Background: The HIF-1 and HIF-2 transcription factors coordinate adaptive responses to hypoxia.
Results: Sirt7 decreases HIF-1α and HIF-2α protein levels independent of its deacetylase activity.
Conclusion: Sirt7 inhibits the activity of the HIF-1 and HIF-2 transcription factors.
Significance: This study identifies Sirt7 as a regulator of HIF-1 and HIF-2 signaling.
Keywords: Hypoxia, Hypoxia-inducible Factor (HIF), Sirtuins, Transcription
Abstract
Hypoxia-inducible factor (HIF) 1 and HIF-2 are heterodimeric proteins composed of an oxygen-regulated HIF-1α or HIF-2α subunit, respectively, and a constitutively expressed HIF-1β subunit, which mediate adaptive transcriptional responses to hypoxia. Here, we report that Sirt7 (sirtuin-7) negatively regulates HIF-1α and HIF-2α protein levels by a mechanism that is independent of prolyl hydroxylation and that does not involve proteasomal or lysosomal degradation. The effect of Sirt7 was maintained in the presence of the sirtuin inhibitor nicotinamide and upon deletion or mutation of its deacetylase domain, indicating a non-catalytic function. Knockdown of Sirt7 led to an increase in HIF-1α and HIF-2α protein levels and an increase in HIF-1 and HIF-2 transcriptional activity. Thus, we identify a novel molecular function of Sirt7 as a negative regulator of HIF signaling.
Introduction
Hypoxia-inducible factor (HIF)2 1 is a transcription factor that is essential for mediating a broad repertoire of adaptive responses to hypoxia. First identified in studies of EPO (erythropoietin) gene transcriptional regulation (1), HIF-1 was subsequently shown to coordinate adaptive responses to hypoxia at both the cellular and systemic levels (2–5). HIF-1, which is a heterodimer composed of HIF-1α and HIF-1β subunits (2), has been shown to regulate the expression of hundreds of target genes involved in angiogenesis, such as vascular endothelial growth factor (VEGFA); in erythropoiesis, such as EPO; and in metabolism, autophagy, and other adaptive responses to hypoxia (5). In addition, the HIF-1α subunit has adaptive functions that are independent of transcriptional activity (6). The HIF-2α protein shares sequence similarity with HIF-1α and also promotes adaptive responses to hypoxia but has a more limited tissue distribution, and in some cases, it mediates distinct biological functions, including redox regulation through transactivation of the SOD2 (superoxide dismutase 2) gene, which encodes manganese superoxide dismutase (7).
In recent years, the mechanisms regulating HIF-1 protein stability and transcriptional activity have been extensively investigated. O2-dependent proline hydroxylation marks HIF-1α for ubiquitination by the VHL ubiquitin ligase complex and subsequent proteasomal degradation (8–12), whereas asparagine hydroxylation by FIH-1 (factor inhibiting HIF-1) blocks interaction of HIF-1α with the p300 coactivator (13, 14). During hypoxia, hydroxylation of proline and asparagine residues is inhibited, which provides a molecular basis for the observed increase in HIF-1α protein stability and transcriptional activity (15). The hydroxylases contain Fe(II) in their catalytic centers and use α-ketoglutarate as a co-substrate; therefore, iron chelators, such as desferrioxamine, and competitive antagonists of α-ketoglutarate, such as dimethyloxalylglycine (DMOG), block hydroxylase activity and increase HIF-1α levels (8). Several proteins that interact with HIF-1α and stimulate its proteasomal degradation independent of O2 concentration have also been identified. These include SSAT1 (spermidine/spermine N1-acetyltransferase 1) (16), calcineurin (17), RACK1 (18), hypoxia-associated factor (19), CHIP/HSP70 (heat shock protein of 70 kDa) (20), and SHARP1 (21). We have also recently described a mechanism by which chaperone-mediated autophagy can target HIF-1α for lysosomal degradation, identifying a pathway by which HIF-1α levels can be regulated independent of the proteasome (22).
Sir2, the founding member of the sirtuin family, was identified as a NAD+-dependent histone deacetylase (23), thereby linking cellular redox status to gene transcription. In mammals, there are seven Sir2 homologs (Sirt1–7), which vary in tissue distribution, subcellular localization, and enzymatic targets (23). Physical and functional interactions between several members of the sirtuin family and HIF proteins have recently been identified. As the founding member of the sirtuin family, the effects of Sirt1 have been best characterized. Sirt1 was identified as a HIF-2α deacetylase that augments HIF-2 (but not HIF-1) activity (24, 25). A subsequent study suggested that Sirt1 may repress HIF-1 transactivation ability (26), whereas a third study reported that Sirt1 increased HIF-1α protein levels (27). It is possible that, in different contexts, Sirt1 may have opposing effects on HIF-1 activity. Sirt6 knock-out mice develop lethal hypoglycemia early in life, which is due to the role of Sirt6 as a co-repressor of HIF-1 target genes (28). Finally, Sirt3, which is a mitochondrial deacetylase, may regulate HIF-1 signaling in cancer cell lines (29, 30) by an indirect effect on mitochondrial reactive oxygen species.
Sirt7 is arguably the least studied of the mammalian sirtuins, although it has been reported to activate transcription by RNA polymerase I (31–33) and has been identified recently as a deacetylase of Lys-18 of histone H3 (H3K18) (34). Sirt7-null mice develop cardiac hypertrophy and inflammatory cardiomyopathy, due in part to hyperacetylation of p53 (35).
Here, we report that HIF-1α and HIF-2α protein levels are regulated by Sirt7 via direct physical interactions. Sirt7 overexpression decreased HIF-1α and HIF-2α protein levels, HIF transcriptional activity, and target gene expression, whereas knockdown of Sirt7 had the opposite effect. This inhibitory effect of Sirt7 was preserved in the presence of nicotinamide and with Sirt7 mutants that do not possess deacetylase activity, indicating that Sirt7 inhibits HIF-1α and HIF-2α through a mechanism that is independent of its catalytic activity. Thus, these studies have revealed a novel role for Sirt7 as a negative regulator of the hypoxia response pathway.
EXPERIMENTAL PROCEDURES
Tissue Culture
HeLa, Hep3B, MDA-MB-231, and 293T cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were maintained at 37 °C in a 5% CO2 and 95% air incubator. Cells were subjected to hypoxia by exposure to 1% O2, 5% CO2, and balance N2 at 37 °C in a modulator incubator chamber (Billups-Rothenberg, Inc.).
