Background: In ATP-binding cassette proteins, ATP binding produces association of the two nucleotide-binding domains (NBDs), but the molecular mechanism is unknown.
Results: The NBDs separate following ATP hydrolysis.
Conclusion: NBD dimers dissociate during the hydrolysis cycle supporting monomer/dimer models of operation.
Significance: Knowledge of the molecular mechanism of hydrolysis will help us understand how ATP-binding cassette proteins work.
Keywords: ABC Transporter, ATPases, CFTR, Fluorescence, Kinetics, Multidrug Transporters, LRET, Dimerization, Luminescence Resonance Energy Transfer
Abstract
In ATP-binding cassette proteins, the two nucleotide-binding domains (NBDs) work as dimers to bind and hydrolyze ATP, but the molecular mechanism of nucleotide hydrolysis is controversial. It is still unresolved whether hydrolysis leads to dissociation of the ATP-induced dimers or partial opening of the dimers such that the NBDs remain in contact during the hydrolysis cycle. We studied the bacterial lipid flippase MsbA by luminescence resonance energy transfer (LRET). The LRET signal between optical probes reacted with single-cysteine mutants was employed to follow NBD association/dissociation in real time. The intermonomer distances calculated from LRET data indicate that the NBDs separate completely following ATP hydrolysis, even in the presence of mm MgATP, and that the dissociation occurs following each hydrolysis cycle. The results support association/dissociation, as opposed to constant contact models, for the mode of operation of ATP-binding cassette proteins.
Introduction
ABC2 proteins constitute one of the largest protein superfamilies, extending from bacteria to man, with most members corresponding to membrane proteins that mediate transmembrane substrate transport (1, 2). ABC transporters include importers and exporters, which differ in a number of characteristics (2). Elucidation of the mechanism of ABC exporters is essential to understand processes such as multidrug resistance of cancer cells mediated by P-glycoprotein (MDR1 and ABCB1) (2, 3).
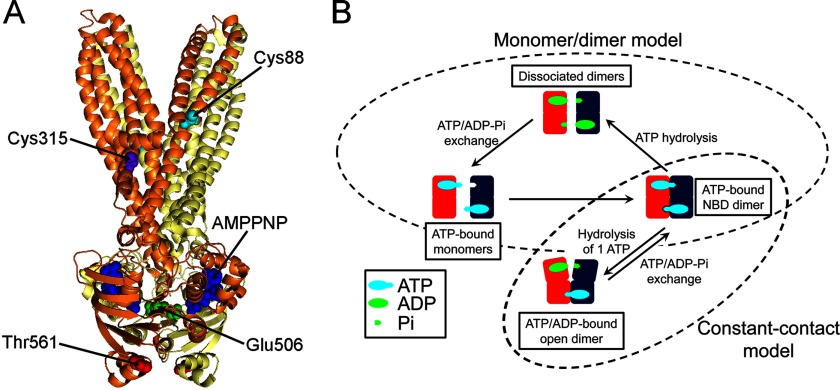
The core structure of ABC proteins comprises two transmembrane domains and two nucleotide-binding domains (NBDs) (see Fig. 1A) (1–3). The NBDs, the engines of ABC proteins, are responsible for nucleotide binding and hydrolysis, and their structure is conserved among ABC proteins with dissimilar functions (1). It has been established that ATP binding induces dimer formation. The head-to-tail orientation of the NBDs in the dimer results in the formation of two nucleotide-binding sites; each site is formed by the conserved motif A (P-loop) of one NBD and the signature motif of the other. In the ATP-bound state (closed conformation), two ATP molecules are “sandwiched” between the NBDs (4, 5). ABC transporters have a central translocation pathway accessible to only one side of the membrane at a time (alternate accessibility model), with the accessibility controlled by the NBDs (2). Despite the detailed structural knowledge of the NBDs, the mechanism by which they control the alternate accessibility is controversial. As a result of experimental discrepancies and a lack of direct information, a number of models have been proposed for the NBD-NBD interactions (see Fig. 1B). These models can be broadly divided in two groups: (a) Monomer/dimer models (4–10). ATP hydrolysis is followed by dissociation of the dimers, and the energy that drives the conformational changes that lead to substrate transport (the power stroke) is provided by dimer association-dissociation. Several variations of this model have been proposed, which differ in the extent of NBD separation, role of substrate binding (e.g., the switch model, in which substrate binding would increase the affinity of the NBDs for ATP) (11, 12), and sequence of ATP hydrolysis at the two sites (e.g., the processive clamp model, where the two ATPs are hydrolyzed sequentially before dimer dissociation (5, 8, 11); and the alternate hydrolysis model, where ATP hydrolysis alternates between the two sites and one hydrolysis event is sufficient for dimer dissociation) (9, 10). (b) Constant contact models (13–16). The NBDs remain in contact during the hydrolysis cycle, and the power stroke results from smaller conformational changes at the NBD-dimer interface. In one model, ATP hydrolysis occurs at one site, which opens after hydrolysis to allow ADP/ATP exchange (13, 15, 17).
FIGURE 1.
Structure and mechanism of MsbA. A, structure of the AMPPNP-bound MsbA. Ribbon representation with the monomers shown in yellow and orange. B, schematic representation of the two potential modes of operation that can explain the catalytic cycle of ABC proteins. Only the NBD-NBD interactions are illustrated. One NBD is shown in red, and the other one is in blue. Dissociated NBDs are depicted bound to ATP and Pi, but they could be nucleotide-free.
To ascertain the NBD movements during the catalytic cycle of a full-length exporter, we studied the bacterial lipid flippase MsbA, a tractable homolog of P-glycoprotein for which extensive structural and functional information is available (18–23). We used luminescence resonance energy transfer (LRET) to measure interdomain distances. LRET has atomic resolution and high sensitivity (24) and can also provide dynamic information on conformational changes in the millisecond to minute time frame (25).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
We studied wild-type MsbA Salmonella typhimurium (WT MsbA), Cys-less MsbA (MsbA CL), the single-Cys active mutant T561C, and the single-Cys inactive mutant E506Q/T561C. WT MsbA with an N-terminal decahistidine tag was generously provided by Dr. Geoffrey Chang (UCSD Skaggs School of Pharmacy and Pharmaceutical Sciences). In MsbA CL, the two native Cys (Cys-88 and Cys-315) were replaced with Ala. The Thr-561 to Cys mutation was introduced into MsbA CL Cys-less background to generate the single-Cys mutant T561C, and the Glu-506 to Gln mutation was introduced into T561C to generate the catalytically inactive E506Q/T561C mutant. The MsbA DNAs cloned into the NdeI/BamHI sites of the pET-19b plasmid (Novagen, Billerica, MA) were used to transform BL21 DE3-RILP Escherichia coli cells (Agilent Technologies, Santa Clara, CA). Expression started from single colonies grown overnight at 37 °C in Terrific Broth with 100 mg/liter ampicillin. Induction was for 4 h at 30 °C with 1 mm isopropyl-β-d-thiogalactopyranoside, and solubilization of crude membranes was performed for 1 h at room temperature in a buffer containing 100 mm NaCl, 20 mm Tris/HCl, pH 8, 15% glycerol, 0.5 mm Tris (2-carboxyethyl) phosphine hydrochloride (TCEP), and 0.5 mm phenylmethanesulfonyl fluoride with 2% n-dodecyl-β-d-maltopyranoside (dodecylmaltoside), and 0.04% sodium cholate. Purification was by metal affinity chromatography using a Talon Co2+ resin (Talon Superflow; Clontech) and gel filtration chromatography on a Superdex 200 column equilibrated with LRET buffer (100 mm NaCl, 20 mm Tris/HCl, pH 7.5, 0.065% dodecylmaltoside, 0.04% sodium cholate, and 0.2 mm TCEP). Protein concentration was determined by the BCA assay (Bio-Rad) and from the absorbance at 280 nm; purity was estimated at >95% from the gel filtration chromatogram and SDS-PAGE gels stained with Coomassie Blue. Monodispersity was assayed by dynamic light scattering as described (28).
ATPase Measurements
ATP hydrolysis was measured using a variant of the linked enzyme assay (38).
