Abstract
Cell division in Escherichia coli begins by assembling three proteins, FtsZ, FtsA, and ZipA, to form a proto-ring at midcell. These proteins nucleate an assembly of at least 35 components, the divisome. The structuring of FtsZ to form a ring and the processes that effect constriction have been explained by alternative but not mutually exclusive mechanisms. We discuss how FtsA and ZipA provide anchoring of the cytoplasmic FtsZ to the membrane and how a temporal sequence of alternative protein interactions may operate in the maturation and stability of the proto-ring. How the force needed for constriction is generated and how the proto-ring proteins relate to peptidoglycan synthesis remain as the main challenges for future research.
Keywords: Bacterial Genetics, Cell Division, Escherichia coli, Membrane Proteins, Protein Complexes, Proto-ring/FtsZ/FtsA/ZipA
Overview of Septation
Cell division is the last event in the bacterial cell cycle. It takes place by producing a rigid transversal septum after the growth processes fully duplicate the contents of the cell. Septation is always preceded by segregation of the duplicated nucleoids, ensuring that each of the daughter cells contains a fully replicated chromosome. Specialized molecules control the positioning of the division machinery at midcell, preventing any harmful fragmentation that septum progression could cause on unsegregated nucleoids.
The division machinery, the divisome, establishes a complex interacting network together with DNA, lipids, and peptidoglycan components. It comprises a variable number of proteins, depending on the bacterial species. These proteins have redundant multiple functions and connections serving as safeguards to complete division and to guarantee the efficiency and versatility of the process. Altogether, an efficient division process allows the proliferation of bacteria under a variety of conditions, and it is one of the reasons for their success in the environment. Free-living bacteria, such as Escherichia coli, have more elaborated divisomes, whereas those that need to grow as intracellular parasites, as such Mycoplasma genitalium, have streamlined ones (1, 2).
The structure of the E. coli divisome as revealed by immunofluorescence images of dividing cells is called the division ring (3). For its assembly, three essential proteins, FtsZ, FtsA, and ZipA, are first gathered at midcell, forming a proto-ring attached to the inner membrane (Fig. 1) (4). In the proto-ring structure, the cytoplasmic GTPase FtsZ is anchored to the membrane by its interaction with ZipA and FtsA. ZipA is a bitopic protein containing a transmembrane domain (5). FtsA is a protein of the actin family that associates with the membrane by a short amphipathic helix (6, 7). The assembly of the FtsZ ring is dependent on a continuous energy supply. The FtsZ ring requires either ZipA or FtsA (8). Although it can be formed in the presence of either of them, division is affected unless both are present (9). A maturation period takes place after the proto-ring is assembled and before the rest of the divisome proteins become incorporated (10). During maturation, the proto-ring assembly is not stable (11). Once the full divisome is assembled, the motor force, likely exerted by the polymerization of FtsZ, results in membrane constriction, followed by the production of a septum.
FIGURE 1.
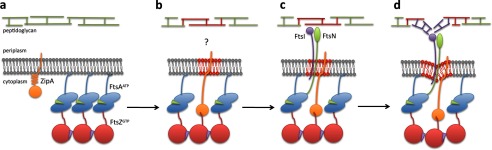
Speculative model showing the proposed stages in the assembly of the E. coli proto-ring. The model focuses on interpretations derived from currently discussed evidence. The initial stage of proto-ring assembly (a) shows FtsZ attached by its C-terminal end to the membrane-associated FtsA protein. The C-terminal ends of FtsZ oligomers (red circles) formed with GTP (purple triangles) contact the ATP-containing FtsA oligomers (green triangles within the blue ovals) bound to the membrane via its amphipathic helix. ZipA is shown in its compact conformation. Rearrangement of the proto-ring components (b) shows the disruption of the FtsA oligomer. This may involve a competition between ZipA and FtsA for the C terminus of FtsZ, in which ZipA may adopt its extended conformation. At this stage, the FtsI-independent preseptal peptidoglycan synthesis (highlighted in red) proceeds by an unknown mechanism requiring FtsZ and ZipA (or FtsA*). The interaction of the ZipA-FtsZ complex simultaneously increases the membrane fluidity (highlighted in red). After the disruption of the FtsA oligomer, its 1C domain is released from the FtsA self-interaction and is free to recruit the late proteins (c): FtsN (green oval) and FtsI (purple circle). Cell constriction and its associated synthesis of septal peptidoglycan (d) may be initiated by the pulling of the membrane. The force, transmitted by ZipA and FtsA, may result from FtsZ polymer dynamics. A transient membrane constriction may become stabilized by septal peptidoglycan (highlighted in purple) newly synthesized by FtsI.
