Abstract
Both caveolin-1 (Cav-1) and Mcl-1 have been implicated in the regulation of cancer cell anoikis, but their relationship and underlying mechanisms of regulation are not known. The present study demonstrated for the first time that Cav-1 regulates Mcl-1 through protein-protein interaction and inhibits its downregulation during cell anoikis in human lung cancer cells. Immunoprecipitation and immunocytochemistry studies showed that Cav-1 interacted with Mcl-1 and prevented it from degradation via the ubiquitin-proteasome pathway. Mcl-1 and Mcl-1-Cav-1 complex were highly elevated in Cav-1-overexpressing cells but were greatly reduced in Cav-1 knockdown cells. Consistent with this finding, we found that Mcl-1 ubiquitination was significantly attenuated by Cav-1 overexpression but increased by Cav-1 knockdown. Together, our results indicate a novel role of Cav-1 in anoikis regulation through Mcl-1 interaction and stabilization, which provides a new insight to the pathogenesis of metastatic lung cancer and its potential treatment.
Keywords: lung cancer, anoikis resistance, metastasis, myeloid cell leukemia-1
resistance to anoikis, a form of apoptotic cell death induced by loss of cell anchorage to extracellular matrixes, has been accepted as a key determinant of cancer cell metastasis (1, 4). Recently, a number of proteins have been identified to facilitate anoikis resistance in various cancer types. Among these, caveolin-1 (Cav-1) has perhaps received the most attention since its expression has been linked to cancer progression and aggressiveness (27). Although some evidence has suggested a tumor suppressing role of Cav-1 (6, 11, 22), in lung cancer, Cav-1 potentiates cancer progression and aggressiveness. Cav-1 expression has been shown to relate to poor prognosis and reduced tumor-free periods in lung cancer patients (10). Moreover, Cav-1 was shown to facilitate metastasis and induce anoikis resistance in lung carcinoma cell lines (2, 23, 29). Not only does Cav-1 play a role in cell death and survival, it also plays a role in cell migration (17), invasion (26), and lipid transportation (21). Cav-1 was reported to exhibit scaffold function and to be essential in regulating several proteins such as endothelial nitric oxide synthase (eNOS), G protein subunit, and nonreceptor tyrosine kinases (14), supporting the wide range of activities of this protein in various cellular processes.1
The prosurvival member of Bcl-2 family protein named myeloid cell leukemia sequence 1 (Mcl-1) has recently been implicated as a key regulator of cell anoikis (24). In melanoma, the depletion of Mcl-1 renders mutant B-RAF melanoma cells sensitive to anoikis (1). Likewise, Mcl-1 degradation and Bim upregulation are a critical determinant of anoikis initiation in wild-type and c-Src-transformed NIH3T3 fibroblast cells. This protein is degraded through the ubiquitin-proteasomal pathway after cell detachment (28). Increasing evidence also indicates the role of Mcl-1 in progressive prostate cancer (31), supporting its clinical significance in cancer metastasis.
The objective of the present study was to investigate the possible relationship between Cav-1 and Mcl-1 and their regulation of anoikis in lung cancer cells. The hypothesis of this study is that Cav-1 mediates its effect on cancer cell anoikis through Mcl-1 regulation. Using gene overexpression and knockdown strategies, we demonstrate this relationship and elucidate the important role of Cav-1 in regulating Mcl-1 through protein interaction and stabilization, thus revealing the existence of a novel mechanism of anoikis regulation which could be important in cancer metastasis.
MATERIALS AND METHODS
Cells and reagents.
Non-small-cell lung cancer (NSCLC)-H460 cells and melanoma G361 cells were obtained from American Type Culture Collection (Manassas, VA). H460 cells were cultured in RPMI 1640 medium, while G361 cells were cultured in DMEM medium. RPMI 1640 was supplemented with 5% fetal bovine serum (FBS), 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin. DMEM was supplemented with 10% FBS, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin. All cell cultures were incubated in a 5% CO2 environment at 37°C. Lactacystin, MG 132, and dimethysulfoxide (DMSO) were obtained from Sigma Chemical (St. Louis, MO); propidium iodide (PI) and Hoechst 33342 were from Molecular Probes, (Eugene, OR); rabbit Cav-1 antibody, rabbit Mcl-1 antibody, mouse monoclonal ubiquitin antibody, mouse monoclonal Cav-1 antibody, and peroxidase-conjugated secondary antibody were from Abcam (Cambridge, MA); MitoTracker Red CMXRos, Alexa Fluor 350 goat anti-mouse IgG (H+L), Alexa Fluor 488 goat anti-rabbit IgG (H+L), and Lipofectamine 2000 were from Invitrogen (Carlsbad, CA). Antibody for ubiquitin, protein G-agarose bead, and β-actin antibody were from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids and transfection.
The Cav-1 expression plasmid [pEX_Cav-1-yellow fluorescent protein (YFP)] and control plasmid (pDS_XB-YFP) were obtained from American Type Culture Collection; the Mcl-1 expression plasmid 25375:pCDNA3.1-hMcl-1 was obtained from Addgene (Cambridge, MA); Cav-1 knockdown plasmid [Cav-1 short hairpin (sh)RNA plasmid] and control plasmid (control shRNA plasmid A) were obtained from Santa Cruz Biotechnology. Stable transfection of cells with Cav-1 expression plasmid or Cav-1 knockdown plasmid was performed by culturing H460 cells in a six-well plate until they reached ∼60% confluence. Lipofectamine reagent (15 μl) and 2 μg of Cav-1, shRNA-Cav-1, or control plasmids were used to transfect the cells in the absence of serum. After 12 h, the medium was replaced with culture medium containing 5% FBS. Approximately 36 h after the beginning of transfection, the cells were digested with 0.03% trypsin, and the cell suspensions were plated onto 75-ml culture flasks and cultured for 24 to 28 days with G418 selection (600 μg/ml). The stable transfectants were pooled and the expression of Cav-1 protein in the transfectants was determined by Western blotting. The cells were cultured in antibiotic-free RPMI 1640 medium for at least two passages before being used in each experiment.