LRET Studies
T561C and E506Q/T561C were labeled with thiol-reactive LRET probes. Glycerol and TCEP were removed from the LRET buffer by gel filtration (Zeba columns; Thermo Fisher Scientific, Rockford, IL), and the protein was immediately labeled with both the thiol-reactive Tb3+-chelate DTPA-cs124-EMPH (Lanthascreen; Invitrogen) as donor (39), and N-(2-aminoethyl)maleimide (Bodipy FL maleimide; Invitrogen) or Cy3 maleimide (GE Healthcare) as acceptor. Donor and acceptor probes were used at 2-fold the molar Cys concentration. MsbA CL was used as control to evaluate nonspecific labeling. The labeled protein was separated from free unreacted probes by gel filtration on a 3-ml Superdex 200 5/150 GL column equilibrated with LRET buffer. To avoid diffusion-enhanced LRET arising from free probes (40), only the peak with higher protein concentration was used. Samples containing 0.25–1 μm MsbA were analyzed in 3-mm-pathlength quartz cuvettes, essentially as described (25). Emission was measured on either a Photon Technology International spectrometer (QM3SS; PTI, London, Canada) or an Optical Building Blocks phosphorescence lifetime photometer (EasyLife L; OBB, Birmingham, NJ). The emission was recorded with a 200-μs delay from the beginning of the ∼1-μs excitation pulse from a xenon flash lamp (gated mode). During those 200 μs, the short lifetime processes decay significantly; these processes include acceptor emission resulting from direct excitation, scattering of the excitation pulse, and autofluorescence. Therefore, in gated mode, the remaining light arises from long lifetime processes such as emission from the donor or sensitized emission from the acceptor. Excitation was set to 335 nm, and emission was collected through bandpass filters (Tb3+: 490/10 nm; Bodipy FL: 520/10 nm; Cy3: 570/10, Omega Optical, Brattleboro, VT). Generally, donor and acceptor emission decays were collected as averages from 1,800 pulses at 100 Hz. For the standard intensity time courses, the excitation frequency was 100 Hz, and the area under each decay curve was integrated between 200 and 300 μs, and averaged for 1 s. Distances between the donor and acceptor probes were calculated according to Equations 1 and 2,
where E is the efficiency of energy transfer, R0 is the Förster distance (the distance at which E = 0.5), and τD and τDA are the lifetimes of the donor in the absence and presence of the acceptor, respectively. The R0 values were determined as previously described (25). In LRET, the lifetime of the donor molecules that participate in energy transfer, τDA, is equal to the sensitized emission lifetime, i.e., the “long” lifetime of the acceptor that arises from energy transfer (6). An additional fast component with a τ of <100 μs is an instrument artifact (scattering of the excitation pulse caused by the “slow” instrument response time). We did not perform corrections because the magnitude of the experimental signal was sufficiently large to start the fits later, when this component was undetectable. Because Tb3+ emission lifetime is long, diffusion-enhanced LRET is expected to elicit very low efficiency energy transfer between MsbA dimers. This energy transfer will produce long lifetimes that approach the donor-only lifetime. Because the average distance between MsbA homodimers in solution >1,000 Å, diffusion-enhanced LRET is not expected to be a significant problem unless there is association between dimers in solution or after reconstitution in liposomes. Its magnitude was estimated by mixing MsbA separately labeled with donor or acceptor, which produced small signals with lifetimes within 5% of the donor-only lifetime. These signals were attributed to diffusion-enhanced LRET (40) and were subtracted for the estimation of the proportion of donor-acceptor pairs at each distance in the different conformational states.
Reconstitution of MsbA into Unilamellar Liposomes
Reconstitution was performed in E. coli lipids by gel filtration. Pilot experiments showed that essentially all T561C inserts into the liposomes with the NBDs on the outside, so labeling was generally performed after reconstitution, following the protocol described above for the protein in detergent. The signal arising from intermolecular LRET was determined in proteoliposomes containing a mixture of T561C prelabeled (in detergent) with donor-only and acceptor-only (see end of previous section). These experiments show minimal signal at lipid:protein reconstitution ratios <1:100 (w/w), and therefore a 1:200 ratio was selected. The average hydrodynamic diameter of the proteoliposomes was 550 Å, as determined by dynamic light scattering measured at 90° on Brookhaven Instruments BI-200SM (Holtsville, NY). Assuming a bilayer thickness of 30 Å and an area of 65 Å2 per phospholipid head (41), the 1:200 reconstitution ratio yields an average of 0.85 MsbA dimers per liposome.
Stopped Flow LRET Studies
The kinetics of the conformational change from ATP-bound T561C to the dynamic equilibrium during continuous ATP hydrolysis (in MgATP) was monitored using a stopped flow mixing accessory (RX2000; Applied Photophysics, Surrey, UK) with a 20-μl cell, placed into the sample compartment of the OBB LRET system. The stopped flow device was driven by pressured N2 and set to mix equal volumes of two syringes in <10 ms. The first syringe contained 1 μm T561C labeled with the Tb3+ chelate and Bodipy FL in LRET buffer containing 1 mm EDTA and 0.5 mm ATP. The second syringe was filled with LRET buffer containing 1 mm EDTA, 0.5 mm ATP, and 20 mm MgSO4. The solutions were equilibrated for 10 min before starting data collection. The data were collected as in the standard intensity time courses, except that the pulse frequency was increased from 100 to 500 Hz, and the averaging was decreased to 5–10 pulses to increase the number of data points captured during the transitions. Excitation and emission filters were as described above. Stopped flow data were fit using SigmaPlot (Systat Software Inc., San Jose, CA). The data recorded at 30 °C or below fit well to a single-exponential decay, whereas a two-exponential decay function was needed to fit data obtained at 37 °C.
Data Presentation and Statistics
The data are shown as means ± S.E. or S.D., as indicated. Statistical comparisons were performed by the Student's t test for paired or unpaired data, or one-way analysis of variance, as appropriate. p < 0.05 in a two-tail analysis was considered significant. The number of experiments (n) corresponds to independent measurements from at least three different protein preparations. The goodness of fit for the analysis of LRET decays and stopped flow LRET fits was determined from the random residual distribution, which showed no structure and chi-squared values near unity.
RESULTS
General Experimental Strategy
There were two important goals for our studies: 1) To assess structure and function under the same conditions on a functional protein. In previous studies of full-length ABC protein exporters, functional and structural studies were performed under different experimental conditions. This is obvious in the case of crystallographic studies, but it is also true in the case of EPR spectroscopic studies, where function was evaluated at “normal” temperatures, but EPR data were obtained at very low temperatures (18, 21, 22). 2) To not only identify conformational states, but to perform kinetic studies of the transitions between those conformational states. Such detailed studies have not been performed in full-length ABC proteins. The only attempt has been a single-molecule FRET study of P-glycoprotein (26). Kinetic studies are essential to define the molecular mechanism of function. LRET is an excellent technique to accomplish these aims, and to apply it to MsbA we generated single-Cys mutants for labeling with thiol-selective donor and acceptor probes. We focused on the NBDs for these initial studies and chose position 561 for the single Cys. This mutant has been shown to have “normal” ATPase activity (23), and EPR data are available for comparison (22). Fig. 1A illustrates the positions of the native MsbA Cys as well as those of the E506Q and T561C mutations. The native cysteines (Cys-88 and Cys-315) were replaced with Ala in the MsbA CL, which was used as background to introduce the single Cys in T561C. A catalytically deficient mutant of T561C, generated by replacement of the catalytic carboxylate Glu-506 with Gln (E506Q/T561C), was used to identify conformational changes dependent on ATP hydrolysis. Glu-506, located at the end of motif B, is the catalytic carboxylate involved in Mg2+ coordination.
Activity of Purified MsbA Mutants
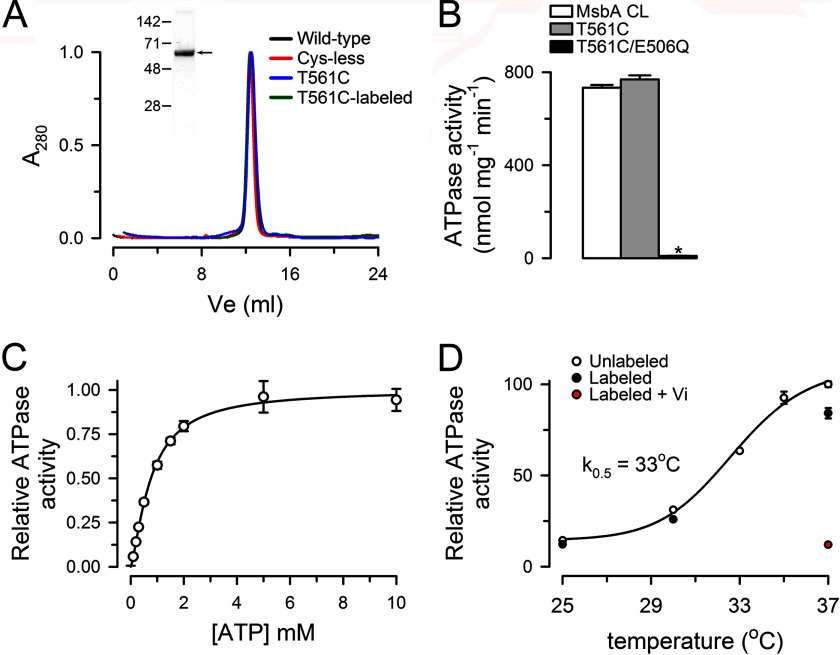
Fig. 2A shows that the hydrodynamic properties of WT MsbA, MsbA CL, and T561C, evaluated by gel filtration chromatography, are essentially identical. The elution is consistent with the expected homodimer of ∼134 kDa because it occurs in close proximity to that of mouse P-glycoprotein (ABCB1, ∼140 kDa) and the connexin 26 (Cx26) hexamer (∼160 kDa), two proteins that we routinely use in our laboratory (27, 28). The Fig. 2A (inset) shows a Coomassie Blue-stained SDS-PAGE of the purified T561C mutant. The expression level and purity of all the MsbAs studied was indistinguishable.
