Abstract
Recent advances in sequencing methods have transformed the field of microbial ecology, making it possible to determine the composition and functional capabilities of uncultured microorganisms. These technologies have been instrumental in the recognition that resident microorganisms can have profound effects on the phenotype and fitness of their animal hosts by modulating the animal signaling networks that regulate growth, development, behavior, etc. Against this backdrop, this review assesses the impact of microorganisms on insect-plant interactions, in the context of the hypothesis that microorganisms are biochemical brokers of plant utilization by insects. There is now overwhelming evidence for a microbial role in insect utilization of certain plant diets with an extremely low or unbalanced nutrient content. Specifically, microorganisms enable insect utilization of plant sap by synthesizing essential amino acids. They also can broker insect utilization of plant products of extremely high lignocellulose content, by enzymatic breakdown of complex plant polysaccharides, nitrogen fixation, and sterol synthesis. However, the experimental evidence for microbial-mediated detoxification of plant allelochemicals is limited. The significance of microorganisms as brokers of plant utilization by insects is predicted to vary, possibly widely, as a result of potentially complex interactions between the composition of the microbiota and the diet and insect developmental age or genotype. For every insect species feeding on plant material, the role of resident microbiota as biochemical brokers of plant utilization is a testable hypothesis.
Keywords: Insect-plant interactions, Microbial brokers, Microbiota, Symbiosis
Introduction
Insect-plant interactions are a major focus in the field of chemical ecology. Some of the most important decisions made by plant-associated insects (e.g., what to eat, where to mate or deposit eggs) are determined by plant chemistry. Commonly, these interactions are interpreted to occur strictly between the insect and plant, although it is recognized that they can be dynamic, including insect-induced changes in plant chemistry (Karban and Baldwin 1997), and can be modified by interactions with other plant-associated organisms, including pathogenic microbes and nematodes and insect-vectored viruses (Ali et al. 2011; Hatcher 1995; Ingwell et al. 2012; Stout et al. 2006; Tack et al. 2012). A central tenet of this powerful paradigm is that the chemically-mediated interaction between the insect and the plant is a major determinant of the fitness of the individual insect and the abundance and distribution of insect populations.
A second, long-standing focus of entomological research is insect-microbial interactions, especially the role of resident microorganisms in enabling insect utilization of nutritionally poor or unbalanced diets (Buchner 1965; Koch 1965; Moran 2007). Although many such studies have concerned insects that feed on plant material, the research effort and literature on insect-microbial interactions have been conducted largely independently on the research community studying insect-plant interactions. Nevertheless, several authors have argued for integration of the two fields, explicitly proposing that these resident microorganisms act as go-betweens, or “biochemical brokers” that enable insects to exploit plants (Douglas 1992; Southwood 1985). The chemical functions mediated by these microorganisms could include detoxification of plant allelochemicals, degradation of plant cell walls, and biosynthesis of nutrients that are essential for the insect but in short supply in the plant food (Douglas 2009).
Recent advances in the life sciences have greatly enhanced the opportunity for integration between the fields of insect-plant interactions and insect-microbial interactions. In particular, understanding of the relationship between animals and microorganisms has changed fundamentally in the last 5-10 years, such that the concept of microbial brokers is fully congruent with the “new” mainstream of microbial ecology. In this review, I first address the technical and conceptual breakthroughs that are enabling the study of the microbiota of plant-feeding insects, and then I review current understanding of these microorganisms as brokers of insect-plant interactions.
Tools to Study the Microbiology of Plant-Associated Insects
Until recently, the greatest barrier to adopting the concept of microbial brokers in insect-plant interactions was technical. Most microorganisms in any environmental sample, including the resident microbial community in insects, are not readily cultured on standard microbiological media, and they are, consequently, not amenable to the traditional discipline of microbiology. Today, it is routine to catalog the composition and activities of complex microbial communities by cultivation-independent methods, making the concept of microbial brokers a readily testable hypothesis.
The field of microbial ecology has been transformed by high-throughput sequencing methods (Chaston and Douglas 2012; Zaneveld et al. 2011). For example, the phylogenetic position of a bacterium can be determined from the sequence of its 16S rRNA gene, obtained from a DNA sample by PCR amplification; and the relative abundance of different bacterial taxa in an environmental sample can be derived by massively-parallel sequencing of PCR-derived 16S amplicons using pyrosequencing or Illumina sequencing, with other technologies offering greater read lengths becoming available. However, the 16S rRNA gene is not a fully reliable predictor of bacterial functional traits because of lateral gene transfer and the bias for gene deletion in bacterial genomes. For example, bacteria with identical or near-identical 16S sequences may differ in their host range (Lerouge et al. 1990; Mandel et al. 2009; Nikoh et al. 2011), and can vary between benign and highly pathogenic phenotypes (Lukjancenko et al. 2010; Rasko et al. 2008). Insight into the functional potential of a single bacterium can be obtained by sequencing its genome, which is now a routine procedure that takes just a few weeks (Loman et al. 2012). In the same way, the genetic capacity for functions can be derived from sequencing the DNA of microbial communities (metagenomics), and the realized functional capacity obtained by sequencing the transcripts (metatranscriptomics) (Hess et al. 2011; Qin et al. 2010; Warnecke et al. 2007).
These high-throughput sequencing methods are best considered to provide a community level insight into microbial function. They have only a limited capacity to assign genes of interest to particular bacterial taxa, but generate hypotheses of taxonomic/functional relationships (Temperton and Giovannoni 2012). These hypotheses are increasingly becoming testable through advanced microscopical methods. It has long been feasible to identify individual microbial cells by hybridization of fluorescently-labeled 16S rDNA or other sequences to cells in whole mounts or sectioned material (Amann et al. 1995), and microscopical advances are increasingly enabling the gene expression profile and metabolic traits of individual microorganisms to be quantified in situ (Musat et al. 2012; Pernice et al. 2012). Methods to sequence the genome or transcriptome of single cells are being developed, providing valuable complementary strategies to interrogate the functional capability of individual microbial cells (Lasken 2013; Pamp et al. 2012).
