Abstract
Genomic regions with replicated linkage to asthma-related phenotypes likely harbor multiple susceptibility loci with relatively minor effects on disease susceptibility. The 11q13 chromosomal region has repeatedly been linked to asthma with five genes residing in this region with reported replicated associations. Cortactin, an actin-binding protein encoded by the CTTN gene in 11q13, constitutes a key regulator of cytoskeletal dynamics and contractile cell machinery, events facilitated by interaction with myosin light chain kinase; encoded by MYLK, a gene we recently reported as associated with severe asthma in African Americans. To evaluate potential association of CTTN gene variation with asthma susceptibility, CTTN exons and flanking regions were re-sequenced in 48 non-asthmatic multiethnic samples, leading to selection of nine tagging polymorphisms for case-control association studies in individuals of European and African descent. After ancestry adjustments, an intronic variant (rs3802780) was significantly associated with severe asthma (odds ratio [OR]: 1.71; 95% confidence interval [CI]: 1.20-2.43; p = 0.003) in a joint analysis. Further analyses evidenced independent and additive effects of CTTN and MYLK risk variants for severe asthma susceptibility in African Americans (accumulated OR: 2.93, 95% CI: 1.40-6.13, p = 0.004). These data suggest that CTTN gene variation may contribute to severe asthma and that the combined effects of CTTN and MYLK risk polymorphisms may further increase susceptibility to severe asthma in African Americans harboring both genetic variants.
Keywords: CTTN, MLCK, cytoskeleton, SNP, asthma
Introduction
Asthma is a common airway inflammatory disorder affecting 7% of the US population [Mannino et al., 2002]) characterized by airflow obstruction and bronchial hyper-responsiveness, often associated with atopy. Incomplete twin concordance and patterns of inheritance in families have suggested that asthma and related phenotypes have considerable and complex genetic components [Ober, 2005]. Genome-wide linkage analyses have revealed at least 20 broadly defined major asthma susceptibility regions, several of which have been replicated [Wills-Karp and Ewart, 2004]. Candidate gene association studies have indicated that many regions contain multiple susceptibility loci with relatively small effects on disease risk [Ober and Hoffjan, 2006]. Chromosome 11q13, a region replicated across various studies [Wills-Karp and Ewart, 2004], contains at least five asthma-related genes identified in two or more association studies.
Histopathological studies of asthmatic airway tissue samples typically demonstrate persistent airway tissue inflammation, irreversible airway structural changes [Moore and Peters, 2006] and vascular permeability consistent with tissue inflammatory changes and both angiogenic and microvascular remodeling processes [Orsida et al., 1999; McDonald, 2001]. Although the majority of putative asthma susceptibility genes are related to immune function or therapeutic target pathways [Ober and Hoffjan, 2006], it has become apparent that vascular genes (involved in regulation of both angiogenesis and inflammation) as well as genes governing pro-fibrotic processes (cell hypertrophy, cell adhesion and migration), are also relevant to asthmatic inflammatory lung injury, tissue remodeling and pathogenesis. Asthma susceptibility genes implicated in airway tissue remodeling include ADAM33 [Van Eerdewegh et al., 2002; Kedda et al., 2006], EGFR [Wang et al., 2006], ITGB3 [Weiss et al., 2005; Thompson et al., 2007], SELP [Bourgain et al., 2003], TGFB1 [Pulleyn et al., 2001; Hoffjan et al., 2004; Silverman et al., 2004], and VEGFR2 [Park et al., 2006]. Cytoskeletal genes provide dynamic regulation of cell shape, cell motility and adhesion and are involved in remodeling processes during asthma and are up-regulated in asthmatic peripheral blood lymphocytes [Hansel et al., 2005] and in asthmatic airway smooth muscle cells [Benayoun et al., 2003]. We recently reported that a non-synonymous variation in the myosin light chain kinase gene (MYLK), a central regulator of cellular contraction, is strongly associated with severe asthma in African Americans [Flores et al., 2007], a finding subsequently validated by our report in an Afro-Caribbean asthmatic cohort from Barbados [Gao et al., 2007].
Cortactin, an F-actin binding multidomain scaffold protein, is involved in cortical actin assembly and dynamic actin cytoskeleton rearrangement [Cosen-Binker and Kapus, 2006]. We have previously described the interaction between cortactin and MLCK [Dudek et al., 2002], and cortactin participates in diverse cellular processes, including lung vascular barrier regulation [Dudek et al., 2004], cell motility (overexpression enhances migration of both fibroblasts and endothelial cells and promotes metastasis in human cancers [Daly, 2004]), intercellular junction assembly [Cosen-Binker and Kapus, 2006], and leukocyte adhesion and transmigration [Tilghman and Hoover, 2002; Johansson et al., 2004; Yang et al., 2006]. Additionally, current evidences suggest that cortactin is critical for the angiogenic lipid factor sphingolipid sphingosine 1-phosphate (S1P), found in elevated concentrations in the airways of asthmatic (but not control) subjects after segmental antigen challenge [Ammit et al., 2001], in mediating lymphocyte chemotaxis and transmigration [Roviezzo et al., 2004; Brinkmann and Baumruker, 2006], and for the enhancement of endothelial and vascular smooth muscle cell proliferation and migration [Black and Johnson, 2002; Sawicka et al., 2003; Lee et al., 2006a].
Cortactin is encoded by the CTTN gene (∼ 38 kilobases [kb]), located in close proximity to previously identified asthma-related genes (FCER1B, CC16, GSTP1, GPR44 and IL18) in the 11q13 linkage region [Ober and Hoffjan, 2006]. Based on the linkage evidence and the biological plausibility for its implication in immunoregulation and angiogenesis, we explored the potential association of CTTN gene variants with susceptibility to asthma. We sequenced CTTN to identify novel polymorphisms, selected nine SNPs for genotyping a multiethnic case-control sample collected in Chicago, and identified a significant association with severe asthma with an intronic CTTN polymorphism. In addition, we observed that a recently reported susceptibility variant of MYLK, the gene encoding MLCK, coupled to the CTTN polymorphism increases the risk for severe asthma in African Americans (∼ three fold).
