128)].
Metabolic Signatures of the GI Environment
The intestinal environment including the gut luminal content is defined by the colliding processes of nutrient absorption, host defense, and host-microbial interactions. The GI milieu is highly variable across individuals because of inherent interindividual variation and environmental factors including diet and lifestyle. High-resolution magic-angle spinning (HRMAS) 1H NMR spectroscopy in combination with orthogonal projection to latent structure-discriminant analysis (O-PLS-DA) was recently used to investigate metabolic profiles of human intestinal tissue samples (276). The typical 1H Carr-Purcell-Meiboom-Gill HRMAS NMR spectra of intact human biopsies of antrum, duodenum, jejunum, ileum, and transverse colon showed 35 common metabolites including amino acids, carboxylic acids, pyrimidines, membrane component metabolites, creatine, glucose, inositols, lipids, and triglycerides. These metabolites were found in the various human GI tract samples irrespective of the sampling position. However, variation in the relative signal intensities indicated different concentrations of components in different intestinal locations. O-PLS-DA of NMR spectra of samples indicated that 1) there was no sex variation in any part of the GI tract mucosa; 2) variations between duodenum and jejunum were negligible, 3) the antrum contained higher levels of choline, glycogen, phosphorylethanolamine, taurine, and acetyl glycoproteins, and lower levels of amino acids, lipids, glucose, and lactate compared with other gut regions; 4) the duodenum and jejunum were similar in terms of their biochemical composition and were rich in choline, glutathione, glycerophosphocholine (GPC), lipids, and glycoproteins but contained relatively lower levels of acetate, betaine, glycogen, inositols, and phosphorylethanolamine; 5) the ileum was rich in amino acids, but low in GPC; and 6) the transverse colonic mucosa contained higher levels of acetate, glutamate, inositols, lactate, with lower levels of creatine, GPC, and taurine (Fig. 2) (276).
Fig. 2.
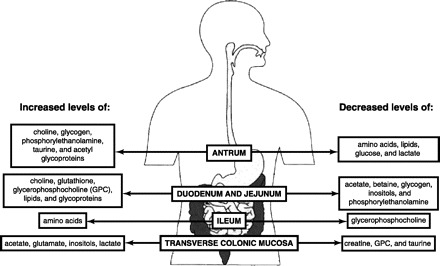
Biochemical composition of the different gastrointestinal regions [Adapted from Wang et al. (276)].
A similar global biochemical composition of the different GI tract regions was reported for conventional and conventionalized mice in a study by Martin et al. (166). This study aimed to investigate the impact of the intestinal microbiota on the biochemical composition of intact tissues obtained from conventional mice, conventionalized mice, and mice transplanted with a simplified human baby microbiota supplemented with probiotics and supplemented with synbiotics.
Modification of the GI Environment by CRC
Research studies have shown that the tumor microenvironment, the environment within colorectal neoplastic lesions, is significantly different to the normal intestine. The modified tumor environment can have adverse effects on drug uptake and the intrinsic sensitivity of malignant cells to treatment (175, 176). Early 1H NMR spectroscopy studies of colon tumors vs. normal mucosal biopsies demonstrated a significant increase in the concentration of the endogenous compounds lactate, glutamate, aspartate, taurine, spermine, glutathione and glycerophosphoethanolamine, and a significant decrease of myo- and scyllo-inositol (183, 184). A recent quantitative metabolome profiling of the colon cancer microenvironment by Hirayama et al. (114) confirmed increased concentrations of most amino acids in colon tumors with the exception of glutamine. Additionally, decreased levels of glucose were detected in tumor tissue, which can be explained by the fact that increased glycolitic activities in cancer cells deplete glucose in a hypovascular microenvironment. This fact also explains an increased concentration of lactate and a decreased concentration of pyruvate in tumor tissue. This study also identified higher levels of the tricarboxylic acid (TCA) metabolites succinate, fumarate, and malate in tumor samples, which contradicts the studies by Chan et al. (53) and Denkert et al. (76). Both studies reported lower levels of metabolites of the TCA cycle, which confirm previous proteomic reports that show an impaired TCA cycle in colon carcinoma tissues (6, 27). Chan et al. (53), using HR-MAS NMR, identified 10 marker metabolites that separated normal from tumor samples. Lipids, polyethylene glycol, and glucose were identified at higher levels in normal mucosa compared with tumor tissues, whereas choline-containing compounds, taurine, scyllo-inositol, glycine, phosphoethanolamine, lactate, and phosphocholine were present at higher levels in the CRC samples. Twenty-four additional metabolites were identified in the same study by gas chromatography mass spectrometry, of which only fumarate, malate, mannose, galactose, glucose, 1-hexadecanol, and arachidonic acid showed increased levels in normal mucosa compared with tumor samples. Increased concentrations of lactate, phosphate, l-glycine, l-proline, l-phenylalanine, palmitic acid, marganic acid, oleic acid, stearic acid, uridine, 11,14-eicosadienoic acid, 11-eicosenoic acid, 1-O-heptadecylglycerol, 1-monooleoylglycerol, propyl octadecanoate, and cholesterol were found in tumor samples (53).
Immune Surveillance of the GI Tract
Because humans and their microbiota have coevolved, the human immune system is designed to generally tolerate commensals while retaining the capacity to rapidly mobilize against invading pathogens. Not surprisingly, the gut immune system is distinct in both form and function from other components of human immunity, with the extensive involvement of normal microbiota in maintaining an effective epithelial barrier and with lymphocyte induction occurring at sites disseminated throughout the GI lamina propria (95). The barrier capacity of the mucosal epithelium derives from an extensive network of tight junctions (TJ) that stitch the epithelial cells together. Pathogenic disruption of TJs can induce inflammation, which then further exacerbates TJ leakiness, as seen when enteropathogenic Escherichia coli (E. coli) triggers dephosphorylation and disassociation of the TJ protein, occludin (247). Toll-like receptor (TLR)-dependent activation of epithelial cells, macrophages, and dendritic cells (DCs) proximal to the pathogenic lesion initiates an inflammatory response characterized by release of proinflammatory cytokines including IL-1β, IL-6, IL-8, IL-12, IL-13, IL-17, IL-23, IFN-γ and TNF-α that exert wide-ranging effects on the mucosa [for example, IL-13 along with the chemokine CXCL10 induces epithelial cell cycling (61)] and recruit immune effector cells. Inflammatory signals are balanced by tolerogenic cytokines, particularly IL-10 produced by TRegs to promote oral tolerance of food compounds and normal bacterial microbiota.
A robust gut commensal microbiota can provide an effective barrier against pathogenic invasion. The gut epithelial cells of mice raised in a germ-free environment show reduced levels of ATP, major histocompatibility complex II, and TLR-9 and are attached to a lamina propria that is thinner and less cellular than in normally colonized mice (reviewed in Ref. 228). At the same time, the gut microbiota guides both development and function of the gut immune system. Before colonization, inductive sites are reduced in both number and size in the guts of germ-free animals (33). Likewise, dimeric IgA production and effector T cell numbers are reduced in the small intestine of germ-free animals (122, 125, 182). Select commensals including species of Bifidobacterium, Bacteroides, and Lactobacillus have been linked to various immune-restoring activities including development of inductive tissue, restoration of numbers, and balance between various TH and TReg subsets (170) and epithelial immune functions (2, 49). At the same time, commensals exert proinflammatory signals including activation of TLR9 (109, 125) and IL-17 (125).
Chronic inflammation has been linked to CRC (68, 84, 135, 260). In pathological conditions like inflammatory bowel disease (IBD), homeostasis gives way to a chronic inflammatory state characterized by massive immune infiltration, immune-mediated tissue destruction, and attendant disruption of epithelial function and morphology. Gut bacteria such as Bacteroides fragilis and Streptococcus bovis have been linked to CRC because of their ability to activate immune cells to release promitogenic and proangiogenic cytokines like IL-6 and IL-17 (85, 283). Consequently, IBD conditions including ulcerative colitis and Crohn's disease dramatically increase a patient's risk of developing CRC. Indeed, epidemiological data suggest that up to 15% of human cancer incidence is associated with inflammation (142, 161).
Evidence suggests that IBD develops at least in part as a response to changes in the normal microbiota (dysbiosis) rather than from pathogenic invasion (170, 203, 234). For example, the abundance of Faecalibacerium prausnitzii, which has been shown to increase anti-inflammatory IL-10 and reduce proinflammatory TNF-α levels in the mouse colon following oral administration, is reduced in a significant percentage of patients suffering from Crohn's disease (251). The dysbiosis model of IBD proposes that genetic or environmental changes alter gut homeostasis and shift the microbial balance away from symbiotic species (those with known health-promoting effects) and toward pathobiotic species (organisms with pathogenic potential such as Clostridium and Helicobacter that are resident but not normally pathogenic) (228). This shift in turn leads to the induction of an inflammatory state that becomes chronic and significantly increases the risk for CRC.
A growing body of evidence suggests that probiotics can alter both the basal and induced states of gut immunity. Lactic acid bacteria (LAB) in particular have been shown to induce DC maturation and subsequent TReg activation in the gut (83) and also to induce natural killer cell cytotoxicity and cytokine secretion in peripheral blood mononuclear cells (74). Additionally, Lactobacillus sobrius (L. sobrius) has been shown to stabilize epithelial TJs in the gut, counteracting the occluding depletion caused by E. coli K88 (5). Activation of mesenteric lymph node TReg upon ovalbumin injection correlated with elevated production of IL-10 and transforming growth factor-β when rats were pretreated with L. rhamnosus GG (227). LAB treatment of the gut epithelial cell line, Caco-2, induces production of the human β-defensin-2 defensin and inhibits LPS-induced IL-23 secretion via TLR-2-dependent signaling (200). Finally, consistent with their anti-inflammatory effect on cultured epithelial cells, LAB strains including L. casei, L. delbrueckii, and L. acidophilus fed to Balb/c mice enhanced production of IgG1 and IL-4, indicative of a Th2-dominated immune response to injected ovalbumin (202).
Although probiotic bacterial interactions with the gut (and systemic) immune system are extremely complex, the data clearly suggest a modulatory effect on gut inflammation, with LAB particularly stabilizing epithelial TJs, inducing epithelial defensin production, and inducing the anti-inflammatory and immunomodulatory capacity of TReg and their DCs. Much work remains to determine how individual modulations impact overall gut health and development, which probiotics exert particular effects, and how probiotics can best be used to shape the gut immune state.
The Normal Intestinal Microbiota and the Microbiota Associated with CRC
Nonpathogenic commensal microbiota have a profound impact on normal GI physiology. They ensure effective intestinal mucosal motility, growth, and immunity as well as nutrient digestion, absorption, angiogenesis, and fortification of the mucosal barrier. Additionally, bacteria promote host epithelial cell production of fucosylated glycans (on which many gut bacteria feed) (228). Other functions of the GI microbiota include energy recovery from poorly digestible nutrients, modification of bile acids (BAs), and the nutritional supplementation of additional auxotrophic compounds not obtained through diet such as folate and biotin (129, 268).
The majority of the bacteria comprising the microbiota are uncultivable, which meant in the past that researchers were unable to characterize them. Since Antony van Leeuwenhoek observed and described what he called “living animalcules”, traditional microbiology has focused on the study of individual species as isolated units. However, many species have never been successfully isolated as viable specimens for analysis, presumably because we have not been capable of reproducing their optimal growth conditions experimentally. High-throughput sequencing and molecular taxonomic technologies are filling the gaps left by conventional microbiology techniques to provide a more comprehensive catalog of the normal microbiota. Phylogenetic analysis of bacterial 16S rRNA genes, amplified directly from complex communities, identified 82 distinct operational taxonomic units in human fecal samples with the vast majority (95%) distributed among three major monophyletic groups: the Bacteroides group, the Clostridium coccoides group, and the Clostridium leptum subgroup (253). Another comprehensive culture-independent phylogenetic analysis of microorganisms from 107 samples of resected tissue from Crohn's disease, ulcerative colitis, and control patients without patients showed that, regardless of disease state or anatomical site of sampling, the majority of sequences were associated with the same four phyla of the bacteria: Firmicutes (49% of clones), Bacteroidetes (23%), Proteobacteria (21%), and Actinobacteria (5%), with almost half of the sequences (45%) belonging to two subgroups, the order Bacteroidales and the family Lachnospiraceae (which comprises the Clostridium XIVa and IV groups within the order Clostridiales) (89). Comparison of small-subunit rRNA sequences from non-IBD colon and small intestine samples indicated differences in microbial populations with respect to the sampling site (Fig. 3). The colon contained more Bacteroidetes and more Actinobacteria species compared with the small intestine, which contained more Firmicutes. Within this phylum, the small intestine was enriched in species of the family Streptococcacea, and a higher relative percentage of species of Lactobacillaceae was observed in the colon (89). The most recent data generated by deep sequencing of the human intestinal microbiota has revealed that most bacterial species are present at low abundance (species defined as organisms sharing ≥97% sequence identity in their 16S rRNA genes) (212). Qin et al. (212) concluded that humans possess ∼1,000 different bacterial species in the GI tract, close to the 878 and 768 species detected in genetically identical twins by Turnbaugh et al. (269). Although most authors agree on a human intestinal core microbiota at the phylum level composed of Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria (59, 60, 212, 258, 266), the presence of a core microbiota at the genus or species level is still under debate. Qin et al. (212) identified a common core with 75 species common to >50% of individuals and 57 species common to >90%, whereas Turnbaugh et al. (269) concluded that only 35.9% and 49.1% of the species-level phylotypes found in the fecal communities of the two twins were shared between the two fecal samples, indicating the absence of a core set of abundantly represented universally shared phylotypes. The analysis by Claesson et al. (60) revealed a core gut microbiota across four samples (4 elderly subjects ages 60–87 yr) that consisted of species from Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria, and at the genus level it consisted of Alistipes, Anaerostipes, Anaerotruncus, Anaerostipes, Anaerotruncus, Bacteroides, Bifidobacterium, Blautia, Clostridium, Coprococcus, Dorea, Eubacterium, Faecalibacterium, Holdemania, Leuconostoc, Oscillospira, Peptostreptococcus, Roseburia, Ruminococcus, and Streptococcus. The study by Qin et al. (212) added the following genera to the previous list: Collinsella, Parabacteroides, Akkermansia, Prevotella, Butyrivibrio, Escherichia, Gordonibacter, Mollicutes, and Bryantella.
Fig. 3.
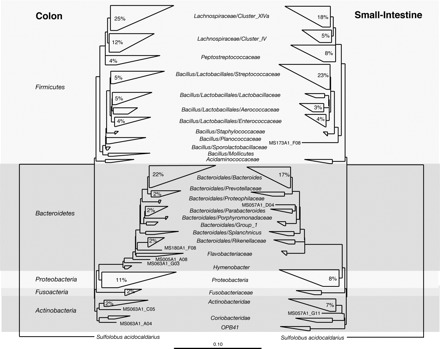
Comparison of SSU rRNA sequences isolated from colon and small intestine specimens. The phylogenetic trees depict classification of sequences isolated from non-inflammatory bowel disease (IBD) colon (left) and non-IBD small intestine (right) samples. Numbers within wedges represent the proportion of sequences within each sample set (i.e., colon or small intestine) that were assigned to a particular clade (values <2% are omitted). Wedge widths represent the taxa with the longest (top) and shortest (bottom) distances within the clade. Wedge areas represent the number of taxa in each clade. The scale bar represents base changes per site. [Reproduced with permission from Frank et al. (89)].
A number of research studies have catalogued the composition of the gut microbiome of healthy humans (212, 267) and the microbiome associated with IBD (105, 156, 280) and obesity (147, 265, 266); however, characterization of the CRC-associated gut microbiota is still behind. Recent deep-sequencing analysis (250) conducted on 12 stool samples (6 with CRC and 6 with normal colonoscopy) selected from 179 patients revealed a total number of 819 genera distributed among cancerous and normal samples with few variations between replicates. A significant difference between CRC and normal samples was observed for the Bacteroides/Prevotella group, with higher bacterial counts in cancer samples. Interestingly, the levels of Bacteroides/Prevotella were not influenced by age, BMI, family history, or size and location of tumors. In contrast, another study showed that the composition of the mucosa-associated microbiota differed significantly between colon adenoma cases and controls, with increased abundance of Proteobacteria and decreased levels of Bacteroidetes in adenoma samples (245). This study also demonstrated a higher overall diversity in adenoma samples, which was previously shown for two specific Clostridium species in an earlier study conducted in fecal samples of patients with CRC (236). A culture-dependent study of the microbiota of patients with polyps and CRC identified 15 bacterial taxa significantly associated with high risk of colon cancer (including species of Bacteroides, Eubacterium, and Bifidobacterium) and five associated with low risk of colon cancer (including species of Lactobacillus) in Japanese-Hawaiians, North American Caucasians, and rural native Japanese populations (181).
Biological Activities of the Intestinal Microbiota That Contribute to CRC
Factors involved in the modulation of the intestinal epithelial cell function present in the lumen may include dietary compounds, products of alimentary secretions from salivary glands, the stomach, pancreas, or intestinal glandular cells, secreted regulatory peptides, and constituents or products of the microbiota (77). It has been proposed that a steady state exists, where the end products of carbohydrate metabolism in the intestine (butyrate, acetate, and propionate) have a beneficial effect because they can act as colonic nutrients, whereas the products of the metabolism of proteins (phenolic compounds, amines, ammonia, N-nitroso compounds, and indoles) might have detrimental effects on the host (69). Although extreme, this concept is interesting because it provides a basis for the study of beneficial vs. detrimental bacteria and their metabolites.