Anoikis assay.
For anoikis evaluation, six-well tissue culture plates were coated with 200 μl (6 mg/ml in 95% ethanol) of poly 2-hydroxyethylmethacrylate (poly-HEMA; Sigma) and left for 10 h in a laminar flow hood. Cells in a single-cell suspension were seeded in poly-HEMA-coated plates at a density of 1 × 105 cells/ml and incubated for various times up to 24 h at 37°C. Cells were harvested, washed, and incubated with 20 μM 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) for 4 h at 37°C. Optical density was then determined using V-max photometer (Molecular Devices, Menlo Park, CA) at a 450-nm wavelength. Absorbance ratio of treated to nontreated cells was calculated and is presented as relative cell viability. For Hoechst 33342 and PI assays, cells were incubated with 10 μM Hoechst 33342 or 15 μM PI for 30 min at 37°C. Apoptotic cells having condensed chromatin and/or fragmented nuclei and PI-positive necrotic cells were scored under a fluorescence microscope (Olympus IX51 with DP70).
Western blot analysis.
After specific treatments, cells were incubated in lysis buffer containing 20 mM Tris·HCl (pH 7.5), 1% Triton X-100, 150 mM sodium chloride, 10% glycerol, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 100 mM phenylmethylsulfonyl fluoride, and a commercial protease inhibitor cocktail (Roche Molecular Biochemicals, Basel, Switzerland) for 30 min on ice. Cell lysates were collected and determined for protein content using the Bradford method (Bio-Rad, Hercules, CA). Equal amount of proteins of each sample (40 μg) were denatured by heating at 95°C for 5 min with Laemmli loading buffer and were subsequently loaded on 10% SDS-polyacrylamide gel electrophoresis. After separation, proteins were transferred onto 0.45-μm nitrocellulose membranes (Bio-Rad). The transferred membranes were blocked for 1 h in 5% nonfat dry milk in TBST [25 mM Tris·HCl (pH 7.5), 125 mM NaCl, 0.05% Tween-20] and incubated with the appropriate primary antibodies at 4°C overnight. Membranes were washed twice with TBST for 10 min and incubated with horseradish peroxidase-coupled isotype-specific secondary antibodies for 1 h at room temperature. The immune complexes were detected by enhanced with chemiluminescence substrate (Supersignal West Pico; Pierce, Rockford, IL) and quantified using analyst/PC densitometry software (Bio-Rad).
Immunoprecipitation.
Cells were washed after treatment and lysed in lysis buffer at 4°C for 20 min. After centrifugation at 14,000 g for 15 min at 4°C, the supernatants were collected and determined for protein content. Cell lysates were normalized, and equal amounts of protein per sample (60 μg) were incubated with anti-Cav-1 antibody conjugated to protein G plus-agarose beads (Santa Cruz) for 6 h at 4°C. The immune complexes were washed five times with ice-cold lysis buffer, resuspended in 2 × Laemmli sample buffer, and boiled at 95°C for 5 min. Immune complexes were separated by 10% SDS-PAGE and detected for Cav-1 and Mcl-1 complexes by Mcl-1 antibody. For detection of the ubiquitin-Mcl-1 complex, the anti-Mcl-1 antibody was incubated with the cell lysate in the immunoprecipitation step followed by Western blot analysis using anti-ubiquitin antibody.
Quantitative real-time RT-PCR.
One microgram of TRIzol-extracted RNA was reverse-transcribed in a 100-μl reaction mixture containing 500 μM dNTP, 125 units of MultiScribe Reverse Transcriptase (Applied Biosystems, Foster City, CA), 40 units of RNase inhibitor, 2.5 μM oligo(dT), 1 × TaqMan reverse transcriptase buffer, and 5 mM MgCl2 at 48°C for 40 min. The primers for Mcl-1 (Hs03043899_m1*) and 18s rRNA (Hs99999901_s1) were obtained from Applied Biosystems. Amplification was performed at the following cycling conditions: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. A SYBR Green PCRMasterMix (Applied Biosystems) was used with 1 ng of cDNA and with 100–400 nM primers. A negative control without any cDNA template was run with every assay. All PCR reactions were performed by using ABI PRISM7900 Sequence Detection System (Applied Biosystems). Relative mRNA levels were determined by using the comparative CT (threshold cycle) method (16), where the Mcl-1 target is normalized to the control and compared with a reference sample (assigned a relative value of 1) by the equation: 2−ΔΔCT.
Immunofluorescence.
Cells (0.5×106/well) were seeded in six-well plates for 24 h to allow the cell to completely adhere to the surface. Then, the cells were fixed in 3.7% formaldehyde for 10 min at room temperature and were then permeabilized and blocked in a solution containing 0.5% saponin, 1% FBS, and 1.5% goat serum for 30 min. After primary antibody incubation with both Cav-1 mouse monoclonal antibody (Abcam) at 1:100 dilution and Mcl-1 rabbit polyclonal antibody (Abcam) at 1:100 dilution for 1 h, cells were washed and incubated together with Alexa Fluor 350 goat anti-mouse IgG (H+L) conjugated secondary antibody (Invitrogen) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) conjugated secondary antibody (Invitrogen) for 30 min. Mitochondria were stained with MitoTracker Red CMXRos (Invitrogen). Cells were cytospun onto a glass slide and mounted using the anti-fade reagent Fluoromont-G (Southern Biotech, Birmingham, AL). Images were acquired by confocal laser scanning microscopy (Zeiss LSM 510).