Methods
Study populations and clinical evaluation
Unrelated asthma cases (110 European Americans and 192 African Americans) were recruited in the Chicago area as part of the Collaborative Study on the Genetics of Asthma protocol [CSGA, 1997]. A brief clinical description of these samples has been reported [Ober et al., 2000; Lester et al., 2001]. Controls (209 European-Americans and 193 African Americans) consisted of adult individuals with negative personal and first-degree relative family history for asthma. Cases and controls reported at least three grandparents who were either of European or African American ancestry. No medical history was taken, and no medical testing was performed on control individuals. Twenty seven European Americans and 103 African Americans asthmatics were diagnosed with severe asthma on the basis of nocturnal symptoms, prescribed use of either inhaled or oral steroids, a forced expiratory volume measurements (FEV1) < 60% predicted value at any time and time related reversibility (≥ 15% increase in baseline FEV1 after treatment). This study was approved by the Institutional Review Board and written informed consent was obtained from all participants. Five samples (three European American controls and two African American cases, one of them with severe asthma) were excluded from subsequent analysis [Flores et al., 2007] since their self-reported ancestry did not match molecular ancestry determined by the genotypes of 30 microsatellite markers selected to be informative on individual European, Asian and African ancestries [Amundadottir et al., 2006].
CTTN Polymorphism Discovery
DNA samples from 48 unrelated non-asthmatic individuals (19 African and 29 European Americans) were used to search for common variations in the CTTN gene. DNA sequencing protocols and polymorphism identification were performed as previously described [Gao et al., 2006]. Primer pairs used are described in Table AI (Appendix A). The genomic sequence NM_005231 corresponding to CTTN isoform a, the longest transcript of the gene, was used as the reference sequence. Sequence variation is reported following established guidelines [Antonarakis, 1998].
Genotyping
Genotyping was conducted by means of TaqMan™ allelic discrimination assays on a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) except for SNPs rs7131282 and rs34959377, that were genotyped using the SNaPshot® multiplex kit (Applied Biosystems, Foster City, CA). Briefly, amplification of SNP-containing fragments were performed in two separate PCR reactions (AmpliTaq Gold™ DNA Polymerase with 2.5 mM Mg2+, 10% DMSO and 0.1 μM of primers 5′-TGGAGTTATGTGGTGGAAAC-3′ and 5′-GGGAAGAGAACACAGAGAAA-3′ for rs7131282, and 5′-GTATTCTCTGAACCCTTGGA-3′ and 5′-CTGAGGCTGCTCTTAAACTG-3′ for rs34959377 respectively). The SNaPshot® single base extension reaction was carried out simultaneously for both SNPs using 1 μM of primers 5′-CCCTGCAGAGATGCGG-3′ for rs7131282 and 5′-CCCCCCGGGAACTCAGGGGAACG-3′ for rs34959377 based on manufacturer's recommendations. All genotype calls, excepting for rs7131282 and rs34959377 (genotyped by SNaPshot® reactions), were performed automatically by the SDS software (Applied Biosystems, Foster City, CA) based on discriminating plots (95% confidence) from the ratio between the allele probe fluorescent intensities after normalization. Genotyping was blind to the ethnic background of the sample or case and control status. Approximately 10% of the samples were genotyped in duplicate to monitor genotyping quality.
Statistical Analysis
Departures from Hardy-Weinberg equilibrium (HWE) were tested by means of an exact test. The multiple-marker selection algorithm, haplotype r2, included in TagIT 3.03 software [Weale et al., 2003] was used to select a set of tagging SNPs (tSNPs) satisfying a minimum performance of r2 > 0.90. This algorithm is known to maintain the tSNPs prediction accuracy while tending to increase tagging efficiency by selecting fewer tSNPs than other existing methods [Ding and Kullo, 2007]. A SNP-dropping-with-re-sampling method [Ahmadi et al., 2005] was used to verify that the expected properties of the tSNP set ensured a minimum performance of r2 > 0.85 in both populations. SNP associations were tested by means of an additive model using the Armitage trend test. SPSS 14.0 (SPSS Inc., Chicago, IL) was used for multiple logistic regression analysis, including use of individual ancestries based on 30 microsatellites as covariates [Flores et al., 2007] when appropriate, to estimate the odds ratios (OR) with their respective 95% confidence interval (CI). EPIDAT 3.0 was used for the Mantel-Haenszel stratified analysis and to estimate the effects of the two risk variants in CTTN and MYLK genes, the latter using dominant models for both SNPs due to the small sample sizes. A non-additive effect for gene-gene interaction was tested by means of SNPstats [Sole et al., 2006]. Individual SNP associations were adjusted for multiple testing as implemented in the Simple Interactive Statistical Analysis web site [Sankoh et al., 1997], using the Šidák procedure taking into account the composite linkage disequilibrium (LD), which works well in capturing the LD correlation among SNPs [Zaykin et al., 2006], as a correlation estimator. In addition, we used two replication samples to reduce the likelihood of type I error. P-values were not corrected for the two conditions tested due to the non-independent nature of both phenotypes. The patterns of LD were explored using Haploview 3.32 [Barrett et al., 2005].
Results
SNP discovery and tagging SNP (tSNP) selection
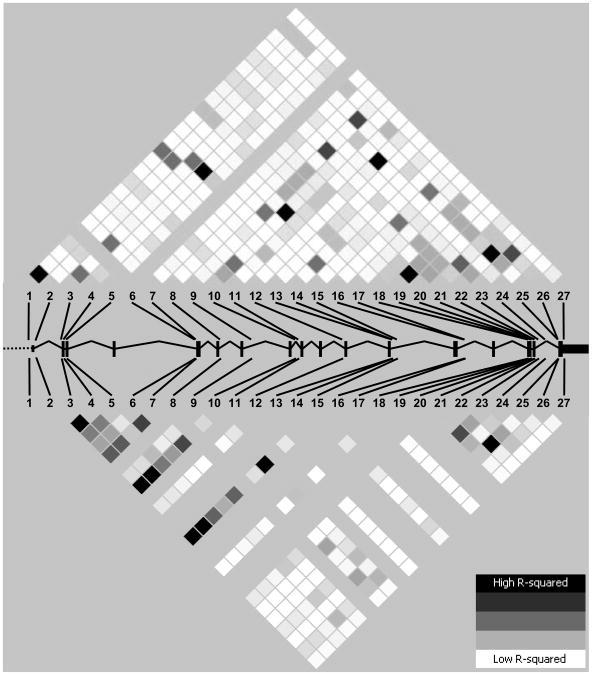
A total of 27 CTTN variants, all in Hardy-Weinberg equilibrium, were identified by re-sequencing 9,746 base pairs in each sample with each variant consisting of a single base change (SNP), with the exception of variation number 27 (ss76859860) which demonstrated a single base insertion (Table I). These SNPs generate an overall occurrence of one SNP every ∼375 bp, similar to the average rate (one per 348 bp) reported in a re-sequencing survey of 106 genes [Cargill et al., 1999]. Ten SNPs were entirely novel, two SNPs predicted an amino acid change, and 4 SNPs were not previously validated by frequency. CTTN SNPs 1, 2, 8, 13 and 27 showed a MAF < 5% in both populations whereas the previously described CTTN SNPs 1 (rs11825246) and 2 (rs11825335) demonstrated allelic frequencies of ∼2-3% in Yoruba Nigerians (YRI) in the newly released HapMap [The International HapMap Consortium, 2005]. Linkage disequilibrium (LD) plots suggested a broad heterogeneity of LD in the region with strong and moderate values clustering in the 3′ end of the gene in both populations, also extending to the 5′ end in European Americans (Fig. 1). After excluding SNPs with MAF < 5%, a total of 6 and 7 SNPs were sufficient to provide an adequate coverage of CTTN gene variation in European and African Americans, respectively. Nine cosmopolitan tSNPs, providing adequate coverage in both populations, were selected for association studies. These tSNPs tagged common SNPs (MAF ≥ 5%) in YRI subjects and in samples from Utah residents with ancestry from northern and Western Europe (CEU) with slightly increased performances than those achieved in the re-sequenced samples. Additionally, a total of 4 and 7 tSNPs from HapMap provided adequate coverage of CTTN gene variation in CEU and YRI, respectively, congruent with our estimates for European- and African Americans.