The intestinal microbiota has been linked to CRC by production of toxic and genotoxic bacterial metabolites that can lead to mutations by binding specific cell surface receptors and affecting intracellular signal transduction. Using germ-free and gnotobiotic technology, Uronis and collaborators (270) recently showed that the intestinal microbiota has a direct impact on the development of colitis-associated colon cancer. In this study, azoxymethane-treated Il10−/− mice develop colitis-associated colon cancer in the presence of Bacteroides vulgatus, whereas germ-free mice remain disease free. The presence of colitis directly correlates with tumor multiplicity and acts as a promoter of CRC. Additionally, disruption of MyD88 signaling, a key integrator of multiple Toll-like receptors, prevented the development of colorectal tumors in Il10−/− mice, indicating a role of the microbiota in triggering intestinal inflammation and neoplastic changes in a susceptible host. The following subsections present bacterial activities presumed to generate metabolites involved in colorectal carcinogenesis. Table 3 lists the probiotic features of potential importance in CRC prevention and the bacterial activities previously linked to increased risk of CRC.
Table 3.
Features in colorectal cancer prevention and increased risk
| Characteristics |
|---|
Probiotic features of potential importance in colorectal cancer prevention
|
Bacterial activities that have been linked to increased risk of colorectal cancer
|
ROS, reactive oxygen species.
Secondary bile salt transformations.
An interesting study by O'Keefe et al. (192) highlighted that colon cancer is extremely rare in native African populations (less than 1/100,000 compared with 65/100,000 in African Americans) and correlated the higher incidence of colon cancer in African Americans with a diet high in meat and animal fat and with higher counts of 7α-dehydroxylating colonic bacteria. Bacterial biotransformations of conjugated BAs begin with bile salt hydrolases (BSH) that deconjugate liver-derived conjugated BAs to liberate primary BA, cholic acid (CA), chenodeoxycholic acid (CDA), and amino acids, which are further modified through dehydroxylation, dehydrogenation, and sulfation. On the basis of the composition of BA in feces of healthy individuals, 7α-dehydroxylation is quantitatively the most important secondary bacterial bile salt transformation in the human large intestine, yielding deoxycholic acid (DCA) and lithocholic acid (LCA) (221). High concentrations of secondary BAs in feces, blood, and bile have been linked to cholesterol gallstone disease and colon cancer (173). There is considerable experimental evidence that secondary BAs, such as DCA, are cytotoxic to colonic epithelial cells, as well as mutagenic with antiapoptotic properties (25). DCA and other hydrophobic BAs increase oxidative stress through generation of reactive oxygen species (ROS), which in turn cause oxidative DNA damage, in part, through the iron-catalyzed Fenton reaction (31), and impair the human mismatch repair system (54). Figure 4 shows a schematic to illustrate potential roles of BA in DNA damage and GI cancer (25).
Fig. 4.
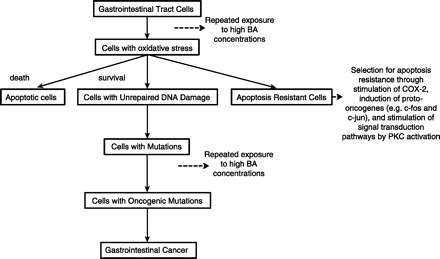
Pathway of induction of DNA damage and gastrointestinal cancer by bile acid (BA). COX-2, cyclooxygenase-2. [Modified from Bernstein et al. (25)].
BSHs are widely distributed in the genera Lactobacillus and Bifidobacterium, members of the lactic acid bacteria, most of which are considered probiotics (23). In contrast, 7α-dehydroxylation activity is prevalent in species of the genus Clostridium, including C. absonum, C. scindens, C. bifermentans, C. limosum, and C. hylemonae (221).
Production of hydrogen sulfide.
A diet high in meat has been shown to significantly increase the levels of taurine conjugation to bile acids. Unlike glycine, taurine contains a sulfonic acid moiety that is reduced and dissimilated to hydrogen sulfide after deconjugation (221). Meat is a rich source of dietary sulfur (through sulfur-amino acids), which promotes the growth of sulfur-reducing bacteria (SRB). SRB have the ability to use sulfate (SO42−) as an oxidant to degrade organic matter. An equivalent amount of hydrogen sulfide (H2S) is formed per mole of sulfate reduced (120, 191). Hydrogen sulfide is a toxic compound that generates free radicals (18), impairs cytochrome oxidase, suppresses butyrate utilization, and inhibits the synthesis of mucus and the methylation of DNA (57). In fact, a study by Ramasamy et al. (214) concluded that ineffective detoxification of hydrogen sulfide by dysregulation of the two rhodanese isoenzymes thiosulfate sulfurtransferase and mercaptopyruvate sulfurtransferase may result in mucosal insult, inflammation, and ultimately CRC. The diversity and ecology of human colonic SRB is largely unknown although one study identified the main SRB as lactate- and H2-utilizing Desulfovibrio spp. (64–81%), acetate-utilizing Desulfobacter spp. (9–16%), propionate- and H2-utilizing Desulfobulbus spp. (5–8%), lactate-utilizing Desulfomonas spp. (3–10%), and acetate- and butyrate-utilizing Desulfotomaculum spp. (2%) (97).
Production of aglycones.
Glycoside hydrolases (glucosidases, EC 3.2.1.-) are a widespread group of enzymes that hydrolyze the glycosidic bond between two or more carbohydrates or between a carbohydrate and a noncarbohydrate moiety. They remove one monosaccharide from the nonreducing end of their substrates in each catalytic cycle with release of aglycones (162). McBain and Macfarlane (172) showed that most of the glucosidase activities were associated with the bacterial fraction of colon contents. Older reports showed that cycasin (methylazoxymethanol-β-d-glucoside), a toxic aglycone released from the nuts, roots, and leaves of Cycas circinalis and Cycas revoluta by the action of β-glucosidases present in the plant or produced by the intestinal microbiota, was carcinogenic in rodents and has hepatotoxic, neurotoxic, teratogenic, and radiomimetic (imitates the effects of radiation) properties (115).
Bacterial β-glucuronidases.
The colonic microbial community may convert innocuous compounds into carcinogenic metabolites through a number of enzymatic activities, of which β-glucuronidation has been the most extensively investigated as a biomarker of CRC risk. A number of compounds, including carcinogens formed in food during cooking, drugs, etc., are metabolized in the liver, conjugated to glucuronic acid, and excreted into the small intestine via the bile duct (118, 229, 252). Once these components reach the intestinal tract, they act as substrates for host and bacterial enzymes. Specifically, the β-glucuronidase activity in the GI tract is influenced by diet and composition of the microbiota because only some of the members of the microbiota possess this enzyme. A study of 40 intestinal strains of the phylogenetic groups Clostridium (clusters IV, XIVa, and XVI), Bifidobacterium sp., and Bacteroides sp. showed that ∼30% of the clostridia but none of the Bacteroides and Bifidobacterium species had β-glucuronidase activity (66). Additionally, fecal specimens of patients with colon cancer were reported to have significantly higher β-glucuronidase activity compared with healthy subjects (132).
With regard to diet, the carcinogenic properties of 2-amino-3-methylimidazo(4,5-f)quinoline (IQ), which is one of the heterocyclic amines formed when various meats and fish are cooked, have been directly correlated with the presence of the bacterial β-glucuronidase encoded by the uidA gene in E. coli. In gnotobiotic rats monoassociated with a wild-type strain of E. coli (carrying the gene uidA) or a mutant strain (with the insertionally inactivated version of the gene), the presence of β-glucuronidase in the digestive lumen dramatically increased the genotoxicity of IQ in the colon (118). Some dietary components have been associated with a decrease in β-glucuronidase activity, including decreased dietary oligosaccharide diet content in turkeys (127) and hesperetin supplementation in rats. Hesperetin is a citrus flavonoid, abundant in orange and grape juices (12). However, a crossover feeding trial that studied the effect of fruit and vegetable intake in 63 healthy women and men ages 20 to 40 yr (167) and a study that evaluated the impact of a regular consumption of yogurt on the composition and metabolism of the human intestinal microbiota did not show a significant effect of either fruit/ vegetables or yogurt on β-glucuronidase activity of the intestinal microbiota (8).
Production of aromatic amines by azoreductases.
Bacterial azoreductases metabolize a number of compounds including azo colorants and drugs (for example, Sulfazalazine, a drug used for the treatment of inflammatory bowel diseases) to produce aromatic amines, which may have carcinogenic activities (106). Water-soluble azo dyes are metabolized by the intestinal microbiota, whereas water-insoluble azo dyes are metabolized by reductases in the liver (58). Azoreductases catalyze the reductive cleavage of azo groups (-N=N-). Different types of bacterial azoreductases have been isolated and characterized. Their size range varies from 28 to 62 kDa; they can use NADH or NADPH as electron donor, may or may not require flavin mononucleotide as cofactor, and can be aerobic or oxygen sensitive (55, 124). Intestinal bacteria reported to have azoreductase activity include Acidaminococcus fermentans, Enterobacter aerogenes, Bacillus sp., Bacteroides sp., Bacteroides fragilis, B. thetaiotaomicron, Bifidobacterium adolescentis, B. infantis, Butyrivibrio sp., Citrobacter sp., Clostridium nexile, C. clostridiiforme, C. paraputrificum, C. ramosum, C. sporogenes, Coprococcus catus, Enterococcus faecalis, E. coli, Eubacterium sp., Eubacterium aerofaciens, E. biforme, E. hadrum, Fusobacterium sp., Faecalibacterium prausnitzii, Klebsiella aerogenes, Lactobacillus sp., Lactobacillus catenaforme, Peptococcus prevotii, Peptostreptococcus productus, Pneumococcus sp., Proteus sp., Proteus vulgaris, Pseudomonas sp., Pseudomonas aeruginosa, P. pyrocyanea, Ruminococcus bromii, Salmonella paratyphi, S. typhimurium, Shigella dysenteriae (Type 1), Staphylococcus aureus, Streptococcus faecalis, Enterococcus faecalis, E. faecium, S. haemolyticus, and Veillonella parvula (reviewed by Chung and Stevens, Ref. 58). A more recent study confirmed azoreductase activity in 17 out of 40 microbial intestinal species using microarrays. Among these 17 species, Clostridium perfringens, Clostridium clostridioforme, Enterococcus faecalis, Ruminococcus obeum, and Bifidobacterium adolescentis showed the highest azo dye reduction activity (275). Although azoreductase activity has been linked to cancer and to members of the intestinal microbiota, one study showed that there was no correlation between azoreducer strains isolated from healthy adults vs. strains of the same species isolated from patients with colon cancer (186). This study highlights the need for further characterization of intestinal bacteria and their metabolic activities.
Production of aromatic amines by nitroreductases.
Another source of aromatic amines results from the metabolism of aromatic nitro compounds like dinitro toluene, nitrobenzenes, and nitropyrenes by nitroreductases. Two types of bacterial nitroreductases have been described on the basis of reduction of the nitro groups of polynitroaromatic compounds through the one- or the two-electron mechanism. Type I nitroreductases are oxygen-insensitive and catalyze the sequential reduction of nitro groups through the addition of electron pairs from NAD(P)H to produce the nitroso, hydroxylamino, and amino derivatives. Type II nitroreductases are oxygen sensitive and catalyze the single-electron reduction of the nitro group to produce a nitro anion radical, which can be reoxidized aerobically to the original structure with the concomitant production of the superoxide anion in a futile cycle (reviewed by Roldan et al., Ref. 225). The type I nitroreductase NfsB has been described in a strain of E. coli isolated from the jejunum of rats. NfsB catalyzes the nitroreduction of nitrobenzodiazepines, which are sedative-hypnotic drugs used for the treatment of anxiety (151). Homologs of NfsB can be found in species of Neisseria, Desulfovibrio, Shigella, Salmonella, Enterobacter, Citrobacter, Klebsiella, Vibrio, Pseudomonas, Shewanella, Flavobacteria, and Shigella. Also, a P-nitrobenzoate reductase, PnbA, has been recently characterized in Lactobacillus plantarum WCFS1. This enzyme showed low identity with nitroreductases from enterobacteria such as E. cloacae, but homologs were found in Lactococcus lactis, L. gasseri, L. antri, L. sakei, L. fermentum, and L. vaginalis, among others (107). The significance and role of different types of nitroreductases in the generation of carcinogenic compounds cannot be assessed at this time. As more intestinal bacterial genomes are sequenced, it should be possible to test whether there is a relationship between specific nitroreductases and the generation of toxic compounds.
Generation of acetaldehyde.
The first step in the metabolism of ethanol is the oxidation to acetaldehyde by alcohol dehydrogenases (ADHs). Acetaldehyde is then metabolized to mainly acetate and NADH by acetaldehyde dehydrogenases (ALDHs). Acetaldehyde is considered a potent carcinogenic compound, and alcohol consumption has been linked to cancer of the oral cavity, pharynx, esophagus, liver, colon, rectum, and breast in women (29, 238). Human ADHs are encoded by at least seven genes and comprise five different classes (116). The different isozymes are differentially active in different tissues. A study in rodents detected expression of ADH1, ADH3, and ADH4 as well as ALDH2 in the mucosal layer of the GI tract, with characteristic regional differences (279). Given the contribution of human ADHs to acetaldehyde production, it is not entirely possible to correlate microbial producers of acetaldehyde in the GI tract with CRC. However, a study by Seitz et al. (239) showed that mucosal acetaldehyde levels in rodents were significantly lower in germ-free compared with conventional animals. In fact, a search for enzymes with the EC number 1.1.1.1(ADH) in the Integrated Microbial Genomes-Human Microbiome Project (IMG/HMP) system (163) indicated the presence of this domain in the majority of annotated microbial genomes of GI origin in the database (125/185) with several organisms encoding more than one copy of the gene. The genera of GI origin encoding putative ADHs are Akkermansia, Anaerofustis, Anaerostipes, Anaerotruncus, Bacillus, Bacteroides, Bifidobacterium, Blautia, Bryantella, Burkholderia, Citrobacter, Clostridiales, Clostridium, Collinsella, Coprococcus, Desulfovibrio, Dorea, Enterobacter, Enterococcus, Escherichia, Eubacterium, Faecalibacterium, Finegoldia, Helicobacter, Klebsiella, Lactobacillus, Laribacter, Leuconostoc, Mitsuokella, Parabacteroides, Prevotella, Proteus, Providencia, Ruminococcus, Streptococcus, Subdoligranulum, Vibrio, Victivallis, and Yersinia.
Desulfation of BAs.
Sulfation of BAs is an important metabolic pathway to detoxify and promote fecal secretion of BAs because BA-sulfates are more water soluble and undergo limited enterohepatic recirculation (224). Sulfation is catalyzed by a group of enzymes called sulfotransferases being monosulfation at the 3-OH position predominant in humans (7, 101). The role of the commensal microbiota in BA desulfation was suggested by Robben et al. (223) in a study that showed in rats that microbial desulfation of intraperitoneally injected (24–14C)taurolithocholate-3-sulfate caused a fivefold decrease in the fecal plus urinary excretion rate of the isotope to approximately that found for sulfated (24–14C)taurolithocholate. A study conducted to investigate hydrolytic and reductive enzymes involved in formation of genotoxic metabolites in the large intestine identified C. perfingens and species of bacteroides and bifidobacteria as members of the commensal microbiota with aryslsulfatase activity (172). The presence of arylsulfatases in the following genera was confirmed by searching the IMG/HMP system: Anaerotruncus, Bacteroides, Bifidobacterium, Blautia, Bryantella, Citrobacter, Clostridium, Collinsella, Escherichia, Holdemania, Klebsiella, Parabacteroides, Proteus, Providencia, Roseburia, and Subdoligranulum.
Generation of ROS.
The results of recent studies suggest that the NADPH oxidase (NOX) family, notably NOXs and dual oxidases (DUOXs), are primarily involved in the generation of ROS by various nonphagocytic cells, including those of the gut epithelia (224). Oxidative stress damages cell components including proteins, lipids, membranes, and DNA. Types of DNA damage include depurination and depyrimidination, single- and double-stranded DNA breaks, base and sugar modifications, and DNA-protein crosslinks. The permanent modification of DNA resulting from the oxidative damage is one of the most important factors involved in mutagenesis that leads to carcinogenesis (3). In addition, large quantities of H2O2 are produced and excreted by human tumor cells (255), a mechanism that may enhance tumor invasion and proliferation. On the other hand, because ROS have a role in promoting cell apoptosis, a number of anticancer agents act by generating ROS-induced tumor cell death (38, 143, 286). Studies have shown that commensal bacteria induce generation of ROS in gut epithelial cells and that these metabolites act as key messengers that modulate the protein degradation machinery of several essential signaling components, which in turn influence diverse physiological processes of the host cells including cell proliferation and inflammation (141, 224). Therefore, ROS-generating enzyme systems have been studied as potential approaches to both prevent and treat cancer (188).