FIGURE 2.
ATPase activity of purified MsbA. A, hydrodynamic properties of purified MsbA in dodecylmaltoside. Gel filtration analysis of purified MsbA run as described under “Experimental Procedures.” A280 is the absorbance measured at 280 nm, normalized to the total absorbance area. T561C was labeled with the Tb3+ chelate and Bodipy FL maleimides as described under “Experimental Procedures.” The inset shows a Coomassie Blue-stained gel of purified T561C MsbA. The positions of molecular mass markers (in kDa) are indicated on the left. B, ATPase activity of purified MsbA mutants. The asterisk denotes p < 0.01 compared with MsbA CL values. The values are presented as means ± S.D. (n = 6 for each condition). C, dependence of the T561C ATPase activity on ATP concentration. Mg2+ was kept constant at 12 mm. The values are presented as means ± S.D. (n = 4). D, dependence of the T561C ATPase activity on temperature. The concentration of MgATP was 10 mm (12 mm Mg2+). The effects of labeling with Tb3+ and Bodipy FL on T561C ATPase activity are also shown. Vi refers to inhibition by orthovanadate, achieved by 10-min preincubation with 250 μm Vi in the presence of 4 mm MgATP. The values normalized to the activity of unlabeled T561C at 37 °C are presented as means ± S.D. (n = 6 for unlabeled T561C at 37 °C and n = 3 for all other conditions). See “Experimental Procedures” for details.
Fig. 2B shows that T561C has an ATPase activity similar to that of the MsbA CL, whereas the catalytically deficient mutant E506Q/T561C has a very low activity (∼1% of the T561C activity). The dependence of the ATPase activity on ATP concentration in the presence of 12 mm MgSO4 is illustrated in Fig. 2C. The apparent Km for ATP was 763 ± 43 μm with a Hill coefficient of 1.4 ± 0.1 (n = 4), similar to previous reports (19, 23, 29). Fig. 2D shows that the ATPase activity of T561C is highly dependent on temperature and that reaction of Cys-561 with Bodipy FL maleimide or the Tb3+ chelate produced only a small reduction in activity (∼15%). Consistent with the preserved activity of the labeled protein, labeling had no effect on the elution from the gel filtration column, showing absence of effects on the hydrodynamic properties of T561C (Fig. 2A).
Although the rates of ATP hydrolysis from purified MsbA are among the highest reported, we have measured significantly higher rates in buffers of different composition (i.e., in 50 mm Tris/HCl, pH 7.5). All activities reported here were measured at the same temperature and in the same buffer used for the LRET experiments (100 mm NaCl, 20 mm Tris/HCl, pH 7.5, 0.065% dodecylmaltoside, 0.04% sodium cholate, and 0.2 mm TCEP); this was essential to directly compare the functional and structural data in a quantitative manner.
LRET in MsbA-T561C
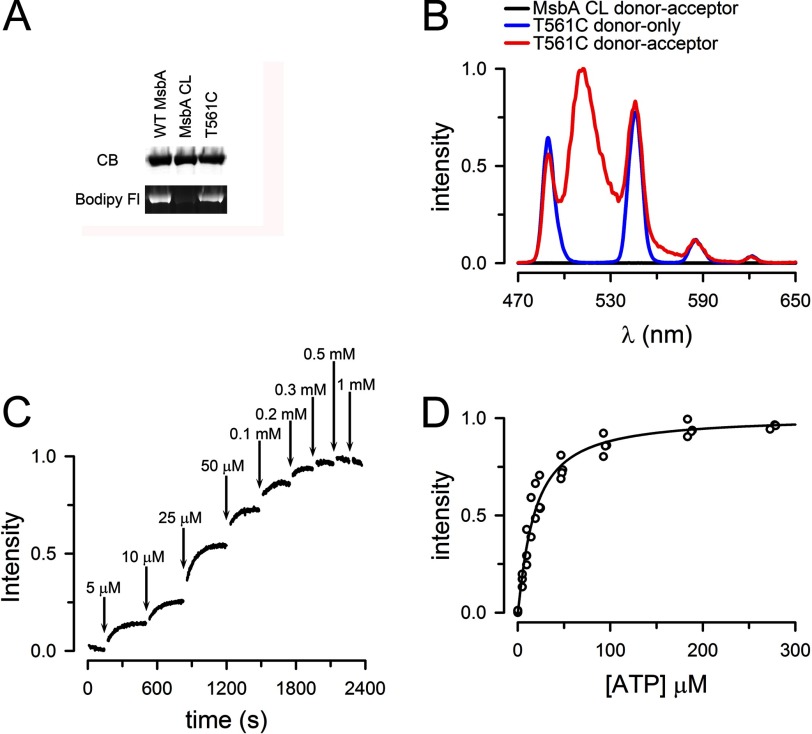
Fig. 3A shows that reaction of the fluorescent thiol-reagent Bodipy FL maleimide with MsbA CL is minimal, whereas both WT MsbA and the single-Cys T561C mutant are well labeled. The labeling intensity was similar in WT MsbA and T561C, suggesting that only one of the two native Cys is labeled, consistent with the structure that shows Cys-315 buried (19).
FIGURE 3.
Sensitized emission from T561C labeled with LRET probes. A, selective labeling of T561C with Bodipy FL maleimide. CB, Coomassie Blue-stained SDS-PAGE gel; Bodipy Fl, Bodipy FL fluorescence from the same gel. B, emission spectra from ATP-bound T561C labeled with Tb3+ only (T561C donor only, blue trace) or Tb3+ and Bodipy FL (T561C donor-acceptor, red trace). The emission spectra of MsbA CL subjected to the Tb3+/Bodipy FL labeling procedure is also shown (MsbA CL donor-acceptor, black trace). Protein concentration was ∼1 μm, and 0.5 mm ATP was present for at least 10 min before collecting the spectra. MsbA CL values were normalized to the T561C donor-acceptor sensitized emission maximum (∼510 nm). T561C donor-only data were scaled to the T561C donor-acceptor 585-nm peak. C, typical T561C LRET intensity changes in response to increasing ATP concentration. The sensitized Bodipy FL emission was measured at ATP concentrations ranging from 0 to 1 mm. The data were normalized to the 0.5 mm peak value. The solutions were nominally divalent cation-free and contained 1 mm EDTA to prevent ATP hydrolysis. D, summary of the dependence of the LRET signal on ATP concentration. The sensitized emission data were obtained from four independent experiments, such as that in C, and the data were normalized to the 0.5 mm peak value.
Fig. 3B illustrates the emission spectra of labeled Thr-561 MsbA in the presence of 0.5 mm ATP. The Tb3+-labeled spectrum (blue trace, T561C donor only) shows sharp peaks separated by dark regions. The emission spectrum from T561C labeled with the Tb3+-chelate and Bodipy FL (red trace, T561C donor-acceptor) shows the Tb3+ peaks superimposed upon a broad peak (500–535 nm) that corresponds to the sensitized emission of Bodipy FL; the sensitized emission arises from excitation by energy transfer from Tb3+. The emission spectra for MsbA CL subjected to the same Tb3+/Bodipy FL-labeling protocol (black trace, MsbA CL donor-acceptor) resulted in minimal LRET emission. Therefore, the Bodipy FL-sensitized emission comes from energy transfer between donor and acceptor LRET probes bound to Cys-561. In most experiments, we labeled T561C with a 2-fold molar excess of both donor and acceptor simultaneously. Because the same residue (Cys-561) is labeled in each T561C monomer in the MsbA homodimer, assuming random labeling, 50% of the homodimers will be labeled with one donor and one acceptor, whereas the homodimers labeled with donor-only (25%) or acceptor-only (25%) will not contribute to the signal. Because the long lifetime acceptor emission can only arise from LRET, the stoichiometry of labeling affects the intensity of the signal, but not the distance calculations or the Kd determinations, which depend on probes lifetimes and relative changes, respectively (24, 25). In any case, we estimated the labeling stoichiometry at ∼0.6 mol probe/mol MsbA.