Immunoprecipitation and Immunoblot Assays
Cells were lysed in PBS with 0.1% Tween 20, 1 mm DTT, protease inhibitor mixture, 1 mm Na3VO4, and 10 mm NaF, followed by gentle sonication. For immunoprecipitation assays, a 2-μg aliquot of anti-Myc epitope antibody (Novus Biologicals) was incubated overnight with 2.5 mg of cell lysate at 4 °C, followed by a 3-h incubation with 30 μl of protein G-Sepharose (GE Healthcare). Beads were washed four times with lysis buffer. Proteins were eluted in SDS sample buffer and fractionated by SDS-PAGE. The following antibodies were used in immunoblot and immunoprecipitation assays: anti-β-actin (Santa Cruz Biotechnology), anti-HIF-1α (BD Biosciences), anti-FLAG (Sigma), anti-HIF-2α and anti-Myc epitope (Novus Biologicals), and anti-V5 epitope (Invitrogen).
Luciferase Reporter Assay
HeLa or Hep3B cells were seeded onto 24-well plates at 20,000 cells/well, and 48 h after seeding, the cells were transfected with plasmid DNA using PolyJet (SignaGen Laboratories). Reporters pSV-RL (10 ng) and p2.1 (120 ng) were cotransfected with expression vectors (31). Cells were lysed, and luciferase activities were determined with a multiwell luminescence reader (PerkinElmer Life Sciences) using the Dual-Luciferase reporter assay system (Promega).
Plasmids
The coding sequences of human full-length Sirt5, Sirt6, and Sirt7 and sequences encoding deletion mutants of Sirt7 were inserted into the pCMV-Myc vector (Clontech) and verified by DNA sequence analysis. Catalytically inactive mutants of Sirt7 were generated using the QuikChange site-directed mutagenesis kit (Stratagene). Validated shRNA vectors against Sirt7 were purchased from Sigma. Other constructs were described previously (24, 36).
Statistical Analysis
All data are presented as means ± S.E., except where indicated otherwise. Differences between two conditions were analyzed using Student's t test.
RESULTS
Sirt7 Interacts with HIF-1α and HIF-2α
We investigated whether members of the sirtuin family are capable of interacting with HIF-1α. Co-immunoprecipitation experiments demonstrated that Myc epitope-tagged Sirt6 interacted with coexpressed FLAG epitope-tagged HIF-1α (Fig. 1A), which was consistent with a recent report (29) and served as a positive control. Myc-tagged Sirt5 did not interact with FLAG-HIF-1α or HIF-2α, but interaction of Myc-tagged Sirt7 with both FLAG-HIF-1α (Fig. 1A) and HIF-2α (Fig. 1B) was detected.
FIGURE 1.
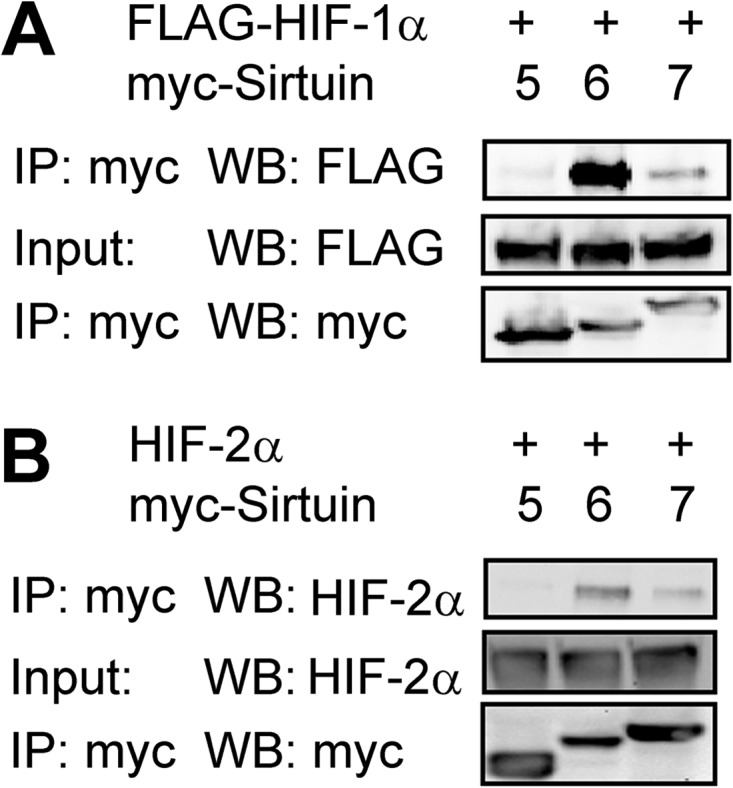
Sirt7 binds to HIF-1α and HIF-2α. A, 293T cells were cotransfected with FLAG epitope-tagged HIF-1α expression vector and expression vector encoding Myc epitope-tagged Sirt5, Sirt6, or Sirt7. 24 h post-transfection, cell lysates were immunoprecipitated (IP) with anti-Myc epitope antibody, and each Western blot (WB) was probed with the indicated antibodies. B, 293T cells were cotransfected with HIF-2α expression vector and expression vector encoding Myc-tagged Sirt5, Sirt6, or Sirt7. 24 h post-transfection, lysates were immunoprecipitated with anti-Myc antibody and probed with the indicated antibodies.