The rich datasets and precise hypotheses that can now be obtained by culture-independent methods have generated the impetus to re-visit microbial cultivation technologies, especially to develop methods suitable for taxa that are adapted to low-nutrient conditions or are nutritionally fastidious (Carini et al. 2013; Goodman et al. 2011; Singh et al. 2013). Genome-based methods, such as metabolic modeling, can assist in identifying specific nutritional requirements, and have been used successfully to construct suitable culture media (Renesto et al. 2003). Culture-independent and culture-dependent approaches are increasingly seen as complementary, and not alternative, tools to investigate microbial diversity and function.
From the perspective of the microbiology of plant-associated insects, the important point is that these approaches are not dependent on extensive prior genomic information about the specific insect species. Genomics is no longer restricted to a few model species, such as Drosophila melanogaster and Bombyx mori. The DNA from any insect can be sequenced and our capacity to interpret the sequence data is made ever easier by the exponentially-increasing sequence data deposited in the publicly-available databases. Genomics is now a democratic endeavor.
The New Biology of Resident Microbiota In Animals
The capacity to identify and analyze the function of uncultured microorganisms (see above) has precipitated large consortial initiatives to study the resident microorganisms in humans, e.g., The Human Microbiome Project (commonfund.nih.gov), MetaHIT (metahit.eu), and these initiatives have further accelerated the sequencing technologies and bioinformatics tools for analysis. They also have contributed to a profound change in our understanding of the relationship between animals and their resident microbiota. Throughout much of the twentieth century, microorganisms were divided into three categories: disease-causing pathogens, commensals with no discernible impact on host health or fitness, and beneficial mutualists. Most microorganisms associated with most animals were treated as commensals. Plant-feeding insects provided some classic examples of specialized mutualists, such as the cellulose-degrading protists in the paunch (proximal hindgut) of lower termites, the dedicated organs (bacteriomes) housing bacteria in many plant sap-feeding insects, and the fungi maintained in the nests of leaf-cutting Attinine ants (Buchner 1965). These associations were considered as special cases, of significance only to the insect groups that bear them.
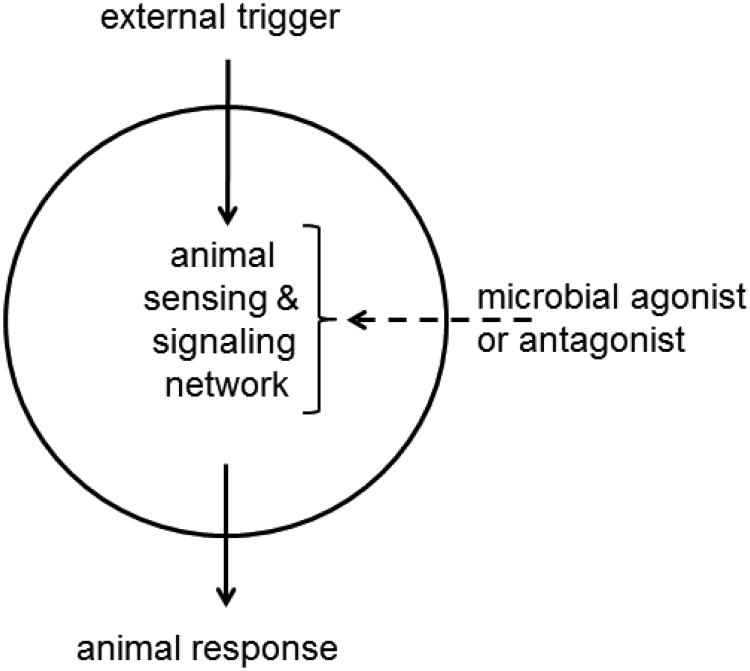
We now realize that many of the so-called commensal microorganisms that colonize animals are not commensal at all, but critically important to their animal host. Germ-free Drosophila display delayed development, altered nutrient allocation and metabolic rates, and depressed gut immunity (Ridley et al. 2012; Ryu et al. 2008; Storelli et al. 2011); and germ-free mice are profoundly altered in multiple physiological systems, including intestinal function and nutrition, the respiratory and vascular systems, immunity, and global metabolism (Smith et al. 2007). There is an emerging recognition that the health and fitness of animals has a microbial basis (McFall-Ngai et al. 2013). Although the detail of the mechanisms are still unclear, it appears that the signaling networks that coordinate the growth, development, and maintenance of animals are structured to function in the context of interactions with the resident microbiota (Fig. 1). When the microorganisms are removed or their composition or activities dramatically altered, the signaling networks become dysfunctional, leading to depressed animal vigor and fitness. This condition is known as dysbiosis (Stecher et al. 2013).
Fig. 1.
The sensing and signaling networks that regulate animal response to external factors are modulated by chemical signals from resident microorganisms. For example, acetic acid released from Acetobacter bacteria in the Drosophila gut amplifies Drosophila insulin signaling (see text), the levels of biologically-active dopamine and serotonin in the mouse gut are microbiota-dependent (Asano et al., 2012), and plant growth can be stimulated by the release of plant auxin hormones from rhizosphere bacteria (Lugtenberg and Kamilova, 2009).
Most research on dysbiosis has focused on humans and laboratory mice, but there are clear indications that the phenomenon extends to insects. Specifically, a recent study of Shin et al. (2011) on Drosophila offers a vivid illustration of microbial modulation of animal signaling networks. The blood glucose level in Drosophila is regulated by insulin-like peptides, in a fashion comparable to the insulin hormone of mammals. Germ-free Drosophila display reduced expression of key insulin-like peptide genes (specifically, dilp-3 and dilp-5), and a consequent elevation of their blood glucose levels. The normal phenotype is rescued by just one member of the gut microbiota, Acetobacter pomorum. Shin et al. (2011) demonstrated that A. pomorum with a null mutation in one gene, coding for the enzyme pyrroloquinoline quinone-dependent alcohol dehydrogenase (PQQ-ADH), display reduced expression of dilp-3/5 and elevated glucose levels. The conventional phenotype can be rescued by adding acetic acid to the diet, indicating that the bacterial-derived acetic acid increases the amplitude of insulin signaling in Drosophila.
In summary, it appears that the signaling networks that shape the function of animals, including insects, are not insulated from the influence of resident microorganisms. This condition may reflect the deep evolutionary history of associations with microorganisms, such that the signaling networks regulating animal function evolved in the context of pre-existing interactions with microorganisms (McFall-Ngai et al. 2013).