The dependence of the Bodipy FL-sensitized emission on ATP concentration is shown in Fig. 3C. MsbA T561C was labeled with the Tb3+-chelate and Bodipy FL, and the sensitized emission intensity was measured as a function of ATP concentration in the absence of Mg2+ (1 mm EDTA was added to chelate trace divalent cations). The LRET signal is expected to increase if the NBDs approach each other in the presence of ATP. Fig. 3C shows that this was indeed the case and that the apparent affinity for ATP of this conformational change was ∼20 ± 2 μm, with a Hill coefficient of 1.2 ± 0.1 (n = 4; Fig. 3D). This value is significantly lower than the Km for the ATPase activity of ∼750 μm. The difference between the Kd (ATP) for NBD dimerization and the Km (ATP) for the ATPase activity suggests that the dimerization is not the rate-limiting step of the hydrolysis cycle and that there a significant contribution of the ATPase cycle to the kinetics of the functional enzyme. As is the case for many enzymes, steps that follow ATP-induced NBD association determine the apparent affinity of the complete ATP hydrolysis cycle (i.e., contribution of the turnover rate or ADP dissociation following ATP hydrolysis).
Changes in LRET during the MsbA Catalytic Cycle
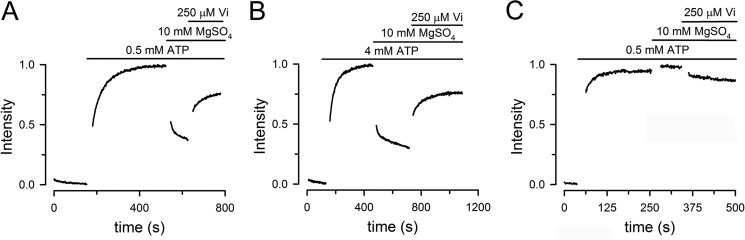
The ATP dependence of the Bodipy FL-sensitized emission suggests that the LRET signal can be used to follow conformational changes during the ATP-hydrolysis cycle. Such changes are illustrated in Fig. 4A. The addition of ATP to 0.5 mm increased the LRET signal intensity. This increase in intensity is the result of a conformational change that brings the donor-acceptor LRET pair closer, and it is due to ATP binding because in the absence of Mg2+ (nominally divalent-free LRET buffer with 1 mm EDTA to chelates trace divalent cations) ATP hydrolysis was null. Subsequent addition of 10 mm MgSO4 to initiate ATP hydrolysis triggered additional conformational changes that reduced the signal intensity. The decrease in intensity after 1 min in MgATP was 55 ± 2% (n = 6). Under hydrolysis conditions, addition of orthovanadate (Vi) increased the signal intensity. The inhibition of the T561C ATPase activity by Vi under these conditions was 88 ± 1% (n = 6). The Vi-inhibited state corresponds to a high energy posthydrolysis state also referred to as a posthydrolysis transition state. Experiments at varying ATP concentrations indicated that the slower decrease is due to signal quenching by ATP, independent of conformational changes. For that reason, we preferred to use 0.5 mm ATP for most studies. However, experiments with 4 mm ATP showed very similar results (Fig. 4B).
FIGURE 4.
Time course of NBDs association/dissociation monitored by LRET. A, changes in Bodipy FL-sensitized emission in T561C MsbA. Response to sequential additions of ATP, MgSO4, and orthovanadate. Traces show signals normalized to the total change. B, same as A, but ATP concentration was 4 mm instead of 0.5 mm. C, changes in Bodipy FL-sensitized emission in E506Q/T561C MsbA. See A for details. For all panels, the protein concentration was ∼1 μm, and the traces are representative of data from more than six similar experiments.
The dependence of the conformational changes on ATP hydrolysis was further examined with the catalytically deficient mutant E506Q/T561C. Fig. 4C shows an experiment identical to that in Fig. 4A, but on the E506Q/T561C mutant. The addition of ATP elicited an increase in LRET signal similar to that seen in the catalytically active T561C, but the responses to the subsequent additions of Mg2+ and Vi were essentially absent. These results support the assumption that the intensity changes in MgATP and after Vi addition in T561C are the result of conformational changes that require ATP hydrolysis. Comparison of panels A and C in Fig. 4 shows that the increase in LRET signal in response to ATP is faster in E506Q/T561C than in T561C. We have not explored this difference in detail, but we have previously shown a similar effect in the catalytic carboxylate mutant in the isolated NBD MJ0796, and we ascribed it to electrostatic effects at the dimer interface (30); the reduced negative charge in the E506Q mutant would increase the speed of dimerization of ATP-bound monomers.
Donor-Acceptor Distances in MsbA T561C during the Catalytic Cycle
The changes in LRET intensity shown in Fig. 4A can result from a “long” donor-acceptor distance changing to a new “short” distance or from changes in the proportions of existing conformations. In the first case, the intensity increase in response to ATP addition would result from dissociated NBDs coming into contact and producing a new shorter donor-acceptor distance. In the second case, ATP would displace the equilibrium between associated and dissociated NBDs toward MsbA with associated NBDs, and although the distances would not change, the fraction of molecules displaying the shorter distances would increase. To discriminate between these possibilities, we calculated donor-acceptor distances from the sensitized emission lifetimes.
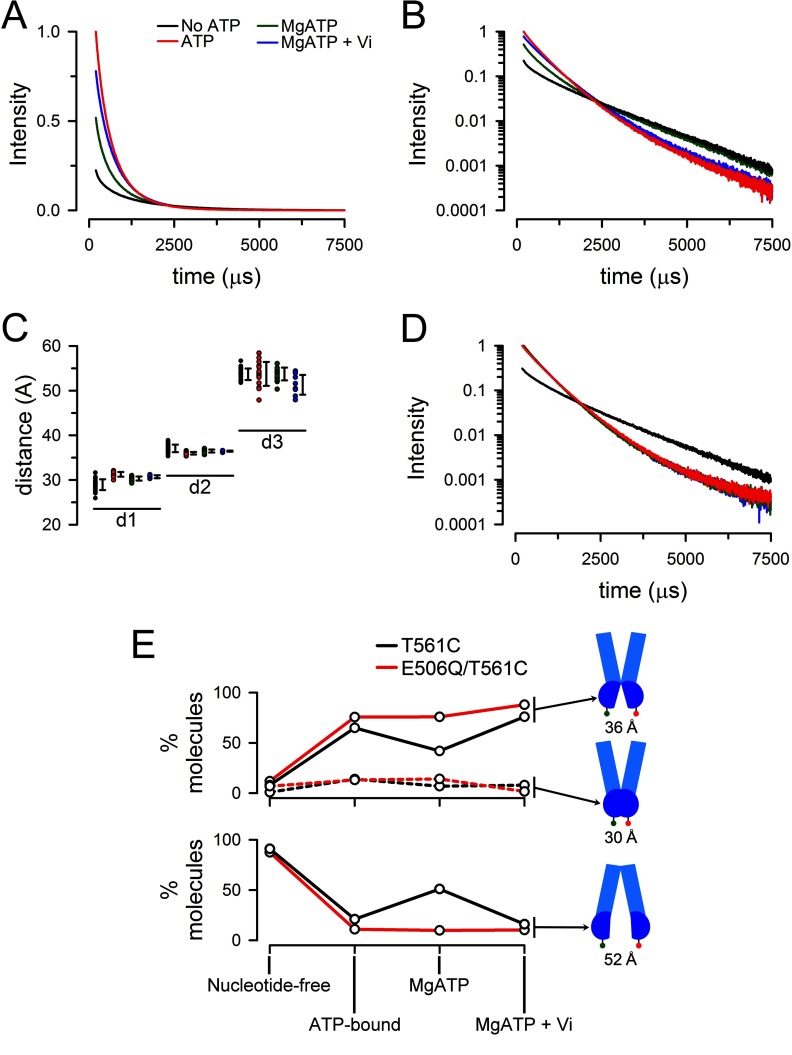
Fig. 5 shows typical intensity decays from T561C labeled with the Tb3+/Bodipy FL pair. As expected from the intensity time course in Fig. 4A, addition of ATP increased the signal intensity severalfold, whereas the addition of Mg2+ decreased the LRET signal intensity of the ATP-bound T561C, and subsequent addition of Vi increased the signal back toward the ATP-bound state. For a defined donor-acceptor distance like that seen in isolated NBDs (25), the intensity decay follows a single-exponential function of time and therefore will appear as a straight line when the log of the intensity is plotted. The observation of such a semilog plot in Fig. 5B clearly shows that the intensity decay does not follow a single exponential, implying the presence of multiple donor-acceptor distances. The decay curves illustrate that the ATP-bound and Vi-inhibited states have a faster overall decay and that they are more similar to each other than to the nucleotide-free (apo) state and the equilibrium state during continuous hydrolysis (MgATP).
FIGURE 5.