Sirt7 Inhibits HIF-1 and HIF-2 Transcriptional Activity
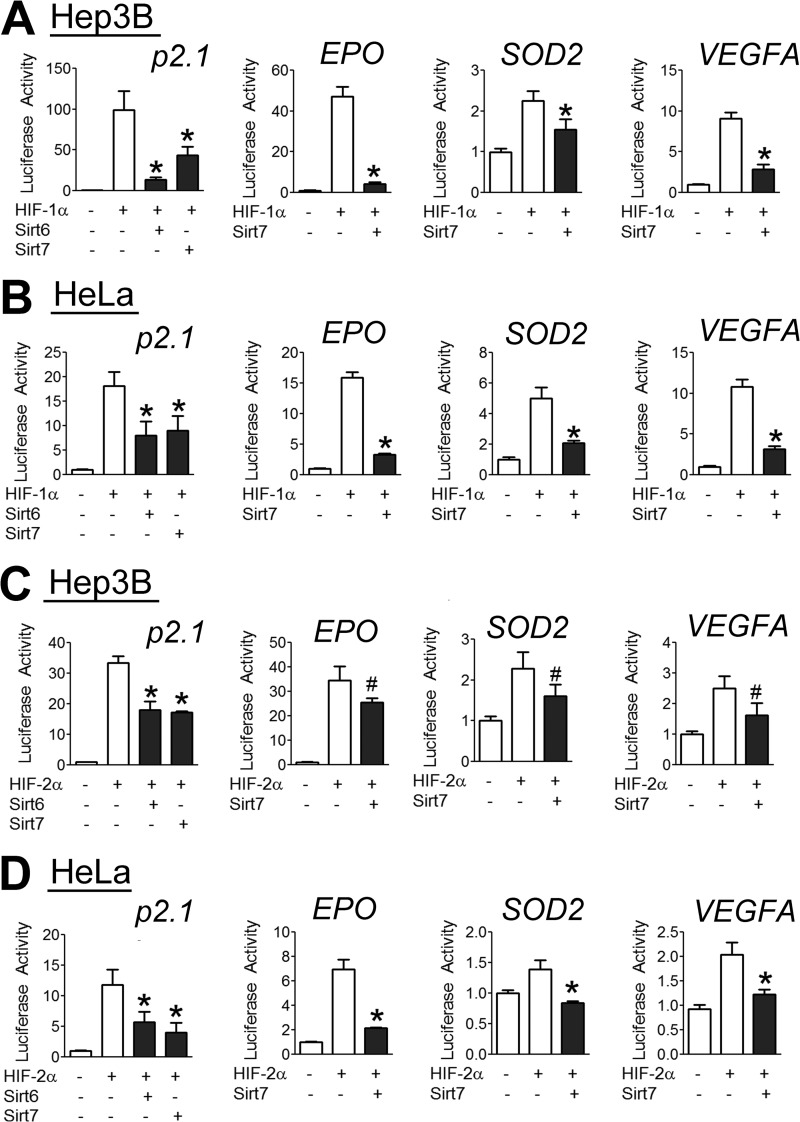
To examine the effect of Sirt7 on the transcriptional activity of HIF-1 and HIF-2, we utilized several previously described reporter plasmids (24, 36). First, Hep3B human hepatocellular carcinoma cells were cotransfected with p2.1, a reporter plasmid that contains a 68-bp hypoxia response element (from the human ENO1 gene) upstream of an SV40 promoter and firefly luciferase coding sequences, and pSV-RL, a control reporter that contains Renilla luciferase coding sequences downstream of the SV40 promoter only. The ratio of firefly to Renilla luciferase activity served as a measure of HIF transcriptional activity. In addition, we used three reporter plasmids containing the promoter regions of the EPO, VEGFA, and SOD2 genes, respectively, upstream of firefly luciferase coding sequences. We found increased p2.1 reporter activity and promoter activity of all three HIF target genes in response to HIF-1α overexpression in Hep3B cells (Fig. 2A). However, coexpression of Sirt7 inhibited this effect (Fig. 2A). Similar results were obtained in HeLa human cervical carcinoma cells (Fig. 2B), indicating that Sirt7 can regulate HIF-1 activity in multiple cell types. Likewise, overexpression of HIF-2α led to an increase in reporter activity, which was blocked by overexpression of Sirt7, in both Hep3B (Fig. 2C) and HeLa (Fig. 2D) cells. Despite the weaker interaction of Sirt7 with HIF-1α or HIF-2α compared with Sirt6 (Fig. 1, A and B), the inhibition of HIF-1 (Fig. 2, A and B) and HIF-2 (Fig. 2, C and D) activity by Sirt7 was comparable in magnitude to that by Sirt6 in both HeLa and Hep3B cells. The differences between the co-immunoprecipitation and reporter assays suggest that additional proteins may stabilize the interaction of Sirt7 with HIF-1α or HIF-2α at HIF-binding sites in chromatin. In any case, the results demonstrate that Sirt7 is a negative regulator of HIF-1α- and HIF-2α-dependent transcriptional activity.
FIGURE 2.
Sirt7 inhibits HIF transcriptional activity. A and B, Hep3B (A) or HeLa (B) cells were cotransfected with the indicated firefly luciferase reporter gene, which contained the ENO1 hypoxia response element upstream of a basal SV40 promoter (p2.1) or an intact promoter from the EPO, SOD2, or VEGFA gene; control Renilla luciferase reporter gene pSV-RL; HIF-1α expression vector; and either an empty vector (EV) or Sirt7 expression vector. 24 h post-transfection, cells were lysed, and the ratio of firefly to Renilla luciferase activity was determined. C and D, Hep3B (C) or HeLa (D) cells were cotransfected with the indicated luciferase promoter construct, HIF-2α expression vector, and either EV or Sirt7 expression vector. 24 h post-transfection, cells were lysed, and luciferase activity was determined. Results are shown as means ± S.E. #, p < 0.05; *, p < 0.01 versus HIF-1α or HIF-2α alone.