Statistical analysis.
Mean data from independent experiments were normalized with control treatment groups. All of the experiments were repeated at least three times. A statistical analysis between treatments versus control was verified by Student's t-test. The strength of relationships, correlation coefficient (r), between each protein level after detachment was determined with SPSS software (version 16; SPSS, Chicago, IL). P < 0.05 was considered as statistically significant.
RESULTS
Caveolin-1 inhibits anoikis of H460 cells.
We and others have previously reported the role of Cav-1 in anoikis regulation in various cell types (7, 23). To assure the role of this protein in anoikis regulation of the test cell system, we first characterized the effect of different ectopic Cav-1 expression levels on cell anoikis of H460 cells. Through stable gene transfection, we generated Cav-1-overexpressing (HCav-1) cells, shRNA knockdown (shCav-1) cells, and vector (pDS_XB-YFP and control shRNA plasmid A) control cells, as described in materials and methods. These mutant clones were analyzed for Cav-1 expression by Western blotting (Fig. 1A). To study anoikis, Cav-1 overexpressed, Cav-1 knockdown, and vector control cells were detached and incubated in adhesion-resistant poly-HEMA-coated plates. Cell survival was then determined at various times by XTT assay. Analysis of cell viability showed that detachment of the cells caused a time-dependent decrease in cell survival, with approximately 80%, 50%, and 30% of the overexpressed, vector control, and knockdown Cav-1 cells, respectively, remaining viable after 6 h (Fig. 1B). At 24 h postdetachment, HCav-1 cells exhibited ∼60% viability, whereas both control and shCav-1 cells showed a survival rate of <40%. Control experiments, in which cells were allowed to attach in normal tissue culture plates, showed no significant change in cell viability over the 24-h test period (data not shown). Analysis of cell apoptosis by Hoechst 33342 assay showed that shCav-1 cells were most susceptible to apoptosis induced by cell detachment, whereas HCav-1 cells were least susceptible (Fig. 1C). This finding is consistent with the cell viability data showing the highest rate of survival of HCav-1 cells after detachment. Morphological analyses of apoptotic and necrotic cell death by Hoechst 33342 and PI assays showed that apoptosis was the primary mode of cell death induced by cell detachment in H460 cells (Fig. 1D).
Fig. 1.
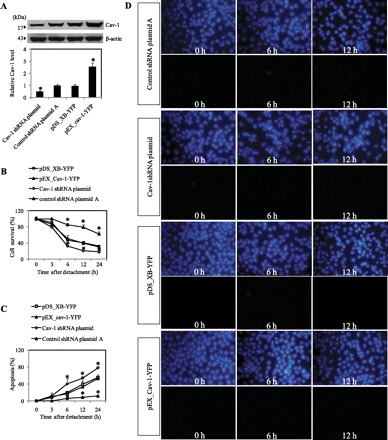
Caveolin-1 (Cav-1) overexpression increases anoikis resistance in H460 cells. A: control, HCav-1 [expression plasmid (pEX)_Cav-1 plasmid transfectant H460], or short hairpin (sh)Cav-1-transfected H460 cells were constructed and grown in culture that was then analyzed for Cav-1 expression by Western blotting. Blots were reprobed with β-actin antibody to confirm equal loading of samples. The immunoblot signals were quantified by densitometry, and mean data from independent experiments were normalized to the results. Columns are means ± SD (n = 3). *P < 0.05 vs. control transfected cells. B: subconfluent (90%) monolayers of control transfected, Cav-1-overexpressing cells, and Cav-1 knockdown cells were detached and suspended in poly-HEMA-coated plates for various times (0–24 h). At the indicated times after detachment, the cells were collected and their survival was determined by XTT assay. Viability of detached cells at time 0 was considered as 100%. C: percentage of cell detachment-induced apoptosis was analyzed by Hoechst 33342 nuclear fluorescence. Data points represent means ± SD (n = 3). *P < 0.05 vs. control transfected cells. D: detachment-induced apoptosis and necrosis in control transfected cells, Cav-1-overexpressing, and Cav-1 knockdown cells. Detached cells were suspended in poly-HEMA-coated plates for 0–12 h, and cell apoptosis and necrosis were determined by Hoechst 33342 and propidium iodide (PI) fluorescence measurements, respectively. pDS_XB-YFP, yellow fluorescent protein Cav-1 control plasmid.
Cell detachment induces Cav-1 and Mcl-1 downregulation.
The role of Cav-1 and Mcl-1 in cancer cell anoikis is unclear. To provide evidence for the role of these proteins, we evaluated the expression profiles of Cav-1 and Mcl-1 after cell detachment in lung cancer H460 cells. The cells were detached, suspended in adhesion-resistant plates, and analyzed for Cav-1 and Mcl-1 protein expression by Western blotting. Figure 2, A and B, shows that after cell detachment, both Cav-1 and Mcl-1 expression gradually decreased over time concomitant with cell viability and death, suggesting their potential relationship and role in anoikis regulation. Like Cav-1, the role of Mcl-1 in anoikis regulation was studied using stable gene transfection. Figure 2C shows that stably transfected Mcl-1 (HMcl-1) cells expressed a high level of Mcl-1 protein as compared with vector-transfected control cells. The HMcl-1 cells also showed resistance to anoikis as indicated by their increased viability after cell detachment over control cells (Fig. 2D).