Emission decays from MsbA labeled with Tb3+ and Bodipy FL at Cys-561. A, sensitized Bodipy FL emission decays from T561C. No ATP, nucleotide-free apo state (black trace) obtained in nominally divalent cation-free buffer with 1 mm EDTA to prevent ATP hydrolysis; ATP, ATP-bound state (red trace) obtained 5 min after addition of ATP to 0.5 mm; MgATP, continuous hydrolysis state (green trace), obtained 3 min after addition of MgSO4 to 10 mm; MgATP + Vi, vanadate-inhibited or posthydrolysis transition state (blue trace) obtained 3 min after addition of 250 μm sodium orthovanadate. Intensities were normalized to the ATP intensity at 200 μs. The traces are representative of more than seven similar experiments. B, sensitized Bodipy FL emission decays from T561C. Semilog plot of the data in A. C, distribution of calculated distances from the LRET data. Distances in angstroms calculated from the exponential components d1, d2, and d3. The black symbols to the right of each individual set of experimental points denotes ± 1 S.D. Color coding is as in A, from left to right: black, no ATP; red, ATP; green, MgATP; and blue, MgATP + Vi. D, semilog plot of the sensitized Bodipy FL emission decays from E506Q/T561C. See A for details. The traces are representative of more than six similar experiments. E, donor-acceptor LRET pair distributions during the hydrolysis cycle. The mean percentages of molecules in each conformation were calculated from the average fractional intensity contribution of each component (Table 1) divided by the rate of energy transfer (k = 1/τDA − 1/τD), after subtracting the long lifetime background component arising from intermolecular LRET. The latter was estimated from the LRET signal measured between T561C labeled separately only with donor or acceptor.
The sensitized emission lifetimes and calculated donor-acceptor distances for T561C labeled with Tb3+ and Bodipy FL are shown in Table 1. The donor-only decay (τD, measured at 490 nm) at 37 °C was well fitted by a two-exponential decay function that yielded lifetimes of 657 ± 104 and 1939 ± 15 μs (n = 7). The slower component accounted for 98.2 ± 0.3% of the signal and was used for the distance calculations. A three-exponential function was needed to fit τDA. Calculations yielded three distinct distances that were present and essentially identical under all studied conditions. The differences in sensitized emission decays among conformational states (e.g., nucleotide-free versus ATP-bound) were the result of variations in the proportion of those three distances. We identified two shorter distances of ∼30 and ∼36 Å and a longer distance of ∼53 Å. The percentage of the LRET signal originating from the longer (∼53 Å) donor-acceptor distance decreased from ∼50% in the nucleotide-free state to ∼5% in the ATP-bound state. It doubled to ∼10% during hydrolysis in MgATP and decreased back to the ATP-bound state level of ∼5% in the Vi-inhibited protein. The calculated distances for individual experiments are shown in Fig. 5C, which illustrates the presence of the three distinct distances under all experimental conditions and also gives an indication of variability. Although differences in variability among conformational states (e.g., nucleotide-free versus ATP-bound) could reflect changes in the “compactness” of MsbA, they can also be explained by differences in signal intensity that affect the experimental error. For example, in the nucleotide-free state, the low intensity shorter distance components are less well defined than the more intense longer distance component, but this pattern is reversed for the ATP-bound state.
TABLE 1.
Tb3+-Bodipy FL distances during the catalytic cycle in dodecylmaltoside in catalytically active MsbA T561C and catalytically deficient E506Q/T561C
The three identified exponential components of the sensitized emission decay are presented as d1, d2, and d3. τDA, Bodipy FL-sensitized emission lifetime; R, calculated donor-acceptor distance; Signal, contribution of each pre-exponential component. Apo, nucleotide-free state in 1 mm EDTA/magnesium-free solution; ATP-bound, ATP-bound state in 0.5 mm ATP; MgATP, continuous hydrolysis state in 0.5 mm MgATP; Vi-inhibited, high energy posthydrolysis state or transition state, obtained after addition of 250 μm Vi in the presence of 0.5 mm MgATP. The data are presented as means ± S.D., and n is the number of independent experiments in each conformational state (first T561C and then E506Q/T561C). The d1, d2, and d3 values for τDA, R, and Signal were statistically different from each other under all conditions (p < 0.01). The d1, d2, and d3 values for τDA and R among the different conformational states were statistically indistinguishable from each other.
| State | T561C |
E506Q/T561C |
||||
|---|---|---|---|---|---|---|
| τDA | R | Signal | τDA | R | Signal | |
| μs | Å | % | μs | Å | % | |
| Apo (n = 24; n = 10) | ||||||
| d1 | 218 ± 47 | 29.0 ± 1.2 | 18.3 ± 3.2 | 166 ± 43 | 27.5 ± 1.3 | 12.4 ± 2.4 |
| d2 | 685 ± 61 | 37.1 ± 0.9 | 33.1 ± 1.9 | 656 ± 45 | 36.7 ± 0.6 | 41.4 ± 1.1 |
| d3 | 1,615 ± 38 | 53.7 ± 1.3 | 48.6 ± 3.2 | 1,536 ± 39 | 51.3 ± 1.0 | 46.2 ± 2.6 |
| ATP-bound (n = 15; n = 7) | ||||||
| d1 | 319 ± 26 | 31.3 ± 0.5 | 31.1 ± 4.6a | 341 ± 25 | 31.7 ± 0.5 | 28.2 ± 5.1a |
| d2 | 609 ± 20 | 36.0 ± 0.3 | 65.1 ± 4.2a | 630 ± 17 | 36.3 ± 0.2 | 69.3 ± 4.4a |
| d3 | 1,610 ± 86 | 53.7 ± 2.7 | 3.8 ± 0.7a | 1,529 ± 101 | 51.3 ± 2.4 | 2.5 ± 0.8a |
| MgATP (n = 15; n = 7) | ||||||
| d1 | 274 ± 21 | 30.3 ± 0.5 | 31.7 ± 3.9a | 325 ± 12 | 31.4 ± 0.2 | 28.4 ± 2.1a |
| d2 | 643 ± 24 | 36.5 ± 0.3 | 58.3 ± 4.0a | 595 ± 7 | 35.8 ± 0.1 | 69.3 ± 2.1a |
| d3 | 1,617 ± 46 | 53.7 ± 1.5 | 10.0 ± 2.9a | 1,540 ± 43 | 51.4 ± 1.1 | 2.3 ± 0.2a |
| Vi-inhibited (n = 8; n = 7) | ||||||
| d1 | 293 ± 16 | 30.7 ± 0.3 | 20.4 ± 2.9 | 334 ± 22 | 31.6 ± 0.4 | 28.4 ± 2.7a |
| d2 | 640 ± 9 | 36.4 ± 0.1 | 74.5 ± 3.4a | 607 ± 12 | 36.0 ± 0.2 | 69.3 ± 2.5a |
| d3 | 1,531 ± 83 | 51.3 ± 2.2 | 5.1 ± 1.4a | 1,542 ± 120 | 52.0 ± 3.8 | 2.3 ± 0.4a |
a Statistical differences in the Signal of d1, d2, and d3 components compared with the corresponding components in the Apo state (p < 0.05).
The sensitized emission intensity from a single donor-acceptor pair depends on the donor-acceptor distance; that is, the sensitized emission from a donor-acceptor pair where the donor and acceptor are in close proximity is brighter than that from a pair where there is more separation between donor and acceptor. Consequently, the relative intensities of each exponential component are not linearly related to the number of donor-acceptor pairs that they originate from. However, the rate of energy transfer (k = 1/τDA − 1/τD) can be used in combination with the intensity to calculate the number donor-acceptor pairs that contribute to a distance component (31). By correcting for the energy transfer rate and also for intermolecular LRET caused by diffusion-enhanced LRET, we estimated the percentage of MsbA molecules in each conformation (Fig. 5E, black lines). Basically, the ∼10% of NBDs that are associated in the nucleotide-free state climbs to 80–85% in the ATP-bound and Vi-inhibited states. During continuous hydrolysis, there is a dynamic equilibrium with ∼50% of the NBDs dissociated. The latter indicates that during continuous hydrolysis, under the conditions of our experiments, neither NBD association nor dissociation is clearly favored.
To confirm that the distance measured for the dissociated NBDs was ∼53 Å and not ∼85 Å, as estimated from the apo MsbA crystal structure (19), we used T561C labeled with the Tb3+/Cy3 LRET pair. This LRET pair has a Förster distance of 62 Å, and it is therefore more sensitive to changes in longer distances than the Tb3+/Bodipy FL pair. The analysis of Cy3-sensitized emission decays obtained in T561C labeled with Tb3+ chelate and Cy3 showed a long lifetime component (1,849 ± 48 μs, mean ± S.D., n = 26) that did not change in intensity in the different conformational states and was therefore attributed to intermolecular LRET. The lifetimes of the two additional decay components were 124 ± 33 and 590 ± 40 μs (means ± S.D., n = 7) in the apo state. These lifetimes did not differ in the ATP-bound state, but their intensities were conformational state-sensitive, with the 590-μs lifetime component accounting for 50 ± 2% (n = 7) of the signal in the nucleotide-free state and 21 ± 2% (n = 7) in the ATP-bound state. The distances calculated from the Tb3+/Cy3 were 37.4 ± 1.7 and 53.2 ± 1.5 Å (means ± S.D., n = 26). These distances are very similar to the ∼36 and 53 Å determined for the Tb3+/Bodipy FL pair (Table 1). The shorter distance of ∼30 Å is expected to produce an average lifetime of ∼25 μs, which is buried within the instrument response time and therefore cannot be detected for the Tb3+/Cy3 pair.