Sirt7 Down-regulates HIF-1α and HIF-2α Protein Levels
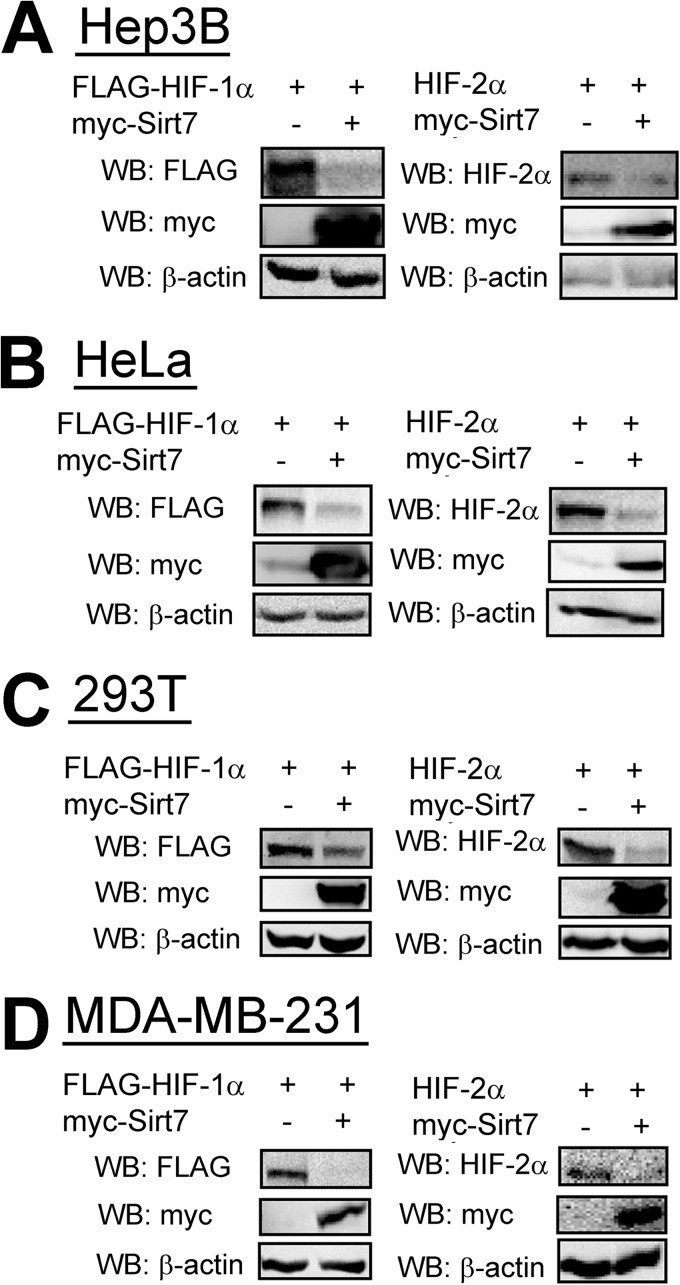
To investigate the mechanism by which Sirt7 regulates HIF-1 and HIF-2 activity, we analyzed HIF-1α levels by immunoblot assay. Overexpression of Sirt7 led to a large decrease in HIF-1α protein levels in Hep3B (Fig. 3A), HeLa (Fig. 3B), 293T human embryonic kidney (Fig. 3C), and MDA-MB-231 human breast cancer (Fig. 3D) cells. Sirt7 overexpression had a similar inhibitory effect on HIF-2α protein levels in all four cell lines (Fig. 3, A–D).
FIGURE 3.
Sirt7 overexpression decreases HIF-1α and HIF-2α protein levels. Hep3B (A), HeLa (B), 293T (C), and MDA-MB-231 (D) cells were cotransfected with HIF-1α or HIF-2α expression vector and either EV or Sirt7 expression vector. 24 h post-transfection, cell lysates were subjected to Western blot assay (WB) with the indicated antibodies.
The Effect of Sirt7 on HIF-1 and HIF-2 Is Independent of Deacetylase Activity
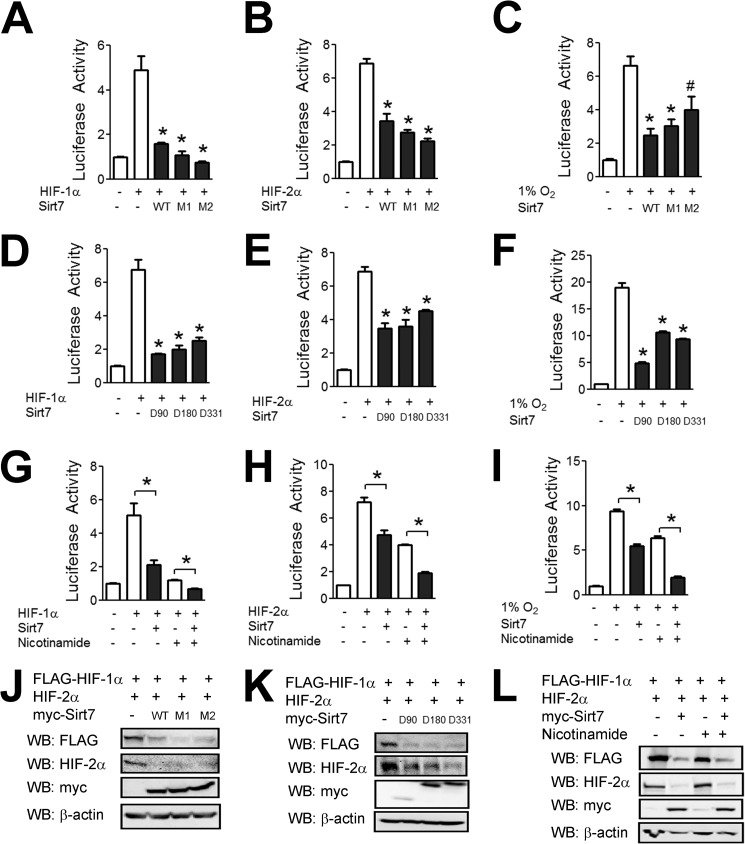
Sirt7 has been reported to act as a protein deacetylase, and we investigated whether its enzymatic activity is required for the effect on HIF-1 and HIF-2 activity. Two previously described catalytically inactive mutants of Sirt7 (31, 34) retained the ability to inhibit HIF transcriptional activity induced by HIF-1α overexpression (Fig. 4A), HIF-2α overexpression (Fig. 4B), or exposure of cells to hypoxia (Fig. 4C). The deacetylase domain of Sirt7 resides in residues 90–330, and deletion of this entire region had no effect on the ability of Sirt7 to inhibit HIF transcriptional activity induced by HIF-1α overexpression (Fig. 4D), HIF-2α overexpression (Fig. 4E), or exposure of cells to hypoxia (Fig. 4F). Finally, the response of cells to treatment with nicotinamide, a broad-spectrum sirtuin inhibitor that blocks NAD+-dependent deacetylation (23), was analyzed. Nicotinamide treatment itself had a large effect on HIF activity in Hep3B cells, which reflects the role of other sirtuins in regulating HIF-1 (24–30), but had no effect on the ability of Sirt7 to decrease HIF-1 or HIF-2 activity (Fig. 4, G–I).
FIGURE 4.