The recognition of the microbial basis of animal health has profound implications for insect-plant interactions, including a clarification of the defining features of microbial brokers of insect-plant interactions. Some microorganisms associated with the insects may be of no material significance to the insect host, bearing in mind that every microbial taxon identified in an insect need not “have a function”. Others may affect the fitness of the insect through their interactions with the insect regulatory networks, irrespective of the insect diet. Finally, there are the candidate microbial brokers: those microorganisms that may enable insect utilization of plant material by their capacity to synthesize nutrients in short supply in the ingested plant food, or to degrade plant cell wall material or plant toxins (Table 1). Below, I review the evidence for microbial brokers.
Table 1. survey of microbial brokers of insect-plant interactions (see text for details).
| Metabolic Capability of | Microbial brokers and their insect | Key references |
|---|---|---|
| Microorganisms | Hosts | |
| Essential amino acid synthesis | Various bacteria and fungi (“yeast-like symbionts”) in Hemiptera, e.g. Buchnera in aphids, Sulcia and Baumannia in leafhoppers | Buchner (1965); Douglas(2003); McCutcheon et al. (2009a) |
| Vitamin synthesis | Various bacteria in Coleoptera (especially stored product beetles, anobiid beetles) and Hemiptera | Nakabachi and Ishikawa, (1999); McCutcheon and Moran (2007); Buchner (1965) |
| Nitrogen fixation | Various termites | Ohkuma et al. (1999) |
| Sterol synthesis | “Yeast-like symbionts” in some planthoppers and some xylophagous beetles | Noda and Koizumi (2003); Norris et al., (1969) |
| Degradation of complex plant polysaccharides | Protists in lower termites, bacteria in scarab beetles | Bugg et al. (2011); Calderon-Cortes et al. (2012) |
| Detoxification of allelochemicals | Attinine ants, Dendroctonus beetles | De Fine Licht et al. (2013); Adams et al. (2013) |
Microorganisms that Provide Insects with Essential Nutrients
The insects are, as a group, metabolically impoverished. They share with other animals the inability to synthesize at least 9 protein-amino acids (the essential amino acids) and various co-factors required for the function of many metabolic enzymes (some vitamins). Insects and other arthropods additionally lack the capacity to synthesize sterols, which contribute to membrane architecture and are the basis for ecdysteroid hormones (Behmer and Nes 2003). Most insects derive these essential nutrients from their diet, but some plant products are grossly deficient in these compounds.
The most persuasive evidence for microbial brokers contributing to the synthesis of essential nutrients relates to insects feeding on plant (phloem or xylem) sap. The capacity to utilize plant sap through the life cycle has evolved multiple times within the order Hemiptera, but apparently in no other animal (Douglas 2003). Most of the nitrogen in plant sap is in the form of free amino acids (phloem sap additionally contains peptides and proteins, but these compounds generally account for a small proportion of the total nitrogen). Although phloem sap and xylem sap differ in their total concentration of amino acids (50-800 mM and 0.5-3 mM amino acids, respectively), they share the common property of a grossly unbalanced amino acid composition, containing less than 20% essential amino acids (Brodbeck et al. 1993; Christensen and Fogel 2011; Douglas 2003). Budget analysis revealed that the observed increase in the protein content of the final instar pea aphid Acyrthosiphon pisum could not be supported by the essential amino acids ingested from in the host plant Vicia faba (Akman Gunduz and Douglas 2009).
All Hemipterans that feed on plant sap through the life cycle bear large populations of specific microorganisms that are restricted to a specific location in the insect body (Buchner 1965). Although the details of the anatomical location and the identity of the microorganisms vary among the hemipteran groups, most associations involve specialized insect cells called bacteriocytes that house and maintain the microorganisms. The anatomical organization of bacteriocytes varies, including a single aggregation of cells in the body cavity (e.g., in aphids and whiteflies) or paired structures associated with the lateral wall of the abdomen (in leafhoppers and cicadas) (Buchner 1965). Alternative structural arrangements include the localization of bacteria to the distal midgut of plataspid stinkbugs and of yeasts to the fat body of some planthoppers (Dong et al. 2011; Nikoh et al. 2011). These microorganisms are vertically transmitted, usually via the female ovaries, such that the cytoplasm of each egg bears a maternal inoculum of symbionts at oviposition; gut symbionts are deposited externally, and acquired by the feeding offspring (Buchner 1965).
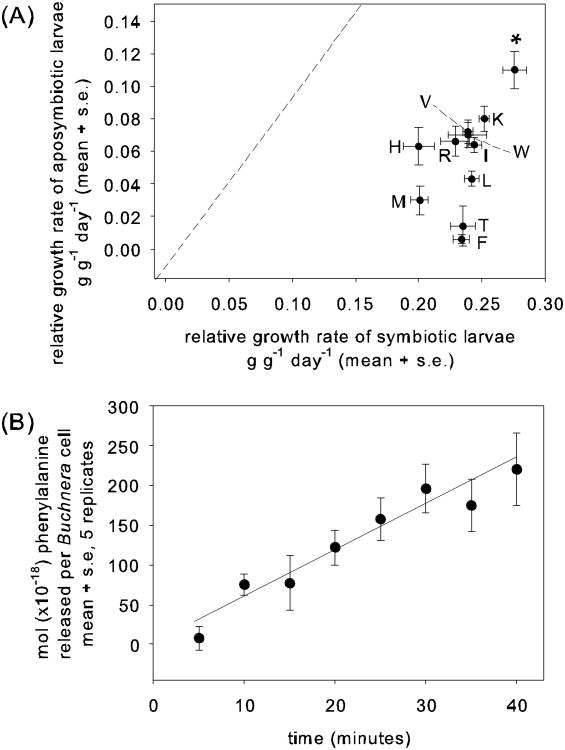
There is overwhelming evidence that the microbial symbionts provide the insect with essential amino acids, supplementing the plant sap. The evidence is genomic, dietary, and physiological. The genomes of many of these bacteria are very small (<1 Mb) and have a much reduced gene content relative to free-living bacteria (McCutcheon and Moran 2012). Despite the loss of many metabolic capabilities, these bacteria have retained genetic capacity for essential amino acid synthesis (McCutcheon et al. 2009a, b; McCutcheon and Moran 2010; Nikoh et al. 2011; Shigenobu et al. 2000;Sloan and Moran 2012). The dietary and physiological evidence comes almost entirely from research on aphids, simply because these insects can readily be maintained on chemically-defined diets. Unlike animals, generally, aphids do not have a dietary requirement for all the essential amino acids (Fig. 2a), and they can synthesize essential amino acids from dietary sucrose, aspartate, and glutamate; and when the bacteria are eliminated by antibiotic treatment, these capabilities are lost (Douglas et al. 2001; Febvay et al. 1999). Confirming the interpretation of these data that the bacterial endosymbiont Buchnera provides essential amino acids, metabolically-active Buchnera cells isolated from the association release essential amino acids at appreciable rates (Fig. 2b).