Efficacy of Probiotics of the LAB in CRC and CRC Prevention
The complex interaction between diet, normal intestinal microbiota, and health has promoted the development of strategies that allow for the selective growth of beneficial microorganisms or probiotics. These bacteria include those of the Bifidobacterium and Lactobacillus genera, which are used as markers of stability of the normal human intestinal microbiota. Probiotics have drawn attention both as a way of managing disorders of the GI tract and as a result of their role in the modulation of the immune system (98, 152, 259). Probiotics are “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” (219). The potential role of probiotics has been extensively reviewed (99, 198), and their beneficial effects include reinforcement of the natural defense mechanisms and protection against GI disorders. Probiotics have been successfully used to manage infant diarrhea (56, 93), food allergies (205), and IBD (32). A number of publications have reviewed the role of probiotics in chemoprevention of CRC (88, 153, 165). There is not a general consensus on the role of probiotics in CRC protection. In many cases this lack of agreement is due to the fact that researchers refer to probiotics as a group without considering 1) whether the probiotic tested is composed of one or more strains, 2) the species included in the probiotic preparation, 3) the preparation of the probiotic blend, e.g., the physiological state of the bacterial cultures at the moment of lyophilization or drying, 4) the age of the probiotic preparation (was the preparation kept cool, dry, and protected from the light?), and 5) whether the doses administered (CFU/ml) were adequate and comparable. There is, however, a general agreement that specific probiotic strains can beneficially affect metabolic activities that occur in the GI tract and that they enhance the host's immune response. Evidence for the effect of probiotics in animal and human studies as well as in vitro studies is presented below.
Animal studies.
Table 1 reviews the effect of probiotic intervention in animal models of CRC. Chemically induced (autochthonous) tumors in rodents are considered good test models to obtain results transferable to the clinical situation (10) with 1,2-dimethylhydrazine (DMH) being the most common carcinogen used in colon cancer induction. DMH is extensively metabolized in vivo, and toxicity has been ascribed to metabolism-generated reactive intermediates, such as alkyldiazonium ions, carbon-centered radicals, and ROS (92). From the studies detailed in Table 1, we can generally conclude that the beneficial effects of probiotics are species and strain dependent and that the LAB have to be alive. In addition, research suggests that probiotics are more effective in high-fat diets, an interesting fact considering that CRC has been linked to high consumption of red and processed meats and indirectly to high consumption of fat. A clear reduction in aberrant crypt foci (ACF) was mostly observed with prebiotic and synbiotic preparations (nutritional supplements combining probiotics and prebiotics); however, the effect of probiotics alone is not as clear. Finally, beneficial effects were observed when probiotics were fed before and early during carcinogen treatment but not later in the studies.
Table 1.
Effect of probiotic intervention in animal models of colorectal cancer
| Model | Treatment | Results | Reference |
|---|---|---|---|
| Sprague-Dawley rats, AOM-induced colon cancer model | Bifidobacterium lactis (DSM Food Specialties, Australia) plus “resistant starch” (RS) | RS plus B. lactis significantly protect against the development of colorectal cancer in the rat-AOM model. | 145 |
| Male Wistar SPF rats, DMH-induced colon cancer model | Moderate or intense physical exercise, alone or in combination with the consumption of a soy product fermented with Enterococcus faecium | Soy product and the practice of physical exercise (intense or moderate) were incapable, separately or combined, of inhibiting the formation of ACF in DMH-induced rats. | 246 |
| Rat AOM-induced colon cancer model | Lactobacillus rhamnosus GG and Bifidobacterium lactis Bb12, alone or plus inulin enriched with oligofructose (SYN) | SYN supplementation in carcinogen-treated rats primarily modulated immune functions in the gut-associated lymphoid tissue, coinciding with a reduced number of colon tumors. | 226 |
| Sprague-Dawley rats, AOM-induced colon cancer model | Not specified “novel synbiotic preparation”. | Significantly reduced ratio of ACF/colon and of ACF per colon and per each single focus | 64 |
| Rat AOM-induced colon cancer model | Lactobacillus casei strain Shirota (LcS) | The number of rats with colon cancers and the number of colon cancers per rat were significantly decreased in the rats that had consumed the LcS diet. LcS inhibited chemically induced colon carcinogenesis in the rat. | 284 |
| Male Fisher 344 weanling rats, AOM-induced colon cancer model | Bifidobacterium longum BB536 (Morinaga Milk Industry, Japan) alone or plus lactulose (SYN). | There was a significant reduction in the total number of ACF in the colons of the rats consuming B. longum, lactulose, or SYN diets compared with the control. There was no significant difference between the B. longum and lactulose groups in total number of ACF. | 52 |
| Male Fisher 344 weanling rats, AOM-induced colon cancer model | Bifidobacterium longum | Lyophilized cultures of B. longum significantly inhibited the ACF formation and the crypt multiplicity in the colon. A significant decrease in the fecal bacterial β-glucuronidase was also observed. | 137 |
| Male Wistar SPF rats, DMH-induced colon cancer model | Lactococcus lactis NZ9000 | No differences in tumor incidence, multiplicity, dimensions, and stage in the colonic mucosa were observed among the groups. | 148 |
| Male Fisher 344 rats, AOM-induced colon cancer model | Lactobacillus GG, L. delbrueckii subsp. “rhamnosus”, and Bifidobacterium lactis Bb12 (Chr. Hansen, Horsholm, Denmark), alone or plus raftilose (SYN) | After 16 wk of feeding, SYN significantly increased ACF multiplicity. After 32 wk, SYN significantly decreased intestinal tumors. Crypts with scarce or absent mucins were identified (MDF), visible in all AOM-treated rats, and correlated with tumor induction. There were fewer MDF/colon than ACF, and they were histologically more dysplastic than mucinous lesions identified as ACF in high-iron diamine Alcian blue-stained colon. | 44 |
| Male Fisher 344 rats, AOM-induced colon cancer model | Bifidobacterium lactis (Bb12) and Lactobacillus rhamnosus (LGG), alone or plus raftilose | 31 wk after AOM, rats treated with ratfilose had a significantly lower number of tumors (adenomas and cancers). Nonsignificant effect of probiotics in reducing tumors was also observed. Cecal short-chain fatty acids (SCFA) were higher in the groups treated with raftilose. Apoptosis was increased in the normal mucosa of the probiotic group, whereas no variation was observed in the tumors. Colonic proliferation was lower in the raftilose group. | 86 |
| Sprague-Dawley rats, AOM-induced colon cancer model | Bifidobacterium longum, Lactobacillus acidophilus, and Lactobacillus casei selected to inhibit MNNG and DMH-induced genetic damage to the colon | A significant decrease in AOM-induced colonic ACF was observed in rats fed a high-fat diet (corn oil) given L. acidophilus or inulin. In a concurrent group of animals fed a low-fat diet, no significant decrease in ACF was observed. | 30 |
| Sprague-Dawley rats, DMH-induced colon cancer model Diet: high-fat semipurified (AIN-93) | Lactobacillus acidophilus Delvo Pro LA-1, Lactobacillus rhamnosus GG, Bifidobacterium animalis CSCC1941, and Streptococcus thermophilus DD145 | Large intestinal tumor burden and mass were significantly lower for rats treated with L. acidophilus. L. acidophilus and L. rhamnosus GG but not B. animalis and S. thermophilus were reisolated from feces, indicating their survival trough the GI tract. | 174 |
| Male Fisher 344 rats, AOM-induced colon cancer model | Lactobacillus acidophilus NCFM | NCFM significantly suppressed AOM-induction of colonic ACF, in terms of total number, as well as crypt multiplicity and number of ACF/cm2 colon. NCFM inhibited AOM-induced colonic ACF formation in a dose-dependent manner. A significant dose-dependent reduction of cecal β-glucuronidase activities was observed. | 216 |
| Male Fisher 344 rats, AOM-induced colon cancer model | Bifidobacterium longum 25 (Unilever Research Laboratory, Vlaardingen) alone or plus inulin (SYN) | B. longum or inulin were associated with a decrease in AOM-induced small ACF (1-3 aberrant crypts per focus). SYN administration resulted in more potent inhibition of ACF (80% inhibition of small ACF) and decreased the incidence of large ACF (>4 aberrant crypts per focus). | 230 |
| Rats, DMH-induced colon cancer model | Lactobacillus acidophilus, Lactobacillus casei (NDRI, Karnal), and curd culture Lactococcus lactis biovar. Diacetylactis DRC-1 | A significant reduction in DNA damage assessed by the comet assay was observed in the probiotic curd group. | 139 |
| Male Fisher 344 rats, AOM-induced colon cancer model | Nonspecified EPS-producing and non-EPS-producing cultures | Rats fed diets supplemented with fermented milk made with 2 EPS-positive and 1 EPS-negative strains had significantly lowered incidence of colon tumor and colon tumor multiplicity. Cyclooxygenase-2 enzyme activity was significantly lower in the colon tissue of rats fed diets containing milk fermented with 4 EPS-producing and 1 nonproducing cultures than that in rats fed diets supplemented with acidified milk. No relationship was found between rheological properties or level of ropiness of fermented milk and its chemopreventive effect. | 210 |
| Rats, MNNG- and DMH-induced colon cancer model | Lactobacillus acidophilus (from commercially available yogurt), Lactobacillus gasseri P79, Lactobacillus confusus DSM20196, Streptococcus thermophilus NCIM 50083, Bifidobacterium breve and Bifidobacterium longum (from human infant stool) | Pretreatment orally with lactic acid bacteria (LAB) on 4 consecutive mornings before DMH gavage (8 h after the last LAB application) revealed that L. acidophilus, L. confusus, L. gasseri, B. longum, and B. breve inhibited the genotoxic effect of DMH. One of four S. thermophilus and one of three Lactobacillus delbrueckeii ssp. bulgaricus strains were also protective. Heat-treated L. acidophilus did not inhibit DMH-induced genotoxicity. | 206 |
| Male Fischer 344, DMH-induced colon cancer model 20% or 5% corn oil diet | Lactobacillus rhamnosus GG | Significant decrease in the incidence of colon tumors and the number of small intestinal and colon tumors per tumor-bearing animal for rats fed a 20% corn oil diet. Animals fed a 5% corn oil diet had a lower tumor incidence and number of tumors resulting from the decrease in dietary fat. Decrease in tumor incidence or number of tumors was not seen when animals were fed the Lactobacillus after the ninth week of carcinogen treatment. | 104 |
| Male Sprague-Dawley rats, AOM-induced colon cancer model | Lactobacillus acidophilus, Bifidobacterium adolescentis, Bacteroides fragilis, Escherichia coli and Clostridium perfringens | The number of ACF 5 wk after the start of the experiment was decreased in the rats treated with L. acidophilus and C. perfringens. The inhibitory effect of L. acidophilus is due to the enhanced removal of O6-methylguanine from the colon mucosal DNA. | 15 |
| Rats, DMH-induced colon cancer model | Bifidobacterium sp Bio (Danone strain 173010) | Significant reduction of aberrant crypts | 1 |
| Male Wistar SPF rats, DMH-induced colon cancer model | Bifidobacterium sp. (Chr. Hansen, Inc., Milwaukee, WI), L. acidophilus, alone, in combination and plus fructooligosaccharides (FOS) | Bifidobacteria + FOS administration significantly decreased the number of aberrant crypts that developed. Bifido-FOS treatment led to significantly fewer aberrant crypts and ACF than the Bifido-Basal treatment. The Skim-FOS group had significantly more cecal bifidobacteria than the Skim-Basal group and significantly fewer C. perfringens than the Skim-Basal and Bifido-Basal. The number of aberrant crypts was not significantly different among the groups when L. acidophilus was added. However, the number of C. perfringens was significantly decreased by the addition of bifidobacteria, L. acidophilus, or the combination of the two. | 91 |
| F344 rats fed the high-fat diet containing 2-amino-3-methylimidazo(4,5-f)quinoline (IQ) | Lyophilized cultures of Bifidobacterium longum | B. longum significantly inhibited the IQ-induced incidence (percentage of animals with tumors) of colon (100% inhibition) and liver (80% inhibition) tumors and multiplicity (tumors/animal) of colon, liver, and small intestinal tumors in male rats. Dietary supplementation of B. longum resulted in a significant inhibition of colon, small intestine, and liver tumor incidences in male rats (P < 0.05). In female rats, dietary supplementation of B. longum also decreased mammary carcinogenesis to 50% and liver carcinogenesis to 27% of those observed in animals fed the control diet, but the differences did not reach a statistical significance. | 218 |
| Male Fischer 344, DMH-induced colon cancer model | Viable Lactobacillus acidophilus | The colon cancer incidence after a 20-wk induction period was lower in the animals receiving L. acidophilus (40% vs. 77% in controls), but no difference in incidence was discerned after a 36-wk period. | 103 |
| Fischer 344, DMH-induced colon cancer model | Milk fermented with Streptococcus thermophilus or Lactobacillus bulgaricus | Feeding fermented milks altered the metabolism of 1,2-dimethylhydrazine and shifted the target organ from the ear duct to the small intestine. In addition, the colon tumor distribution for the fermented-milk groups appeared to shift toward the anus. | 244 |
| Male Fisher 344 rats, AOM-induced colon cancer model modified AIN-76A diet | Lyophilized cultures of B. longum | Significant suppression of colon tumor incidence and tumor multiplicity and also reduced tumor volume were seen. Results also revealed that ingestion of B. longum significantly inhibited AOM-induced cell proliferation, ODC activity, and expression of ras-p21 oncoprotein. | 248 |
| ICR mice inoculated with sarcoma-180 intraperitoneally and BDF1 mice inoculated with L1210 leukemia intraperitoneally | Lactobacillus casei YIT 9018 (LC 9018) | Intravenous injection of LC 9018 markedly inhibited the growth of subcutaneously inoculated sarcoma-180. This organism was also effective against methylcholanthrene-induced syngeneic MCA K-1 tumor in BALB/c mice. The antitumor activity of LC 9018 was reduced by treatment with carrageenan, an antimacrophage agent and was also observed in T-cell-deficient athymic nude mice. | 130 |
| Germfree (GF) mice human-flora-associated (HFA) mouse treated with AAC | Streptococcus faecalis T-110, Clostridium butyricum TO-A, and Bacillus mesentericus TO-A mixture (Biothree, TOA Pharmaceutical Tokyo, Japan) | The mean level of the DNA adducts in the colonic epithelium of the probiotic group was significantly lower than that of control group. | 117 |
| DMH-treated BALB/c mice | Lactococcus lactis expressing catalase | Animals that received the catalase-producing L. lactis had a significantly lesser extent of colonic damage and inflammation. | 72 |
| DMH-treated Male albino rats of Wistar strain | Lactobacillus acidophilus, Lactobacillus casei ssp. casei, and dahi culture DRC-1 (Lactococcuslactis ssp. biovar. diacetylactis NCDC-60) | Addition of Ac Dahi to wheat bran acted synergistically against intestinal carcinogenesis. | 140 |
AOM, azoxymethane; ACF, aberrant crypt foci; DMH, 1,2-dimethylhydrazine; MDF, mucin-depleted foci; MNNG, N-methyl-N′-nitro-N-nitrosoguanidine; IQ, 2-amino-3-methylimidazo[4,5-f]quinoline; AAC, 2-amino-9H-pyrido[2,3-b]indole (2-amino-α-carboline); EPS, exopolysaccharide; ICR, imprinting control region.
Studies in humans.
A number of publications report beneficial (75, 197) or detrimental (100) effects of consuming probiotics to treat or prevent diarrhea induced by cancer therapies. However, the number of clinical trials involving the use of probiotics for prevention or treatment of CRC is scarce (Table 2). With the overall goal of contributing to the formulation of improved dietary advice, the EU-sponsored project SYNCAN aimed to evaluate the potential CRC-preventing activity of a synbiotic product in human volunteers. Preliminary in vitro studies first identified synbiotic combinations offering the greatest competitive advantages in the colon ecosystem and established the anticarcinogenic effect of the most promising synbiotic combination in a rat colon cancer model (207, 272). From these studies, Pool-Zobel et al. (207) concluded that the fermentation products of inulin-type fructans by the gut microbiota, specifically butyrate and propionate, are capable of inhibiting histone deacetylases and growth of colon tumor cells. Despite this finding, the mechanisms involved in a potential effect of probiotics in colon cancer are not yet fully characterized.
Table 2.
Clinical trials of probiotic intervention for prevention or treatment of colorectal cancer
| Model | Treatment | Results | Reference |
|---|---|---|---|
| 20 human volunteers | R/DB/PC/C. Resistant starch (RS) containing high-amylose maize starch and Bifidobacterium lactis (alone or in combination). | The synbiotic intervention fostered a unique fecal stream bacterial community, with greater proportion of patients harboring fecal Lachnospiraceae spp. No significant effect on epithelial proliferation or crypt height was observed, and fecal SCFA concentrations did not change between interventions or when compared with baseline (probably due to low doses of RS). | 282 |
| 38 healthy men | R/DB/PC/C. Lactobacillus rhamnosus LC705 (LC705) together with Propionibacterium freudenreichii ssp shermanii JS (PJS) | Administration of LC705 and PJS was followed by an increase in the fecal counts of lactobacilli and propionibacteria and a decrease in the activity of β-glucosidase. | 111 |
| 150 patients diagnosed with colorectal cancer | Lactobacillus GG | Lactobacillus GG supplementation was well tolerated and may reduce the frequency of severe diarrhea and abdominal discomfort related to 5-FU-based chemotherapy. | 197 |
| 398 men and women presently free from tumor who had had at least 2 colorectal tumors removed | Wheat bran and/or L. casei | L. casei prevented atypia of colorectal tumors. | 123 |
| 31 subjects undergoing elective colorectal resection for cancer | Mixture of Bifidobacterium longum (BB536) and Lactobacillus johnsonii (La1) | La1, but not BB536, adheres to the colonic mucosa and affects intestinal microbiota by reducing the concentration of pathogens and modulates local immunity. | 96 |
| 37 patients with colon cancer and 43 polypectomized patients | R/DB/PC. A synbiotic preparation-oligofructose-enriched inulin (SYN1) + Lactobacillus rhamnosus GG (LGG) and Bifidobacterium lactis Bb12 (BB12) | Numbers of Bifidobacterium and Lactobacillus increased, and Clostridium perfringens decreased. There was a reduction of colorectal proliferation, and the capacity of fecal water to induce necrosis in colonic cells and improve epithelial barrier function in polypectomized patients. Synbiotic consumption prevented an increased secretion of interleukin-2 by peripheral blood mononuclear cells in polypectomized patients and increased the production of interferon-γ in cancer patients. | 213 |
| 100 patients with colorectal carcinoma scheduled for radical colorectomy randomly divided into control (n = 50) and probiotics groups (n = 50) | Probiotics containing Lactobacillus plantarum (CGMCC No. 1258, 1011 CFU/g), Lactobacillus acidophilus (LA-11, 1010 CFU/g), and Bifidobacterium longum (BL-88, 1010 CFU/ g) | Intervention lasted 16 days (6 days preoperatively and 10 days postoperatively). The total probiotic daily dose was 2.6 × 1014 CFU/g). The treatment improved the integrity of the gut mucosal barrier and balance of the gut microbiota and decreased rate of post surgical infection. | 154 |
R/DB/PC/C: Randomized, double-blind, placebo-controlled, crossover trial.