Effects of Sirt7 are independent of deacetylase activity. A and B, Hep3B cells were cotransfected with p2.1, pSV-RL, HIF-1α or HIF-2α expression vector, and either EV or expression vector encoding wild-type Sirt7 (WT) or catalytically inactive missense mutant Sirt7-S112A (M1) or Sirt7-H188Y (M2). 24 h post-transfection, cells were lysed, and the ratio of firefly to Renilla luciferase activity was determined. C, Hep3B cells were cotransfected with p2.1; pSV-RL; and EV or vector encoding wild-type Sirt7, Sirt7-S112A, or Sirt7-H188Y. 24 h post-transfection, cells were exposed to 1% O2 for an additional 24 h and lysed, and luciferase activity was determined. D and E, Hep3B cells were cotransfected with p2.1; pSV-RL; HIF-1α or HIF-2α expression vector; and either EV or expression vector encoding Sirt7 amino acids 1–90 (D90), 1–180 (D180), or 1–331 (D331). 24 h post-transfection, cells were lysed, and luciferase activity was determined. F, Hep3B cells were cotransfected with p2.1; pSV-RL; and either EV or vector encoding Sirt7 amino acids 1–90, 1–180, or 1–331. 24 h post-transfection, cells were exposed to 1% O2 for an additional 24 h and lysed, and luciferase activity was determined. G and H, Hep3B cells were cotransfected with p2.1, pSV-RL, HIF-1α or HIF-2α vector, and either EV or Sirt7 vector. 24 h post-transfection, cells were left untreated or treated with nicotinamide (20 mm) for an additional 24 h, after which cells were lysed, and luciferase activity was determined. I, Hep3B cells were cotransfected with p2.1, pSV-RL, and either EV or Sirt7 vector. 24 h post-transfection, cells were exposed to 1% O2 for an additional 24 h in the presence or absence of nicotinamide (20 mm) as indicated. J, Hep3B cells were cotransfected with vector encoding FLAG-HIF-1α or HIF-2α and either EV or vector encoding wild-type Sirt7, Sirt7-S112A, or Sirt7-H188Y. 24 h post-transfection, cell lysates were subjected to Western blotting (WB). K, Hep3B cells were cotransfected with vector encoding FLAG-HIF-1α or HIF-2α and either EV or vector encoding Sirt7 amino acids 1–90, 1–180, or 1–331. 24 h post-transfection, cell lysates were subjected to Western blotting. L, Hep3B cells were cotransfected with vector encoding FLAG-HIF-1α or HIF-2α and either EV or Sirt7 vector. Cells were left untreated or treated with nicotinamide (20 mm) for 24 h, after which cell lysates were subjected to Western blotting. Results are shown as means ± S.E. #, p < 0.05; *, p < 0.01 versus HIF-1α or HIF-2α alone.
Consistent with these results, we found that catalytically inactive Sirt7 mutants retained the ability to decrease HIF-1α and HIF-2α protein levels (Fig. 4J), as did Sirt7 mutants with the deacetylase domain deleted (Fig. 4K). Similarly, nicotinamide had no effect on the ability of Sirt7 to decrease HIF-1α and HIF-2α protein levels (Fig. 4L). Taken together, the data indicate that the deacetylase activity of Sirt7 is not required for negative regulation of HIF-1 and HIF-2.
The Effect of Sirt7 Is Hydroxylase-, Proteasome-, and Lysosome-independent
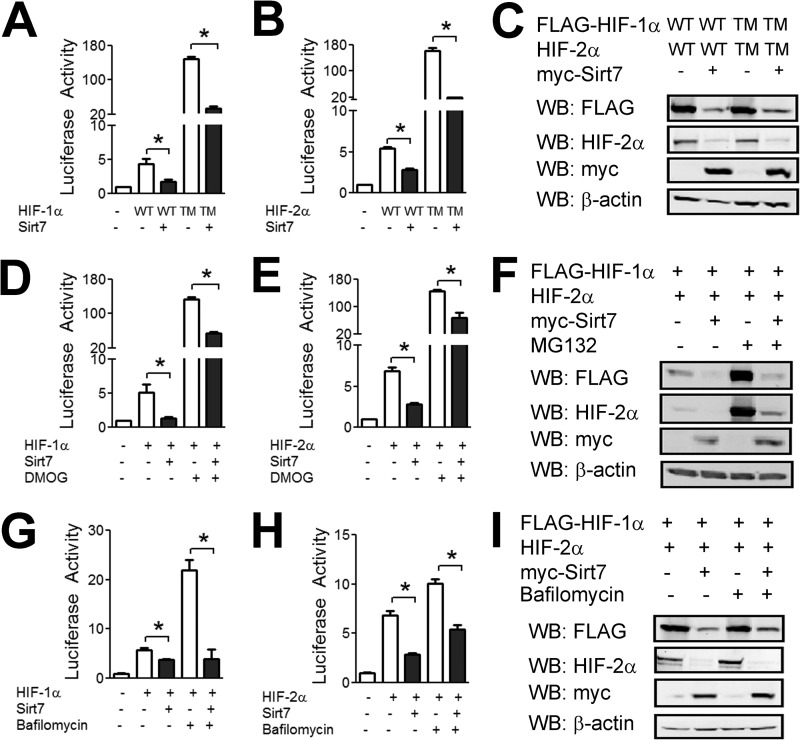
Hydroxylation of HIF-1α at Pro-402 and Pro-564 leads to subsequent ubiquitination and proteasomal degradation, whereas hydroxylation at Asn-803 inhibits HIF-1α transactivation domain function. To determine whether hydroxylation is necessary for the effect of Sirt7, we utilized a triple-mutant HIF-1α construct harboring P402A, P564A, and N803A mutations. Overexpression of Sirt7 inhibited reporter activity induced by overexpression of either wild-type or triple-mutant HIF-1α to a comparable degree (Fig. 5A). Similar results were observed upon overexpression of wild-type or triple-mutant HIF-2α (Fig. 5B). Immunoblot assays demonstrated that Sirt7 decreased the protein levels of triple-mutant HIF-1α and HIF-2α to a similar degree as wild-type HIF-1α and HIF-2α (Fig. 5C). Treatment with the hydroxylase inhibitor DMOG led to an increase in reporter activity induced by overexpression of HIF-1α (Fig. 5D) or HIF-2α (Fig. 5E), but DMOG did not block the inhibitory effect of Sirt7 overexpression. Likewise, treatment with the proteasome inhibitor MG132 did not block the effect of Sirt7 (Fig. 5F).
FIGURE 5.