Localization of the Division Site
Two negative regulatory systems, nucleoid occlusion and the Min system, have been described to determine the position of the division site at midcell. Their combined action inhibits polymerization of FtsZ, blocking the assembly of the proto-ring at places other than the cell center. Nucleoid occlusion blocks septation across the nucleoid, therefore excluding a potentially harmful guillotine effect on the chromosome. In E. coli, it is accomplished by SlmA, a DNA-associated division inhibitor directly involved in preventing FtsZ polymerization in the vicinity of the nucleoid (12). A dimer of SlmA binds simultaneously to two molecules of FtsZ and to specific DNA sequences distributed all over the chromosome except at the Ter region. Prior to division, Ter localizes at midcell, coinciding with the site in which the FtsZ ring is assembled, and it is the last chromosomal region to segregate (13, 14).
The E. coli Min system comprises three proteins, MinC, MinD, and MinE. Together, they inhibit the polymerization of FtsZ at the poles and therefore prevent the production of nonviable anucleated minicells. Although with a low activity, MinC is sufficient to inhibit the assembly of FtsZ polymers (15). It has been proposed that the C-terminal domain of MinC interacts directly with FtsZ filaments, preventing FtsZ-FtsZ lateral interactions and disrupting the interactions with FtsA and ZipA, which are essential for Z ring formation (15–17). MinD is an ATPase that activates and anchors MinC to the membrane. MinE regulates the localization of the MinCD complex by restricting it to the cell poles, thus allowing the FtsZ ring assembly only at midcell (for review, see Ref. 18). However, even in the absence of MinE, FtsZ rings located at the poles are more sensitive to MinCD-induced disassembly than those present at non-polar positions (19). In addition, the functionality of the Min system may be further enhanced at the poles, probably because MinCD senses membrane curvature (20, 21). The ability of the MinCDE system to modify the placement of proteins is not limited to septation, as it can also influence the localization into polar foci of proteins unrelated to cell division, such as the tryptophanase TnaA or the chaperonin GroES (22).
The Proto-ring: The Scaffold of the Divisome
FtsZ polymers need two more proto-ring components, the FtsA and ZipA proteins, both associated with the inner membrane, to localize in a ring in close proximity to it. Although nonessential for the division progress, the Zap (FtsZ-associated protein) proteins may be considered as accessory components of the proto-ring because they affect the assembly and dynamics of FtsZ.
Assembly, Organization, and Stabilization of FtsZ Polymers in the Proto-ring
The best described and phylogenetically most conserved component of the proto-ring is the cytoplasmic GTPase FtsZ, a structural homolog of eukaryotic tubulin (23–25). Highly dynamic protofilaments of FtsZ are reorganized in the vicinity of the cell membrane at the division site. Their continuous rearrangement during the cell division process is postulated to be the main force driving membrane constriction, a process that starts when the divisome is fully assembled (reviewed in Refs. 26 and 27).