Table I.
Summary of variation (tSNPs in bold) in the CTTN gene exons and flanking intronic regions in European and African Americans.
| SNP # | Positiona | Location (effect)b | rs/ss # | Flanking (5′) | Flanking (3′) | Valc |
|---|---|---|---|---|---|---|
| 1 | 69928894 | IVS1-125C>G | rs11825246 | - | - | ‡ |
| 2 | 69929306 | IVS2+191C>C | rs11825335 | - | - | ‡ |
| 3 | 69931037 | IVS2-15C>T | rs2298396 | - | - | ‡ |
| 4 | 69931123 | c.72C>T (p.Thr24Thr) | rs2298397 | - | - | ‡ |
| 5 | 69931234 | IVS3+96C>T | rs569732 | - | - | ‡ |
| 6 | 69938246 | IVS5-50C>T | rs34053053 | - | - | † |
| 7 | 69938257 | IVS5-39T>C | rs7932550 | - | - | † |
| 8 | 69939481 | IVS7+10G>A | ss76859851 | GGTAAGACGC | AAAGGTGCAG | - |
| 9 | 69940999 | IVS8+122G>A | rs1198234 | - | - | ‡ |
| 10 | 69943894 | IVS9+284G>A | rs35617256 | - | - | † |
| 11 | 69944090 | IVS9-62A>G | ss76859852 | TTGAAAACAT | CTTTCCACCT | - |
| 12 | 69946558 | IVS11-136C>T | rs12802851 | - | - | ‡ |
| 13 | 69949352 | IVS13+197C>T | ss76859853 | ACCTTTTAGA | GCAGCATGGT | - |
| 14 | 69949368 | IVS13+213C>T | ss76859854 | ATGGTGCAGC | GTTTCAGTGG | - |
| 15 | 69949575 | IVS13+420G>A | rs3781650 | - | - | ‡ |
| 16 | 69952996 | IVS14+43C>A | ss76859855 | TCCCTGGGAC | TGTGCCGAGG | - |
| 17 | 69953003 | IVS14+50G>A | ss76859856 | GACCTGTGCC | AGGGGATTGG | - |
| 18 | 69956879 | c.1291C>G (p.Leu431Val) | ss76859857 | CAAGGCAGAG | TGAGCTACAG | - |
| 19 | 69957113 | IVS16+81G>T | rs11825631 | - | - | ‡ |
| 20 | 69957275 | IVS16+243G>A | ss76859858 | GCAGAGGAAG | GAGGGTTTCA | - |
| 21 | 69957407 | c.1451G>A (p.Ser484Asn) | ss76859859 | TTAGAGGACA | CACCTACGAT | - |
| 22 | 69957414 | c.1458C>T (p.Tyr486Tyr) | rs643301 | - | - | ‡ |
| 23 | 69957575 | IVS17+103G>A | rs17160866 | - | - | ‡ |
| 24 | 69957890 | IVS17+418A>G | rs34959377 | - | - | † |
| 25 | 69958640 | IVS17-140G>A | rs3802780 | - | - | ‡ |
| 26 | 69958655 | IVS17-125T>C | rs7131282 | - | - | ‡ |
| 27 | 69958933 | g.*18_19insC | ss76859860 | GCCCCCCCCC | GGAGCTGCGC | - |
Chromosome position according to NCBI build 36;
As in CTTN isofrom a;
Validation status:
frequency available,
unknown or by 2-hit as in dbSNP build 126.
Figure 1.
Linkage disequilibrium patterns (measured by r2) in African (upper panel) and European Americans (lower panel). Boxes denote the approximate location of CTTN gene exons. Polymorphisms are numbered as in Table I.
CTTN gene variants in disease susceptibility
All SNPs displayed average automatic quality call values over 99% confidence. Among the successfully retyped samples, no discrepancies were found with the original genotypes for any of the SNPs, giving an estimated overall discordance rate of 0.0% (95% CI: 0.0-0.9%). Additionally, all CTTN tSNPs were in HWE in both panels (Table BI, Appendix B). The minor allele frequencies by population, SNP information, and p-values for association with asthma and severe asthma are shown in Table II. No SNP was associated with susceptibility to asthma in either European or African Americans, however, an intronic SNP (rs3802780) showed a significant uncorrected association with severe asthma in African Americans (OR: 1.62, 95% CI: 1.08-2.42). When adjusted for multiple testing, this association trended towards significance (p ∼ 0.08) and strikingly, this was the only SNP with a significant uncorrected association in European Americans (OR: 2.21, 95% CI: 1.07-4.57), despite the small number of severe asthmatics in this panel. Nevertheless, an adjustment for multiple testing also drove this association towards borderline significance (p ∼ 0.09). We noted that HWE p-value for this SNP was nearly significant in European American controls. However, quality measures for this SNP revealed satisfactory results since genotyping was simultaneous and blind to affectation status or ethnicity and non-significant HWE p-values were found for other sample groups, automatic quality call was 99.45%, completion rate was 97%, and no differences for the rate of completion proportions were found between controls and severe asthmatics (p = 0.32). Thus, systematic errors in genotyping do not appear to account for these results. To rule out potentially spurious associations due to population stratification, proportions of ancestry estimated from 30 unlinked microsatellites available from these samples [Amundadottir et al., 2006] were considered for adjustments. Estimated individual ancestries were not significantly different between African American controls and severe asthmatics [Flores et al., 2007], and the results remained unchanged when included in a logistic regression model (OR: 1.61, 95% CI: 1.07-2.42, p = 0.022). In European Americans, both European (0.98 ± 0.054 in controls vs. 0.99 ± 0.021 in severe asthmatics, t-test p = 0.015) and Asian (0.014 ± 0.049 in controls vs. 0.00 ± 0.006 in severe asthmatics, t-test p = 0.001) ancestries differed slightly but significantly between controls and severe asthmatics. However, the observed SNP association in European Americans remained unchanged in a logistic regression model (OR: 2.62, 95% CI: 1.22-5.63, p = 0.014). A stratified Mantel-Haenszel test (p = 0.0022) or a joint logistic regression model for the two populations taking into account the estimated ancestries (p = 0.003) further showed a strong association of this SNP with severe asthma. The rs3802780 A allele was associated with risk in both populations (joint OR: 1.71, 95% CI: 1.20-2.43) (Table III). A sliding window haplotype analysis performed as we previously described [Flores et al., 2007] failed to reveal significant results for any sample comparison (not shown).