Designing Tailored Probiotics
From the many studies published in the field we could conclude that, when it comes to probiotics there is no “one size fits all”. The vast majority of studies involving probiotics suggests a role for probiotics mainly to prevent CRC. However, there is gray area between prevention and early stages of CRC that could benefit from treatment with selected probiotic strains. From animal studies it is evident that species of Bifidobacterium and Lactobacillus, specifically B. lactis, B. longum, L. acidophilus, L. casei, and L. rhamnosus have a beneficial impact on preventing and reducing the number of ACFs in mice (see Table 1 for references), whereas Enterococcus and Lactococcus are mostly ineffective. However, specific activities such as butyrate production are present in E. durans (217), and in one study DMH-treated mice that received catalase-producing L. lactis had a lower extent of colonic damage and inflammation (72). This one example indicates that there might be two paths to achieve a full probiotic effect; one is to genetically modify one or more than one strains to combine and include multiple beneficial activities. This strategy is definitely showing some promise, as a number of studies using genetically modified probiotic strains have demonstrated beneficial effects. A Phase I clinical trial was conducted with a Lactococcus lactis strain in which the thymidylate synthase gene was replaced with a synthetic sequence encoding mature human interleukin-10. Ten patients with Crohn's disease were included and showed a decrease in disease activity (34). The fact that the recombinant bacterium requires thymidine to grow prevents its widespread distribution in nature and provides an effective containment strategy. Other studies have shown that expression of the MnSOD in L. plantarum, L. lactis, and L. gasseri improved probiotic strain survival (39, 41) and reduced colitis symptoms (48). B. breve and Lactococcus lactis expressing the bilE gene from Listeria monocytogenes showed increased resistance to bile and were also recovered at significantly higher levels than control strains from the feces and intestines of mice (278). Alternatively, a carefully selected blend of strains may be the more appropriate approach given the controversy that might arise from the use of genetically modified organisms (42, 271). From the topics covered in this review we have identified microbial features with an effect, beneficial or otherwise, in CRC as well as essential characteristics needed by probiotic strains to survive and thrive in the GI tract.
Survival and colonization of the specific intestinal microenvironment.
The GI tract has the dual tasks of absorbing nutrients and protecting the body from potentially harmful microorganisms. It possesses a unique structure with a massive surface area that enhances absorption and also houses the largest numbers of immune cells in the body (168). These characteristics create an exceptional environment that is able to host and shape the vast human intestinal microbiota. It is not surprising that the disruption of the intestinal environment by disease, diet, physical trauma, antibiotics, and other agents has a profound effect on the residing microorganisms and can create a microbial dysbiosis, which negatively affects the host's overall health.
ACID RESISTANCE.
Chan et al. (53) showed that tumorogenic colon microenvironments had increased concentrations of lactate, phosphate, l-glycine, l-proline, l-phenylalanine, palmitic acid, marganic acid, oleic acid, stearic acid, uridine, 11,14-eicosadienoic acid, 11-eicosenoic acid, 1-O-heptadecylglycerol, 1-monooleoylglycerol, propyl octadecanoate, and cholesterol. The high lactate production rate of malignant cells was documented as early as 1956 (277) when Warburg proposed an energy deficiency in normal cells after suppression of their respiration, which “forces the cells to replace the irretrievably lost respiration energy in some way,” thus increasing fermentation. The production of increased concentrations of lactate persists even in the presence of oxygen, a phenomenon occasionally called “aerobic glycolysis” or the “Warburg effect”. On the contrary, production of lactate is repressed, and glycolysis is slowed down by environmental oxygen in most normal mammalian cells, which is referred to as the “Pasteur effect” (274). Consequently, resistance to lactate is an important feature when selecting for probiotic strains to increase their chances for survival in an inflamed/tumorogenic GI environment. The mechanisms involved in acid resistance by lactic acid bacteria include 1) proton pumps, of which the most prevalent are F1F0ATPases; 2) amino acid decarboxylation reactions, which result in the formation of biogenic amines; and 3) the urease and arginine deiminase pathways, which produce NH3 that combines with protons in the cytoplasm to produce NH4+, raising the internal pH (20, 64, 240). Additionally, we have recently shown that superoxide dismutases, involved in resistance to oxidative stress, can also protect some organisms against acid stress (40). Other systems involved in the resistance to acidic environments include 1) chaperones, proteases, and heat shock proteins, which protect or degrade proteins if damaged; 2) DNA repair systems that excise errors or restart stalled replication forks; 3) transcriptional regulators such as two-component regulatory systems and σ-factors that induce minor or global responses; and 4) quorum-sensing-driven events. In addition, cells respond to acidic environments by altering architecture, composition, stability, and activity of their envelope (21, 36, 64, 240).
RESISTANCE TO OXIDATIVE STRESS.
A number of studies have shown a significant role of oxidative stress in the initiation and growth of colorectal adenomas and demonstrated adaptation of CRC cells to oxidative stress (189, 194, 249). Consequently, enzymes that can detoxify ROS are desirable in probiotic strains. Oxidative stress can cause several types of damage to bacterial cells and has bacteriostatic and bactericidal effects (90, 287). Although oxygen by itself is unable to cause any damage to the cell, ROS are generated during cellular processes where O2 is partially reduced to water. These intermediates have a high oxidizing potential and thus are responsible for cellular oxygen toxicity (90). To offset the harmful effects of ROS, some LAB have evolved protective mechanisms that utilize antioxidant enzymes, such as superoxide dismutases and hydroperoxidases (i.e., catalases and peroxidases; or KatE and KatG), intracellular enzymes that scavenge superoxide radicals and hydrogen peroxide, respectively, preventing the formation of HO via Fenton chemistry (87). In Streptococcus and Lactococcus the elimination of ROS conform to a general bacterial antioxidative defense system because both genera posses MnSOD (209, 235). With a few exceptions (70, 138), most lactobacilli lack this general defense system. Some lactobacilli species developed an alternative nonenzymatic defense system that involves the accumulation of high intracellular concentrations of Mn(II), which can scavenge O2− (13, 14). Additionally, to minimize the potentially toxic effects of the oxygen, Lactobacillus contains enzymes such NADH oxidases, NADH peroxidases, or pyruvate oxidases (102, 257) and the competitive advantage of very low iron growth requirement (82, 121).
RESISTANCE TO BILE.
The role of bile acids as carcinogens in the GI tract has been recently recognized (see above). However, not all BAs are considered cytotoxic. Secondary BAs are the products of bacterial dehydroxylation, dehydrogenation, and sulfation, or 7α-dehydroxylation of deconjugated bile salts and have been shown to stimulate colorectal epithelial proliferation in animals (9) and, in high concentrations in feces, blood, and bile, have been linked to cholesterol gallstone disease and colon cancer (173). A higher conversion of primary to secondary BAs in the GI tract is strongly stimulated by a diet high in red meats and fat (24, 25). A general mechanism by which secondary bile acids may act to promote tumorigenesis is by generating ROS and reactive nitrogen species (RNS), which can lead to increased DNA damage and then an increase in the mutation rates (80). As discussed above, 7α-dehydroxylation activity is present is several species of the genus Clostridium (C. absonum, C. scindens, C. bifermentans, C. limosum, and C. hylemonae) (221), whereas BSHs are widely distributed in the genera Lactobacillus and Bifidobacterium, members of the LAB, most of which are considered probiotics (23, 204). The importance of a probiotic blend resistant to BAs and capable of decreasing gut microbiota-mediated generation of secondary BAs is evident.
ADHESION.
The ability of probiotics to adhere to GI mucus is of considerable importance in their ability to exert a modulatory effect in situ. Moreover, naturally adhesive strains are of relevance for continuous production of probiotics using biofilm technology (discussed below). Mucus adherence has been extensively studied in several strains of probiotic LAB (22, 131, 256) and has revealed a broad repertoire of proteinaceous and nonproteinaceous surface components involved in adhesion to epithelial cells through passive or steric forces and electrostatic and hydrophobic interactions. These components include lipoteichoic acids and specific structures such as external appendages covered by lectins (reviewed in Ref. 241). The adhesion of probiotic bacteria to epithelial cells has been shown to prevent the establishment of pathogens; however, adhesion per se does not warrant persistence in the GI tract (150). Several studies that characterized LAB from different origins have shown that the ability to adhere to epithelial cells is strain dependent. The human cell lines Caco-2 and a mucus-secreting derivative (HT29-MTX cells) have been extensively and successfully used to identify adherent strains (47, 62, 63, 199, 232, 233).
COMPETITIVE EXCLUSION.
As stated above, probiotic strains with adhesive properties can compete with and prevent establishment of pathogenic bacteria by competitive exclusion. Although the mechanisms by which probiotics inhibit pathogenic activity are not entirely known, it has been shown that it goes beyond a mere physical barrier. Resta-Lenert and Barret (220) showed that exposure of cell monolayers to live but not heat inactivated probiotic S. thermophilus and L. acidophilus strains significantly limited adhesion, invasion, and physiological dysfunction induced by exposure to an enteroinvasive strain of E. coli. As stated in the previous section, several proteinaceous and nonproteinaceous surface components have been involved in adhesion to epithelial cells (see references above). The probiotic strains increased transepithelial resistance, which was accompanied by maintained (actin, zonulin-1) or enhanced (actinin, occludin) phosphorylation of cytoskeletal and TJ proteins. A similar effect has been demonstrated for a probiotic strain of L. plantarum, which had a protective effect against damage to the integrity of Caco-2 monolayers and the structure and distribution of TJ proteins by enteroinvasive E. coli (211). In addition to the above described mechanism of competitive exclusion, studies have indicated that specific probiotic strains can promote mucus secretion. Given that the mucus layer is the first line of defense against pathogens, increased mucus thickness can be beneficial in cases where the mucus layer has been compromised as a result of inflammation or disease (193).
Activities that reduce generation of mutagenic or genotoxic compounds.
A role for the comensal gut microbiota in the generation of mutagenic, carcinogenic, and/or genotoxic compounds in the GI tract has been established (see The Normal Intestinal Microbiota and the Microbiota Associated with CRC). In contrast, some commensal and probiomic bacteria have the potential to reduce such compounds and may diminish cancer risk (112, 149) via enzymatic activities that produce bioactive molecules like short-chain fatty acids, which are the endproducts of carbohydrate fermentation (specifically resistant starches and dietary fiber) by anaerobic bacteria. Fecal concentrations of different short-chain fatty acids include acetate > propionate > butyrate in a molar ratio of ∼60:20:20, respectively (262). Butyrate has been regarded as the most important nutrient for colonocytes and also has a major role in regulation of cell proliferation and differentiation (reviewed in Ref. 281). Butyrate is produced mainly in the proximal colon, where substrate availability is higher, and hence this region of the colon has a lower pH, which is significant because the lower pH enriches this region with the more acid-resistant LAB strains. A number of studies have shown that butyrate inhibits human colon carcinoma cell proliferation and induces apoptosis in human colon carcinoma cells (79, 195, 231). Additionally, butyrate possesses possible anticarcinogenic effects by suppressing the COX-2 expression in HT-29 and in Caco-2 cancer cell lines (190, 261) and promoting expression of differentiation markers such as alkaline phosphatase in colonic cell lines (78, 196). Butyrate also induces expression of the host's glutathione-S-transferases (GST) and other stress response genes (recently reviewed in Ref. 237). GSTs catalyze the reaction between glutathione and lipophilic compounds with electrophilic centers, which leads to the deactivation of toxic compounds, xenobiotics, and products of oxidative stress (81). Bacterial pathways of butyrate production have been characterized in Clostridium acetobutylicum, a solventogenic bacteria of technological importance, and more recently in Butyrivibrio fibrisolvens, a butyrate producer that has been found in the GI tract of humans, dogs, and cats (16, 17). The pathway for butyrate production involves two phases; the first converts acetyl-CoA to butyryl-CoA and requires four enzymes, thiolase, β-hydroxybutyryl-CoA dehyrogenase, crotonase, and butyryl-CoA dehyrogenase. Subsequently, butyryl-CoA is converted into butyrate via two alternative mechanisms, either a phosphotransbutyrylase and butyrate kinase convert butyryl-CoA to butyrate with the intermediate formation of butyryl-phosphate, or a butyryl-CoA:acetate CoA-transferase transfers the CoA moiety to external acetate, which leads to the formation of acetyl-CoA and butyrate (177, 185). Among LAB, no butyrate producers have been isolated in the genera Lactobacillus; however, genome sequencing of probiotic lactobacilli strains has shown the presence of the butyrate kinase gene in L. casei (46, 169). Additionally, a bacterial gut isolate identified as Enterococcus durans has been shown to be able to synthesize and secrete butyrate (217). Other bioactive compounds with cancer-preventive properties generated by microbial enzymes are equol (produced from the main isoflavin in soy, daidzein, see above) and conjugated linoleic acids (recently reviewed in Ref. 69).
BSHs deconjugate bile salts by the enzymatic hydrolysis of the C-24 N-acyl amide bond linking bile acids to their amino acid conjugates. They are in the choloylglycine hydrolase family (EC 3.5.1.24) and are widely distributed in gut microbial species (see above). The presence of this enzyme is correlated with resistance to bile in the GI tract and, hence, persistence and activity of probiotics in situ although some authors have questioned the beneficial effects of bacterial BSHs on the host (23, 201).
Probably the single most important bacterial enzymes correlated with cancer prevention are the ones with antioxidative activities because it has been extensively proven that exposure to ROS, RNS, or reactive lipid peroxidation products damages cellular DNA, causing oncogenic mutations or epigenetic changes (189). Hence, bacterial antioxidant enzymes are involved in both persistence of the probiotic in GI tract (see resistance to oxidative stress) and prevention of DNA damage by products of the oxidative metabolism. In this respect, studies have showed anti-inflammatory properties of Lactobacillus gasseri, L. plantarum, and L. lactis expressing manganese superoxide dismutase using the interleukin 10-deficient mouse or trinitrobenzene sulfonic acid rat models of colitis (48, 110).
Although specific aglycones have been reported to be carcinogenic in mice (see Production of aglycones), equol [7-hydroxy-3-(4′-hydroxyphenyl)-chroman] is an example of health-promoting aglycone. It is generated by the microbiota in response to soy isoflavone intake in some but not all humans and exhibits a wide range of biological properties (126, 242). In soybeans and nonfermented soy products, daidzein (one of the main isoflavonoids) occurs only in small amounts as aglycone (less than 5% of total daidzein), whereas the rest is mainly constituted of nonabsorbable, biologically inactive glycosides, acetylglycosides, or malonylglycosides (243). After ingestion, malonyglycosides are partially hydrolyzed in the small intestine, but a considerable portion of isoflavonoids reach the colon to be metabolized by specific bacterial groups. The bioactive compound equol, aglycone derived from daidzein (4′,7-dihydroxyisoflavone) exclusively by bacterial enzymatic activities in the colon, has been found to exhibit antiandrogenic activities, inhibiting prostate cancer development (155). Conversely, a recent study reported that in mice early exposure to equol was not chemopreventive against mammary tumors induced by 7,12-dimethylbenz(a)anthracene although for R-(+)equol there was a trend toward delaying tumor formation (37). It is not clear which bacterial species are involved in equol production in the colon. One study identified a mixed microbial culture consisting of Enterococcus faecium, Lactobacillus mucosae, Finegoldia magna, and Veillonella sp. (73) and, more recently, a new species of the genus Eggerthella (285) as equol producers. Another example of aglycones with potential anticancer activities is resveratrol-aglycone, which reduced the number of aberrant crypt foci/mouse in CF-1 mice (134). Table 3 lists the probiotic features of potential importance in CRC prevention and the bacterial activities previously linked to increased risk of CRC.
Production of probiotics: improving the survival and efficacy of strains.