Fig. 2.
The symbiotic bacterium Buchnera aphidicola as a source of essential amino acids in the pea aphid Acyrthosiphon pisum. A. Relative growth rates of larval aphids that are symbiotic (untreated, bearing Buchnera bacteria) and aposymbiotic (experimentally deprived of Buchnera) on chemically-defined diets containing all 10 essential amino acids (*) or lacking one amino acid (F phenylalanine, H histidine, I isoleucine, K lysine, L leucine, M methionine, R arginine, T threonine, V valine, W tryptophan). The dashed diagonal is the line of equivalent growth rates [E. Gunduz and A. E. Douglas, unpub data: methods exactly as in Douglas et al. (2001)]. (B) Buchnera cells isolated from the aphids continue to release some essential amino acids into the medium [C W Russell and A E Douglas, unpub data. Buchnera preparations were obtained from dissected pea aphid bacteriocytes as described in MacDonald et al. (2012) and resuspended in reaction medium comprising 28 mM glucose, 8.6 mM NaCl, 1 mM MgSO4, 1 mM glutamate, 1 mM glutamine, 1 mM serine, 1 mM aspartate, 0.1 mM CaCl2, 0.25 M sucrose, 50 mM NaH2PO4, 13 mM K2H2PO4, pH 7.5 at a density 4 × 108 cells ml−1. Timecourse reactions were stopped by centrifugation at 1000 g for 70 seconds, and the supernatant was immediately flash-frozen in liquid nitrogen and stored at −80°C. The phenylalanine content of the supernatant was quantified using the AccQ Tag derivatization kit (Waters) by UPLC with PDA detector (Waters Acquity), as described in MacDonald et al. (2012).]
An alternative route by which microorganisms can act as brokers in relation to nitrogen nutrition is by the fixation of atmospheric nitrogen. This capability is of the greatest potential value to an insect feeding on low total nitrogen content, such as sound wood, which contains <0.1% nitrogen. There is persuasive evidence for bacterial nitrogen fixation in some termites (Ohkuma et al. 1999). Bacteria with a predicted capacity for nitrogen fixation or nitrogen-fixation genes have been identified in some analyses preparations of gut contents, for example in ants, the Hessian fly and tephritid flies (Aharon et al. 2013; Bansal et al. 2011; Russell et al. 2009). We should, however, be cautious in interpreting these data for several reasons. First, nitrogen fixation genes are not universally detected in bacteria associated with phytophagous insects. For example, the metagenome study of Shi et al. (2013) detected nitrogenase genes only in the gut contents of a termite Nasutitermes sp., and not the grasshopper Acrida cinerea or cutworm Agrotis ipsilon, despite deep sequence coverage. Second, the detection of the genetic capacity for nitrogen fixation is not sufficient evidence for nitrogen fixation. Gene expression is tightly regulated (for example, nitrogen fixation genes are repressed in the presence of combined nitrogen sources in most bacteria), and nitrogenase enzyme activity is irreversibly inhibited by molecular oxygen prevailing in the gut lumen of many insects. Finally, nitrogen fixation is advantageous to the insect only where the rates are substantial, requiring the nitrogen-fixing bacteria to represent at least 1-10% the total microbiota. As a reminder, nitrogen-fixing bacteria and genes are routinely detected in the human microbiome, but they are not abundant, and there is no suggestion that they contribute to the human nitrogen economy (Human Microbiome Project 2012). One intriguing instance of bacterial nitrogen fixation in an insect on a less extreme diet than wood products relates to the fruit-feeding tephritid fly Ceratitis capitatis, with nitrogen-fixing forms contributing an estimated 8% of the culturable bacteria and readily detectable nitrogen fixation rates, as quantified by the acetylene reduction assay (Behar et al. 2005).
Microorganisms associated with insects can act as brokers of plant utilization through additional biosynthetic capabilities, especially in relation to vitamin and sterol synthesis. Classic dietary studies revealed that elimination of the symbiotic microorganisms results in a dietary requirement for one or several B vitamins in planthoppers, stored product beetles, e.g., Oryzaephilus and Sitophilus species, xylophagous anobiid beetles [reviewed by Douglas (1989)]. Despite recent studies providing dietary and genomic evidence that several bacterial symbionts of small genome size have retained the pathways for the synthesis of some vitamins (McCutcheon and Moran 2007; Nakabachi and Ishikawa 1999), vitamin transfer from bacteria to the insect has not been demonstrated directly.
Only eukaryotic microorganisms can act as brokers of the sterol nutrition of plant-feeding insects because bacteria lack the capacity for sterol synthesis. Living plant tissue contains sterols. Although definitive sterol budget analyses of phytophagous insects remain to be conducted, there is a general consensus that plant sterol content is quantitatively sufficient for insect growth. The sterol mismatch between plants and most insects is that plants contain a diverse array of sterols, and the sterol metabolism of most insects is founded on a single sterol, cholesterol that is generally barely detectable in plant tissues. This mismatch is resolved by the intrinsic metabolic capabilities of the insect to convert the phytosterols to cholesterol (Behmer and Nes 2003). Fungal symbionts capable of sterol synthesis likely are crucially important to some xylophagous beetles, such as the ambrosia beetle Xyleborus ferrugineus (Norris et al. 1969) and to stored product insects because these substrates commonly contain barely detectable sterols. In particular, the elegant experiments of Pant and Fraenkel (1954) demonstrated a strong dietary requirement for sterols when the fungal symbionts were eliminated from the stored product beetles Lasioderma serricone and Sitophilus paniceum (=Stegobium paniceum). However, the relationship between symbiotic sources of sterols and low sterol content of the insect diet is not consistent. Contrary to the expectation that fungal symbionts are an important source of sterols for xylophagous insects, sterol analysis of the wood wasp Sirex noctilio suggests that this xylophage derives its sterols from the diet and not the fungal symbiont (Thompson et al. 2013). Also, there is persuasive evidence that the yeast-like symbionts provide sterols to the phloem-feeding planthoppers (Noda and Koizumi 2003), even though plant phloem sap is a sufficient source of sterols for other phloem-feeding insects with bacterial symbionts (Behmer et al. 2011). These results indicate that it would be premature to draw general conclusions linking microbial brokers of insect sterol nutrition to diet.