Table II.
Genotyped SNPs, MAFs and unadjusted p-values for association with asthma and severe asthma in the two populations.
| SNPa | Location (effect)b | rs/ss # | Position (Build 36) | African Americans | European Americans | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| MAF | p-value | MAF | p-value | ||||||||||
|
|
|
||||||||||||
| Controls (N=193) | Asthma (N=190)c | Severe (N=102)c | CAd | CSe | Controls (N=206)c | Asthma (N=110) | Severe (N=27) | CAd | CSe | ||||
| 5 | Intron 3 | rs569732 | 69931234 | 0.19 | 0.20 | 0.19 | 0.663 | 0.962 | 0.40 | 0.47 | 0.50 | 0.086 | 0.157 |
| 7 | Intron 5 | rs7932550 | 69938257 | 0.16 | 0.17 | 0.14 | 0.684 | 0.488 | 0.22 | 0.24 | 0.18 | 0.491 | 0.524 |
| 11 | Intron 9 | ss76859852 | 69944090 | 0.28 | 0.25 | 0.27 | 0.412 | 0.766 | - | - | - | - | - |
| 12 | Intron 11 | rs12802851 | 69946558 | 0.07 | 0.05 | 0.04 | 0.112 | 0.133 | 0.19 | 0.20 | 0.16 | 0.649 | 0.632 |
| 19 | Intron 16 | rs11825631 | 69957113 | 0.34 | 0.30 | 0.31 | 0.377 | 0.591 | - | - | - | - | - |
| 21 | Exon 17 (Ser484Asn) | ss76859859 | 69957407 | 0.19 | 0.15 | 0.16 | 0.219 | 0.415 | - | - | - | - | - |
| 24 | Intron 17 | rs34959377 | 69957890 | 0.11 | 0.11 | 0.09 | 0.871 | 0.426 | 0.18 | 0.20 | 0.17 | 0.639 | 0.904 |
| 25 | Intron 17 | rs3802780 | 69958640 | 0.22 | 0.27 | 0.30 | 0.118 | 0.020 | 0.21 | 0.26 | 0.35 | 0.172 | 0.028 |
| 26 | Intron 17 | rs7131282 | 69958655 | 0.47 | 0.44 | 0.43 | 0.433 | 0.282 | 0.20 | 0.23 | 0.15 | 0.516 | 0.395 |
As in Table I;
As in CTTN isofrom a;
Excluding outlier individuals determined in Flores et al. [2007];
Control vs. asthma comparison;
Control vs. severe asthma comparison. Statistically significant p-values in bold.
Table III.
Effects of rs3802780 in severe asthma adjusted for individual ancestries in European and African Americans individually and in a joint analysis.
| Samplea | Genotype | Sample OR (95% CI) | p-value | Joint OR (95% CI) | p-value | ||
|---|---|---|---|---|---|---|---|
| GG | GA | AA | |||||
| African Americans | |||||||
| Severe asthma | 48 (48.5%) | 42 (42.4%) | 9 (9.1%) | ||||
| Controls | 112 (60.5%) | 66 (35.7%) | 7 (3.8%) | 1.61 (1.07-2.42) | 0.022 | ||
| European Americans | |||||||
| Severe asthma | 10 (43.5%) | 10 (43.5%) | 3 (13.0%) | ||||
| Controls | 116 (59.5%) | 75 (38.5%) | 4 (2.0%) | 2.62 (1.22-5.63) | 0.014 | 1.71 (1.20-2.43) | 0.003 |
Excluding outlier individuals determined in Flores et al. [2007]
Combined effects of CTTN and MYLK risk variants in severe asthma
The CTTN SNP rs3802780 was found to be significantly associated with severe asthma in the same African American case-control samples in which we previously reported the association (p = 0.005) of a non-synonymous MYLK coding SNP (rs9840993) with severe asthma [Flores et al., 2007]. As we have previously described strong biological interaction between these gene products [Dudek et al., 2002; Dudek et al., 2004], we explored the combined effects of both genes on severe asthma in African Americans. Other than an additive accumulation of risk from the alleles at both genes (Figure 2), interaction effects were not detected under any model tested (p-value > 0.10). However, African Americans carriers of at least one risk allele at both genes had almost a three-fold increase risk for severe asthma (OR: 2.93, 95% CI: 1.40-6.13, p = 0.004) compared to non-risk allele carriers at both genes.
Figure 2.

Combined effects of CTTN (rs3802780) and MYLK (rs9840993) genotypes in severe asthma in African Americans. Histograms indicating the odds ratios for the different genotype combinations for the CTTN rs3802780 G/A and MYLK rs9840993 T/C compared to the reference non risk genotypes for both SNPs. Genotype counts in cases (above the diagonal) and controls (below the diagonal) are also shown.