In addition to desirable metabolic characteristics, probiotic strains or blends of strains should also be selected for high stability, viability, and survival to ensure that a sufficient number of viable bacteria will reach the GI tract (108). Improving probiotic cell survival during the production process itself is also important to reduce the uncertainty and variability in probiotic viability and improve consistence in biological effects. It is also essential to define the dose of probiotics that guarantees at least a transient modification of the homeostatic equilibrium of the gut microbiota. Variables like strain viability, associated strains, survival in the GI tract, and interactions with other bacteria and with the host must be assessed during experimental development and validation of probiotic blends, while the dose to administer will be a function of the probiotic behavior after assessment of the mentioned variables. Traditionally, the production approach to generate starter cultures has been to control and maintain a specific physiological state of the strains. The physiological state concept was introduced by Malek (160) and has been debated for decades. Nowadays, it is accepted that terms as “young cells”, “old cells”, “midlog phase cells”, and “stationary-phase cells” do not adequately define the age of bacterial cultures because the age of the cells can only be defined in relation to their inherent rates of cell division. The age of bacterial cultures during batch cultivation cannot be controlled, and consequently the environmental conditions are always transient. An improved approach to study and produce probiotics is to unequivocally control their physiological state by growing strains in continuous system under steady-state or pseudo-steady-state conditions. In this condition, rates of cell division are maintained constant, which enables the study of specific growth conditions in an environmental equilibrium. Therefore, continuous open culture systems must be selected for production of probiotics to eliminate variations in biological performance. Continuous open culture systems include biofilm operational systems that reproduce more closely the natural behavior of bacterial strains. Biofilm formation is a natural process in which microbial cells of single or multiple species adsorb to support without chemicals or polymers to entrap the bacteria. Microbial biofilms normally exhibit modified patterns of growth rate and gene expression, and their physiology and architecture have been extensively reviewed (67, 136, 144).
GI in vitro systems to evaluate new probiotics.
The process of validating new probiotic alternatives to manage growing pathologies associated to Western diets includes preclinical assessments (biochemical and cellular assays) and animal toxicity testing before validation in human clinical trials. An increasing number of promising probiotic candidates fail in phase III clinical trials, which highlights the need for a better method to accurately evaluate the absorption, distribution (stability or viability in case of living bacteria), metabolism, elimination, and toxicity (ADMET) of new probiotic candidates early in the validation process (4). Two preliminary methods commonly used to determine the toxicological and pharmacological profiles of potential drugs are in vitro cell cultures and animal models. Single cell type monolayer cell cultures cannot measure the systemic changes caused by the compound of interest and its metabolites, and animal experiments can take months to complete and require large quantities of product. New testing tools such as bioassays, computer modeling techniques, biomarkers, and validated surrogate endpoints are required to improve validation of new probiotic strains or combination of strains and to permit earlier termination of ineffective preparations and thereby reduce research costs.
Traditionally mouse intestinal models and genetic intervention to generate knockout mouse models have been the approach of choice to test and validate the physiological properties of probiotics in vivo (Table 1). Increasing evidence that the microbiota help maintain a healthy GI homeostasis has been derived from studies in germ-free rodents. These models have clearly demonstrated that, in the absence of an intestinal microbiota, intestinal motility is disrupted and also that normalization of motor patterns does not occur with all species of the intestinal microbiota. These studies have also introduced the concept of species specificity, demonstrating that effects on gut function obtained with one bacterial species cannot be extrapolated to another (45, 119). Recent transplant experiments using germ-free mouse recipients of microbiota from obese or lean mice demonstrated the clear influence of the microbiome populations on host obesity. This pioneer study confirmed how different energy-harvesting abilities of the adapted microbiota can influence food assimilation by the host (264). Following this approach, we should be able to find microbial populations more adapted to environmental toxins, specific diets, etc.. Novel in vitro models should be able to accurately assess the ADMET steps for new probiotic candidates early in the development process, allowing researchers to reduce costs and ethical concerns when screening characteristics that are host independent and replicable in vitro. In vitro systems cannot fully simulate the physiological processes of the gut wall, such as active or facilitated transport and local cellular and humoral immune system and feedback mechanisms. However, they are essential to better understand the characteristics of individual members of the microbiome as well as the molecular basis of the interaction between microorganisms and to explore practical applications. Consequently, to investigate the intestinal microbiota as an ecosystem, several in vitro systems have been developed. Examples of such models are the single bioreactor GI tract simulator, the Human Intestinal Microbial Ecosystem (SHIME) (180, 222, 254), a three-step colon model (157), a semicontinuous four-channel colon simulator (158, 159), and an in vitro microscale cell culture analog of the GI tract (171). Other in vitro systems designed to study the intestinal microbiota are the multicompartmental, dynamic, computer-controlled systems that simulate the human GI tract, TIM-1 (178), and large intestinal system TIM-2 (28, 178), which were used to investigate specific drug dosage forms for colonic or rectal delivery and the effect of the complex microbiota on the stability of drug compounds.
Conclusions
The bacteria associated with the human body outnumber our own cells by at least one order of magnitude, and the majority reside within our GI tract. The GI microbiota allows us to defend ourselves from pathogens and digest nutrients, but it is also implicated in human disease susceptibility including risk of CRC. The rapid evolution of “omics” technologies is beginning to provide researchers the information needed to modulate that immense bacterial ecosystem by introducing a carefully selected blend of beneficial organisms capable of survival, persistence, and delivery of a wide range of bioactive compounds. Certainly, the beneficial effects of probiotics, which are greatly influenced by our individual and varied genetic makeup and environment, including diet, have been long debated because of conflicting research. However, present and future studies will help overcome these barriers to enable development and more rigorous testing of probiotics as more natural and less disruptive treatments to prevent CRC and other GI-related malignancies.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
The authors thank P. K. Lund and E. Altermann for their insightful comments and critical reading of this review.
REFERENCES
- 1. Abdelali H, Cassand P, Soussotte V, Daubeze M, Bouley C, Narbonne JF. Effect of dairy products on initiation of precursor lesions of colon cancer in rats. Nutr Cancer 24: 121–132, 1995 [DOI] [PubMed] [Google Scholar]
- 2. Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest 12: 355–364, 1963 [PubMed] [Google Scholar]
- 3. Acharya A, Das I, Chandhok D, Saha T. Redox regulation in cancer: a double-edged sword with therapeutic potential. Oxid Med Cell Longev 3: 23–34, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Agres T. Finding blockbusters a struggle. Drug Discovery & Development. 2005 [Google Scholar]
- 5. Akdis M, Akdis CA. Mechanisms of allergen-specific immunotherapy. J Allergy Clin Immunol 119: 780–791, 2007 [DOI] [PubMed] [Google Scholar]
- 6. Alfonso P, Nunez A, Madoz-Gurpide J, Lombardia L, Sanchez L, Casal JI. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics 5: 2602–2611, 2005 [DOI] [PubMed] [Google Scholar]
- 7. Alnouti Y. Bile acid sulfation: a pathway of bile acid elimination and detoxification. Toxicol Sci 108: 225–246, 2009 [DOI] [PubMed] [Google Scholar]
- 8. Alvaro E, Andrieux C, Rochet V, Rigottier-Gois L, Lepercq P, Sutren M, Galan P, Duval Y, Juste C, Dore J. Composition and metabolism of the intestinal microbiota in consumers and non-consumers of yogurt. Br J Nutr 97: 126–133, 2007 [DOI] [PubMed] [Google Scholar]
- 9. Amaral JD, Viana RJ, Ramalho RM, Steer CJ, Rodrigues CM. Bile acids: regulation of apoptosis by ursodeoxycholic acid. J Lipid Res 50: 1721–1734, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amberger H. Different autochthonous models of colorectal cancer in the rat. J Cancer Res Clin Oncol 111: 157–159, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aoki K, Taketo MM. Adenomatous polyposis coli (APC): a multi-functional tumor suppressor gene. J Cell Sci 120: 3327–3335, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Aranganathan S, Selvam JP, Nalini N. Effect of hesperetin, a citrus flavonoid, on bacterial enzymes and carcinogen-induced aberrant crypt foci in colon cancer rats: a dose-dependent study. J Pharm Pharmacol 60: 1385–1392, 2008 [DOI] [PubMed] [Google Scholar]
- 13. Archibald FS, Fridovich I. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J Bacteriol 145: 442–451, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Archibald FS, Fridovich I. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J Bacteriol 146: 928–936, 1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arimochi H, Kinouchi T, Kataoka K, Kuwahara T, Ohnishi Y. Effect of intestinal bacteria on formation of azoxymethane-induced aberrant crypt foci in the rat colon. Biochem Biophys Res Commun 238: 753–757, 1997 [DOI] [PubMed] [Google Scholar]
- 16. Asanuma N, Ishiwata M, Yoshii T, Kikuchi M, Nishina Y, Hino T. Characterization and transcription of the genes involved in butyrate production in Butyrivibrio fibrisolvens type I and II strains. Curr Microbiol 51: 91–94, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Asanuma N, Kawato M, Ohkawara S, Hino T. Characterization and transcription of the genes encoding enzymes involved in butyrate production in Butyrivibrio fibrisolvens. Curr Microbiol 47: 203–207, 2003 [DOI] [PubMed] [Google Scholar]
- 18. Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res 5: 455–459, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Austin GL, Adair LS, Galanko JA, Martin CF, Satia JA, Sandler RS. A diet high in fruits and low in meats reduces the risk of colorectal adenomas. J Nutr 137: 999–1004, 2007 [DOI] [PubMed] [Google Scholar]
- 20. Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL, Cano RJ, Klaenhammer TR. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl Environ Microbiol 70: 5315–5322, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Azcarate-Peril MA, McAuliffe O, Altermann E, Lick S, Russell WM, Klaenhammer TR. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl Environ Microbiol 71: 5794–5804, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Azcarate Peril MA, Tallon R, Klaenhammer TR. Temporal gene expression and probiotic attributes of Lactobacillus acidophilus during growth in milk. J Dairy Sci 92: 870–886, 2009 [DOI] [PubMed] [Google Scholar]
- 23. Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol 72: 1729–1738, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol 15: 3329–3340, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 589: 47–65, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Berquin IM, Edwards IJ, Chen YQ. Multi-targeted therapy of cancer by omega-3 fatty acids. Cancer Lett 269: 363–377, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bi X, Lin Q, Foo TW, Joshi S, You T, Shen HM, Ong CN, Cheah PY, Eu KW, Hew CL. Proteomic analysis of colorectal cancer reveals alterations in metabolic pathways: mechanism of tumorigenesis. Mol Cell Proteomics 5: 1119–1130, 2006 [DOI] [PubMed] [Google Scholar]
- 28. Blanquet S, Zeijdner E, Beyssac E, Meunier JP, Denis S, Havenaar R, Alric M. A dynamic artificial gastrointestinal system for studying the behavior of orally administered drug dosage forms under various physiological conditions. Pharm Res 21: 585–591, 2004 [DOI] [PubMed] [Google Scholar]
- 29. Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncol 7: 149–156, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Bolognani F, Rumney CJ, Pool-Zobel BL, Rowland IR. Effect of lactobacilli, bifidobacteria and inulin on the formation of aberrant crypt foci in rats. Eur J Nutr 40: 293–300, 2001 [DOI] [PubMed] [Google Scholar]
- 31. Booth LA, Gilmore IT, Bilton RF. Secondary bile acid induced DNA damage in HT29 cells: are free radicals involved? Free Radic Res 26: 135–144, 1997 [DOI] [PubMed] [Google Scholar]
- 32. Bourlioux P, Koletzko B, Guarner F, Braesco V. The intestine and its microflora are partners for the protection of the host: report on the Danone Symposium “The Intelligent Intestine,” held in Paris, June 14, 2002. Am J Clin Nutr 78: 675–683, 2003 [DOI] [PubMed] [Google Scholar]
- 33. Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456: 507–510, 2008 [DOI] [PubMed] [Google Scholar]
- 34. Braat H, Rottiers P, Hommes DW, Huyghebaert N, Remaut E, Remon JP, van Deventer SJ, Neirynck S, Peppelenbosch MP, Steidler L. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn's disease. Clin Gastroenterol Hepatol 4: 754–759, 2006 [DOI] [PubMed] [Google Scholar]
- 35. Bray F, Guerra Yi M, Parkin DM. The comprehensive cancer monitoring programme in Europe. Eur J Public Health 13: 61–66, 2003 [DOI] [PubMed] [Google Scholar]
- 36. Broadbent JR, Larsen RL, Deibel V, Steele JL. Physiological and transcriptional response of Lactobacillus casei ATCC 334 to acid stress. J Bacteriol 192: 2445–2458, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown NM, Belles CA, Lindley SL, Zimmer-Nechemias L, Witte DP, Kim MO, Setchell KD. Mammary gland differentiation by early life exposure to enantiomers of the soy isoflavone metabolite equol. Food Chem Toxicol 48: 3042–3050, 2010 [DOI] [PubMed] [Google Scholar]
- 38. Brozovic A, Ambriovic-Ristov A, Osmak M. The relationship between cisplatin-induced reactive oxygen species, glutathione, and BCL-2 and resistance to cisplatin. Crit Rev Toxicol 40: 347–359, 2010 [DOI] [PubMed] [Google Scholar]
- 39. Bruno-Barcena JM, Andrus JM, Libby SL, Klaenhammer TR, Hassan HM. Expression of a heterologous manganese superoxide dismutase gene in intestinal lactobacilli provides protection against hydrogen peroxide toxicity. Appl Environ Microbiol 70: 4702–4710, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bruno-Barcena JM, Azcarate-Peril MA, Hassan HM. Role of antioxidant enzymes in bacterial resistance to organic acids. Appl Environ Microbiol 76: 2747–2753, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bruno-Barcena JM, Azcarate-Peril MA, Klaenhammer TR, Hassan HM. Marker-free chromosomal integration of the manganese superoxide dismutase gene (sodA) from Streptococcus thermophilus into Lactobacillus gasseri. FEMS Microbiol Lett 246: 91–101, 2005 [DOI] [PubMed] [Google Scholar]
- 42. Bruno-Barcena JM, Azcarate Peril MA, Sineriz F. Problems with genetically modified foods. In: Methods in Biotechnology, Vol 14: Food Microbiology Protocols, edited by Spencer JT, Ragout de Spencer AL. Totowa, NJ: Humana, 2001, p. 481–484 [Google Scholar]
- 43. Butt MS, Sultan MT. Green tea: nature's defense against malignancies. Crit Rev Food Sci Nutr 49: 463–473, 2009 [DOI] [PubMed] [Google Scholar]
- 44. Caderni G, Femia AP, Giannini A, Favuzza A, Luceri C, Salvadori M, Dolara P. Identification of mucin-depleted foci in the unsectioned colon of azoxymethane-treated rats: correlation with carcinogenesis. Cancer Res 63: 2388–2392, 2003 [PubMed] [Google Scholar]
- 45. Caenepeel P, Janssens J, Vantrappen G, Eyssen H, Coremans G. Interdigestive myoelectric complex in germ-free rats. Dig Dis Sci 34: 1180–1184, 1989 [DOI] [PubMed] [Google Scholar]