To confirm the identity of the processes dependent on ATP hydrolysis, we calculated donor-acceptor distances in the catalytically deficient mutant E506Q/T561C. Fig. 5D shows that addition of ATP produced the same effects as in the catalytically active T561C (increased intensity and faster decay), but subsequent addition of Mg2+ and Vi had no additional effects, consistent with the results in Fig. 4C. The sensitized emission lifetimes and calculated distances between the donor and acceptor probes are summarized in Table 1. Fig. 5E (red lines) shows that the percentage of molecules in each conformation matches very closely those in the active T561C protein under nucleotide-free and ATP-bound states. However, as expected from the deficiency in ATP hydrolysis, subsequent additions of Mg2+ and Vi had no significant effects on the distribution of conformations. These results indicate that the changes elicited by MgATP and Vi depend on ATP hydrolysis. Also, the calculated distances and sizes of the exponential components were similar to those of the T561C in the nucleotide-free and ATP-bound states, but they confirmed that no additional conformational changes took place after addition of Mg2+ and Vi.
Donor-Acceptor Distances in MsbA T561C Reconstituted in Liposomes
The studies presented above were performed on MsbA in detergent because the experiments are simpler, and MsbA is active in detergent. However, the native environment of membrane proteins is the lipid bilayer, and structural and functional differences between MsbA in solution and incorporated into a lipid bilayer are possible. The general behavior of T561C was similar in liposomes and detergent in the sense that the sensitized Bodipy FL emission increased from the apo to the ATP-bound state. The analysis of the sensitized emission decays in Table 2 shows three distances that are very similar to those found for T561C in detergent. The only noticeable difference was a small (∼3 Å) decrease in the longest distance (dissociated NBDs). However, it is apparent that the increase in intensity elicited by ATP is smaller in the liposomes (Fig. 6) and that this is partially due to an approximate doubling of the ∼36 Å component intensity in the nucleotide-free state at the expense of the longest distance component. This corresponds to a 4-fold increase in the percentage of associated NBDs in the nucleotide-free state in liposomes, from ∼9 to ∼35%. Unfortunately, the large increase in light scattering elicited by the aggregation of liposomes in the presence of Mg2+ precluded us from obtaining meaningful intensity changes between the ATP-bound and continuous hydrolysis states.
TABLE 2.
Tb3+-Bodipy FL distances in liposomes
The values are means ± S.D. (n = 7). The d1, d2, and d3 values for τDA and R were statistically different from each other under all conditions (p < 0.01). See Table I for details.
| Conformational state | τDA | R |
|---|---|---|
| μs | Å | |
| Apo | ||
| d1 | 187 ± 25 | 28.2 ± 0.7 |
| d2 | 639 ± 19 | 36.4 ± 0.3 |
| d3 | 1454 ± 44 | 49.3 ± 1.1 |
| ATP-bound | ||
| d1 | 208 ± 31 | 28.4 ± 0.8 |
| d2 | 598 ± 26 | 35.8 ± 0.4 |
| d3 | 1468 ± 85 | 49.7 ± 2.0 |
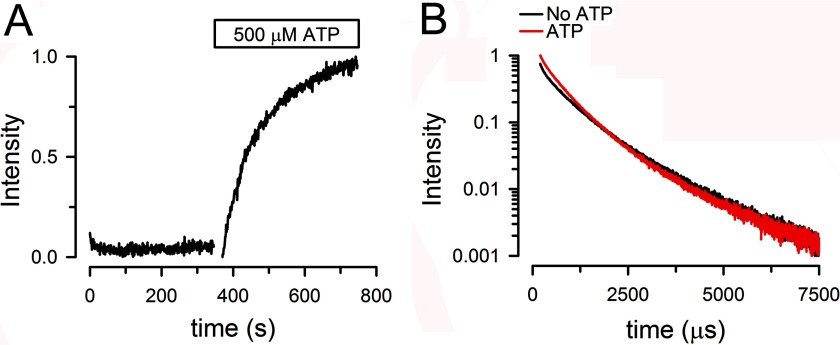
FIGURE 6.
NBD association monitored by LRET in T561C MsbA reconstituted in liposomes. A, effect of ATP. Changes in Bodipy FL-sensitized emission in response to addition of ATP. The data were normalized to the total change. B, effect of ATP on sensitized Bodipy FL emission decays. No ATP, nucleotide-free apo state (black trace); ATP, ATP-bound state (red trace) obtained 5 min after addition of ATP to 0.5 mm. The intensity was normalized to the ATP value at 200 μs. The data are representative of seven similar experiments. See legends to Figs. 4 and 5 for additional details.
Kinetics of the Conformational Changes
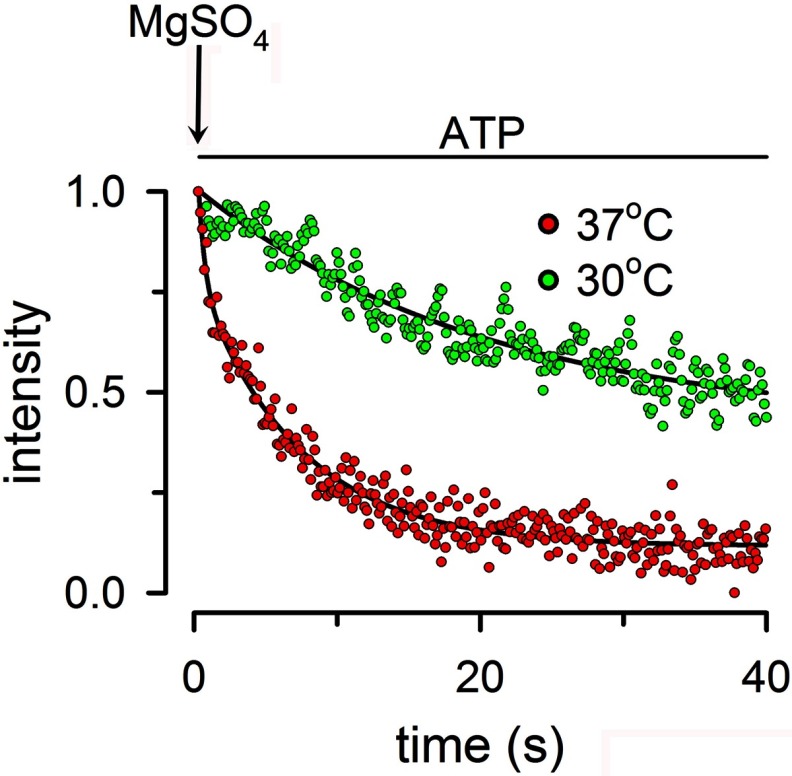
The T561C distance changes clearly indicate that significant separation occurs between the NBDs during the MsbA hydrolysis cycle. Our data and most previous distance estimates in full-length ABC proteins show significant NBD movements between conformational states (18, 19, 22). However, these data do not provide kinetic information on the transitions between states, which is essential to understand the ABC catalytic cycle. For example, detection of an ATP-bound state with associated NBDs and a posthydrolysis state with dissociated NBDs does not provide information on whether NBD dissociation occurs after hydrolysis of 1 or 100 ATPs; it just indicates that the two states exist under defined experimental conditions. To address this issue, we determined the rate of the conformational change elicited by ATP hydrolysis. For these experiments, we incubated Tb3+/Bodipy FL-labeled T561C with ATP in the absence of Mg2+ (nominally Mg2+-free solution with 1 mm EDTA) to produce the ATP-bound state. Then we mixed in a stopped flow cell the ATP-bound protein with a buffer that differed in the presence of MgSO4. Rapid mixing of the solutions initiated ATP hydrolysis, and we followed the rate of decrease in the LRET signal as a function of time. The experiments are essentially identical to the ATP to MgATP transition in Fig. 4A, but manual Mg2+ addition and mixing prevented recording of the “fast” transition in those experiments. Fig. 7 illustrates a typical experiment showing the transition from the ATP-bound to the MgATP state during continuous hydrolysis at 37 °C (red circles) and 30 °C (green circles). The initial rate of decrease in the LRET signal at 37 °C was 2.5 ± 0.2 s−1 (n = 27, five independent experiments), within a factor of 2 of the rate of ATP hydrolysis under similar conditions 1.4 ± 0.2 s−1 (n = 8, 2 independent experiments). Although we are measuring the rate of equilibration (the rate of NBD association is not zero because the “monomeric” NBD concentration becomes sizable), the initial rate of decrease in LRET is likely to be close to the NBD dissociation rate. This approximation is supported by the similarity between this equilibration rate and the hydrolysis rate, which supports a monomer/dimer mode of operation. At 30 °C, the dissociation rate was reduced to 0.053 ± 0.002 s−1 (n = 17), a more pronounced decrease than that of the rate of hydrolysis (0.43 ± 0.05 s−1). These results correspond to one NBD dissociation approximately every eight ATP hydrolysis events and suggest that at lower temperatures, MsbA operates in a predominantly constant contact mode.