Fig. 2.
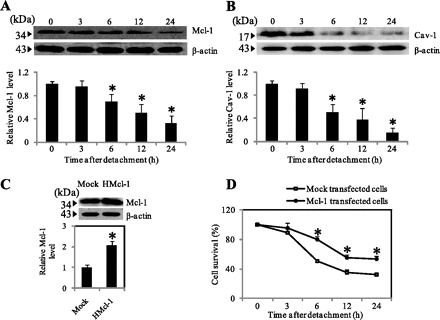
Cav-1 and myeloid cell leukemia sequence 1 (Mcl-1) expression after cell detachment. A and B: H460 cells were detached and suspended in poly-HEMA-coated plates for various times (0–24 h). Blots were probed with antibodies specific to Mcl-1 and Cav-1 and were reprobed with β-actin antibody. Columns are means ± SD (n = 3). *P < 0.05 vs. control at time 0. C: mock and HMcl-1 cells were grown in culture and analyzed for Mcl-1 expression by Western blotting. Blots were reprobed with β-actin antibody to confirm equal loading of samples. The immunoblot signals were quantified by densitometry, and mean data from independent experiments were normalized to the results. Columns are means ± SD (n = 3). *P < 0.05 vs. control transfected cells. D: subconfluent (90%) monolayers of Mock and HMcl-1 cells were detached and suspended in poly-HEMA-coated plates for various times (0–24 h). At the indicated times, the cells were collected and their survival was determined by XTT assay. Viability of detached cells at time 0 was considered as 100%. Data points represent means ± SD (n = 3). *P < 0.05 vs. control transfected cells.
Mcl-1 downregulation during cell anoikis is regulated by Cav-1 interaction.
Cav-1 has been shown to function as a scaffold protein regulating the stability and function of several proteins (14). The observations that both Cav-1 and Mcl-1 have a similar effect on anoikis and their expression is similarly downregulated during anoikis suggest the possible linkage and shared mechanism of anoikis regulation. Since Mcl-1 is recognized as a relatively short half-life protein (19) and its scaffolding interaction with Cav-1 has not been reported, we explored their possible interaction by generating a correlation plot between Cav-1 and Mcl-1 expression during cell anoikis (Fig. 3A). Not only did the reduction of these proteins correlate well with the induction of cell anoikis, but the plot also revealed a highly correlated profile of Cav-1 and Mcl-1 downregulation with the correlation coefficient of 0.98.
Fig. 3.
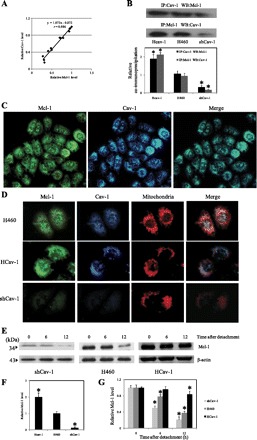
Interaction and localization of Cav-1 and Mcl-1. A: correlation analysis of the expression of Cav-1 and Mcl-1 after detachment of H460 cells. B: immunoprecipitation (IP) experiments were performed using specific anti-Cav-1 antibody; immunoblots were probed with anti-Mcl-1 antibody and vice versa. Equal amounts of protein (25 μg) were loaded in each lane. WB, Western blotting. Columns are means ± SD (n = 3). *P < 0.05 vs. control transfected cells. C: H460 cells were analyzed for localization of Cav-1 and Mcl-1 by immunofluorescence microscopy. Immunofluorescence was performed using mouse anti-Cav-1 monoclonal antibody and rabbit anti-Mcl-1 polyclonal antibody, followed by appropriate secondary antibodies labeled with Alexa Fluor 350 and Alexa Fluor 488 to visualize Cav-1 and Mcl-1, respectively. Cells were also stained with MitoTracker Red CMXRos (300 nM) to aid visualization of mitochondria. D: differential expression of Cav-1 and Mcl-1 in HCav-1, shCav-1, and H460 cells. Cells were fixed and processed for immunofluorescence staining. E: dependence of Mcl-1 reduction after cell detachment on Cav-1 expression. HCav-1, shCav-1, and H460 cells were detached and suspended in poly-HEMA-coated plates for various times (0–12 h). Blots were probed with specific antibody to Mcl-1 and were reprobed with β-actin antibody to confirm equal loading of samples. F: relative Mcl-1 levels in attached cells. G: relative Mcl-1 levels in shCav-1, HCav-1, and H460 cells after detachment for 0, 6, and 12 h. Columns are means ± SD (n = 3). *P < 0.05 vs. control at detachment time = 0 h.
Next, we used immunoprecipitation techniques to determine the direct interaction between the two proteins. Cell lysates of HCav-1, shCav-1, and H460 cells were then prepared, immunoprecipitated using Cav-1 antibody, and analyzed for Cav-1-Mcl-1 complex by Western blotting using Mcl-1 antibody as a probe. The results showed that Cav-1-Mcl-1 complex formation was most pronounced in HCav-1 cells, which express the highest level of Cav-1, and least expressed in Cav-1 knockdown (shCav-1) cells (Fig. 3B). Moreover, consistent results were observed in the immunoprecipitation experiment using Mcl-1 antibody followed by Western blot analysis using Cav-1 antibody. These results suggest that Cav-1 plays a role in scaffolding Mcl-1 protein and that its interaction with Mcl-1 may play a role in regulating Mcl-1 level. To provide supporting evidence for the Cav-1 and Mcl-1 interaction, immunocytochemical studies were performed to evaluate the intracellular localization of the two proteins. Figure 3C shows immunofluorescent staining of Mcl-1 and Cav-1, which are strikingly similar and supportive of the protein colocalization.