- 46. Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL. Genome sequence and comparative genome analysis of lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol 2009: 239–257, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Gionchetti P, Rizzello F, Campieri M, Brigidi P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol 125: 286–292, 2008 [DOI] [PubMed] [Google Scholar]
- 48. Carroll IM, Andrus JM, Bruno-Barcena JM, Klaenhammer TR, Hassan HM, Threadgill DS. Anti-inflammatory properties of Lactobacillus gasseri expressing manganese superoxide dismutase using the interleukin 10-deficient mouse model of colitis. Am J Physiol Gastrointest Liver Physiol 293: G729–G738, 2007 [DOI] [PubMed] [Google Scholar]
- 49. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313: 1126–1130, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castells A, Castellvi-Bel S, Balaguer F. Concepts in familial colorectal cancer: where do we stand and what is the future? Gastroenterology 137: 404–409, 2009 [DOI] [PubMed] [Google Scholar]
- 51. CDCUS United States Cancer Statistics: 1999–2005 Incidence and Mortality Web-based Report Atlanta [Online]. Cancer Statistics Working Group, Department of Health and Human Services, Centers for Disease Control and Prevention, and National Cancer Institute; http://apps.nccd.cdc.gov/uscs/[2009] [Google Scholar]
- 52. Challa A, Rao DR, Chawan CB, Shackelford L. Bifidobacterium longum and lactulose suppress azoxymethane-induced colonic aberrant crypt foci in rats. Carcinogenesis 18: 517–521, 1997 [DOI] [PubMed] [Google Scholar]
- 53. Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J Proteome Res 8: 352–361, 2009 [DOI] [PubMed] [Google Scholar]
- 54. Chang CL, Marra G, Chauhan DP, Ha HT, Chang DK, Ricciardiello L, Randolph A, Carethers JM, Boland CR. Oxidative stress inactivates the human DNA mismatch repair system. Am J Physiol Cell Physiol 283: C148–C154, 2002 [DOI] [PubMed] [Google Scholar]
- 55. Chen H. Recent advances in azo dye degrading enzyme research. Curr Protein Pept Sci 7: 101–111, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chouraqui JP, Van Egroo LD, Fichot MC. Acidified milk formula supplemented with bifidobacterium lactis: impact on infant diarrhea in residential care settings. J Pediatr Gastroenterol Nutr 38: 288–292, 2004 [DOI] [PubMed] [Google Scholar]
- 57. Christl SU, Gibson GR, Cummings JH. Role of dietary sulphate in the regulation of methanogenesis in the human large intestine. Gut 33: 1234–1238, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chung KT, Stevens SE, Jr, Cerniglia CE. The reduction of azo dyes by the intestinal microflora. Crit Rev Microbiol 18: 175–190, 1992 [DOI] [PubMed] [Google Scholar]
- 59. Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, van Sinderen D, O'Connor M, Harnedy N, O'Connor K, Henry C, O'Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O'Toole PW. Microbes and health sackler colloquium: Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA 108: 4586–4591, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Claesson MJ, O'Sullivan O, Wang Q, Nikkila J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O'Toole PW. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLos One 4: e6669, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cliffe LJ, Humphreys NE, Lane TE, Potten CS, Booth C, Grencis RK. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science 308: 1463–1465, 2005 [DOI] [PubMed] [Google Scholar]
- 62. Collado MC, Grzeskowiak L, Salminen S. Probiotic strains and their combination inhibit in vitro adhesion of pathogens to pig intestinal mucosa. Curr Microbiol 55: 260–265, 2007 [DOI] [PubMed] [Google Scholar]
- 63. Collado MC, Isolauri E, Salminen S. Specific probiotic strains and their combinations counteract adhesion of Enterobacter sakazakii to intestinal mucus. FEMS Microbiol Lett 285: 58–64, 2008 [DOI] [PubMed] [Google Scholar]
- 64. Cotter PD, Hill C. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev 67: 429–453, table of contents, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. da Silva FC, Valentin MD, Ferreira Fde O, Carraro DM, Rossi BM. Mismatch repair genes in Lynch syndrome: a review. Sao Paulo Med J 127: 46–51, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol 66: 487–495, 2008 [DOI] [PubMed] [Google Scholar]
- 67. Dagher SF, Ragout AL, Sineriz F, Bruno-Barcena JM. Cell immobilization for production of lactic acid biofilms do it naturally. Adv Appl Microbiol 71: 113–148, 2010 [DOI] [PubMed] [Google Scholar]
- 68. Danese S. Inflammatory bowel disease and inflammation-associated colon cancer: partners in crime (Abstract). Curr Drug Targets 9: 360, 2008 [DOI] [PubMed] [Google Scholar]
- 69. Davis CD, Milner JA. Gastrointestinal microflora, food components and colon cancer prevention. J Nutr Biochem 20: 743–752, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. De Angelis M, Gobbetti M. Lactobacillus sanfranciscensis CB1: manganese, oxygen, superoxide dismutase and metabolism. Appl Microbiol Biotechnol 51: 358–363, 1999 [DOI] [PubMed] [Google Scholar]
- 71. de Miranda NF, Nielsen M, Pereira D, van Puijenbroek M, Vasen HF, Hes FJ, van Wezel T, Morreau H. MUTYH-associated polyposis carcinomas frequently lose HLA class I expression—a common event amongst DNA-repair-deficient colorectal cancers. J Pathol 219: 69–76, 2009 [DOI] [PubMed] [Google Scholar]
- 72. de Moreno de LeBlanc A, LeBlanc JG, Perdigon G, Miyoshi A, Langella P, Azevedo V, Sesma F. Oral administration of a catalase-producing Lactococcus lactis can prevent a chemically induced colon cancer in mice. J Med Microbiol 57: 100–105, 2008 [DOI] [PubMed] [Google Scholar]
- 73. Decroos K, Vanhemmens S, Cattoir S, Boon N, Verstraete W. Isolation and characterisation of an equol-producing mixed microbial culture from a human faecal sample and its activity under gastrointestinal conditions. Arch Microbiol 183: 45–55, 2005 [DOI] [PubMed] [Google Scholar]
- 74. Delcenserie V, Martel D, Lamoureux M, Amiot J, Boutin Y, Roy D. Immunomodulatory effects of probiotics in the intestinal tract. Curr Issues Mol Biol 10: 37–54, 2008 [PubMed] [Google Scholar]
- 75. Delia P, Sansotta G, Donato V, Messina G, Frosina P, Pergolizzi S, De Renzis C, Famularo G. Prevention of radiation-induced diarrhea with the use of VSL#3, a new high-potency probiotic preparation. Am J Gastroenterol 97: 2150–2152, 2002 [DOI] [PubMed] [Google Scholar]
- 76. Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, Niesporek S, Noske A, Buckendahl A, Dietel M, Fiehn O. Metabolite profiling of human colon carcinoma—deregulation of TCA cycle and amino acid turnover (Abstract). Mol Cancer 7: 72, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dignass AU. Mechanisms and modulation of intestinal epithelial repair. Inflamm Bowel Dis 7: 68–77, 2001 [DOI] [PubMed] [Google Scholar]
- 78. Domokos M, Jakus J, Szeker K, Csizinszky R, Csiko G, Neogrady Z, Csordas A, Galfi P. Butyrate-induced cell death and differentiation are associated with distinct patterns of ROS in HT29-derived human colon cancer cells. Dig Dis Sci 55: 920–930, 2010 [DOI] [PubMed] [Google Scholar]
- 79. Dronamraju SS, Coxhead JM, Kelly SB, Mathers JC. Differential antineoplastic effects of butyrate in cells with and without a functioning DNA mismatch repair. Nutr Cancer 62: 105–115, 2010 [DOI] [PubMed] [Google Scholar]
- 80. Dvorak K, Payne CM, Chavarria M, Ramsey L, Dvorakova B, Bernstein H, Holubec H, Sampliner RE, Guy N, Condon A, Bernstein C, Green SB, Prasad A, Garewal HS. Bile acids in combination with low pH induce oxidative stress and oxidative DNA damage: relevance to the pathogenesis of Barrett's oesophagus. Gut 56: 763–771, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Economopoulos KP, Sergentanis TN. GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: A comprehensive meta-analysis. Eur J Cancer 46: 1617–1631, 2010 [DOI] [PubMed] [Google Scholar]
- 82. Elli M, Zink R, Rytz A, Reniero R, Morelli L. Iron requirement of Lactobacillus spp. in completely chemically defined growth media. J Appl Microbiol 88: 695–703, 2000 [DOI] [PubMed] [Google Scholar]
- 83. Elmadfa I, Meyer A, Nowak V, Hasenegger V, Putz P, Verstraeten R, Remaut-DeWinter AM, Kolsteren P, Dostalova J, Dlouhy P, Trolle E, Fagt S, Biltoft-Jensen A, Mathiessen J, Velsing Groth M, Kambek L, Gluskova N, Voutilainen N, Erkkila A, Vernay M, Krems C, Strassburg A, Vasquez-Caicedo AL, Urban C, Naska A, Efstathopoulou E, Oikonomou E, Tsiotas K, Bountziouka V, Benetou V, Trichopoulou A, Zajkas G, Kovacs V, Martos E, Heavey P, Kelleher C, Kennedy J, Turrini A, Selga G, Sauka M, Petkeviciene J, Klumbiene J, Holm Totland T, Andersen LF, Halicka E, Rejman K, Kowrygo B, Rodrigues S, Pinhao S, Ferreira LS, Lopes C, Ramos E, Vaz Almeida MD, Vlad M, Simcic M, Podgrajsek K, Serra Majem L, Roman Vinas B, Ngo J, Ribas Barba L, Becker V, Fransen H, Van Rossum C, Ocke M, Margetts B. European Nutrition and Health Report 2009. Forum Nutr 62: 1–405, 2009 [DOI] [PubMed] [Google Scholar]
- 84. Erdman SE, Poutahidis T. Roles for inflammation and regulatory T cells in colon cancer. Toxicol Pathol 38: 76–87, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, Matthews VB, Armes J, Bhathal PS, Hughes NR, Marcusson EG, Karras JG, Na S, Sedgwick JD, Hertzog PJ, Jenkins BJ. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest 118: 1727–1738, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Femia AP, Luceri C, Dolara P, Giannini A, Biggeri A, Salvadori M, Clune Y, Collins KJ, Paglierani M, Caderni G. Antitumorigenic activity of the prebiotic inulin enriched with oligofructose in combination with the probiotics Lactobacillus rhamnosus and Bifidobacterium lactis on azoxymethane-induced colon carcinogenesis in rats. Carcinogenesis 23: 1953–1960, 2002 [DOI] [PubMed] [Google Scholar]
- 87. Fenton HJH. Oxidation of tartaric acid in the presence of iron. J Chem Soc (Lond) 65: 897–910, 1984 [Google Scholar]
- 88. Fotiadis CI, Stoidis CN, Spyropoulos BG, Zografos ED. Role of probiotics, prebiotics and synbiotics in chemoprevention for colorectal cancer. World J Gastroenterol 14: 6453–6457, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fridovich I. Oxygen toxicity: a radical explanation. J Exp Biol 201: 1203–1209, 1998 [DOI] [PubMed] [Google Scholar]
- 91. Gallaher DD, Stallings WH, Blessing LL, Busta FF, Brady LJ. Probiotics, cecal microflora, and aberrant crypts in the rat colon. J Nutr 126: 1362–1371, 1996 [DOI] [PubMed] [Google Scholar]
- 92. Gamberini M, Leite LC. Proliferation of mouse fibroblasts induced by 1,2-dimethylhydrazine auto-oxidation: role of iron and free radicals. Biochem Biophys Res Commun 234: 44–47, 1997 [DOI] [PubMed] [Google Scholar]
- 93. Gaon D, Garcia H, Winter L, Rodriguez N, Quintas R, Gonzalez SN, Oliver G. Effect of Lactobacillus strains and Saccharomyces boulardii on persistent diarrhea in children. Medicina (B Aires) 63: 293–298, 2003 [PubMed] [Google Scholar]
- 94. Gargiulo S, Torrini M, Ollila S, Nasti S, Pastorino L, Cusano R, Bonelli L, Battistuzzi L, Mastracci L, Bruno W, Savarino V, Sciallero S, Borgonovo G, Nystrom M, Bianchi-Scarra G, Mareni C, Ghiorzo P. Germline MLH1 and MSH2 mutations in Italian pancreatic cancer patients with suspected Lynch syndrome. Fam Cancer 8: 547–553, 2009 [DOI] [PubMed] [Google Scholar]
- 95. Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell 140: 859–870, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gianotti L, Morelli L, Galbiati F, Rocchetti S, Coppola S, Beneduce A, Gilardini C, Zonenschain D, Nespoli A, Braga M. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J Gastroenterol 16: 167–175, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gibson GR, MacFarlane S, Macfarlane GT. Metabolic interactions involving sulphate-reducing and methanogenic bacteria in the human large intestine. FEMS Microbiol Ecol 12: 117–125, 2006 [Google Scholar]
- 98. Gill H, Prasad J. Probiotics, immunomodulation, and health benefits. Adv Exp Med Biol 606: 423–454, 2008 [DOI] [PubMed] [Google Scholar]
- 99. Gill HS, Guarner F. Probiotics and human health: a clinical perspective. Postgrad Med J 80: 516–526, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Giralt J, Regadera JP, Verges R, Romero J, de la Fuente I, Biete A, Villoria J, Cobo JM, Guarner F. Effects of probiotic Lactobacillus casei DN-114 001 in prevention of radiation-induced diarrhea: results from multicenter, randomized, placebo-controlled nutritional trial. Int J Radiat Oncol Biol Phys 71: 1213–1219, 2008 [DOI] [PubMed] [Google Scholar]
- 101. Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact 129: 141–170, 2000 [DOI] [PubMed] [Google Scholar]
- 102. Goffin P, Muscariello L, Lorquet F, Stukkens A, Prozzi D, Sacco M, Kleerebezem M, Hols P. Involvement of pyruvate oxidase activity and acetate production in the survival of Lactobacillus plantarum during the stationary phase of aerobic growth. Appl Environ Microbiol 72: 7933–7940, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Goldin BR, Gorbach SL. Effect of Lactobacillus acidophilus dietary supplements on 1,2-dimethylhydrazine dihydrochloride-induced intestinal cancer in rats. J Natl Cancer Inst 64: 263–265, 1980 [DOI] [PubMed] [Google Scholar]
- 104. Goldin BR, Gualtieri LJ, Moore RP. The effect of Lactobacillus GG on the initiation and promotion of DMH-induced intestinal tumors in the rat. Nutr Cancer 25: 197–204, 1996 [DOI] [PubMed] [Google Scholar]
- 105. Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol 44: 4136–4141, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gorbach SL, Goldin BR. The intestinal microflora and the colon cancer connection. Rev Infect Dis 12, Suppl 2: S252–S261, 1990 [DOI] [PubMed] [Google Scholar]
- 107. Guillen H, Curiel JA, Landete JM, Munoz R, Herraiz T. Characterization of a nitroreductase with selective nitroreduction properties in the food and intestinal lactic acid bacterium lactobacillus plantarum WCFS1. J Agric Food Chem 57: 10457–10465, 2009 [DOI] [PubMed] [Google Scholar]
- 108. Gupta V, Garg R. Probiotics. Indian J Med Microbiol 27: 202–209, 2009 [DOI] [PubMed] [Google Scholar]
- 109. Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29: 637–649, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Han W, Mercenier A, Ait-Belgnaoui A, Pavan S, Lamine F, van S, Kleerebezem M, 2nd, Salvador-Cartier C, Hisbergues M, Bueno L, Theodorou V, Fioramonti J. Improvement of an experimental colitis in rats by lactic acid bacteria producing superoxide dismutase. Inflamm Bowel Dis 12: 1044–1052, 2006 [DOI] [PubMed] [Google Scholar]
- 111. Hatakka K, Holma R, El-Nezami H, Suomalainen T, Kuisma M, Saxelin M, Poussa T, Mykkanen H, Korpela R. The influence of Lactobacillus rhamnosus LC705 together with Propionibacterium freudenreichii ssp. shermanii JS on potentially carcinogenic bacterial activity in human colon. Int J Food Microbiol 128: 406–410, 2008 [DOI] [PubMed] [Google Scholar]
- 112. Hayatsu H, Hayatsu T. Suppressing effect of Lactobacillus casei administration on the urinary mutagenicity arising from ingestion of fried ground beef in the human. Cancer Lett 73: 173–179, 1993 [DOI] [PubMed] [Google Scholar]
- 113. Herrera L, Kakati S, Gibas L, Pietrzak E, Sandberg AA. Gardner syndrome in a man with an interstitial deletion of 5q. Am J Med Genet 25: 473–476, 1986 [DOI] [PubMed] [Google Scholar]
- 114. Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res 69: 4918–4925, 2009 [DOI] [PubMed] [Google Scholar]
- 115. Hirono I. Natural carcinogenic products of plant origin. Crit Rev Toxicol 8: 235–277, 1981 [DOI] [PubMed] [Google Scholar]
- 116. Holmes RS. Alcohol dehydrogenases: a family of isozymes with differential functions. Alcohol Alcohol Suppl 2: 127–130, 1994 [PubMed] [Google Scholar]
- 117. Horie H, Zeisig M, Hirayama K, Midtvedt T, Moller L, Rafter J. Probiotic mixture decreases DNA adduct formation in colonic epithelium induced by the food mutagen 2-amino-9H-pyrido(2,3-b)indole in a human-flora associated mouse model. Eur J Cancer Prev 12: 101–107, 2003 [DOI] [PubMed] [Google Scholar]
- 118. Humblot C, Murkovic M, Rigottier-Gois L, Bensaada M, Bouclet A, Andrieux C, Anba J, Rabot S. Beta-glucuronidase in human intestinal microbiota is necessary for the colonic genotoxicity of the food-borne carcinogen 2-amino-3-methylimidazo(4,5-f)quinoline in rats. Carcinogenesis 28: 2419–2425, 2007 [DOI] [PubMed] [Google Scholar]
- 119. Husebye E, Hellstrom PM, Sundler F, Chen J, Midtvedt T. Influence of microbial species on small intestinal myoelectric activity and transit in germ-free rats. Am J Physiol Gastrointest Liver Physiol 280: G368–G380, 2001 [DOI] [PubMed] [Google Scholar]
- 120. Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med (Maywood) 229: 586–597, 2004 [DOI] [PubMed] [Google Scholar]
- 121. Imbert M, Blondeau R. On the iron requirement of lactobacilli grown in chemically defined medium. Curr Microbiol 37: 64–66, 1998 [DOI] [PubMed] [Google Scholar]