Effects of Sirt7 are independent of proteasomal and lysosomal degradation. A, Hep3B cells were cotransfected with p2.1, pSV-RL, EV or Sirt7 expression vector, and vector encoding wild-type HIF-1α (WT) or triple-mutant HIF-1α (TM; P402A/P564A/N803A). 24 h post-transfection, cells were lysed, and luciferase activity was determined. B, Hep3B cells were cotransfected with p2.1, pSV-RL, EV or Sirt7 expression vector; and vector encoding wild-type or triple-mutant HIF-2α. 24 h post-transfection, cells were lysed, and luciferase activity was determined. C, Hep3B cells were cotransfected with either 1 μg of wild-type HIF-1α or HIF-2α vector or 100 ng of triple-mutant HIF-2α vector and either EV or Sirt7 vector. Total plasmid transfected was equalized with EV. 24 h post-transfection, cell lysates were analyzed by Western blotting (WB). D and E, Hep3B cells were cotransfected with p2.1, pSV-RL, HIF-1α (D) or HIF-2α (E) vector, and either EV or Sirt7 vector. 24 h post-transfection, cells were left untreated or treated with DMOG (1 mm) for an additional 24 h, after which cells were lysed, and luciferase activity was determined. F, Hep3B cells were cotransfected with HIF-1α or HIF-2α vector and either EV or Sirt7 vector. 24 h post-transfection, cells were treated with MG132 (20 μm) for an additional 8 h, after which cells were lysed, and Western blot assays were performed. G and H, Hep3B cells were cotransfected with p2.1, pSV-RL, expression vector encoding HIF-1α (G) or HIF-2α (H), and either EV or Sirt7 vector. 24 h post-transfection, cells were left untreated or treated with bafilomycin (5 nm) for an additional 24 h, after which cells were lysed, and luciferase activity was determined. I, Hep3B cells were cotransfected with HIF-1α or HIF-2α vector and either EV or Sirt7 vector. 24 h post-transfection, cells were treated with bafilomycin for an additional 8 h, after which Western blot assays were performed. Results are shown as means ± S.E. *, p < 0.01 for the indicated comparisons.
We recently reported that HIF-1α is subject to lysosomal degradation through the process of chaperone-mediated autophagy (22). Treatment with the lysosomal inhibitor bafilomycin led to an expected increase in HIF-1 (Fig. 5G) and HIF-2 (Fig. 5H) transcriptional activity but had no effect on inhibition of HIF activity that was mediated by Sirt7. Similarly, bafilomycin had no effect on the inhibition of HIF-1α and HIF-2α protein levels by Sirt7 (Fig. 5I). We conclude that Sirt7 exerts effects on HIF-1α and HIF-2α protein levels that are independent of both the proteasomal and lysosomal degradation pathways.
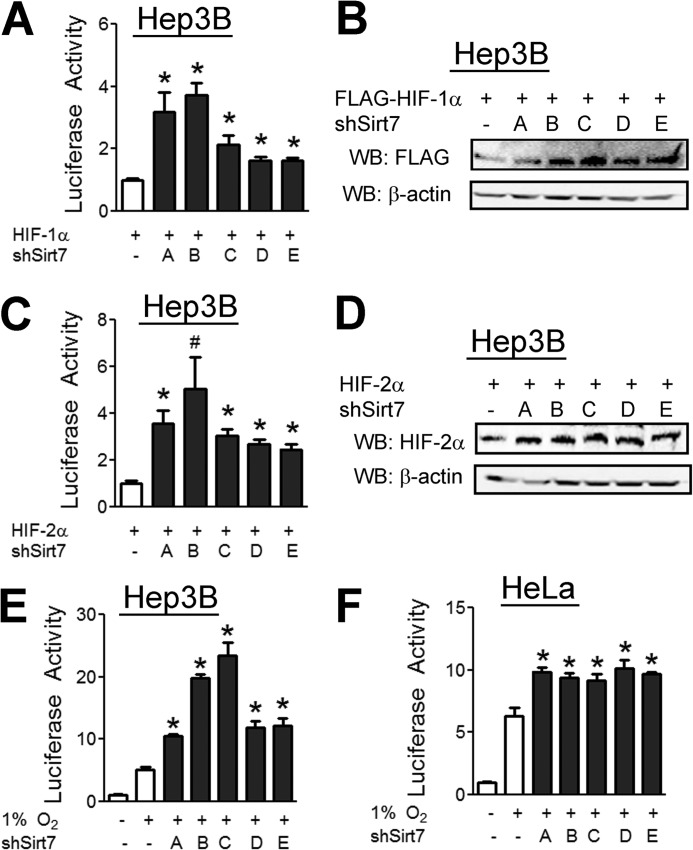
Knockdown of Sirt7 Expression Increases HIF-1 and HIF-2 Activity
To verify that Sirt7 regulates HIF-1 and HIF-2 at endogenous levels, we utilized five validated shRNA vectors targeting different sequences in Sirt7 mRNA. Knockdown of Sirt7 with each of the Sirt7 shRNA vectors led to an increase in HIF-1 reporter activity (Fig. 6A) and HIF-1α protein levels (Fig. 6B). Sirt7 knockdown likewise led to an increase in HIF-2 reporter activity (Fig. 6C) and HIF-2α protein levels (Fig. 6D). Finally, knockdown of Sirt7 led to an increase in endogenous HIF transcriptional activity induced by hypoxia in both Hep3B (Fig. 6E) and HeLa (Fig. 6F) cells.
FIGURE 6.
Knockdown of Sirt7 expression increases HIF-1α and HIF-2α protein levels and HIF transcriptional activity. A, Hep3B cells were cotransfected with p2.1, pSV-RL, HIF-1α expression vector; and either empty shRNA vector (−) or vector encoding one of five shRNA sequences targeting Sirt7 mRNA (shSirt7; labeled A–E). 48 h post-transfection, cell lysates were subjected to Western blot assay. B, Hep3B cells were cotransfected with FLAG-HIF-1α vector and either empty shRNA vector or vector encoding shRNA sequences targeting Sirt7, followed by Western blotting (WB). C, Hep3B cells were treated as described for A except that HIF-2α expression vector was used instead of HIF-1α. D, Hep3B cells were cotransfected with HIF-2α vector and either empty shRNA vector or vector encoding shRNA sequences targeting Sirt7, followed by Western blotting. E and F, Hep3B (E) and HeLa (F) cells were cotransfected with p2.1, pSV-RL, and either empty shRNA vector or vector encoding shRNA sequences targeting Sirt7. 24 h post-transfection, cells were exposed to 1% O2 for an additional 24 h and then lysed, and luciferase activity was determined. Results are shown as means ± S.E. #, p < 0.05; *, p < 0.01 versus HIF-1α, HIF-2α, or 1% O2 alone.