Available techniques still do not allow an exact visualization of the detailed arrangement of FtsZ polymers in the proto-ring. Two models, ribbon and scattered, have been proposed to describe the arrangement of FtsZ polymers in the ring. The ribbon model proposes that FtsZ would form sufficiently long filaments to coat the internal surface of the inner membrane and assemble side-by-side as a single band. A compact ring would then be maintained by lateral contacts between filaments. The proof to support this model is that in vitro conditions favoring the elongation of and the lateral associations between protofilaments lead to the assembly of FtsZ into filaments longer than 500 nm (28). Such filaments have been observed when FtsZ is adsorbed onto mica surfaces, when calcium is present, or at low pH (<6.0) (29–31). Long FtsZ filaments are also formed under more physiological conditions in the presence of accessory proteins or crowding agents that mimic the in vivo conditions (32–34). In solution, long FtsZ protofilaments have also been found assembled as ribbons on the outer surface of artificial membrane tubules (35).
The scattered model proposes that short protofilaments of FtsZ are arranged in a disperse and divergent manner. It is supported by images obtained in vivo using high-resolution microscopy. In Bacillus subtilis and E. coli, the half-time turnover of FtsZ molecules within the division ring when estimated by fluorescence recovery after photobleaching is 9 s, supporting that short protofilaments (45–90 monomers, ∼200 nm long) are formed during the recovery time (36). Using wide-field single molecule fluorescence imaging, the depolymerization of FtsZ from Caulobacter crescentus was determined to occur on the nanosecond timescale (37). FtsZ ring images have been obtained from C. crescentus by electron cryotomography and by two- and three-dimensional super-resolution imaging and from E. coli using photoactivated localization microscopy. In both cases, they are compatible with a scattered arrangement of short filaments that accumulate within a limited space, forming a loose ring assembly (37–39).
The assembly and stabilization of FtsZ polymers in the proto-ring are regulated by the Zap proteins (ZapA, ZapB, ZapC, and ZapD), which exhibit functionally redundant roles in binding and bundling polymeric FtsZ (32, 40–46). These proteins act at midcell, promoting the transition of FtsZ polymers from a helical band into a compact ring by cooperatively stimulating the lateral association of protofilaments (47, 48). The Zap proteins bind to a conserved motif of 10–16 residues found within the C-terminal end of FtsZ (49–52). The motif mediates the binding to FtsA, ZipA, MinC, and ClpX. MinC is a negative regulator of FtsZ polymerization, and ClpX is the recognition moiety of the ClpXP protease. In consequence, a role as a central hub to integrate signals that modulate divisome assembly in E. coli has been assigned to this C-terminal end of FtsZ (53). In B. subtilis, in which Zap proteins are not present, a positively charged variable region found downstream of this conserved sequence mediates electrostatic interactions between FtsZ protofilaments. The equivalent region of the E. coli FtsZ protein is not charged, requiring the Zap proteins to stabilize FtsZ lateral interactions (54).
FtsA and ZipA: The Anchors of FtsZ Polymers to the Membrane
FtsA is an ATP-binding protein and belongs to the actin/Hsp70 superfamily (55). The self-interaction of FtsA is essential for the E. coli division process (56, 57). The ATPase activity of FtsA has not been firmly established, perhaps because its hydrolytic activity is low and an unidentified cofactor is required in vivo for its full activation. The ATP-binding activity is nevertheless associated with polymerization. FtsA polymers are formed in vitro either in solution or when attached to lipids and in vivo when overproduced (58–60). When ATP is available, the Streptococcus pneumoniae FtsA protein interacts in a head-to-tail manner, growing bidirectionally to form protofilaments until no monomers are available (59). Given the negligible ATPase activity, FtsA polymers remain as helical protofilaments, showing a tendency to form bundles. When attached to a lipid monolayer, FtsA protofilaments from Thermotoga maritima (60) and E. coli (61) undergo a conformational change, forming straight tubular structures. A short C-terminal amphipathic helix in FtsA is responsible for its attachment to the membrane (6), and it also diminishes self-interaction (56).