- 122. Inagaki H, Suzuki T, Nomoto K, Yoshikai Y. Increased susceptibility to primary infection with Listeria monocytogenes in germfree mice may be due to lack of accumulation of L-selectin+ CD44+ T cells in sites of inflammation. Infect Immun 64: 3280–3287, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ishikawa H, Akedo I, Otani T, Suzuki T, Nakamura T, Takeyama I, Ishiguro S, Miyaoka E, Sobue T, Kakizoe T. Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int J Cancer 116: 762–767, 2005 [DOI] [PubMed] [Google Scholar]
- 124. Ito K, Nakanishi M, Lee WC, Sasaki H, Zenno S, Saigo K, Kitade Y, Tanokura M. Three-dimensional structure of AzoR from Escherichia coli. An oxidereductase conserved in microorganisms. J Biol Chem 281: 20567–20576, 2006 [DOI] [PubMed] [Google Scholar]
- 125. Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4: 337–349, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Izumi T, Piskula MK, Osawa S, Obata A, Tobe K, Saito M, Kataoka S, Kubota Y, Kikuchi M. Soy isoflavone aglycones are absorbed faster and in higher amounts than their glucosides in humans. J Nutr 130: 1695–1699, 2000 [DOI] [PubMed] [Google Scholar]
- 127. Jankowski J, Juskiewicz J, Gulewicz K, Lecewicz A, Slominski BA, Zdunczyk Z. The effect of diets containing soybean meal, soybean protein concentrate, and soybean protein isolate of different oligosaccharide content on growth performance and gut function of young turkeys. Poult Sci 88: 2132–2140, 2009 [DOI] [PubMed] [Google Scholar]
- 128. Johnson IT, Lund EK. Review article: nutrition, obesity and colorectal cancer. Aliment Pharmacol Ther 26: 161–181, 2007 [DOI] [PubMed] [Google Scholar]
- 129. Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA 105: 13580–13585, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kato I, Kobayashi S, Yokokura T, Mutai M. Antitumor activity of Lactobacillus casei in mice. Gann 72: 517–523, 1981 [PubMed] [Google Scholar]
- 131. Khalil R. Evidence for probiotic potential of a capsular-producing Streptococcus thermophilus CHCC 3534 strain. Pol J Microbiol 58: 49–55, 2009 [PubMed] [Google Scholar]
- 132. Kim DH, Jin YH. Intestinal bacterial beta-glucuronidase activity of patients with colon cancer. Arch Pharmacol Res (Seoul) 24: 564–567, 2001 [DOI] [PubMed] [Google Scholar]
- 133. Kim JY, Kwon O. Garlic intake and cancer risk: an analysis using the Food and Drug Administration's evidence-based review system for the scientific evaluation of health claims. Am J Clin Nutr 89: 257–264, 2009 [DOI] [PubMed] [Google Scholar]
- 134. Kineman BD, Au A, Paiva NL, Kaiser MS, Brummer EC, Birt DF. Transgenic alfalfa that accumulates piceid (trans-resveratrol-3-O-beta-D-glucopyranoside) requires the presence of beta-glucosidase to inhibit the formation of aberrant crypt foci in the colon of CF-1 mice. Nutr Cancer 58: 66–74, 2007 [DOI] [PubMed] [Google Scholar]
- 135. Klampfer L. Cytokines, inflammation and colon cancer. Curr Cancer Drug Targets. 11: 451–64, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kolter R. Biofilms in lab and nature: a molecular geneticist's voyage to microbial ecology. Int Microbiol 13: 1–7, 2010 [DOI] [PubMed] [Google Scholar]
- 137. Kulkarni N, Reddy BS. Inhibitory effect of Bifidobacterium longum cultures on the azoxymethane-induced aberrant crypt foci formation and fecal bacterial beta-glucuronidase. Proc Soc Exp Biol Med 207: 278–283, 1994 [DOI] [PubMed] [Google Scholar]
- 138. Kullisaar T, Zilmer M, Mikelsaar M, Vihalemm T, Annuk H, Kairane C, Kilk A. Two antioxidative lactobacilli strains as promising probiotics. Int J Food Microbiol 72: 215–224, 2002 [DOI] [PubMed] [Google Scholar]
- 139. Kumar A, Singh NK, Sinha PR. Inhibition of 1,2-dimethylhydrazine induced colon genotoxicity in rats by the administration of probiotic curd. Mol Biol Rep 37: 1373–1376, 2009 [DOI] [PubMed] [Google Scholar]
- 140. Kumar A, Singh NK, Sinha PR, Kumar R. Intervention of Acidophilus-casei dahi and wheat bran against molecular alteration in colon carcinogenesis. Mol Biol Rep 37: 621–627, 2010 [DOI] [PubMed] [Google Scholar]
- 141. Kumar A, Wu H, Collier-Hyams LS, Hansen JM, Li T, Yamoah K, Pan ZQ, Jones DP, Neish AS. Commensal bacteria modulate cullin-dependent signaling via generation of reactive oxygen species. EMBO J 26: 4457–4466, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kuper H, Adami HO, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med 248: 171–183, 2000 [DOI] [PubMed] [Google Scholar]
- 143. Landriscina M, Maddalena F, Laudiero G, Esposito F. Adaptation to oxidative stress, chemoresistance, and cell survival. Antioxid Redox Signal 11: 2701–2716, 2009 [DOI] [PubMed] [Google Scholar]
- 144. Lazar V, Chifiriuc MC. Architecture and physiology of microbial biofilms. Roum Arch Microbiol Immunol 69: 95–107, 2010 [PubMed] [Google Scholar]
- 145. Le Leu RK, Hu Y, Brown IL, Woodman RJ, Young GP. Synbiotic intervention of Bifidobacterium lactis and resistant starch protects against colorectal cancer development in rats. Carcinogenesis 31: 246–251, 2009 [DOI] [PubMed] [Google Scholar]
- 146. Leedham SJ, Schier S, Thliveris AT, Halberg RB, Newton MA, Wright NA. From gene mutations to tumours—stem cells in gastrointestinal carcinogenesis. Cell Prolif 38: 387–405, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102: 11070–11075, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Li W, Li CB. Lack of inhibitory effects of lactic acid bacteria on 1,2-dimethylhydrazine-induced colon tumors in rats. World J Gastroenterol 9: 2469–2473, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Lidbeck A, Nord CE, Gustafsson JA, Rafter J. Lactobacilli, anticarcinogenic activities and human intestinal microflora. Eur J Cancer Prev 1: 341–353, 1992 [DOI] [PubMed] [Google Scholar]
- 150. Lievin-Le, Moal V, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev 19: 315–337, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Linwu SW, Syu CJ, Chen YL, Wang AH, Peng FC. Characterization of Escherichia coli nitroreductase NfsB in the metabolism of nitrobenzodiazepines. Biochem Pharmacol 78: 96–103, 2009 [DOI] [PubMed] [Google Scholar]
- 152. Liong MT. Probiotics: a critical review of their potential role as antihypertensives, immune modulators, hypocholesterolemics, and perimenopausal treatments. Nutr Rev 65: 316–328, 2007 [DOI] [PubMed] [Google Scholar]
- 153. Liong MT. Roles of probiotics and prebiotics in colon cancer prevention: Postulated mechanisms and in-vivo evidence. Int J Mol Sci 9: 854–863, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Liu Z, Qin H, Yang Z, Xia Y, Liu W, Yang J, Jiang Y, Zhang H, Wang Y, Zheng Q. Randomised clinical trial: the effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery - a double-blind study. Aliment Pharmacol Ther 33: 50–63, 2011 [DOI] [PubMed] [Google Scholar]
- 155. Lund TD, Munson DJ, Haldy ME, Setchell KD, Lephart ED, Handa RJ. Equol is a novel anti-androgen that inhibits prostate growth and hormone feedback. Biol Reprod 70: 1188–1195, 2004 [DOI] [PubMed] [Google Scholar]
- 156. Macfarlane GT, Blackett KL, Nakayama T, Steed H, Macfarlane S. The gut microbiota in inflammatory bowel disease. Curr Pharm Des 15: 1528–1536, 2009 [DOI] [PubMed] [Google Scholar]
- 157. Macfarlane GT, Macfarlane S, Gibson GR. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35: 180–187, 1998 [DOI] [PubMed] [Google Scholar]
- 158. Mahler GJ, Esch MB, Glahn RP, Shuler ML. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol Bioeng 104: 193–205, 2009 [DOI] [PubMed] [Google Scholar]
- 159. Makivuokko HA, Saarinen MT, Ouwehand AC, Rautonen NE. Effects of lactose on colon microbial community structure and function in a four-stage semi-continuous culture system. Biosci Biotechnol Biochem 70: 2056–2063, 2006 [DOI] [PubMed] [Google Scholar]
- 160. Malek I. The physiological state of microorganisms during continuous culture. In: Continuous Cultivation of Microorganisms: A Symposium. Prague, Czech Republic: Academis Publishing House Czechoslovak Academy of Sciences, 1958 [Google Scholar]
- 161. Mantovani A, Garlanda C, Allavena P. Molecular pathways and targets in cancer-related inflammation. Ann Med 42: 161–170, 2010 [DOI] [PubMed] [Google Scholar]
- 162. Marana SR. Molecular basis of substrate specificity in family 1 glycoside hydrolases. IUBMB Life 58: 63–73, 2006 [DOI] [PubMed] [Google Scholar]
- 163. Markowitz VM, Szeto E, Palaniappan K, Grechkin Y, Chu K, Chen IM, Dubchak I, Anderson I, Lykidis A, Mavromatis K, Ivanova NN, Kyrpides NC. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res 36: D528–D533, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Marotta F, Naito Y, Minelli E, Tajiri H, Bertuccelli J, Wu CC, Min CH, Hotten P, Fesce E. Chemopreventive effect of a probiotic preparation on the development of preneoplastic and neoplastic colonic lesions: an experimental study. Hepatogastroenterology 50: 1914–1918, 2003 [PubMed] [Google Scholar]
- 165. Marshall JR. Prevention of colorectal cancer: diet, chemoprevention, and lifestyle. Gastroenterol Clin North Am 37: 73–82, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Martin FP, Wang Y, Yap IK, Sprenger N, Lindon JC, Rezzi S, Kochhar S, Holmes E, Nicholson JK. Topographical variation in murine intestinal metabolic profiles in relation to microbiome speciation and functional ecological activity. J Proteome Res 8: 3464–3474, 2009 [DOI] [PubMed] [Google Scholar]
- 167. Maruti SS, Chang JL, Prunty JA, Bigler J, Schwarz Y, Li SS, Li L, King IB, Potter JD, Lampe JW. Serum beta-glucuronidase activity in response to fruit and vegetable supplementation: a controlled feeding study. Cancer Epidemiol Biomarkers Prev 17: 1808–1812, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Mason KL, Huffnagle GB, Noverr MC, Kao JY. Overview of gut immunology. In: GI Microbiota and Regulation of the Immune System, edited by Huffnagle GB, Noverr MC. Austin, TX: Landes Bioscience and Springer Science+Business Media, 2008, p. 1–14 [Google Scholar]
- 169. Maze A, Boel G, Zuniga M, Bourand A, Loux V, Yebra MJ, Monedero V, Correia K, Jacques N, Beaufils S, Poncet S, Joyet P, Milohanic E, Casaregola S, Auffray Y, Perez-Martinez G, Gibrat JF, Zagorec M, Francke C, Hartke A, Deutscher J. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J Bacteriol 192: 2647–2648, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453: 620–625, 2008 [DOI] [PubMed] [Google Scholar]
- 171. McAuliffe GJ, Chang JY, Glahn RP, Shuler ML. Development of a gastrointestinal tract microscale cell culture analog to predict drug transport. Mol Cell Biomech 5: 119–132, 2008 [PubMed] [Google Scholar]
- 172. McBain AJ, Macfarlane GT. Ecological and physiological studies on large intestinal bacteria in relation to production of hydrolytic and reductive enzymes involved in formation of genotoxic metabolites. J Med Microbiol 47: 407–416, 1998 [DOI] [PubMed] [Google Scholar]
- 173. McGarr SE, Ridlon JM, Hylemon PB. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J Clin Gastroenterol 39: 98–109, 2005 [PubMed] [Google Scholar]
- 174. McIntosh GH, Royle PJ, Playne MJ. A probiotic strain of L. acidophilus reduces DMH-induced large intestinal tumors in male Sprague-Dawley rats. Nutr Cancer 35: 153–159, 1999 [DOI] [PubMed] [Google Scholar]
- 175. McLean MH, Murray GI, Stewart KN, Norrie G, Mayer C, Hold GL, Thomson J, Fyfe N, Hope M, Mowat NA, Drew JE, El-Omar EM. The inflammatory microenvironment in colorectal neoplasia. PLos One 6: e15366, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer 9: 665–674, 2009 [DOI] [PubMed] [Google Scholar]
- 177. Miller TL, Jenesel SE. Enzymology of butyrate formation by Butyrivibrio fibrisolvens. J Bacteriol 138: 99–104, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Minekus M, Smeets-Peeters M, Bernalier A, Marol-Bonnin S, Havenaar R, Marteau P, Alric M, Fonty G, Huis in't Veld JH. A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl Microbiol Biotechnol 53: 108–114, 1999 [DOI] [PubMed] [Google Scholar]
- 179. Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet 1: 229–233, 1992 [DOI] [PubMed] [Google Scholar]
- 180. Molly K, Van de Woestyne M, Verstraete W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol 39: 254–258, 1993 [DOI] [PubMed] [Google Scholar]
- 181. Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol 61: 3202–3207, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun 21: 532–539, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Moreno A, Arus C. Quantitative and qualitative characterization of 1H NMR spectra of colon tumors, normal mucosa and their perchloric acid extracts: decreased levels of myo-inositol in tumours can be detected in intact biopsies. NMR Biomed 9: 33–45, 1996 [DOI] [PubMed] [Google Scholar]
- 184. Moreno A, Rey M, Montane JM, Alonso J, Arus C. 1H NMR spectroscopy of colon tumors and normal mucosal biopsies; elevated taurine levels and reduced polyethyleneglycol absorption in tumors may have diagnostic significance. NMR Biomed 6: 111–118, 1993 [DOI] [PubMed] [Google Scholar]
- 185. Mullany P, Clayton CL, Pallen MJ, Slone R, al-Saleh A, Tabaqchali S. Genes encoding homologues of three consecutive enzymes in the butyrate/butanol-producing pathway of Clostridium acetobutylicum are clustered on the Clostridium difficile chromosome. FEMS Microbiol Lett 124: 61–67, 1994 [DOI] [PubMed] [Google Scholar]
- 186. Nakamura J, Kubota Y, Miyaoka M, Saitoh T, Mizuno F, Benno Y. Comparison of four microbial enzymes in Clostridia and Bacteroides isolated from human feces. Microbiol Immunol 46: 487–490, 2002 [DOI] [PubMed] [Google Scholar]
- 187. Nielsen M, Joerink-van de Beld MC, Jones N, Vogt S, Tops CM, Vasen HF, Sampson JR, Aretz S, Hes FJ. Analysis of MUTYH genotypes and colorectal phenotypes in patients With MUTYH-associated polyposis. Gastroenterology 136: 471–476, 2009 [DOI] [PubMed] [Google Scholar]
- 188. Nishikawa M, Tamada A, Hyoudou K, Umeyama Y, Takahashi Y, Kobayashi Y, Kumai H, Ishida E, Staud F, Yabe Y, Takakura Y, Yamashita F, Hashida M. Inhibition of experimental hepatic metastasis by targeted delivery of catalase in mice. Clin Exp Metastasis 21: 213–221, 2004 [DOI] [PubMed] [Google Scholar]
- 189. Nowsheen S, Wukovich RL, Aziz K, Kalogerinis PT, Richardson CC, Panayiotidis MI, Bonner WM, Sedelnikova OA, Georgakilas AG. Accumulation of oxidatively induced clustered DNA lesions in human tumor tissues. Mutat Res 674: 131–136, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Nurmi JT, Puolakkainen PA, Rautonen NE. Bifidobacterium Lactis sp. 420 up-regulates cyclooxygenase (Cox)-1 and down-regulates Cox-2 gene expression in a Caco-2 cell culture model. Nutr Cancer 51: 83–92, 2005 [DOI] [PubMed] [Google Scholar]
- 191. O'Keefe SJ. Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol 24: 51–58, 2008 [DOI] [PubMed] [Google Scholar]
- 192. O'Keefe SJ, Chung D, Mahmoud N, Sepulveda AR, Manafe M, Arch J, Adada H, van der Merwe T. Why do African Americans get more colon cancer than Native Africans? J Nutr 137: 175S–182S, 2007 [DOI] [PubMed] [Google Scholar]
- 193. Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol 298: G807–G819, 2010 [DOI] [PubMed] [Google Scholar]
- 194. Oka K, Tanaka T, Enoki T, Yoshimura K, Ohshima M, Kubo M, Murakami T, Gondou T, Minami Y, Takemoto Y, Harada E, Tsushimi T, Li TS, Traganos F, Darzynkiewicz Z, Hamano K. DNA damage signaling is activated during cancer progression in human colorectal carcinoma. Cancer Biol Ther 9: 246–252, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Ooi CC, Good NM, Williams DB, Lewanowitsch T, Cosgrove LJ, Lockett TJ, Head RJ. Efficacy of butyrate analogues in HT-29 cancer cells. Clin Exp Pharmacol Physiol 37: 482–489, 2010 [DOI] [PubMed] [Google Scholar]
- 196. Orchel A, Dzierzewicz Z, Parfiniewicz B, Weglarz L, Wilczok T. Butyrate-induced differentiation of colon cancer cells is PKC and JNK dependent. Dig Dis Sci 50: 490–498, 2005 [DOI] [PubMed] [Google Scholar]
- 197. Osterlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P, Kouri M, Elomaa I, Joensuu H. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: a randomised study. Br J Cancer 97: 1028–1034, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Ouwehand AC, Salminen S, Isolauri E. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82: 279–289, 2002 [PubMed] [Google Scholar]
- 199. Ouwehand AC, Tuomola EM, Tolkko S, Salminen S. Assessment of adhesion properties of novel probiotic strains to human intestinal mucus. Int J Food Microbiol 64: 119–126, 2001 [DOI] [PubMed] [Google Scholar]
- 200. Paolillo R, Romano Carratelli C, Sorrentino S, Mazzola N, Rizzo A. Immunomodulatory effects of Lactobacillus plantarum on human colon cancer cells. Int Immunopharmacol 9: 1265–1271, 2009 [DOI] [PubMed] [Google Scholar]
- 201. Patel AK, Singhania RR, Pandey A, Chincholkar SB. Probiotic bile salt hydrolase: Current developments and perspectives. Appl Biochem Biotechnol 162: 166–180, 2010 [DOI] [PubMed] [Google Scholar]