Microorganisms that Degrade Plant Cell Walls
Plant cell walls comprise a diverse mix of polysaccharides, dominated by cellulose and hemicellulose, which are embedded in a pectin matrix (Palin and Geitmann 2012; Pettolino et al. 2012). In secondary cell walls, the polysaccharides are further covalently linked by ester and ether linkages to the aromatic polymer, lignin, to form the structurally robust and chemically resistant lignocellulose. The difficulty in depolymerizing the lignin and extracting soluble carbohydrate from this recalcitrant structure is illustrated by the biofuel industry's urgent need for novel strategies to extract energy from plant biomass (Pauly and Keegstra 2010). Yet, various animals extract energy from plant cell walls, including lignocellulose, without the intervention of extreme temperature or pressure. The biological degradation of plant cell wall material is mediated by a battery of hydrolytic enzymes, including three classes of cellulases (endo-β-1,4-glucanases, exo-β-1,4-glucanases, and β-glucanases), xylanases that break down hemicellulose, ligninases (principally a laccase activity and peroxidase activity), pectinases, and pectin methylesterases, and multiple of other hydrolases and esterases with diverse specificities for different substituted carbohydrates (Calderon-Cortes et al. 2012; Gilbert 2010)
Vertebrate herbivory is dependent on the complementary functions of the animal and members of its gut microbiota: the teeth, gizzard etc. of the animal mediate mechanical disruption, making the polysaccharides available to enzymes of various gut bacteria that catalyze the chemical degradation of the cellulose, hemicellulose, etc. The enzymatic reactions take place slowly, and foregut or hindgut regions of vertebrate herbivores are modified to form large fermentation chambers in which the plant polysaccharides are transformed into sugars, and then fermented to short chain fatty acids that are released to the host. The bacteria are microbial brokers of vertebrate utilization of plant material, with vertebrates as a group lacking the enzymatic capacity for cellulose degradation.
The contribution of microorganisms to the utilization of plant products is more variable in insects than in vertebrates (Table 2). The concept of microbial brokers is fully relevant to many insects feeding on sound wood and other plant products with a high lignocellulose content, but less important for insects feeding on living plant material, which includes appreciable amounts of soluble carbohydrates and proteins (in the plant cell contents) and often little or no lignin.
Table 2. distribution of cellulases of endogenous and microbial origin in plant-feeding insects. [data and primary references in calderon-cortes et al. 2012].
| Insect | Cellulase Endogenous origin | Microbial origin |
|---|---|---|
| Orthoptera | ||
| multiple species | + | |
| Isoptera (termites) | + | + |
| multiple species | ||
| Coleoptera | ||
| Cerambycidae | + | + |
| Chrysomelidae | + | |
| Curculionidae | + | + |
| Scarabeidae | + | |
| Tenebronidae | + | |
| Hymenoptera | ||
| Siricid wasp (Amylostereum | + | |
| chailletii) | ||
| Apidae (Apis mellifera, | + | |
| Bombus terrestris) | ||
| Formidae (ants)a | + | |
| Lepidopterab | ||
| (Bombycidae) Bombyx mori | + | |
| Sessidae (Melittia | + | |
| satyriniformis, Synanthedon | ||
| scitula) | ||
| Diptera | ||
| Tipulidae (Tipula | + | |
| abdominalis) |
The microbiota in ants tested lack cellulase activity
Most species tested, including Plutella xylostella (Plutellidae) and representatives of Crambidae, Pyralidae, Nuctuidae and Pieridae, lack detectable cellulase activity of endogenous or microbial origin
A major contributory factor to the independence of many plant-feeding insects from microbial degradation of plant cell walls is that the genomes of many insects, unlike vertebrates, code for glucosyl hydrolases with predicted capacity to degrade cellulose and other plant cell wall polysaccharides. In various insects that feed on plant material, including Orthoptera, Lepidoptera, Coleoptera, and Isoptera (termites), these genes are expressed in the midgut, and their biochemical activity against complex carbohydrates, including cellulose, has been demonstrated (Calderon-Cortes et al. 2012; Oppert et al. 2010). Compounding this general capacity of insects to degrade complex carbohydrates, the very alkaline midgut (pH 10-12) of some herbivorous insects, notably the Lepidoptera, facilitates the solubilization of plant carbohydrates and proteins, thus further promoting nutrient extraction (Berenbaum 1980).
Nevertheless, two functional groups of plant cell wall-degrading enzymes appear to be lacking from insects: the exo-1,4-β-glucanases (cellobiohydrolases) and ligninases, which facilitate the breakdown of material with a high lignin content and crystalline cellulose content. Bacteria and eukaryotic microorganisms possess these enzymatic functions (Bugg et al. 2011). Microbial capacity for cellulose degradation is exploited by various insects feeding on sound wood and other products with high lignocellulose content, but the evidence for lignin degradation by the gut microbiota in these insects is equivocal (Bugg et al. 2011).
The best studied microbial brokers are the cellulolytic protists in lower termites, with accumulating evidence that bacteria contribute to cellulose degradation in higher termites (which lack protists) and some other insects, including scarab beetles and the Thysanura (Calderon-Cortes et al. 2012; Watanabe et al. 2010). As in vertebrates, the microorganisms in many insects are housed in a fermentation chamber, almost invariably an expanded portion of the hindgut. Consequently, the plant cell wall material delivered to the microbiota is the residue that is recalcitrant to the enzymatic activities of the digestive enzymes in the insect midgut.