Cav-1 stabilizes Mcl-1 in H460 cells.
Having shown that Cav-1 interacts with Mcl-1, we further investigated whether such interaction is essential for Mcl-1 stability after cell detachment. Adhered HCav-1, shCav-1, and H460 cells were stained with antibodies for Mcl-1, Cav-1, and MitoTracker, and their fluorescent signals were observed by microscopy. While the MitoTracker signals are relatively constant in these cells, the intensities of Cav-1 and Mcl-1 signals in these cells vary greatly (Fig. 3D). Interestingly, cells that express a high level of Cav-1 (HCav-1) also exhibit a high level of Mcl-1, while those that express a low level of Cav-1 (shCav-1) also show a low level of Mcl-1, suggesting the stabilizing effect of Cav-1 on Mcl-1. To further study this effect, HCav-1, shCav-1, and H460 cells were detached and incubated in adhesion-resistant plates for 0–12 h. Western blot analysis of Mcl-1 was then performed at 0, 6, and 12 h postdetachment. Figure 3E shows that at various times of the detachment, Mcl-1 levels in these cells varied depending on the expression levels of Cav-1 in each cell type. These findings strengthen the above finding that Cav-1 interacts with Mcl-1 and stabilizes the protein under different attachment conditions.
Mcl-1 reduction after cell detachment is mediated through ubiquitin-proteasomal degradation.
Although Mcl-1 has been reported to be degraded via the proteasomal pathway (28), we suspected both transcription and degradation to play a role in Mcl-1 downregulation during cell anoikis. To test this, we performed quantitative real-time RT-PCR and proteasome inhibition studies in detached H460 cells. Mcl-1 mRNA level was significantly reduced as early as 1 h (data not shown) and remained unchanged up to 24 h after detachment (Fig. 4A). This finding excluded the possibility that Cav-1 could stabilize Mcl-1 through a transcription-dependent mechanism. Therefore, we tested the involvement of ubiquitin-proteasomal system on Mcl-1 downregulation after cell detachment. Figure 4B shows that cell detachment caused a substantial reduction in Mcl-1 protein level and that treatment of the cells with specific proteasomal inhibitors, lactacystin and MG132, completely inhibited the Mcl-1 reduction. These results indicate that Mcl-1 downregulation after cell detachment is mediated mainly by the proteasome degradation pathway.
Fig. 4.
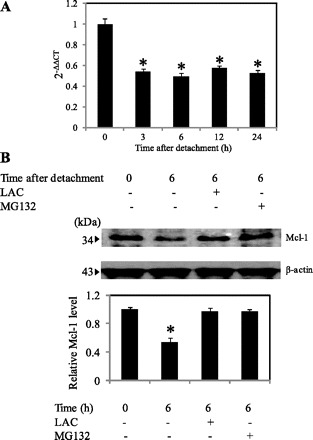
Transcription and degradation of Mcl-1 after cell detachment. A: real-time PCR analysis of Mcl-1 mRNA expression after cell detachment. The relative mRNA expression was determined by using the comparative CT method as described in materials and methods. Columns are means ± SD (n = 3). *P < 0.05 vs. control at detachment time = 0 h. B: relative Mcl-1 expression after detachment for 0–6 h in the presence or absence of lactacystin (LAC; 20 μM) or MG132 (10 μM) in H460 cells. Columns are means ± SD (n = 3). *P < 0.05 vs. control at detachment time = 0 h.
Cav-1 stabilizes Mcl-1 by attenuating Mcl-1 ubiquitination.
Proteasomal degradation of a protein is triggered by protein ubiquitination. To test the potential involvement of ubiquitination in Mcl-1 stability and its regulation by Cav-1, Mcl-1 immunoprecipitation and ubiquitination studies were performed in various Cav-1 expressing cells. In normal H460 cells, the formation of ubiquitin-Mcl-1 complexes gradually increased as early as 1 h after cell detachment and peaked at ∼6 h (Fig. 5A). The level of ubiquitin-Mcl-1 complex formation was minimal in Cav-1 overexpressing (HCav-1) cells and maximal in Cav-1 knockdown (shCav-1) cells as compared with normal H460 cells (Fig. 5B). These results indicate that Cav-1 attenuated the ubiquitination of Mcl-1 and stabilized the protein after cell detachment.
Fig. 5.
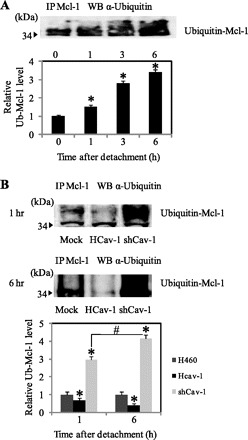
Effect of Cav-1 expression on Mcl-1 ubiquitination. A: H460 cells were detached and suspended in poly-HEMA-coated plates for various times. Cell lysates were prepared and immunoprecipitated (IP) with anti-Mcl-1 antibody. The resulting immune complexes were analyzed for ubiquitin by Western blotting (WB) using anti-ubiquitin antibody. Maximum Mcl-1 ubiquitination was observed at 6 h after cell detachment. The immunoblot signals were quantified by densitometry. Columns are means ± SD (n = 3). *P < 0.05 vs. control at detachment time = 0 h. B: HCav-1, shCav-1, and H460 cells were detached and suspended in poly-HEMA-coated plates for 1 and 6 h. Cell lysates were immunoprecipitated with anti-Mcl-1 antibody, and the resulting immune complexes were analyzed for ubiquitin (Ub) by Western blotting. Columns are means ± SD (n = 3). *P < 0.05 vs. control transfected at detachment time = 1 h; #P < 0.05 vs. the indicated control.