Gene constructs engineered in E. coli to produce FtsA-FtsA in tandem result in FtsZ polymers being retained at the cell poles after septation, suggesting that the dimerization or oligomerization of FtsA modifies the stability of FtsZ polymers and promotes Z ring integrity (62). FtsZ is proposed to be involved in FtsA dimerization given that FtsA crystallizes as a dimer when the FtsZ C-terminal peptide is bound to FtsA but as a monomer when the FtsZ peptide is absent (55, 60). These data suggest a cooperative interaction of three components, i.e. FtsZ, FtsA, and the cytoplasmic membrane, to assemble in a stable proto-ring composed of FtsZ and FtsA polymers arranged in the correct orientation at the inner membrane (Fig. 1a).
FtsA is also a key connector between the proto-ring components and some downstream proteins, such as FtsN. The 1C subdomain of FtsA is necessary and sufficient to interact with the cytoplasmic domain of FtsN (57, 63, 64), and it is also involved in the interaction between two FtsA molecules (57, 59, 60). It follows that both FtsA functions, self-interaction and recruitment of the late divisome proteins, may be mutually exclusive (65). The oligomerization state of FtsA may then have a role in recruiting the late-assembling divisome proteins.
An additional essential component of the Gammaproteobacteria proto-ring is ZipA, a transmembrane protein found in the E. coli cytoplasmic membrane as a monomer or homodimer. The monomer provides an attachment of FtsZ, its only known protein partner, to the membrane. The homodimeric form may activate the production or stability of FtsZ polymers during the assembly of the division ring (66). ZipA may provide a stronger physical link of FtsZ to the membrane compared with FtsA, as both interactions between ZipA and the membrane and between ZipA and FtsZ are stronger than the equivalent interactions provided by FtsA. The already mentioned phylogenetically conserved C-terminal peptide of FtsZ fits into a pocket located in the globular domain of ZipA and establishes a hydrophobic interaction (67), whereas the same FtsZ peptide, adopting a different conformation, interacts with the surface of the 2B domain of FtsA through a weaker electrostatic force (60). The association of FtsA with the membrane would also be weaker, as it is established through a short C-terminal amphipathic helix (6), whereas ZipA is physically anchored through a N-terminal transmembrane domain (5). The essential role of ZipA in septation can be replaced by FtsA*, a gain-of-function mutant (68, 69). However, division driven by FtsA* is not completely normal, suggesting that ZipA is relevant to some extent in the process. ZipA can protect FtsZ from degradation by ClpP, an activity that FtsA* cannot perform; and therefore, it may play some role in division in addition to the anchoring of FtsZ to the membrane (53).
Proto-ring Maturation
The proto-ring components remain assembled at midcell during part of the cell cycle (15–35% depending on the growth rate) before the recruitment of the rest of the divisome components (10). This is a surprisingly long time particularly when considering that, at fast growth rates, the initial assembly of the proto-ring that will work in the division of one cell occurs in its mother (70).
During this period, the proto-ring may be engaged in redirecting peptidoglycan synthesis to initiate transversal growth, leading to the production of the septum. This period may also involve rearrangements of the stoichiometry and/or of the interactions of the proto-ring elements between themselves or with the membrane (10). After the addition of GTP, the polymerization of FtsZ bound to a ZipA protein attached to a lipid bilayer (Fig. 1b) is not immediate and requires an incubation interval to occur (29). Polymerization under these conditions is accompanied by a change in the fluidity of the membrane (71, 72). In addition, FtsZ and ZipA, but not FtsA, are required for preseptal peptidoglycan synthesis, which is independent of FtsI (also called PBP3), an enzyme involved in the synthesis of the septum (68). Among the rest of the proteins involved in peptidoglycan synthesis and maturation, none has been shown to be essential for preseptal peptidoglycan synthesis. However, the role of ZipA in preseptal peptidoglycan synthesis in cells containing FtsA* is dispensable (68, 69). FtsZ may then be the key actor, through its indirect association with the membrane, in effecting preseptal peptidoglycan synthesis, although a molecular mechanism to describe this activity remains to be discovered (Fig. 1b).