Discussion
Although the TENOR [Dolan et al., 2004] and the ENFUMOSA [2003] multi-centric studies have provided new epidemiological information regarding severe or difficult-to-treat asthma, the pathogenic mechanisms leading to this condition remain unclear. The structural changes in the airways accompanying asthma, a consequence of an airway injury/repair driven by chronic inflammation [Orsida et al., 1999; McDonald, 2001; Black and Johnson, 2002], are collectively termed airway remodeling with epithelial changes, mucus gland and goblet cell hyperplasia, subepithelial fibrosis, airway smooth muscle cell hypertrophy and hyperplasia, and vascular remodeling. Remodeling is most prominent in severe asthma and contributes to the development of persistent bronchial hyperresponsiveness, chronic airflow obstruction and lack of responsiveness to corticosteroid treatment [Holgate and Polosa, 2006]. Twin studies have suggested that asthma severity exhibits a hereditary component [Sarafino and Goldfedder, 1995] and, congruently, several genes have been associated with disease severity [Ober and Hoffjan, 2006]. Firmly replicated susceptibility genes, such as ADAM33 and TGFB1, encode molecules which participate in the remodeling processes (rather than in immunological aspects), are highly expressed in airway tissues/cells of severe asthmatics [Minshall et al., 1997; Balzar et al., 2005; Lee et al., 2006b; Foley et al., 2007], and demonstrate significant association with severe asthma exacerbations [Pulleyn et al., 2001; Jongepier et al., 2004; Kedda et al., 2006; Mak et al., 2006].
Despite the evidence provided by biochemical studies, the exact role of cortactin in asthma remains ill-defined but likely involve participation in processes such as cell migration, angiogenesis and immunoregulation thereby contributing to the complex cellular and molecular events involved in remodeling [Jeffery, 2001]. Consistent with this concept, re-sequencing CTTN gene and genotyping 9 tSNPs in case-control samples of diverse ancestry revealed a SNP (rs3802780) significantly associated with severe asthma, where remodeling is suggested to be the major determinant for the persistent and uncontrolled phenotypic characteristic [Holgate and Polosa, 2006]. Despite this persuasive pathobiologic rationale, we fully recognize that individually, our results are only modestly significant for CTTN effects in asthma and did not survive strict correction for multiple testing (adjusted p-values ≥ 0.09). However, the consistent association of the identical SNP with the same phenotype in asthma in two independent case-control samples of African and European American descent provides strong evidence for a modest but potentially significant role of CTTN gene in severe asthma. We also recognize the limitations of our reduced sample size studied. As a result, our results must be interpreted with caution since statistical power was not adequate to detect associations of variants with weak-to-moderate effects in severe asthma, particularly in European Americans (power < 30%). Additional limitations of our study is the utilization of controls in which asthma was not clinically excluded thereby failing to allow for evaluation of potential confounding factors that may alternatively explain the association. Moreover, it can be argued that the inclusion of control samples not clinically characterized would tend to favor null hypothesis (no association between variants and phenotypes) due to the potential inclusion of subjects with undiagnosed asthma, reducing the power of the study even further (on average <4% as estimated by Kurz et al., [2006]). It remains unlikely, however, that any of controls subjects had undiagnosed asthma (undiagnosed severe asthma is unquestionably highly unlikely), particularly given that no first degree relative had asthma in this over 18 years old cohort. We reduced the likelihood of false positives by adjusting for the proportions of ancestry estimated from 30 unlinked microsatellites that were selected for being informative for distinguishing between European, African and Asian ancestries [Amundadottir et al., 2006]. Although not empirically evaluated for their individual assignment ability, similar numbers of less informative microsatellites have demonstrated an assignment accuracy of >99% among European and African Americans [Yang et al., 2005]. However, since these markers were not selected for their ability to detect substructure within continental regions [Price et al., 2008], another limitation of the study is that modest levels of population stratification cannot be discarded that may have biased the results.
Although rs3802780 is intronic, the G allele is conserved across several (but not all) mammalian species (rhesus monkey, dog, armadillo and opossum) as assessed by the UCSC Genome Browser. In order to explore if other variant with apparent functional consequences in CTTN or in flanking genes may explain the association of this SNP with severe asthma, we examined the LD patterns of common (MAF ≥ 5%) variation in our re-sequenced samples and in the ∼400 kb region around CTTN (including the two flanking genes, PPFIA1 and SHANK2, the latter encoding a multidomain scaffolding protein which, similar to MLCK, binds directly to the SH3 domain of cortactin [Redecker et al., 2001; McWilliams et al., 2004]) in CEU and YRI HapMap samples. Since rs3802780 did not exhibit moderate-to-strong LD levels to SNPs outside of CTTN gene, this suggested that the association with severe asthma was most likely to be directly due to the CTTN gene (Figure C1, Appendix C). Maximum r2 values with flanking regions were 0.26 with rs3781649 in YRI and 0.68 with rs3781646 in CEU, both SNPs located in PPFIA1 gene. Seven additional CTTN SNPs in high LD with rs3802780 (r2 ≥ 0.85) and common to both populations were: rs643301, rs482438, rs1198236, rs548687, rs611216, rs592501, and rs1198234. Of these, only rs643301 was a coding SNP but only predicted a synonymous change (Tyr486Tyr). The other three coding SNPs in CTTN showed weak levels of LD with the associated SNP in both HapMap samples (rs2298397 was the only other coding SNP available and showed MAF ≥ 5% in CEU; r2 = 0.07) and the re-sequencing samples (maximum r2 values were 0.26 with the novel SNP ss76859857 in African Americans, 0.18 with the novel SNP ss76859859 in European Americans). Thus, non-synonymous CTTN SNPs were weakly correlated with rs3802780, and functional consequences of the SNPs in strong LD with it in either the CTTN gene itself or in flanking genes were not apparent.
Finally, we also demonstrated that severe asthma risk for African Americans increases by three-fold in carriers of at least one risk allele at both rs3802780 and rs9840993, a previously described severe asthma susceptibility polymorphism in MYLK gene [Flores et al., 2007]. Although these two proteins physically interact in cellular environments [Dudek et al., 2002], an effect due to gene-gene interaction was not detected. Rather, our results suggest that variation in multiple genes in this pathway have an additive effect on risk for severe asthma. However, according to our own estimations following Gauderman [2002], larger sample sizes would be required to derive a firm conclusion pertaining to the gene-gene interaction.