Cav-1 regulates Mcl-1 expression and anoikis in human melanoma G361 cells.
An upregulation of Cav-1 and Mcl-1 has been found not only in non-small-cell lung cancer but also in other forms of cancer such as human melanoma (13, 20). To test whether Cav-1 might have a similar regulatory role on Mcl-1 and anoikis in other cancer cells, melanoma G361 cells were stably transfected with Cav-1, shCav-1, or control plasmids. After clonal selection, the cells were analyzed for Mcl-1, Cav-1, and anoikis. Figure 6A shows that the Cav-1 transfected cells (G361-Cav-1) expressed the highest level of Cav-1 protein, whereas the shCav-1 transfected cells (G361-shCav-1) exhibited the lowest level.
Fig. 6.
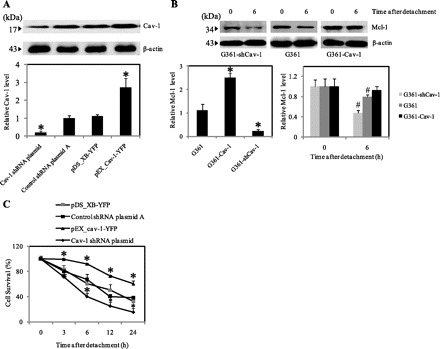
Cav-1 regulates Mcl-1 expression and anoikis in melanoma G361 cells. A: control, G361-Cav-1, and G361-shCav-1 transfected cells were grown in and analyzed for Cav-1 expression by Western blotting. Blots were reprobed with β-actin antibody to confirm equal loading of samples. The immunoblot signals were quantified by densitometry, and mean data from independent experiments were normalized to the results. B: G361, G361-Cav-1 and G361-shCav-1 cells were detached and suspended in poly-HEMA-coated plates for various times (0–6 h). Blots were probed with specific antibody to Mcl-1 and were reprobed with β-actin antibody. Columns are means ± SD (n = 3). *P < 0.05 vs. control G361 cells. #P < 0.05 vs. control at detachment time = 0 h. C: subconfluent (90%) monolayers of mock, G361-Cav-1, and G361-shCav-1 cells were detached and suspended in poly-HEMA-coated plates for various times (0–24 h). At the indicated times, the cells were collected and determined for survival by XTT assay. Viability of detached cells at time 0 was considered as 100%. Data represent means ± SD (n = 3). *P < 0.05 vs. control transfected cells.
To test the interaction between Cav-1 and Mcl-1, cells were harvested at time 0 and at 6 h after detachment. Cell lysates were then prepared and analyzed for Mcl-1 expression by Western blotting. The results show that the levels of Mcl-1 in G361-Cav-1, G361-shCav-1, and G361 cells at 0 and 6 h postdetachment were highly dependent on the cellular level of Cav-1 in each cell (Fig. 6B). Cell anoikis studies also show that Cav-1 functioned as an anoikis inhibitor as evidenced by the inhibitory effect of Cav-1 overexpression and the promoting effect of Cav-1 knockdown on cell anoikis (Fig. 6C). The above results are consistent with the earlier findings in H460 cells and indicate the general role of Cav-1 in anoikis and Mcl-1 regulation.
DISCUSSION
When cells are detached from the extracellular matrix, the loss of anchorage-related signals results in an abrogation of certain cellular processes such as cell survival and growth (12) and consequently initiates the process of anoikis (8). Since anoikis is an important cellular event controlling cancer metastasis, unraveling its underlying mechanisms is critical to the understanding of disease pathogenesis and its treatment. Among the many types of cancer, lung cancer has frequently been found to metastasize at the time of tumor detection. While the exact mechanisms of cancer metastasis have been extensively investigated, an upregulation of Mcl-1 (25) and Cav-1 (23) has been implicated in lung cancer aggressiveness and progression. Mcl-1 was found to overexpress in NSCLC cells and regulate their survival and sensitivity to diverse apoptotic stimuli (25). Apoptotic stimuli such as cell detachment induce Bim (activator of BH3-only protein) expression (3). Recently, Zhang et al. (30) have demonstrated that Mcl-1 can sequester Bim in NSCLC cells which supports the role of Mcl-1 in attenuating anoikis in this cancer cell type. Moreover, amplification or overexpression of Mcl-1 was shown to render cells resistant to detachment-induced apoptosis (24) and the decrease in Mcl-1 level is required in the initiation of cell anoikis (1, 28). Previously, we and others have shown that Cav-1 confers resistance to anoikis in cancer cells (7, 23). Furthermore, the expression of Cav-1 has been used as a biomarker for virulence of some cancers (5). The role of Cav-1 in cancer cell anoikis has been described in many ways such as the induction of survival pathways (15) and the reduction of Cav-1 and Mcl-1 (23, 28). We further demonstrated in this study that, during cell anoikis, Cav-1 and Mcl-1 reduction was tightly correlated. Cav-1 functioned as a scaffolding protein for Mcl-1 binding as demonstrated by immunoprecipitation studies (Fig. 3B). In addition, the Cav-1-Mcl-1 complex significantly increased in the Cav-1 overexpressing (HCav-1) cells, but decreased in the Cav-1 knockdown (shCav-1) cells. Immunocytochemistry studies further confirmed the colocalization of Cav-1 and Mcl-1 in the cells, which was largely associated with the mitochondria (Fig. 3D).