Some insects have been proposed as exploiting the enzymatic capabilities of external microorganisms: the insects feed on microbial cells, which are disrupted in the insect gut, releasing active enzymes that contribute to the digestion of complex plant material. This process has been demonstrated in fungus-growing termites of the subfamily Macrotermitinae (Abo-Khatwa 1978; Martin and Martin 1978), but the quantitative importance of ingested enzymes in the degradation of plant material has been questioned (Slaytor 1992), and remains uncertain.
Do microorganisms associated with insects detoxify plant allelochemicals?
Many secondary metabolites in plants function as defensive compounds that are toxic to insects or deter feeding. These compounds are important chemical determinants of insect-plant interactions. The capacity of insects to transform them to harmless products is an important determinant of the plant range of many phytophagous insects, and has played a crucial role in the evolutionary diversification of various insect groups (Herrera and Pellmyr 2002; Schoonhovenet al. 2005).
Repeatedly, the detoxification of plant allelochemicals has been attributed to microorganisms associated with insects. The authority for these statements is often a mis-cited review article, or a review that has mis-cited the previous literature. The primary literature provides ample evidence for insect-mediated detoxification, involving cytochrome P450 monooxygenases, glutathione S-transferases, and esterases (Despres et al. 2007), but only the most slender evidence for microbial degradation of plant allelochemicals. Nevertheless, two recent publications offer tantalizing evidence for a microbial role. The first study concerns the leaf-cutting ant Acromyrmex echinatior, which maintains and feeds on the symbiotic fungus Leucocoprinus gongylophorus in its nest. De Fine Licht et al. (2013) have shown that enzymatically-active laccase enzyme is passed through the gut of ants feeding on the fungus and released onto plant material, where it can degrade plant compounds, such as tannins and flavonoids. In the second study, Adams et al. (2013) demonstrated that the gut microbiota of the mountain pine beetle Dendroctonus ponderosae include bacteria of the genera Pseudomonas Serratia Rahnella, and Burkholderia that bear genes involved in the degradation of terpenes, the principal defensive compounds of the trees infested by these beetles.
The balance of current evidence would suggest that these two systems may be unusual, with intrinsic detoxification as the norm in plant-feeding insects, but our knowledge of the microbiota of insects is in its infancy. With the availability of high-throughput sequencing methods and ever-improving algorithms and databases for the analysis of sequence data, the scientific community has a unique opportunity to interrogate the detoxification capacity of microorganisms associated with insects, and relate these data to the plant utilization traits of their hosts.
Concluding Comments
Is the concept of microbial brokers useful for the study of insect-plant interactions? I believe that it is valuable as a testable hypothesis. As considered in this article, resident microorganisms play a central role in insect utilization of diets of exceptionally low and unbalanced nutrient contents. This has been demonstrated, in particular, for insects that feed on plant sap or sound wood through the life cycle. For these systems, important unresolved issues include whether natural variation in the composition or activities of the microbiota influence diet choice (including plant range) and fitness of the individual insect, potentially with far-reaching ecological consequences.
In many insects, however, the role of microorganisms as brokers of insect utilization of plants remains as a hypothesis. Without doubt, the techniques now available to study microorganisms will facilitate the testing of this hypothesis. For some systems, the data may prove to be complex, with variation in the relative importance of intrinsic and microbial processes according to the diet, developmental age, genotype, and species of the insect. For example, acridid grasshoppers have been estimated to degrade up to 60% of ingested plant cell wall material (Cazemier et al. 1997): both insect carbohydrases expressed in the midgut and enzymes of bacteria, predominantly γ -proteobacteria, in the hindgut, are likely to contribute to the breakdown of the plant material, and it is extremely unlikely that the relative importance of the two contributors is fixed in either ecological or evolutionary time.
The inclusion of microbial ecology in the study of insect-plant interactions offers great opportunity, but there are two potential pitfalls. The first misconception is conceptual, that resident microorganisms “must have a function”, i.e., they are expected to influence the phenotype of the insect host. Some microorganisms may exploit the insect habitat with minimal effects on insect traits or fitness, even if they have genetic capabilities potentially valuable to the insect. It is important to test for function through analysis of microbial activities in the insect and the insect response to elimination of the microorganism. This leads directly to the second pitfall: interpretation of all microbiota-dependent traits as adaptations to plant utilization. As considered in this article, it is becoming increasingly realized that many fundamental aspects of animal function are microbe-dependent, as a consequence of the deep evolutionary history of animal-microbial relations (McFall Ngai et al. 2013). The role of resident microorganisms as brokers in the interactions between insects and plants overlays their much deeper role as brokers of the function of all animals, including insects.
Acknowledgments
This work was supported by NSF grant BIO 1241099, AFRI grant NYW-2011-04650 and the Sarkaria Institute for Insect Physiology and Toxicology.