In summary, motivated by published positional and biological evidence, we studied the association of 9 carefully selected tSNPs of CTTN gene with susceptibility to asthma in multi-ethnic case-control samples and identified a promising association with the intronic SNP rs3802780 in severe asthma. Differences for the expression level of this gene have been reported between HapMap YRI and CEU cell lines [Storey et al., 2007; Zhang et al., 2008]. However, our data indicates that this SNP per se is not contributing to disparities in asthma severity between ethnic groups [El-Ekiaby et al., 2006]. Additionally, the combined effect of both rs3802780 and rs9840993, a previously described susceptibility SNP in the MYLK gene in African Americans, was much greater than the effects of either individual SNP. These observations, combined with reduced LD in other variants in nearby regions in reference samples [The International HapMap Consortium, 2005], support our hypothesis that the CTTN gene contains severe asthma susceptibility polymorphisms. We can not yet conclude whether this association is truly positive and if it is directly determined by rs3802780 or by other gene variant(s), the focus of additional future studies to confirm this association.
Acknowledgments
This work was supported by NIH grants M01 RR00055, R01 HL72414 and U01 HL49596 (to CO), and HL58064, HL 91889 (JGNG), and HL088144 (SMD).
Footnotes
Electronic Database Information: The URLs for the software used here are as follows:
EPIDAT 3.0, http://dxsp.sergas.es
Simple Interactive Statistical Analysis, http://www.quantitativeskills.com/sisa
UCSC Genome Browser, http://genome.ucsc.edu
References
- Ahmadi KR, Weale ME, Xue ZY, Soranzo N, Yarnall DP, Briley JD, Maruyama Y, Kobayashi M, Wood NW, Spurr NK, Burns DK, Roses AD, Saunders AM, Goldstein DB. A single-nucleotide polymorphism tagging set for human drug metabolism and transport. Nat Genet. 2005;37:84–9. doi: 10.1038/ng1488. [DOI] [PubMed] [Google Scholar]
- Ammit AJ, Hastie AT, Edsall LC, Hoffman RK, Amrani Y, Krymskaya VP, Kane SA, Peters SP, Penn RB, Spiegel S, Panettieri RA., Jr Sphingosine 1-phosphate modulates human airway smooth muscle cell functions that promote inflammation and airway remodeling in asthma. FASEB J. 2001;15:1212–4. doi: 10.1096/fj.00-0742fje. [DOI] [PubMed] [Google Scholar]
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–8. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE. Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Balzar S, Chu HW, Silkoff P, Cundall M, Trudeau JB, Strand M, Wenzel S. Increased TGF-beta2 in severe asthma with eosinophilia. J Allergy Clin Immunol. 2005;115:110–7. doi: 10.1016/j.jaci.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benayoun L, Druilhe A, Dombret MC, Aubier M, Pretolani M. Airway structural alterations selectively associated with severe asthma. Am J Respir Crit Care Med. 2003;167:1360–8. doi: 10.1164/rccm.200209-1030OC. [DOI] [PubMed] [Google Scholar]
- Black JL, Johnson PR. Factors controlling smooth muscle proliferation and airway remodelling. Curr Opin Allergy Clin Immunol. 2002;2:47–51. doi: 10.1097/00130832-200202000-00008. [DOI] [PubMed] [Google Scholar]
- Bourgain C, Hoffjan S, Nicolae R, Newman D, Steiner L, Walker K, Reynolds R, Ober C, McPeek MS. Novel case-control test in a founder population identifies P-selectin as an atopy-susceptibility locus. Am J Hum Genet. 2003;73:612–26. doi: 10.1086/378208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Baumruker T. Pulmonary and vascular pharmacology of sphingosine 1-phosphate. Curr Opin Pharmacol. 2006;6:244–50. doi: 10.1016/j.coph.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–8. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Cosen-Binker LI, Kapus A. Cortactin: the gray eminence of the cytoskeleton. Physiology (Bethesda) 2006;21:352–61. doi: 10.1152/physiol.00012.2006. [DOI] [PubMed] [Google Scholar]
- CSGA A genome-wide search for asthma susceptibility loci in ethnically diverse populations. The Collaborative Study on the Genetics of Asthma (CSGA) Nat Genet. 1997;15:389–92. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- Daly RJ. Cortactin signalling and dynamic actin networks. Biochem J. 2004;382:13–25. doi: 10.1042/BJ20040737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding K, Kullo IJ. Methods for the selection of tagging SNPs: a comparison of tagging efficiency and performance. Eur J Hum Genet. 2007;15:228–36. doi: 10.1038/sj.ejhg.5201755. [DOI] [PubMed] [Google Scholar]
- Dolan CM, Fraher KE, Bleecker ER, Borish L, Chipps B, Hayden ML, Weiss S, Zheng B, Johnson C, Wenzel S. Design and baseline characteristics of the epidemiology and natural history of asthma: Outcomes and Treatment Regimens (TENOR) study: a large cohort of patients with severe or difficult-to-treat asthma. Ann Allergy Asthma Immunol. 2004;92:32–9. doi: 10.1016/S1081-1206(10)61707-3. [DOI] [PubMed] [Google Scholar]
- Dudek SM, Birukov KG, Zhan X, Garcia JG. Novel interaction of cortactin with endothelial cell myosin light chain kinase. Biochem Biophys Res Commun. 2002;298:511–9. doi: 10.1016/s0006-291x(02)02492-0. [DOI] [PubMed] [Google Scholar]
- Dudek SM, Jacobson JR, Chiang ET, Birukov KG, Wang P, Zhan X, Garcia JG. Pulmonary endothelial cell barrier enhancement by sphingosine 1-phosphate: roles for cortactin and myosin light chain kinase. J Biol Chem. 2004;279:24692–700. doi: 10.1074/jbc.M313969200. [DOI] [PubMed] [Google Scholar]
- El-Ekiaby A, Brianas L, Skowronski ME, Coreno AJ, Galan G, Kaeberlein FJ, Seitz RE, Villaba KD, Dickey-White H, McFadden ER., Jr Impact of race on the severity of acute episodes of asthma and adrenergic responsiveness. Am J Respir Crit Care Med. 2006;174:508–13. doi: 10.1164/rccm.200603-431OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENFUMOSA The ENFUMOSA cross-sectional European multicentre study of the clinical phenotype of chronic severe asthma. European Network for Understanding Mechanisms of Severe Asthma. Eur Respir J. 2003;22:470–7. doi: 10.1183/09031936.03.00261903. [DOI] [PubMed] [Google Scholar]