Since Mcl-1 is known to be a short half-life protein due to continuous proteasomal degradation (28), it is possible that its interaction with Cav-1 could affect its stability, which was first demonstrated in this study. Although a rapid decline in Mcl-1 mRNA level was observed at 1 h postdetachment, the mRNA level remained relatively constant during the next 24-h period (Fig. 4A). Because Mcl-1 protein level was significantly decreased at 6 h postdetachment and continued to decline during the 24-h period, this finding ruled out transcriptional regulation as responsible for the Mcl-1 downregulation. Moreover, the observation that proteasome inhibitors completely inhibited detachment-induced Mcl-1 downregulation (Fig. 4B) strongly supported protein degradation and stabilization of Mcl-1 by Cav-1 as a key control mechanism.
Proteasomal degradation of a protein is generally triggered by its ubiquitination (9). We tested and found that Mcl-1 is ubiquitinated during cell detachment and that this process is inhibited by Cav-1. The mechanism by which Cav-1 inhibits Mcl-1 ubiquitination is unclear but likely involves steric hindrance of the ubiquitination sites by Cav-1. The interaction between Cav-1 and Mcl-1 may also affect Mcl-1 phosphorylation which has been linked to its ubiquitination. For example, phosphorylation of Mcl-1 at Ser159 by glycogen synthase kinase-3 was observed during cell anoikis (28) and was found to promote Mcl-1 ubiquitination and subsequent degradation (18).
We extended our finding on the role of Cav-1 in Mcl-1 and anoikis regulation in lung carcinoma H460 cells to melanoma G361 cells due to their reported expression in human melanoma (13, 20). We found that Cav-1-Mcl-1 interaction could be detected in G361 cells and that such interaction is dependent on the cellular level of Cav-1 which determines cellular susceptibility to anoikis (Fig. 6). These results support the general role of Cav-1 as anoikis regulator through Mcl-1 interaction.
In conclusion, we report a novel finding on the role of Cav-1 in anoikis regulation of human lung carcinoma and melanoma cells. While the role of Cav-1 and Mcl-1 in anoikis regulation has been reported, their association and the underlying mechanisms of regulation are unclear. We found that Cav-1 interacts with Mcl-1 and stabilizes the protein by blocking its ubiquitination and subsequent degradation. Because an elevated expression of Cav-1 and Mcl-1 has been linked to the progression of cancer and metastasis, the findings of this study could be beneficial to the understanding of cancer etiology and metastasis mechanisms.
GRANTS
This research was supported by the Higher Education Research Promotion and National Research University Project of Thailand, Ratchadaphiseksomphot Endowment Fund (Chulalongkorn University), Thailand Research Fund (to P. Chanvorachote), and National Institutes of Health (R01-HL076340-04S1).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
P. Chunhacha, V. Pongrakhananon, and P. Chanvorachote performed the experiments; P. Chunhacha and P. Chanvorachote analyzed the data; P. Chunhacha and P. Chanvorachote interpreted the results of the experiments; P. Chunhacha and P. Chanvorachote prepared the figures; P. Chunhacha and P. Chanvorachote drafted the manuscript; P. Chunhacha, V. Pongrakhananon, Y. Rojanasakul, and P. Chanvorachote approved the final version of the manuscript; Y. Rojanasakul and P. Chanvorachote edited and revised the manuscript; P. Chanvorachote, conception and design of research.
ACKNOWLEDGMENTS
The authors thank Mr. Krich Rajprasit for editing assistance.
Footnotes
This article is the topic of an Editorial Focus by Pamela G. Lloyd (16a).
REFERENCES
- 1. Boisvert AK, Longmate W, Abel EV, Aplin AE. Mcl-1 is required for melanoma cell resistance to anoikis. Mol Cancer Res 7: 549– 556, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chanvorachote P, Nimmannit U, Lu Y, Talbott S, Jiang BH, Rojanasakul Y. Nitric oxide regulates lung carcinoma cell anoikis through inhibition of ubiquitin-proteasomal degradation of caveolin-1. J Biol Chem 284: 28476– 28484, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. Bcl-2 and Bcl-xL sequester BH3 domain-only molecule preventing Bax and Bak-mediated mitochondrial apoptosis. Mol Cell 8: 705– 711, 2001 [DOI] [PubMed] [Google Scholar]
- 4. Chiarugi P, Giannoni E. Anoikis: a necessary death program for anchorage-dependent cells. Biochem Pharmacol 76: 1352– 1364, 2008 [DOI] [PubMed] [Google Scholar]
- 5. Corn PG, Thompson TC. Identification of a novel prostate cancer biomarker, caveolin-1: implications and potential clinical benefit. Cancer Manag Res 2: 111– 122, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engelman JA, Chu C, Lin A, Jo H, Ikezu T, Okamoto T, Kohtz DS, Lisanti MP. Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo. A role for the caveolin-scaffolding domain. FEBS Lett 428: 205– 211, 1998 [DOI] [PubMed] [Google Scholar]
- 7. Fiucci G, Ravid D, Reich R, Liscovitch M. Caveolin-1 inhibits anchorage-independent growth, anoikis and invasiveness in MCF-7 human breast cancer cells. Oncogene 21: 2365– 2375, 2002 [DOI] [PubMed] [Google Scholar]