FIGURE 7.
Effect of ATP hydrolysis on the rate of dissociation of NBDs. Rate of equilibration of the LRET signal after rapid mixing of ATP-bound state T561C with Mg2+ in a stopped flow cell. Tb3+/Bodipy FL-labeled T561C was incubated for at least 10 min with 0.5 mm ATP to promote full association of the NBDs. At the arrow labeled MgSO4, the ATP-bound T561C was mixed with buffer containing MgSO4 to increase [Mg2+] from 0 to 10 mm, and the rate of decrease in Bodipy FL-sensitized emission was followed at 30 or 37 °C. The black lines are exponential fits to the data. The data were normalized to the ATP-bound intensity at 37 °C. The final T561C concentration was 0.5 μm. The records are representative from 28 experiments at 37 °C and 8 experiments at 30 °C. See “Experimental Procedures” for details.
DISCUSSION
We recently showed that the ATP hydrolysis cycle of MJ0796, a well characterized thermophile NBD, follows a monomer/dimer mode of operation (25, 30). The NBDs are present as monomers in the absence of ATP and dimerize in the ATP-bound state, in the absence of hydrolysis. Triggering ATP hydrolysis results in NBD dimer dissociation at a rate that matches the rate of ATP hydrolysis (25, 30). These observations indicate that the NBD dimers dissociate after each hydrolysis cycle. There are two main reasons why the behavior of full-length ABC proteins may differ from that of isolated NBDs: 1) The high effective NBD concentration in full-length ABC proteins (caused by limitations in the volume of distribution of the NBDs) could speed-up NBD association. 2) Structural constraints from transmembrane domains/intracellular loops could affect association/dissociation rates. To address the NBD-NBD interactions during the hydrolysis cycle of the ABC protein MsbA, we determined the distances between LRET optical probes attached to the NBDs during the ATP hydrolysis cycle and also evaluated the kinetics of the distance changes in relation to the ATPase activity of the protein.
For the LRET studies, we used the rare element Tb3+ as donor, which has a long lifetime emission (24). LRET has been used to assess intersubunit and intramolecular distances in various proteins (32–36) and has many advantages for our experiments versus traditional fluorescence resonance energy transfer (24). These advantages include a very low background, high signal to noise ratio, and independence of the sensitized emission lifetime from labeling stoichiometry. These features permit the measurement of distances in the 25–100 Å range with little uncertainty because of the probes' orientation factors (24). The residue at position 561 was chosen as target for labeling because it is a surface residue exposed to the aqueous solvent (available for labeling); it is not part of the active site or dimer interface (Fig. 1A), and labeling at Cys-561 has little effect on ATPase activity. In addition, the expected Cys-561 to Cys-561 distance in the closed conformation (associated NBDs) is compatible with LRET. We primarily used the Tb3+/Bodipy FL donor-acceptor pair because its Förster distance is 41 Å, i.e., very sensitive to distance changes in the ∼30–60 Å range). Given the dependence of LRET on the sixth power of the distance between the probes, the use of this LRET pair allows separation changes of only a few angstroms to be detected and minimizes the intermolecular signal between MsbA homodimers.
Our distance estimates from LRET data indicate that the Tb3+-Bodipy FL distances are the same in the nucleotide-free, ATP-bound, and continuous hydrolysis (in MgATP) states. However, the proportion of MsbA in the closed conformation increases in the ATP-bound state, and a dynamic equilibrium between molecules in open and closed conformations occurs during hydrolysis. Two points merit notice. First, there seem to be dominant conformations in each state rather than a continuum of conformations with a broad distance distribution, as suggested from P-glycoprotein single-molecule FRET data (26). In particular, we found a single open state conformation, instead of the multiple conformations proposed from crystal structures (19). Second, although our results support the presence of large movements between the open and closed states, and the LRET distances in the closed state agree with those expected from the crystal structures (19), the longest distance was significantly shorter than that reported for the nucleotide-free MsbA. Our data suggest NBD movements of ∼20 Å between open and closed states versus up to ∼50 Å estimated from the crystal structures (19). Electron spin resonance spectroscopy data suggest a difference in separation between spin labels at position 561 of more than 28 Å between the Apo and Vi-inhibited states (22, 37). In another study, the separation distance at MsbA NBD position 539 for the same states was found to be 33 Å by electron spin resonance spectroscopy and 15 Å from fluorescein homotransfer data (18). It is presently unclear whether the discrepancy is due to differences in temperature, which has a dramatic effect on MsbA function (Fig. 2B), other experimental conditions, or methodological differences such as the size of the probes. It may be argued that longer distances could be missed in our experiments when using an LRET pair with a Förster distance of 41 Å. However, the data obtained with the Tb3+/Cy3 pair (Förster distance of 62 Å) confirmed that the longest measured distance was ∼53 Å. A large systematic underestimation of distances by LRET of tens of angstroms seems unlikely because the dominant distance in the Vi-inhibited state was 36 Å, very close to the AMPPNP-bound MsbA Cα-Cα of 37 Å.
A monomer-dimer mode of operation requires dissociation of the NBDs during the hydrolysis cycle and a close relationship between the rates of hydrolysis and NBDs dissociation. Here, we show data that clearly comply with these two premises. In constant contact models, the NBDs remain in contact during the hydrolysis cycle, and molecular dynamics simulations propose an opening at the ATP hydrolysis site of ∼6 Å (13). Because our probes are positioned away from the active sites, the changes in distance between the probes during the hydrolysis cycle should be even less during opening of one nucleotide-binding site. Our measurements show distances ∼20 Å longer in the nucleotide-free versus nucleotide-bound states, clearly indicating separation of the NBDs. However, only the comparison between the rates of hydrolysis and NBD dimer dissociation can distinguish between monomer/dimer and constant contact modes of operation. Under the more physiological conditions of 37 °C, where the protein is more active, MsbA operates in a monomer/dimer mode. However, the less active protein at 30 °C switches to a constant contact mode of operation.
Our calculated distances in detergent-solubilized MsbA and MsbA reconstituted in liposomes were essentially identical (see Tables 1 and 2). However, there is a significant increase in the proportion of associated NBDs in the nucleotide-free state of reconstituted MsbA. Further studies will be needed to address this difference.
We identified two “short” distances of ∼30 and 36 Å, compatible with associated NBDs in the closed state. One possibility that cannot be discounted without additional studies is that these two distances correspond to different conformations of the probe attached to the Cys side chains. However, it is interesting that the ratio of 36/30 Å distances increases by a factor of ∼2 in the Vi-inhibited MsbA compared with the ATP-bound state. It seems therefore possible that the ∼30 Å distance corresponds to a closed ATP-bound dimer, whereas the ∼36 Å distance arises after a conformational change that follows hydrolysis at one site. As previously discussed, the same T561C to T561C distances were found under all experimental conditions, and the differences between the nucleotide-free, ATP-bound and hydrolysis states are in the proportions of the “short” and “long” distances. This is also true for the Vi-inhibited protein, but because the residual activity in the presence of MgATP plus Vi averaged 12%, the 16% proportion of molecules showing the open conformation (longer distance) (Fig. 5E) is likely an overestimate. It seems probable that Vi locks the protein with essentially all NBDs associated, but these NBDs are present in a more open conformation than that in the ATP-bound state.
In previous studies, including crystallizations and pulse EPR spectroscopy, open and closed states were identified (18, 19, 22, 37), which, as mentioned above, differ from our studies in the magnitude of the presumed NBD movements between these states. However, our studies are the first that assessed the kinetics of the conformational changes. This information is essential to discriminate between monomer/dimer and constant contact modes of operation. Our data strongly support the monomer-dimer model for the catalytic cycle of MsbA at 37 °C. However, the protein operates in a predominantly constant contact mode at lower temperatures, and our data indicate that it is therefore essential to carefully control the experimental conditions. In summary, the behavior of full-length MsbA NBDs at 37 °C is similar to that of the isolated NBDs in detergent but deviates from that at lower temperatures. Although under “physiologic” conditions MsbA operates in a monomer/dimer mode, the results also suggest that a constant contact mode of operation is possible under certain circumstances.
Acknowledgments
We thank Dr. Ina Urbatsch for assistance with ATPase assays and Drs. Blanton, Cuello, Reuss, and Urbatsch for discussions and comments.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM79629 and 3R01GM079629-03S1. This work was also supported by Cancer Prevention & Research Institute of Texas Grant RP101073 (to G. A. A.), American Heart Association Predoctoral Fellowship 11PRE7360046 (to R. S. C.), and a scholarship from the Lubbock Chapter of the Achievement Rewards for College Scientists Foundation (to R. S. C.).