References
- Abo-Khatwa N. Cellulase of fungus-growing termites: a new hypothesis of its origin. Experientia. 1978;34:559–560. [Google Scholar]
- Adams AS, Aylward FO, Adams SM, Erbilgin N, Aukema BH, Currie CR, Suen G, Raffa KF. Mountain pine beetles colonizing historical and naive host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl Environ Microbiol. 2013 doi: 10.1128/AEM.00068-13. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharon Y, Pasternak Z, Ben Yosef M, Behar A, Lauzon C, Yuval B, Jurkevitch E. Phylogenetic, metabolic, and taxonomic diversities shape mediterranean fruit fly microbiotas during ontogeny. Appl Environ Microbiol. 2013;79:303–313. doi: 10.1128/AEM.02761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akman Gunduz E, Douglas AE. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Roy Soc London B. 2009;276:987–991. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali JG, Alborn HT, Stelinski LL. Constitutive and induced subterranean plant volatiles attract both entomopathogenic and plant parasitic nematodes. J Ecol. 2011;99:26–35. [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Revs. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y, Hiramoto T, Nishino R, Aiba Y, Kimura T, Yoshihara K, Koga Y, Sudo N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Amer J Physiol Gastrointest Liver Physiol. 2012;303:G1288–G1295. doi: 10.1152/ajpgi.00341.2012. [DOI] [PubMed] [Google Scholar]
- Bansal R, Hulbert S, Schemerhorn B, Reese JC, Whitworth RJ, Stuart JJ, Chen MS. Hessian fly-associated bacteria: transmission, essentiality, and composition. PLoS One. 2011;6:e23170. doi: 10.1371/journal.pone.0023170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar A, Yuval B, Jurkevitch E. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol Ecol. 2005;14:2637–2643. doi: 10.1111/j.1365-294X.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- Behmer ST, Nes WD. Insect sterol nutrition and physiology: a global overview. Adv Insect Physiol. 2003;31:1–72. [Google Scholar]
- Behmer ST, Grebenok RJ, Douglas AE. Plant sterols and host plant suitability for a phloem-feeding insect. Funct Ecol. 2011;25:484–491. [Google Scholar]
- Berenbaum M. Adaptive significance of midgut pH in larval Lepidoptera. Amer Nat. 1980;115:138–146. [Google Scholar]
- Brodbeck BV, Mizell RF, Andersen PC. Physiological and behavioural adaptations of three species of leafhoppers in response to the dilute nutrient content of xylem sap. J Insect Physiol. 1993;39:73–81. [Google Scholar]
- Buchner P. Endosymbioses of animals with plant microorganisms. Chichester, UK: John Wiley & Sons; 1965. [Google Scholar]
- Bugg TD, Ahmad M, Hardiman EM, Singh R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr Opin Biotechnol. 2011;22:394–400. doi: 10.1016/j.copbio.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Calderon-Cortes N, Quesada M, Watanabe H, Cano-Camacho H, Oyama K. Endogenous plant cell wall digestion: a key mechanism in insect evolution. Annu Revs Ecol Evol Syst. 2012;43:45–71. [Google Scholar]
- Carini P, Steindler L, Beszteri S, Giovannoni SJ. Nutrient requirements for growth of the extreme oligotroph 'Candidatus Pelagibacter ubique' HTCC1062 on a defined medium. ISME J. 2013;7:592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazemir AE, Op Den Camp HJM, Hackstein JHP, Vogels GD. Fibre digestion in arthropods, Comp. Biochem Physiol A Physiol. 1997;118:101–109. [Google Scholar]
- Chaston JM, Douglas AE. Making the most of “omics” for symbiosis research. Biol Bull. 2012;223:21–29. doi: 10.1086/BBLv223n1p21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen H, Fogel ML. Feeding ecology and evidence for amino acid synthesis in the periodical cicada (Magicicada) J Insect Physiol. 2011;57:211–219. doi: 10.1016/j.jinsphys.2010.11.005. [DOI] [PubMed] [Google Scholar]
- De Fine Licht HH, Schiott M, Rogowska-Wrzesinska A, Nygaard S, Roepstorff P, Boomsma JJ. Laccase detoxification mediates the nutritional alliance between leaf-cutting ants and fungus-garden symbionts. Proc Natl Acad Sci USA. 2013;110:583–587. doi: 10.1073/pnas.1212709110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres L, David JP, Gallet C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol Evol. 2007;22:298–307. doi: 10.1016/j.tree.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Dong S, Pang K, Bai X, YU X, Hao P. Identification of two species of yeast-like symbiotes in the brown planthopper, Nilaparvata lugens. Curr Microbiol. 2011;62:1133–1138. doi: 10.1007/s00284-010-9830-z. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 1989;64:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Microbial brokers of insect-plant interactions. In: Menken SBJ, Visser JH, Harrewijn P, editors. Proceedings of the 8th International Insect-Plant Interactions. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1992. pp. 329–336. [Google Scholar]
- Douglas AE. The nutritional physiology of aphids. Adv Insect Physiol. 2003;31:73–140. [Google Scholar]
- Douglas AE. The microbial dimension in insect nutritional ecology. Funct Ecol. 2009;23:38–47. [Google Scholar]
- Douglas AE, Minto LB, Wilkinson TL. Quantifying nutrient production by the microbial symbionts in an aphid. J Exp Biol. 2001;204:349–358. doi: 10.1242/jeb.204.2.349. [DOI] [PubMed] [Google Scholar]
- Febvay G, Rahbe Y, Rynkiewicz M, Guillaud J, Bonnot G. Fate of dietary sucrose and neosynthesis of amino acids in the pea aphid, Acyrthosiphon pisum, reared on different diets. J Exp Biol. 1999;202:2639–2652. doi: 10.1242/jeb.202.19.2639. [DOI] [PubMed] [Google Scholar]
- Gilbert HJ. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 2010;153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher PE. Three-way interactions between plant pathogenic fungi, herbivorous insects and their host plants. Biol Rev Camb Philos Soc. 1995;70:639–694. [Google Scholar]
- Herrera CM, Pellmyr O. Plant-animal interactions. Oxford University Press; 2002. [Google Scholar]
- Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingwell LL, Eigenbrode SD, Bosque-Perez NA. Plant viruses alter insect behavior to enhance their spread. Sci Rep. 2012;2:578. doi: 10.1038/srep00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban R, Baldwin IT. Induced responses to herbivory. University of Chicago Press; Chicago: 1997. [Google Scholar]
- Koch A. Intracellular symbiosis in insects. Annu Rev Microbiol. 1960;14:121–140. doi: 10.1146/annurev.mi.14.100160.001005. [DOI] [PubMed] [Google Scholar]
- Lasken RS. Single-cell sequencing in its prime. Nat Biotechnol. 2013;31:211–212. doi: 10.1038/nbt.2523. [DOI] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Prome JC, Denarie J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–4. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nature Rev Microbiol. 2012;10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- Lukjancenko O, Wassenaar TD, Ussery DW. Comparison of 61 sequenced Escherichia coli genomes. Microb Ecol. 2010;60:708–720. doi: 10.1007/s00248-010-9717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SJ, Lin GG, Russell CW, Thomas GH, Douglas AE. The central role of the host cell in symbiotic nitrogen metabolism. Proc Roy Soc London B. 2012;279:2965–2973. doi: 10.1098/rspb.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–8. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MM, Martin JS. Cellulose digestion in midgut of fungus-growing termite Macrotermes natalensis: role of acquired enzymes. Science. 1978;199:1453–1455. doi: 10.1126/science.199.4336.1453. [DOI] [PubMed] [Google Scholar]