- 8. Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol 124: 619– 626, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373– 428, 2002 [DOI] [PubMed] [Google Scholar]
- 10. Ho CC, Kuo SH, Huang PH, Huang HY, Yang CH, Yang PC. Caveolin-1 expression is significantly associated with drug resistance and poor prognosis in advanced non-small cell lung cancer patients treated with gemcitabine-based chemotherapy. Lung Cancer 59: 105– 110, 2007 [DOI] [PubMed] [Google Scholar]
- 11. Hurlstone AF, Reid G, Reeves JR, Fraser J, Strathdee G, Rahilly M, Parkinson EK, Black DM. Analysis of the CAVEOLIN-1 gene at human chromosome 7q31.1 in primary tumours and tumour-derived cell lines. Oncogene 18: 1881– 1890, 1999 [DOI] [PubMed] [Google Scholar]
- 12. Hynes RO. Cell adhesion: old and new questions. Trends Cell Biol 9: M33– M37, 1999 [PubMed] [Google Scholar]
- 13. Keuling AM, Felton KE, Parker AA, Akbari M, Andrew SE, Tron VA. RNA silencing of Mcl-1 enhances ABT-737-mediated apoptosis in melanoma: role for a caspase-8-dependent pathway. PLoS One 4: e6651, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lajoie P, Nabi IR. Lipid rafts, caveolae, and their endocytosis. Int Rev Cell Mol Biol 282: 135– 136, 2010 [DOI] [PubMed] [Google Scholar]
- 15. Li L, Ren CH, Tahir SA, Ren C, Thompson TC. Caveolin-1 maintains activated Akt in prostate cancer cells through scaffolding domain binding site interactions with and inhibition of serine/threonine protein phosphatases PP1 and PP2A. Mol Cell Biol 23: 9389– 9404, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402– 408, 2001 [DOI] [PubMed] [Google Scholar]
- 16a. Lloyd PG. Caveolin-1, antiapoptosis signaling, and anchorage-independent cell growth. Focus on “Caveolin-1 regulates Mcl-1 stability and anoikis in lung carcinoma cells.” Am J Physiol Cell Physiol (March 14, 2012). doi:10.1152/ajpcell.00075.2012 [DOI] [PubMed]
- 17. Luanpitpong S, Talbott SJ, Rojanasakul Y, Nimmannit U, Pongrakhananon V, Wang L, Chanvorachote P. Regulation of lung cancer cell migration and invasion by reactive oxygen species and caveolin-1. J Biol Chem 285: 38832– 38840, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell 21: 749– 760, 2006 [DOI] [PubMed] [Google Scholar]
- 19. Michels J, Johnson PW, Packham G. Mcl-1. Int J Biochem Cell Biol 37: 267– 271, 2005 [DOI] [PubMed] [Google Scholar]
- 20. Nakashima H, Hamamura K, Houjou T, Taguchi R, Yamamoto N, Mitsudo K, Tohnai I, Ueda M, Urano T, Furukawa K. Overexpression of caveolin-1 in a human melanoma cell line results in dispersion of ganglioside GD3 from lipid rafts and alteration of leading edges, leading to attenuation of malignant properties. Cancer Sci 98: 512– 520, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Quest AF, Leyton L, Párraga M. Caveolins, caveolae, and lipid rafts in cellular transport, signaling, and disease. Biochem Cell Biol 82: 129– 144, 2004 [DOI] [PubMed] [Google Scholar]
- 22. Racine C, Belanger M, Hirabayashi H, Boucher M, Chakir J, Couet J. Reduction of caveolin 1 gene expression in lung carcinoma cell lines. Biochem Biophys Res Commun 255: 580– 586, 1999 [DOI] [PubMed] [Google Scholar]
- 23. Rungtabnapa P, Nimmannit U, Halim H, Rojanasakul Y, Chanvorachote P. Hydrogen peroxide inhibits non-small cell lung cancer cell anoikis through the inhibition of caveolin-1 degradation. Am J Physiol Cell Physiol 300: C235– C245, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett 272: 177– 185, 2008 [DOI] [PubMed] [Google Scholar]
- 25. Song L, Coppola D. Mcl-1 regulates survival and sensitivity to diverse apoptotic stimuli in human non-small cell lung cancer cells. Cancer Biol Ther 4: 267– 276, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Tang Y, Zeng X, He F, Liao Y, Qian N, Toi M. Caveolin-1 is related to invasion, survival, and poor prognosis in hepatocellular cancer. Med Oncol. In press [DOI] [PubMed] [Google Scholar]
- 27. Thompson TC, Tahir SA, Li L, Watanabe M, Naruishi K, Yang G, Kadmon D, Logothetis CJ, Troncoso P, Ren C, Goltsov A, Park S. The role of caveolin-1 in prostate cancer: clinical implications. Prostate Cancer Prostatic Dis 13: 6– 11, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Woods NT, Yamaguchi H, Lee FY, Bhalla KN, Wang HG. Anoikis, initiated by Mcl-1 degradation and Bim induction, is deregulated during oncogenesis. Cancer Res 67: 10744– 10752, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeh D, Chen C, Sun MZ, Shao S, Hao L, Song Y, Gong L, Hu J, Wang Q. Caveolin-1 is an important factor for the metastasis and proliferation of human small cell lung cancer NCI-H446 cell. Anat Rec 292: 1584– 1592, 2009 [DOI] [PubMed] [Google Scholar]
- 30. Zhang H, Guttikonda S, Roberts L, Uziel T, Semizarov D, Elmore SW, Leverson JD, Lam LT. Mcl-1 is critical for survival in a subgroup of non-small-cell lung cancer cell lines. Oncogene 30: 1963– 1968, 2011 [DOI] [PubMed] [Google Scholar]
- 31. Zhang S, Zhau HE, Osunkoya AO, Iqbal S, Yang X, Fan S, Chen Z, Wang R, Marshall FF, Chung LW, Wu D. Vascular endothelial growth factor regulates myeloid cell leukemia-1 expression through neuropilin-1-dependent activation of c-MET signaling in human prostate cancer cells. Mol Cancer 9: 9, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]