DISCUSSION
In this study, we have identified a novel role for Sirt7 as a negative regulator of HIF-1 and HIF-2 transcriptional activity. The inhibitory effect of Sirt7 was demonstrated by experiments analyzing HIF-1α and HIF-2α protein levels, HIF-dependent transcriptional activity of a reporter gene containing a hypoxia response element, and HIF-dependent transcription mediated by the promoters from three different HIF target genes.
The mechanism by which Sirt7 functions to decrease HIF-1α and HIF-2α protein levels remains to be established. The effect was preserved in the presence of hydroxylase inhibitors or mutations of the HIF-1α and HIF-2α hydroxylation sites, indicating that the effect of Sirt7 is independent of the classical oxygen-dependent degradation pathway. There are hydroxylase-independent but proteasomal-dependent pathways by which HIFs are regulated (16–19); however, the effect of Sirt7 was preserved even in the presence of the proteasome inhibitor MG132. Finally, HIF-1α is targeted for lysosomal degradation through chaperone-mediated autophagy (22), but the effect of Sirt7 was also maintained in the presence of the lysosomal inhibitor bafilomycin. Our data suggest that there may be as yet unknown mechanisms by which HIF-1α and HIF-2α can be degraded in the presence of Sirt7. The possibility that Sirt7 regulates translation of the proteins has not been formally excluded but seems unlikely because the inhibitory effects were observed when HIF-1α and HIF-2α were expressed from vectors that lack the 5′- and 3′-untranslated sequences that are present in the native mRNAs.
The mechanism by which Sirt7 regulates HIF activity differs from the other sirtuins because it is independent of its deacetylase activity. The effect of Sirt7 was preserved in the presence of high concentrations of nicotinamide, which is a nonspecific inhibitor of sirtuin catalytic activity. Two point mutations in Sirt7, which have previously been shown to eliminate its enzymatic activity (31, 34), had no effect on its ability to regulate HIF-1. In fact, deletion of the entire deacetylase domain had no effect on the ability of Sirt7 to inhibit HIF signaling. Thus, Sirt7 has cellular functions that are independent of its deacetylase activity. Recent work demonstrated that, in Caenorhabditis elegans, the Sirt7 homolog SIR-2.4 indirectly regulates DAF-16 acetylation and nuclear localization by blocking CBP-mediated acetylation (37). It is possible that Sirt7 regulates HIF-1 and HIF-2 through protein-protein interactions in a similar fashion.
Phylogenetic analysis divides the seven mammalian sirtuins into four classes, with Sirt6 and Sirt7 composing class IV (38). Sirt6 has been reported to function as a transcriptional corepressor of HIF-1α through its activity as an H3K9 deacetylase (28). Sirt6 was found to bind to HIF-1α and associate with the promoter of HIF target genes, and Sirt6 knockdown led to an increase in H3K9 acetylation specifically at those promoters (28). Additionally, Sirt6 appeared to have an effect on transcriptional regulation of HIF-1α itself (28). By contrast, the mechanism utilized by Sirt7 is distinct because its deacetylase activity was not required for its inhibitory effects on HIF-1 and HIF-2. Our results suggest that, despite a high degree of sequence similarity, the sirtuins function through multiple distinct mechanisms to inhibit HIF transcriptional activity. This is similar to previous observations regarding HIF regulation by different members of the SSAT (12, 16), MCM (39) and LIM domain (40–42) protein families and adds to the growing body of evidence identifying the sirtuins as key regulators of the cellular response to hypoxia.
Acknowledgments
We thank Joseph Garcia (University of Texas Southwestern Medical Center) for providing the EPO, SOD2, and VEGF reporter genes and Karen Padgett (Novus Biologicals, Inc.) for providing antibodies against HIF-2α and Myc epitope.
This work was supported, in whole or in part, by National Institutes of Health Contracts N01-HV28180 and HHS-N268201000032c from the United States Public Health Service and Postdoctoral Training Grant T32-HL007525 (to M. E. H.). This work was also supported by the Johns Hopkins Institute for Cell Engineering, American Heart Association Predoctoral Fellowship 10PRE4160120 (to K.), and Komen Foundation Postdoctoral Fellowship KG111254 (to D. M. G.).
- HIF
- hypoxia-inducible factor
- DMOG
- dimethyloxalylglycine
- EV
- empty vector.