- 202. Perdigon G, Maldonado Galdeano C, Valdez JC, Medici M. Interaction of lactic acid bacteria with the gut immune system. Eur J Clin Nutr 56, Suppl 4: S21–S26, 2002 [DOI] [PubMed] [Google Scholar]
- 203. Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3: 417–427, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Pfeiler EA, Azcarate-Peril MA, Klaenhammer TR. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J Bacteriol 189: 4624–4634, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Pohjavuori E, Viljanen M, Korpela R, Kuitunen M, Tiittanen M, Vaarala O, Savilahti E. Lactobacillus GG effect in increasing IFN-gamma production in infants with cow's milk allergy. J Allergy Clin Immunol 114: 131–136, 2004 [DOI] [PubMed] [Google Scholar]
- 206. Pool-Zobel BL, Neudecker C, Domizlaff I, Ji S, Schillinger U, Rumney C, Moretti M, Vilarini I, Scassellati-Sforzolini R, Rowland I. Lactobacillus- and bifidobacterium-mediated antigenotoxicity in the colon of rats. Nutr Cancer 26: 365–380, 1996 [DOI] [PubMed] [Google Scholar]
- 207. Pool-Zobel BL, Sauer J. Overview of experimental data on reduction of colorectal cancer risk by inulin-type fructans. J Nutr 137: 2580S–2584S, 2007 [DOI] [PubMed] [Google Scholar]
- 208. Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. APC mutations occur early during colorectal tumorigenesis. Nature 359: 235–237, 1992 [DOI] [PubMed] [Google Scholar]
- 209. Poyart C, Berche P, Trieu-Cuot P. Characterization of superoxide dismutase genes from gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett 131: 41–45, 1995 [DOI] [PubMed] [Google Scholar]
- 210. Purohit DH, Hassan AN, Bhatia E, Zhang X, Dwivedi C. Rheological, sensorial, and chemopreventive properties of milk fermented with exopolysaccharide-producing lactic cultures. J Dairy Sci 92: 847–856, 2009 [DOI] [PubMed] [Google Scholar]
- 211. Qin H, Zhang Z, Hang X, Jiang YL. plantarum prevents enteroinvasive Escherichia coli-induced tight junction proteins changes in intestinal epithelial cells (Abstract). BMC Microbiol 9: 63, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Rafter J, Bennett M, Caderni G, Clune Y, Hughes R, Karlsson PC, Klinder A, O'Riordan M, O'Sullivan GC, Pool-Zobel B, Rechkemmer G, Roller M, Rowland I, Salvadori M, Thijs H, Van Loo J, Watzl B, Collins JK. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am J Clin Nutr 85: 488–496, 2007 [DOI] [PubMed] [Google Scholar]
- 214. Ramasamy S, Singh S, Taniere P, Langman MJ, Eggo MC. Sulfide-detoxifying enzymes in the human colon are decreased in cancer and upregulated in differentiation. Am J Physiol Gastrointest Liver Physiol 291: G288–G296, 2006 [DOI] [PubMed] [Google Scholar]
- 215. Randi G, Edefonti V, Ferraroni M, La Vecchia C, Decarli A. Dietary patterns and the risk of colorectal cancer and adenomas. Nutr Rev 68: 389–408, 2010 [DOI] [PubMed] [Google Scholar]
- 216. Rao CV, Sanders ME, Indranie C, Simi B, Reddy BS. Prevention of colonic preneoplastic lesions by the probiotic Lactobacillus acidophilus NCFMTM in F344 rats. Int J Oncol 14: 939–944, 1999 [DOI] [PubMed] [Google Scholar]
- 217. Raz I, Gollop N, Polak-Charcon S, Schwartz B. Isolation and characterisation of new putative probiotic bacteria from human colonic flora. Br J Nutr 97: 725–734, 2007 [DOI] [PubMed] [Google Scholar]
- 218. Reddy BS, Rivenson A. Inhibitory effect of Bifidobacterium longum on colon, mammary, and liver carcinogenesis induced by 2-amino-3-methylimidazo(4,5-f)quinoline, a food mutagen. Cancer Res 53: 3914–3918, 1993 [PubMed] [Google Scholar]
- 219. Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, Roberfroid M, Rowland I, Cherbut C, Klaenhammer TR. New scientific paradigms for probiotics and prebiotics. J Clin Gastroenterol 37: 105–118, 2003 [DOI] [PubMed] [Google Scholar]
- 220. Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 52: 988–997, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47: 241–259, 2006 [DOI] [PubMed] [Google Scholar]
- 222. Ritter P, Kohler C, von Ah U. Evaluation of the passage of Lactobacillus gasseri K7 and bifidobacteria from the stomach to intestines using a single reactor model (Abstract). BMC Microbiol 9: 87, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Robben J, Caenepeel P, Van Eldere J, Eyssen H. Effects of intestinal microbial bile salt sulfatase activity on bile salt kinetics in gnotobiotic rats. Gastroenterology 94: 494–502, 1988 [DOI] [PubMed] [Google Scholar]
- 224. Roessner A, Kuester D, Malfertheiner P, Schneider-Stock R. Oxidative stress in ulcerative colitis-associated carcinogenesis. Pathol Res Pract 204: 511–524, 2008 [DOI] [PubMed] [Google Scholar]
- 225. Roldan MD, Perez-Reinado E, Castillo F, Moreno-Vivian C. Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol Rev 32: 474–500, 2008 [DOI] [PubMed] [Google Scholar]
- 226. Roller M, Pietro Femia A, Caderni G, Rechkemmer G, Watzl B. Intestinal immunity of rats with colon cancer is modulated by oligofructose-enriched inulin combined with Lactobacillus rhamnosus and Bifidobacterium lactis. Br J Nutr 92: 931–938, 2004 [DOI] [PubMed] [Google Scholar]
- 227. Roselli M, Finamore A, Britti MS, Mengheri E. Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br J Nutr 95: 1177–1184, 2006 [DOI] [PubMed] [Google Scholar]
- 228. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9: 313–323, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Rowland IR. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des 15: 1524–1527, 2009 [DOI] [PubMed] [Google Scholar]
- 230. Rowland IR, Rumney CJ, Coutts JT, Lievense LC. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 19: 281–285, 1998 [DOI] [PubMed] [Google Scholar]
- 231. Roy MJ, Dionne S, Marx G, Qureshi I, Sarma D, Levy E, Seidman EG. In vitro studies on the inhibition of colon cancer by butyrate and carnitine. Nutrition 25: 1193–1201, 2009 [DOI] [PubMed] [Google Scholar]
- 232. Ruas-Madiedo P, Gueimonde M, Margolles A, de los Reyes-Gavilan CG, Salminen S. Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J Food Prot 69: 2011–2015, 2006 [DOI] [PubMed] [Google Scholar]
- 233. Salazar N, Ruas-Madiedo P, Kolida S, Collins M, Rastall R, Gibson G, de Los Reyes-Gavilan CG. Exopolysaccharides produced by Bifidobacterium longum IPLA E44 and Bifidobacterium animalis subsp. lactis IPLA R1 modify the composition and metabolic activity of human faecal microbiota in pH-controlled batch cultures. Int J Food Microbiol 135: 260–267, 2009 [DOI] [PubMed] [Google Scholar]
- 234. Salzman NH, Bevins CL. Negative interactions with the microbiota: IBD. Adv Exp Med Biol 635: 67–78, 2008 [DOI] [PubMed] [Google Scholar]
- 235. Sanders JW, Leenhouts KJ, Haandrikman AJ, Venema G, Kok J. Stress response in Lactococcus lactis: cloning, expression analysis, and mutation of the lactococcal superoxide dismutase gene. J Bacteriol 177: 5254–5260, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Scanlan PD, Shanahan F, Clune Y, Collins JK, O'Sullivan GC, O'Riordan M, Holmes E, Wang Y, Marchesi JR. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol 10: 789–798, 2008 [DOI] [PubMed] [Google Scholar]
- 237. Scharlau D, Borowicki A, Habermann N, Hofmann T, Klenow S, Miene C, Munjal U, Stein K, Glei M. Mechanisms of primary cancer prevention by butyrate and other products formed during gut flora-mediated fermentation of dietary fibre. Mutat Res 682: 39–53, 2009 [DOI] [PubMed] [Google Scholar]
- 238. Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res Health 30: 38–41; 44–7, 2007 [PMC free article] [PubMed] [Google Scholar]
- 239. Seitz HK, Simanowski UA, Garzon FT, Rideout JM, Peters TJ, Koch A, Berger MR, Einecke H, Maiwald M. Possible role of acetaldehyde in ethanol-related rectal cocarcinogenesis in the rat. Gastroenterology 98: 406–413, 1990 [DOI] [PubMed] [Google Scholar]
- 240. Serrazanetti DI, Guerzoni ME, Corsetti A, Vogel R. Metabolic impact and potential exploitation of the stress reactions in lactobacilli. Food Microbiol 26: 700–711, 2009 [DOI] [PubMed] [Google Scholar]
- 241. Servin AL, Coconnier MH. Adhesion of probiotic strains to the intestinal mucosa and interaction with pathogens. Best Pract Res Clin Gastroenterol 17: 741–754, 2003 [DOI] [PubMed] [Google Scholar]
- 242. Setchell KD, Clerici C. Equol: pharmacokinetics and biological actions. J Nutr 140: 1363S–1368S, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Setchell KD, Cole SJ. Variations in isoflavone levels in soy foods and soy protein isolates and issues related to isoflavone databases and food labeling. J Agric Food Chem 51: 4146–4155, 2003 [DOI] [PubMed] [Google Scholar]
- 244. Shackelford LA, Rao DR, Chawan CB, Pulusani SR. Effect of feeding fermented milk on the incidence of chemically induced colon tumors in rats. Nutr Cancer 5: 159–164, 1983 [DOI] [PubMed] [Google Scholar]
- 245. Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, Jovov B, Abdo Z, Sandler RS, Keku TO. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 1: 138–147, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Silva MF, Sivieri K, Rossi EA. Effects of a probiotic soy product and physical exercise on formation of pre-neoplastic lesions in rat colons in a short-term model of carcinogenic (Abstract). J Int Soc Sports Nutr 6: 17, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Simonovic I, Rosenberg J, Koutsouris A, Hecht G. Enteropathogenic Escherichia coli dephosphorylates and dissociates occludin from intestinal epithelial tight junctions. Cell Microbiol 2: 305–315, 2000 [DOI] [PubMed] [Google Scholar]
- 248. Singh J, Rivenson A, Tomita M, Shimamura S, Ishibashi N, Reddy BS. Bifidobacterium longum, a lactic acid-producing intestinal bacterium inhibits colon cancer and modulates the intermediate biomarkers of colon carcinogenesis. Carcinogenesis 18: 833–841, 1997 [DOI] [PubMed] [Google Scholar]
- 249. Skrzycki M, Majewska M, Podsiad M, Czeczot H. Expression and activity of superoxide dismutase isoenzymes in colorectal cancer. Acta Biochim Pol 56: 663–670, 2009 [PubMed] [Google Scholar]
- 250. Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Van Nhieu JT, Furet JP. Microbial dysbiosis in colorectal cancer (CRC) patients. PLos One 6: e16393, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105: 16731–16736, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Sperker B, Backman JT, Kroemer HK. The role of beta-glucuronidase in drug disposition and drug targeting in humans. Clin Pharmacokinet 33: 18–31, 1997 [DOI] [PubMed] [Google Scholar]
- 253. Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65: 4799–4807, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Sumeri I, Arike L, Adamberg K, Paalme T. Single bioreactor gastrointestinal tract simulator for study of survival of probiotic bacteria. Appl Microbiol Biotechnol 80: 317–324, 2008 [DOI] [PubMed] [Google Scholar]
- 255. Szatrowski TP, Nathan CF. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res 51: 794–798, 1991 [PubMed] [Google Scholar]
- 256. Tallon R, Arias S, Bressollier P, Urdaci MC. Strain- and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J Appl Microbiol 102: 442–451, 2007 [DOI] [PubMed] [Google Scholar]
- 257. Talwalkar A, Kailasapathy K. The role of oxygen in the viability of probiotic bacteria with reference to L. acidophilus and Bifidobacterium spp. Curr Issues Intest Microbiol 5: 1–8, 2004 [PubMed] [Google Scholar]
- 258. Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Munoz-Tamayo R, Paslier DL, Nalin R, Dore J, Leclerc M. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11: 2574–2584, 2009 [DOI] [PubMed] [Google Scholar]
- 259. Teitelbaum JE, Walker WA. Nutritional impact of pre- and probiotics as protective gastrointestinal organisms. Annu Rev Nutr 22: 107–138, 2002 [DOI] [PubMed] [Google Scholar]
- 260. Terzic J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology 138: 2101–2114, 2010 [DOI] [PubMed] [Google Scholar]
- 261. Tong X, Yin L, Giardina C. Butyrate suppresses Cox-2 activation in colon cancer cells through HDAC inhibition. Biochem Biophys Res Commun 317: 463–471, 2004 [DOI] [PubMed] [Google Scholar]
- 262. Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 81: 1031–1064, 2001 [DOI] [PubMed] [Google Scholar]
- 263. Tops CM, Wijnen JT, Hes FJ. Introduction to molecular and clinical genetics of colorectal cancer syndromes. Best Pract Res Clin Gastroenterol 23: 127–146, 2009 [DOI] [PubMed] [Google Scholar]
- 264. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3: 213–223, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol 587: 4153–4158, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature 457: 480–484, 457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature 449: 804–810, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031, 2006 [DOI] [PubMed] [Google Scholar]
- 269. Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, Gordon JI. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA 107: 7503–7508, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLos One 4: e6026, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Van Huynegem K, Loos M, Steidler L. Immunomodulation by genetically engineered lactic acid bacteria. Front Biosci 14: 4825–4835, 2009 [DOI] [PubMed] [Google Scholar]
- 272. Van Loo J, Jonkers N. Evaluation in human volunteers of the potential anticarcinogenic activities of novel nutritional concepts: prebiotics, probiotics and synbiotics (the SYNCAN project QLK1-1999-00346). Nutr Metab Cardiovasc Dis 11: 87–93, 2001 [PubMed] [Google Scholar]
- 273. Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med 319: 525–532, 1988 [DOI] [PubMed] [Google Scholar]
- 274. Walenta S, Mueller-Klieser WF. Lactate: mirror and motor of tumor malignancy. Semin Radiat Oncol 14: 267–274, 2004 [DOI] [PubMed] [Google Scholar]
- 275. Wang RF, Chen H, Paine DD, Cerniglia CE. Microarray method to monitor 40 intestinal bacterial species in the study of azo dye reduction. Biosens Bioelectron 20: 699–705, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Wang Y, Holmes E, Comelli EM, Fotopoulos G, Dorta G, Tang H, Rantalainen MJ, Lindon JC, Corthesy-Theulaz IE, Fay LB, Kochhar S, Nicholson JK. Topographical variation in metabolic signatures of human gastrointestinal biopsies revealed by high-resolution magic-angle spinning 1H NMR spectroscopy. J Proteome Res 6: 3944–3951, 2007 [DOI] [PubMed] [Google Scholar]
- 277. Warburg O. On the origin of cancer cells. Science 123: 309–314, 1956 [DOI] [PubMed] [Google Scholar]
- 278. Watson D, Sleator RD, Hill C, Gahan CG. Enhancing bile tolerance improves survival and persistence of Bifidobacterium and Lactococcus in the murine gastrointestinal tract (Abstract). BMC Microbiol 8: 176, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Westerlund M, Belin AC, Felder MR, Olson L, Galter D. High and complementary expression patterns of alcohol and aldehyde dehydrogenases in the gastrointestinal tract: implications for Parkinson's disease. FEBS J 274: 1212–1223, 2007 [DOI] [PubMed] [Google Scholar]
- 280. Willing B, Dicksved J, Halfvarson J, Andersson A, Lucio M, Zeng Z, Jarnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that GI microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139: 1844–1854, 2010 [DOI] [PubMed] [Google Scholar]
- 281. Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40: 235–243, 2006 [DOI] [PubMed] [Google Scholar]
- 282. Worthley DL, Le Leu RK, Whitehall VL, Conlon M, Christophersen C, Belobrajdic D, Mallitt KA, Hu Y, Irahara N, Ogino S, Leggett BA, Young GP. A human, double-blind, placebo-controlled, crossover trial of prebiotic, probiotic, and synbiotic supplementation: effects on luminal, inflammatory, epigenetic, and epithelial biomarkers of colorectal cancer. Am J Clin Nutr 90: 578–586, 2009 [DOI] [PubMed] [Google Scholar]
- 283. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15: 1016–1022, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Yamazaki K, Tsunoda A, Sibusawa M, Tsunoda Y, Kusano M, Fukuchi K, Yamanaka M, Kushima M, Nomoto K, Morotomi M. The effect of an oral administration of Lactobacillus casei strain shirota on azoxymethane-induced colonic aberrant crypt foci and colon cancer in the rat. Oncol Rep 7: 977–82, 2000 [DOI] [PubMed] [Google Scholar]
- 285. Yokoyama S, Suzuki T. Isolation and characterization of a novel equol-producing bacterium from human feces. Biosci Biotechnol Biochem 72: 2660–2666, 2008 [DOI] [PubMed] [Google Scholar]
- 286. Zheng CL, Che XF, Akiyama S, Miyazawa K, Tomoda A. 2-Aminophenoxazine-3-one induces cellular apoptosis by causing rapid intracellular acidification and generating reactive oxygen species in human lung adenocarcinoma cells. Int J Oncol 36: 641–650, 2010 [DOI] [PubMed] [Google Scholar]
- 287. Zuber P. Management of oxidative stress in Bacillus. Annu Rev Microbiol 63: 575–597, 2009 [DOI] [PubMed] [Google Scholar]