- Flores C, Ma SF, Maresso K, Ober C, Garcia JG. A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet Epidemiol. 2007;31:296–305. doi: 10.1002/gepi.20210. [DOI] [PubMed] [Google Scholar]
- Foley SC, Mogas AK, Olivenstein R, Fiset PO, Chakir J, Bourbeau J, Ernst P, Lemiere C, Martin JG, Hamid Q. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol. 2007;119:863–71. doi: 10.1016/j.jaci.2006.12.665. [DOI] [PubMed] [Google Scholar]
- Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, Shriver MD, Ingersoll R, Scott AF, Beaty TH, Moitra J, Ma SF, Ye SQ, Barnes KC, Garcia JG. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol. 2006;34:487–95. doi: 10.1165/rcmb.2005-0404OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Grant AV, Rafaels N, Stockton-Porter M, Watkins T, Gao P, Chi P, Muñoz M, Watson H, Dunston G, Togias A, Hansel N, Sevransky J, Maloney JP, Moss M, Shanholtz C, Brower R, Garcia JG, Grigoryev DN, Cheadle C, Beaty TH, Mathias RA, Barnes KC. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol. 2007;119:1111–8. doi: 10.1016/j.jaci.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–84. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- Hansel NN, Hilmer SC, Georas SN, Cope LM, Guo J, Irizarry RA, Diette GB. Oligonucleotide-microarray analysis of peripheral-blood lymphocytes in severe asthma. J Lab Clin Med. 2005;145:263–74. doi: 10.1016/j.lab.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Hoffjan S, Ostrovnaja I, Nicolae D, Newman DL, Nicolae R, Gangnon R, Steiner L, Walker K, Reynolds R, Greene D, Mirel D, Gern JE, Lemanske RF, Jr, Ober C. Genetic variation in immunoregulatory pathways and atopic phenotypes in infancy. J Allergy Clin Immunol. 2004;113:511–8. doi: 10.1016/j.jaci.2003.10.044. [DOI] [PubMed] [Google Scholar]
- Holgate ST, Polosa R. The mechanisms, diagnosis, and management of severe asthma in adults. Lancet. 2006;368:780–93. doi: 10.1016/S0140-6736(06)69288-X. [DOI] [PubMed] [Google Scholar]
- Jeffery PK. Remodeling in asthma and chronic obstructive lung disease. Am J Respir Crit Care Med. 2001;164:S28–38. doi: 10.1164/ajrccm.164.supplement_2.2106061. [DOI] [PubMed] [Google Scholar]
- Johansson MW, Lye MH, Barthel SR, Duffy AK, Annis DS, Mosher DF. Eosinophils adhere to vascular cell adhesion molecule-1 via podosomes. Am J Respir Cell Mol Biol. 2004;31:413–22. doi: 10.1165/rcmb.2004-0099OC. [DOI] [PubMed] [Google Scholar]
- Jongepier H, Boezen HM, Dijkstra A, Howard TD, Vonk JM, Koppelman GH, Zheng SL, Meyers DA, Bleecker ER, Postma DS. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy. 2004;34:757–60. doi: 10.1111/j.1365-2222.2004.1938.x. [DOI] [PubMed] [Google Scholar]
- Kedda MA, Duffy DL, Bradley B, O'Hehir RE, Thompson PJ. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet. 2006;14:1027–36. doi: 10.1038/sj.ejhg.5201662. [DOI] [PubMed] [Google Scholar]
- Kurz T, Hoffjan S, Hayes MG, Schneider D, Nicolae R, Heinzmann A, Jerkic SP, Parry R, Cox NJ, Deichmann KA, Ober C. Fine mapping and positional candidate studies on chromosome 5p13 identify multiple asthma susceptibility loci. J Allergy Clin Immunol. 2006;118:396–402. doi: 10.1016/j.jaci.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Lee JF, Ozaki H, Zhan X, Wang E, Hla T, Lee MJ. Sphingosine-1-phosphate signaling regulates lamellipodia localization of cortactin complexes in endothelial cells. Histochem Cell Biol. 2006a;126:297–304. doi: 10.1007/s00418-006-0143-z. [DOI] [PubMed] [Google Scholar]
- Lee JY, Park SW, Chang HK, Kim HY, Rhim T, Lee JH, Jang AS, Koh ES, Park CS. A disintegrin and metalloproteinase 33 protein in patients with asthma: Relevance to airflow limitation. Am J Respir Crit Care Med. 2006b;173:729–35. doi: 10.1164/rccm.200409-1175OC. [DOI] [PubMed] [Google Scholar]
- Lester LA, Rich SS, Blumenthal MN, Togias A, Murphy S, Malveaux F, Miller ME, Dunston GM, Solway J, Wolf RL, Samet JM, Marsh DG, Meyers DA, Ober C, Bleecker ER. Ethnic differences in asthma and associated phenotypes: collaborative study on the genetics of asthma. J Allergy Clin Immunol. 2001;108:357–62. doi: 10.1067/mai.2001.117796. [DOI] [PubMed] [Google Scholar]
- Mak JC, Leung HC, Ho SP, Law BK, Ho AS, Lam WK, Ip MS, Chan-Yeung MM. Analysis of TGF-beta(1) gene polymorphisms in Hong Kong Chinese patients with asthma. J Allergy Clin Immunol. 2006;117:92–6. doi: 10.1016/j.jaci.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Mannino DM, Homa DM, Akinbami LJ, Moorman JE, Gwynn C, Redd SC. Surveillance for asthma--United States, 1980-1999. MMWR Surveill Summ. 2002;51:1–13. [PubMed] [Google Scholar]
- McDonald DM. Angiogenesis and remodeling of airway vasculature in chronic inflammation. Am J Respir Crit Care Med. 2001;164:S39–45. doi: 10.1164/ajrccm.164.supplement_2.2106065. [DOI] [PubMed] [Google Scholar]
- McWilliams RR, Gidey E, Fouassier L, Weed SA, Doctor RB. Characterization of an ankyrin repeat-containing Shank2 isoform (Shank2E) in liver epithelial cells. Biochem J. 2004;380:181–91. doi: 10.1042/BJ20031577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall EM, Leung DY, Martin RJ, Song YL, Cameron L, Ernst P, Hamid Q. Eosinophil-associated TGF-beta1 mRNA expression and airways fibrosis in bronchial asthma. Am J Respir Cell Mol Biol. 1997;17:326–33. doi: 10.1165/ajrcmb.17.3.2733. [DOI] [PubMed] [Google Scholar]
- Moore WC, Peters SP. Severe asthma: an overview. J Allergy Clin Immunol. 2006;117:487–94. doi: 10.1016/j.jaci.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Ober C. Perspectives on the past decade of asthma genetics. J Allergy Clin Immunol. 2005;116:274–8. doi: 10.1016/j.jaci.2005.04.039. [DOI] [PubMed] [Google Scholar]
- Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- Ober C, Leavitt SA, Tsalenko A, Howard TD, Hoki DM, Daniel R, Newman DL, Wu X, Parry R, Lester LA, Solway J, Blumenthal M, King RA, Xu J, Meyers DA, Bleecker ER, Cox NJ. Variation in the interleukin 4-receptor alpha gene confers susceptibility to asthma and atopy in ethnically diverse populations. Am J Hum Genet. 2000;66:517–26. doi: 10.1086/302781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsida BE, Li X, Hickey B, Thien F, Wilson JW, Walters EH. Vascularity in asthmatic airways: relation to inhaled steroid dose. Thorax. 1999;54:289–95. doi: 10.1136/thx.54.4.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HW, Lee JE, Shin ES, Lee JY, Bahn JW, Oh HB, Oh SY, Cho SH, Moon HB, Min KU, Elias JA, Kim YY, Kim YK. Association between genetic variations of vascular endothelial growth factor receptor 2 and atopy in the Korean population. J Allergy Clin Immunol. 2006;117:774–9. doi: 10.1016/j.jaci.2005.12.1328. [DOI] [PubMed] [Google Scholar]
- Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz-Linares A, Groop L, Saetta AA, Korkolopoulou P, Seligsohn U, Waliszewska A, Schirmer C, Ardlie K, Ramos A, Nemesh J, Arbeitman L, Goldstein DB, Reich D, Hirschhorn JN. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyn LJ, Newton R, Adcock IM, Barnes PJ. TGFbeta1 allele association with asthma severity. Hum Genet. 2001;109:623–7. doi: 10.1007/s00439-001-0617-y. [DOI] [PubMed] [Google Scholar]
- Redecker P, Gundelfinger ED, Boeckers TM. The cortactin-binding postsynaptic density protein proSAP1 in non-neuronal cells. J Histochem Cytochem. 2001;49:639–48. doi: 10.1177/002215540104900511. [DOI] [PubMed] [Google Scholar]
- Roviezzo F, Del Galdo F, Abbate G, Bucci M, D'Agostino B, Antunes E, De Dominicis G, Parente L, Rossi F, Cirino G, De Palma R. Human eosinophil chemotaxis and selective in vivo recruitment by sphingosine 1-phosphate. Proc Natl Acad Sci U S A. 2004;101:11170–5. doi: 10.1073/pnas.0401439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoh AJ, Huque MF, Dubey SD. Some comments on frequently used multiple endpoint adjustment methods in clinical trials. Stat Med. 1997;16:2529–42. doi: 10.1002/(sici)1097-0258(19971130)16:22<2529::aid-sim692>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Sarafino EP, Goldfedder J. Genetic factors in the presence, severity, and triggers of asthma. Arch Dis Child. 1995;73:112–6. doi: 10.1136/adc.73.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicka E, Zuany-Amorim C, Manlius C, Trifilieff A, Brinkmann V, Kemeny DM, Walker C. Inhibition of Th1- and Th2-mediated airway inflammation by the sphingosine 1-phosphate receptor agonist FTY720. J Immunol. 2003;171:6206–14. doi: 10.4049/jimmunol.171.11.6206. [DOI] [PubMed] [Google Scholar]
- Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, Faffe DS, Shikanai T, Raby BA, Weiss ST, Shore SA. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med. 2004;169:214–9. doi: 10.1164/rccm.200307-973OC. [DOI] [PubMed] [Google Scholar]
- Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–9. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM. Gene-expression variation within and among human populations. Am J Hum Genet. 2007;80:502–9. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EE, Pan L, Ostrovnaya I, Weiss LA, Gern JE, Lemanske RF, Jr, Nicolae DL, Ober C. Integrin beta 3 genotype influences asthma and allergy phenotypes in the first 6 years of life. J Allergy Clin Immunol. 2007;119:1423–9. doi: 10.1016/j.jaci.2007.03.029. [DOI] [PubMed] [Google Scholar]
- Tilghman RW, Hoover RL. The Src-cortactin pathway is required for clustering of E-selectin and ICAM-1 in endothelial cells. FASEB J. 2002;16:1257–9. doi: 10.1096/fj.01-0969fje. [DOI] [PubMed] [Google Scholar]
- Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, Walsh A, Liu Z, Hayward B, Folz C, Manning SP, Bawa A, Saracino L, Thackston M, Benchekroun Y, Capparell N, Wang M, Adair R, Feng Y, Dubois J, FitzGerald MG, Huang H, Gibson R, Allen KM, Pedan A, Danzig MR, Umland SP, Egan RW, Cuss FM, Rorke S, Clough JB, Holloway JW, Holgate ST, Keith TP. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- Wang X, Saito J, Ishida T, Munakata M. Polymorphism of egfr Intron1 is associated with susceptibility and severity of asthma. J Asthma. 2006;43:711–5. doi: 10.1080/02770900600925247. [DOI] [PubMed] [Google Scholar]
- Weale ME, Depondt C, Macdonald SJ, Smith A, Lai PS, Shorvon SD, Wood NW, Goldstein DB. Selection and evaluation of tagging SNPs in the neuronal-sodium-channel gene SCN1A: implications for linkage-disequilibrium gene mapping. Am J Hum Genet. 2003;73:551–65. doi: 10.1086/378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Lester LA, Gern JE, Wolf RL, Parry R, Lemanske RF, Solway J, Ober C. Variation in ITGB3 is associated with asthma and sensitization to mold allergen in four populations. Am J Respir Crit Care Med. 2005;172:67–73. doi: 10.1164/rccm.200411-1555OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills-Karp M, Ewart SL. Time to draw breath: asthma-susceptibility genes are identified. Nat Rev Genet. 2004;5:376–87. doi: 10.1038/nrg1326. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol. 2005;28:302–12. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- Yang L, Kowalski JR, Yacono P, Bajmoczi M, Shaw SK, Froio RM, Golan DE, Thomas SM, Luscinskas FW. Endothelial cell cortactin coordinates intercellular adhesion molecule-1 clustering and actin cytoskeleton remodeling during polymorphonuclear leukocyte adhesion and transmigration. J Immunol. 2006;177:6440–9. doi: 10.4049/jimmunol.177.9.6440. [DOI] [PubMed] [Google Scholar]
- Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, Dolan ME. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631–40. doi: 10.1016/j.ajhg.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin DV, Meng Z, Ehm MG. Contrasting linkage-disequilibrium patterns between cases and controls as a novel association-mapping method. Am J Hum Genet. 2006;78:737–46. doi: 10.1086/503710. [DOI] [PMC free article] [PubMed] [Google Scholar]