The second event that occurs during maturation of the proto-ring is a rearrangement of the interactions of its components to allow the incorporation of the late divisome proteins. Two interactions between the proto-ring elements and the late proteins have been described: 1) FtsZ with FtsE and 2) the 1C subdomain of FtsA with the cytoplasmic domain of FtsN (57, 63, 64). As both FtsZ and FtsA are likely to be present as oligomers, a mechanism to allow their alternative interaction with their late-assembling divisome protein partners has to be invoked. On the basis of a detailed analysis of FtsA mutants that bypass the need of ZipA, Lutkenhaus and co-workers (65) have proposed a model in which the anchoring of FtsZ to the membrane is initially established through FtsA oligomers. Preliminary evidence suggests that, upon an increase in the density of its association with lipid bilayers, ZipA undergoes a transition from a compact to an extended conformation in which its globular domain can explore a wider volume in its environment (Fig. 1a) (73). ZipA may then compete for the C-terminal tail of FtsZ found inside the FtsA-FtsZ hetero-oligomers at midcell. This would produce a discontinuity in the FtsA oligomers at the sites where a ZipA molecule is inserted and at the same time would gradually replace the anchoring of FtsZ to the membrane provided by FtsA (Fig. 1b). This would then liberate the 1C domain of FtsA from self-interaction, allowing it to engage in recruiting FtsN into the mature divisome (Fig. 1c) (65). The absence of FtsN, the last protein that can be visualized forming a ring at the E. coli divisome, results in the disassembly of proto-rings, with ZipA rings being the most sensitive (11). Moreover, after FtsN deprivation, new proto-rings do not assemble until the levels of FtsN are restored, indicating that the presence of FtsN is required for proto-ring assembly (11). Interestingly, a phylogenetic correspondence between the presence of both genes zipA and ftsN has been observed in Gammaproteobacteria and has been interpreted as an indication of their concurrent role in septation (74).
Proto-ring Stabilization and Divisome Completion
Prior to the initiation of constriction, the remainder of the divisome proteins are recruited, and the proto-ring becomes stabilized into the divisome (Fig. 1d). Besides FtsN (11), a role in maintaining the integrity of the divisome has been suggested for most of the late-assembling proteins, as the number of rings (and even of proto-rings) diminishes in the absence of any of them (75).
Several essential division functions have been ascribed to either individual divisome proteins or their subcomplexes. FtsK is needed to resolve possible chromosome cointegrates resulting from replication (76, 77). FtsEX, an ATP-binding cassette transporter-like complex, acts as a regulator of cell wall hydrolysis at the division site. Upon its interaction with FtsZ, FtsE undergoes conformational changes mediated by ATP hydrolysis to modify the transmembrane FtsX component. The modification favors the recruitment of the amidase activator EnvC by FtsX in the periplasm (78, 79). The small bitopic membrane proteins FtsQ, FtsB, and FtsL form an independent subcomplex able to establish abundant interactions with other divisome proteins (80–82). In the absence of additional evidence, it is possible that they may have either a regulatory or a structural role. FtsW belongs to the SEDS (shape, elongation, division, and sporulation) family of transporters involved in the translocation of lipid-linked peptidoglycan precursors across the cytoplasmic membrane (83). Together with FtsI, its linked class B penicillin-binding protein, FtsW forms a subcomplex specific for septal peptidoglycan synthesis (Fig. 1, c and d) (84). FtsI has a topology similar to FtsN: they contain a transmembrane region, followed by a long linker spanning the periplasm and a C-terminal globular domain contacting the peptidoglycan layer. FtsI and FtsN interact with nonessential divisome proteins that participate in other late septation processes, i.e. with PBP1B (85) or MtgA (86) for peptidoglycan synthesis, with the Tol-Pal complex for the invagination of the cell wall during constriction (87, 88), and with the AmiB and AmiC amidases to effect the hydrolysis of peptidoglycan needed to split the wall of the two daughter cells (89).