REFERENCES
- 1. Semenza G. L., Wang G. L. (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12, 5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. (1998) Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12, 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu A. Y., Shimoda L. A., Iyer N. V., Huso D. L., Sun X., McWilliams R., Beaty T., Sham J. S., Wiener C. M., Sylvester J. T., Semenza G. L. (1999) Impaired physiological responses to chronic hypoxia in mice partially deficient for hypoxia-inducible factor 1α. J. Clin. Invest. 103, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Semenza G. L. (2009) Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology 24, 97–106 [DOI] [PubMed] [Google Scholar]
- 6. Hubbi M. E., Kshitiz, Gilkes D. M., Rey S., Wong C. C., Luo W., Kim D. H., Dang C. V., Levchenko A., Semenza G. L. (2013) A nontranscriptional role for HIF-1α as a direct inhibitor of DNA replication. Sci. Signal. 6, ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prabhakar N. R., Semenza G. L. (2012) Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol. Rev. 92, 967–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 9. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 10. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 11. Yu F., White S. B., Zhao Q., Lee F. S. (2001) HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. U.S.A. 98, 9630–9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baek J. H., Liu Y. V., McDonald K. R., Wesley J. B., Hubbi M. E., Byun H., Semenza G. L. (2007) Spermidine/spermine-N1-acetyltransferase 2 is an essential component of the ubiquitin ligase complex that regulates hypoxia-inducible factor 1α. J. Biol. Chem. 282, 23572–23580 [DOI] [PubMed] [Google Scholar]
- 13. Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 14. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang B. H., Zheng J. Z., Leung S. W., Roe R., Semenza G. L. (1997) Transactivation and inhibitory domains of hypoxia-inducible factor 1α. Modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 272, 19253–19260 [DOI] [PubMed] [Google Scholar]
- 16. Baek J. H., Liu Y. V., McDonald K. R., Wesley J. B., Zhang H., Semenza G. L. (2007) Spermidine/spermine N1-acetyltransferase-1 binds to hypoxia-inducible factor-1α (HIF-1α) and RACK1 and promotes ubiquitination and degradation of HIF-1α. J. Biol. Chem. 282, 33358–33366 [DOI] [PubMed] [Google Scholar]
- 17. Liu Y. V., Hubbi M. E., Pan F., McDonald K. R., Mansharamani M., Cole R. N., Liu J. O., Semenza G. L. (2007) Calcineurin promotes hypoxia-inducible factor 1α expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J. Biol. Chem. 282, 37064–37073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y. V., Baek J. H., Zhang H., Diez R., Cole R. N., Semenza G. L. (2007) RACK1 competes with HSP90 for binding to HIF-1α and is required for O2-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koh M. Y., Darnay B. G., Powis G. (2008) Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1α, leading to its oxygen-independent degradation. Mol. Cell. Biol. 28, 7081–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo W., Zhong J., Chang R., Hu H., Pandey A., Semenza G. L. (2010) Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1α but not HIF-2α. J. Biol. Chem. 285, 3651–3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montagner M., Enzo E., Forcato M., Zanconato F., Parenti A., Rampazzo E., Basso G., Leo G., Rosato A., Bicciato S., Cordenonsi M., Piccolo S. (2012) SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 487, 380–384 [DOI] [PubMed] [Google Scholar]
- 22. Hubbi M. E., Hu H., Kshitiz, Ahmed I., Levchenko A., Semenza G. L. (2013) Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J. Biol. Chem. 288, 10703–10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houtkooper R. H., Pirinen E., Auwerx J. (2012) Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 13, 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dioum E. M., Chen R., Alexander M. S., Zhang Q., Hogg R. T., Gerard R. D., Garcia J. A. (2009) Regulation of hypoxia-inducible factor 2α signaling by the stress-responsive deacetylase sirtuin 1. Science 324, 1289–1293 [DOI] [PubMed] [Google Scholar]
- 25. Chen R., Xu M., Hogg R. T., Li J., Little B., Gerard R. D., Garcia J. A. (2012) The acetylase/deacetylase couple CREB-binding protein/sirtuin 1 controls hypoxia-inducible factor 2 signaling. J. Biol. Chem. 287, 30800–30811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim J. H., Lee Y. M., Chun Y. S., Chen J., Kim J. E., Park J. W. (2010) Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1α. Mol. Cell 38, 864–878 [DOI] [PubMed] [Google Scholar]
- 27. Laemmle A., Lechleiter A., Roh V., Schwarz C., Portmann S., Furer C., Keogh A., Tschan M. P., Candinas D., Vorburger S. A., Stroka D. (2012) Inhibition of SIRT1 impairs the accumulation and transcriptional activity of HIF-1α protein under hypoxic conditions. PLoS One 7, e33433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R. E., Vadysirisack D. D., Guimaraes A., Marinelli B., Wikstrom J. D., Nir T., Clish C. B., Vaitheesvaran B., Iliopoulos O., Kurland I., Dor Y., Weissleder R., Shirihai O. S., Ellisen L. W., Espinosa J. M., Mostoslavsky R. (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bell E. L., Emerling B. M., Ricoult S. J., Guarente L. (2011) SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene 30, 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Finley L. W., Carracedo A., Lee J., Souza A., Egia A., Zhang J., Teruya-Feldstein J., Moreira P. I., Cardoso S. M., Clish C. B., Pandolfi P. P., Haigis M. C. (2011) SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 19, 416–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L. (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grob A., Roussel P., Wright J. E., McStay B., Hernandez-Verdun D., Sirri V. (2009) Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell Sci. 122, 489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsai Y. C., Greco T. M., Boonmee A., Miteva Y., Cristea I. M. (2012) Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol. Cell. Proteomics 11, 60–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barber M. F., Michishita-Kioi E., Xi Y., Tasselli L., Kioi M., Moqtaderi Z., Tennen R. I., Paredes S., Young N. L., Chen K., Struhl K., Garcia B. A., Gozani O., Li W., Chua K. F. (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vakhrusheva O., Smolka C., Gajawada P., Kostin S., Boettger T., Kubin T., Braun T., Bober E. (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703–710 [DOI] [PubMed] [Google Scholar]
- 36. Semenza G. L., Jiang B. H., Leung S. W., Passantino R., Concordet J. P., Maire P., Giallongo A. (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 37. Chiang W. C., Tishkoff D. X., Yang B., Wilson-Grady J., Yu X., Mazer T., Eckersdorff M., Gygi S. P., Lombard D. B., Hsu A. L. (2012) C. elegans SIRT6/7 homolog SIR-2.4 promotes DAF-16 relocalization and function during stress. PLoS Genet. 8, e1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frye R. A. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 39. Hubbi M. E., Luo W., Baek J. H., Semenza G. L. (2011) MCM proteins are negative regulators of hypoxia-inducible factor 1. Mol. Cell 42, 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Foxler D. E., Bridge K. S., James V., Webb T. M., Mee M., Wong S. C., Feng Y., Constantin-Teodosiu D., Petursdottir T. E., Bjornsson J., Ingvarsson S., Ratcliffe P. J., Longmore G. D., Sharp T. V. (2012) The LIMD1 protein bridges an association between the prolyl hydroxylases and VHL to repress HIF-1 activity. Nat. Cell Biol. 14, 201–208 [DOI] [PubMed] [Google Scholar]
- 41. Hubbi M. E., Gilkes D. M., Baek J. H., Semenza G. L. (2012) Four-and-a-half LIM domain proteins inhibit transactivation by hypoxia-inducible factor 1. J. Biol. Chem. 287, 6139–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin J., Qin X., Zhu Z., Mu J., Zhu L., Wu K., Jiao H., Xu X., Ye Q. (2012) FHL family members suppress vascular endothelial growth factor expression through blockade of dimerization of HIF1α and HIF1β. IUBMB Life 64, 921–930 [DOI] [PubMed] [Google Scholar]