- Mccutcheon JP, Mcdonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A. 2009a;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccutcheon JP, Mcdonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009b;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Mcfall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104:8627–33. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musat N, Foster R, Vagner T, Adam B, Kuypers MM. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiol Rev. 2012;36:486–511. doi: 10.1111/j.1574-6976.2011.00303.x. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Ishikawa H. Provision of riboflavin to the host aphid, Acyrthosiphon pisum, by endosymbiotic bacteria, Buchnera. J Insect Physiol. 1999;45:1–6. doi: 10.1016/s0022-1910(98)00104-8. [DOI] [PubMed] [Google Scholar]
- Nikoh N, Hosokawa T, Oshima K, Hattori M, Fukatsu T. Reductive evolution of bacterial genome in insect gut environment. Genome Biol Evol. 2011;3:702–714. doi: 10.1093/gbe/evr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda H, Koizumi Y. Sterol biosynthesis by symbiotes: cytochrome P450 sterol C-22 desaturase genes from yeastlike symbiotes of rice planthoppers and anobiid beetles. Insect Biochem Mol Biol. 2003;33:649–658. doi: 10.1016/s0965-1748(03)00056-0. [DOI] [PubMed] [Google Scholar]
- Norris DM, Baker JM, Chu HM. Symbiotic interrelationships between microbes and ambrosia beetles. III Ergosterol as the source of sterol to the insect. Ann Entomol Soc Am. 1969;62:413–414. [Google Scholar]
- Ohkuma M, Noda S, Kudo T. Phylogenetic diversity of nitrogen fixation genes in the symbiotic microbial community in the gut of diverse termites. Appl Environ Microbiol. 1999;65:4926–4934. doi: 10.1128/aem.65.11.4926-4934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppert C, Klingeman WE, Willis JD, Oppert B, Jurat-Fuentes JL. Prospecting for cellulolytic activity in insect digestive fluids. Comp Biochem Physiol B Biochem Mol Biol. 2010;155:145–154. doi: 10.1016/j.cbpb.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Palin R, Geitmann A. The role of pectin in plant morphogenesis. Biosystems. 2012;109:397–402. doi: 10.1016/j.biosystems.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB) Genome Res. 2012;22:1107–1119. doi: 10.1101/gr.131482.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant NC, Fraenkel G. Studies of the symbiotic yeasts of the two insect species, Lasioderma serricorne F. and Stegobium paniceum. Biol Bull. 1954;107:420–430. doi: 10.1126/science.112.2913.498. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Plant cell wall polymers as precursors for biofuels. Curr Op Plant Biol. 2010;13:305–312. doi: 10.1016/j.pbi.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Pernice M, Meibom A, Van Den Heuvel A, Kopp C, Domart-Coulon I, Hoegh-Guldberg O, Dove S. A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J. 2012;6:1314–1324. doi: 10.1038/ismej.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettolino FA, Walsh C, Fincher GB, Bacic A. Determining the polysaccharide composition of plant cell walls. Nature Protoc. 2012;7:1590–1607. doi: 10.1038/nprot.2012.081. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Rosovitz MJ, Myers GSA, Mogodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bac. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renesto P, Crapoulet N, Ogata H, La Scola B, Vestris G, Claverie JM, Raoult D. Genome-based design of a cell-free culture medium for Tropheryma whipplei. Lancet. 2003;362:447–449. doi: 10.1016/S0140-6736(03)14071-8. [DOI] [PubMed] [Google Scholar]
- Ridley EV, Wong AC, Westmiller S, Douglas AE. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One. 2012;7:e36765. doi: 10.1371/journal.pone.0036765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA. 2009;106:21236–21241. doi: 10.1073/pnas.0907926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonhoven LM, Van Loon JJA, Dicke M. Insect-plant biology. 2nd. Oxford University Press; 2005. [Google Scholar]
- Shi W, Xie S, Chen X, SUN S, Zhou X, Liu L, Geo P, Kyrpides NC, No EG, Yuan JS. Comparative genomic analysis of the microbiome of herbivorous insects revealse eco-environmental adaptations: biotechnology applications. PLoS Genet. 2013;9:e1003131. doi: 10.1371/journal.pgen.1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–6. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- Singh S, Eldin C, Kowalczewska M, Raoult D. Axenic culture of fastidious and intracellular bacteria. Trends Microbiol. 2013;21:92–99. doi: 10.1016/j.tim.2012.10.007. [DOI] [PubMed] [Google Scholar]
- Slaytor M. Cellulose digestion in termites and cockroaches: what role do symbionts play? Compar Biochem Physiol. 1992;103B:775–784. [Google Scholar]
- Sloan DB, Moran NA. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol Lett. 2012;8:986–989. doi: 10.1098/rsbl.2012.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Mckoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Sem Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Southwood TRE. Interactions of plants and animals: pattern and process. Oikos. 1985;44:5–11. [Google Scholar]
- Stecher B, Maier L, Hardt WD. 'Blooming' in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011;14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Stout MJ, Thaler JS, Thomma BP. Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu Rev Entomol. 2006;51:663–689. doi: 10.1146/annurev.ento.51.110104.151117. [DOI] [PubMed] [Google Scholar]
- Tack AJM, Gripenberg S, Roslin T. Cross-kingdom interactions matter: fungal-mediated interactions structure an insect community on oak. Ecol Lett. 2012;15:177–185. doi: 10.1111/j.1461-0248.2011.01724.x. [DOI] [PubMed] [Google Scholar]
- Temperton B, Giovannoni SJ. Metagenomics: microbial diversity through a scratched lens. Curr Opin Microbiol. 2012;15:605–612. doi: 10.1016/j.mib.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Thompson BM, Grebenok RJ, Behmer ST, Gruner DS. Microbial symbionts shape the sterol profile of the xylem-feeding woodwasp, Sirex noctilio. J Chem Ecol. 2013;39:129–139. doi: 10.1007/s10886-012-0222-7. [DOI] [PubMed] [Google Scholar]
- Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, Cayouette M, Mchardy AC, Djordjevic G, Aboushadi N, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Tokuda G. Cellulolytic systems in insects. Annu Rev Entomol. 2010;55:609–632. doi: 10.1146/annurev-ento-112408-085319. [DOI] [PubMed] [Google Scholar]
- Zaneveld JRR, Parfrey LW, Van Treuren W, Lozupone C, Clemente JC, Knights D, Stombaugh J, Kuczynski J, Knight R. Combined phylogenetic and genomic approaches for the high-throughput study of microbial habitat adaptation. Trends Microbiol. 2011;19:472–482. doi: 10.1016/j.tim.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]