- ABC
- ATP-binding cassette
- LRET
- luminescence resonance energy transfer
- MsbA CL
- Cys-less MsbA
- NBD
- nucleotide-binding domain
- Vi
- orthovanadate
- TCEP
- Tris (2-carboxyethyl) phosphine hydrochloride.
REFERENCES
- 1. Bouige P., Laurent D., Piloyan L., Dassa E. (2002) Phylogenetic and functional classification of ATP-binding cassette (ABC) systems. Curr. Protein Pept. Sci. 3, 541–559 [DOI] [PubMed] [Google Scholar]
- 2. Al-Shawi M. K. (2011) Catalytic and transport cycles of ABC exporters. Essays Biochem. 50, 63–83 [DOI] [PubMed] [Google Scholar]
- 3. Sharom F. J. (2008) ABC multidrug transporters. Structure, function and role in chemoresistance. Pharmacogenomics 9, 105–127 [DOI] [PubMed] [Google Scholar]
- 4. Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. (2000) Structural biology of Rad50 ATPase. ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 5. Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., Hunt J. F. (2002) ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen J., Lu G., Lin J., Davidson A. L., Quiocho F. A. (2003) A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell 12, 651–661 [DOI] [PubMed] [Google Scholar]
- 7. Higgins C. F. (2007) Multiple molecular mechanisms for multidrug resistance transporters. Nature 446, 749–757 [DOI] [PubMed] [Google Scholar]
- 8. Janas E., Hofacker M., Chen M., Gompf S., van der Does C., Tampé R. (2003) The ATP hydrolysis cycle of the nucleotide-binding domain of the mitochondrial ATP-binding cassette transporter Mdl1p. J. Biol. Chem. 278, 26862–26869 [DOI] [PubMed] [Google Scholar]
- 9. Oswald C., Holland I. B., Schmitt L. (2006) The motor domains of ABC-transporters. What can structures tell us? Naunyn Schmiedebergs Arch. Pharmacol. 372, 385–399 [DOI] [PubMed] [Google Scholar]
- 10. Vergani P., Lockless S. W., Nairn A. C., Gadsby D. C. (2005) CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature 433, 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jones P. M., O'Mara M. L., George A. M. (2009) ABC transporters. A riddle wrapped in a mystery inside an enigma. Trends Biochem. Sci. 34, 520–531 [DOI] [PubMed] [Google Scholar]
- 12. Moody J. E., Millen L., Binns D., Hunt J. F., Thomas P. J. (2002) Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J. Biol. Chem. 277, 21111–21114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones P. M., George A. M. (2009) Opening of the ADP-bound active site in the ABC transporter ATPase dimer. Evidence for a constant contact, alternating sites model for the catalytic cycle. Proteins 75, 387–396 [DOI] [PubMed] [Google Scholar]
- 14. Dawson R. J., Locher K. P. (2006) Structure of a bacterial multidrug ABC transporter. Nature 443, 180–185 [DOI] [PubMed] [Google Scholar]
- 15. Jones P. M., George A. M. (2007) Nucleotide-dependent allostery within the ABC transporter ATP-binding cassette. A computational study of the MJ0796 dimer. J. Biol. Chem. 282, 22793–22803 [DOI] [PubMed] [Google Scholar]
- 16. Jones P. M., George A. M. (2011) Molecular-dynamics simulations of the ATP/apo state of a multidrug ATP-binding cassette transporter provide a structural and mechanistic basis for the asymmetric occluded state. Biophys. J. 100, 3025–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sauna Z. E., Kim I. W., Nandigama K., Kopp S., Chiba P., Ambudkar S. V. (2007) Catalytic cycle of ATP hydrolysis by P-glycoprotein. Evidence for formation of the E.S reaction intermediate with ATPγS, a nonhydrolyzable analogue of ATP. Biochemistry 46, 13787–13799 [DOI] [PubMed] [Google Scholar]
- 18. Borbat P. P., Surendhran K., Bortolus M., Zou P., Freed J. H., Mchaourab H. S. (2007) Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 5, e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ward A., Reyes C. L., Yu J., Roth C. B., Chang G. (2007) Flexibility in the ABC transporter MsbA. Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dong J., Yang G., McHaourab H. S. (2005) Structural basis of energy transduction in the transport cycle of MsbA. Science 308, 1023–1028 [DOI] [PubMed] [Google Scholar]
- 21. Westfahl K. M., Merten J. A., Buchaklian A. H., Klug C. S. (2008) Functionally important ATP binding and hydrolysis sites in Escherichia coli MsbA. Biochemistry 47, 13878–13886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zou P., Bortolus M., McHaourab H. S. (2009) Conformational cycle of the ABC transporter MsbA in liposomes. Detailed analysis using double electron-electron resonance spectroscopy. J. Mol. Biol. 393, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zou P., McHaourab H. S. (2009) Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J. Mol. Biol. 393, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Selvin P. R. (2002) Principles and biophysical applications of lanthanide-based probes. Annu. Rev. Biophys. Biomol. Struct. 31, 275–302 [DOI] [PubMed] [Google Scholar]
- 25. Zoghbi M. E., Krishnan S., Altenberg G. A. (2012) Dissociation of ATP-binding cassette nucleotide-binding domain dimers into monomers during the hydrolysis cycle. J. Biol. Chem. 287, 14994–15000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verhalen B., Ernst S., Börsch M., Wilkens S. (2012) Dynamic ligand-induced conformational rearrangements in P-glycoprotein as probed by fluorescence resonance energy transfer spectroscopy. J. Biol. Chem. 287, 1112–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bai J., Swartz D. J., Protasevich I. I., Brouillette C. G., Harrell P. M., Hildebrandt E., Gasser B., Mattanovich D., Ward A., Chang G., Urbatsch I. L. (2011) A gene optimization strategy that enhances production of fully functional P-glycoprotein in Pichia pastoris. PLoS One 6, e22577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fiori M. C., Figueroa V., Zoghbi M. E., Saéz J. C., Reuss L., Altenberg G. A. (2012) Permeation of calcium through purified connexin 26 hemichannels. J. Biol. Chem. 287, 40826–40834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Doerrler W. T., Raetz C. R. (2002) ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 [DOI] [PubMed] [Google Scholar]
- 30. Zoghbi M. E., Fuson K. L., Sutton R. B., Altenberg G. A. (2012) Kinetics of the association/dissociation cycle of an ATP-binding cassette nucleotide-binding domain. J. Biol. Chem. 287, 4157–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heyduk T., Heyduk E. (2001) Luminescence energy transfer with lanthanide chelates. Interpretation of sensitized acceptor decay amplitudes. Anal. Biochem. 289, 60–67 [DOI] [PubMed] [Google Scholar]
- 32. Cha A., Snyder G. E., Selvin P. R., Bezanilla F. (1999) Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature 402, 809–813 [DOI] [PubMed] [Google Scholar]
- 33. Knauf P. A., Pal P. (2004) Use of luminescence resonance energy transfer to measure distances in the AE1 anion exchange protein dimer. Blood Cells Mol. Dis. 32, 360–365 [DOI] [PubMed] [Google Scholar]
- 34. Posson D. J., Ge P., Miller C., Bezanilla F., Selvin P. R. (2005) Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature 436, 848–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Posson D. J., Selvin P. R. (2008) Extent of voltage sensor movement during gating of shaker K+ channels. Neuron 59, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rambhadran A., Gonzalez J., Jayaraman V. (2011) Conformational changes at the agonist binding domain of the N-methyl-d-aspartic acid receptor. J. Biol. Chem. 286, 16953–16957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zou P., McHaourab H. S. (2010) Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins. Q-band double electron-electron resonance and nanoscale bilayers. Biophys. J. 98, L18–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Urbatsch I. L., Sankaran B., Weber J., Senior A. E. (1995) P-glycoprotein is stably inhibited by vanadate-induced trapping of nucleotide at a single catalytic site. J. Biol. Chem. 270, 19383–19390 [DOI] [PubMed] [Google Scholar]
- 39. Chen J., Selvin P. R. (1999) Thiol-reactive luminescent chelates of terbium and europium. Bioconjug. Chem. 10, 311–315 [DOI] [PubMed] [Google Scholar]
- 40. Xiao M., Selvin P. R. (2001) Quantum yields of luminescent lanthanide chelates and far-red dyes measured by resonance energy transfer. J. Am. Chem. Soc. 123, 7067–7073 [DOI] [PubMed] [Google Scholar]
- 41. Huang C., Lee L. (1973) Diffusion studies of phosphatydilcholine vesicles. J. Am. Chem. Soc. 95, 234–239 [DOI] [PubMed] [Google Scholar]