Membrane Constriction and Septum Progression
Once the divisome is completely assembled, all of the layers of the cell envelope are connected, and the constriction process (involving all of them) begins. As no contractile elements have been found outside the cell membrane, the constriction force seems to be exerted from the cytoplasm by pulling the envelope inward (Fig. 1d). The available evidence suggests that the main motor exerting the force required for constriction is FtsZ itself.
Two mechanisms have been proposed to describe how FtsZ may exert a pulling force. They involve either bending or condensation of FtsZ polymers and are not mutually exclusive. In the bending model, the motor force is exclusively exerted by FtsZ in a cycle of polymerization, membrane attachment, conformational change, depolymerization, and nucleotide exchange, all driven by GTP hydrolysis (38). Osawa et al. (90) found that a membrane-targeted FtsZ chimera, when self-internalized in flattened tubular liposomes, can assemble into constricted areas that relax as GTP is hydrolyzed. This observation suggests that the bending detected in the FtsZ-rich regions of the liposomes is not stable and that additional components of the septum, such as peptidoglycan, are necessary to produce a stable structure as constriction progresses. In this model, the hydrolysis of GTP would be involved in modifying the curvature of FtsZ polymers (91, 92).
In the condensation model, the constriction force would be generated by the lateral attraction between different FtsZ filaments that would interact to form a compact structure (93). Results from in silico modeling suggest that the FtsZ ring may undergo a transition from a low- to a high-density state by condensation of protofilaments and not by recruiting more FtsZ monomers. In this model, GTP hydrolysis would accommodate a gradual morphological change from a loose helix to a compact FtsZ ring by facilitating monomer turnover during condensation (94, 95). Increasing the number of lateral contacts between protofilaments would compress them, causing a shift from a loose spiral extending along the cell length to a compact ring located at midcell. The FtsZ ring would further condense across the cell diameter to form a thicker annular structure, with its diameter decreasing as constriction progresses. Zap proteins, such as ZapA or ZapB, have been suggested to assist in this mechanism (96). At the end of constriction, the condensed narrow ring quickly disassembles, and the septum closes (39). It is not known if, upon their disassembly, the components of the divisome are degraded or if they can be partially recycled.
Additional Questions
Our knowledge of many aspects of cell division, as summarized in this minireview, provides a detailed account of the components of the divisome, including a description of many of their interactions and their spatial and temporal assembly within the cell. However, only a partial picture of their functions is available. In particular, a description of the detailed molecular mechanisms involved in septation and, moreover, the connection between the biochemical properties of each divisome component and its role in bacterial proliferation has to be completed. A relevant question is how the divisome affects the membrane and modifies the direction of wall growth at the inception of septation. How the divisome components are disposed after the closure of the septum is another question that will also need additional research. Techniques such as high-resolution microscopy (97) and in vitro reconstruction procedures (98) are now available to investigate these questions. The answers will clarify both how the process of septation is initiated and how it finishes. Finally, under ideal growth conditions, E. coli will assemble a divisome to constrict the envelope and, after 20 min, will divide and start again.
Acknowledgments
We thank Marisela Vélez and Paolo Natale for critical reading of this manuscript.
Footnotes
This work was supported by Spanish Ministerio de Educación y Ciencia Grant BIO2011-28941-C03-01, European Union Grant FP7-223431 (DIVINOCELL), and Human Frontier Science Program Grant RGP0050/2010 (all to M. V.).
REFERENCES
- 1. Glass J. I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M. R., Maruf M., Hutchison C. A., 3rd, Smith H. O., Venter J. C. (2006) Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 103, 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Delaye L., Moya A. (2010) Evolution of reduced prokaryotic genomes and the minimal cell concept: variations on a theme. BioEssays 32, 281–287 [DOI] [PubMed] [Google Scholar]
- 3. Vicente M., Rico A. I., Martínez-Arteaga R., Mingorance J. (2006) Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 188, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vicente M., Rico A. I. (2006) The order of the ring: assembly of Escherichia coli cell division components. Mol. Microbiol. 61, 5–8 [DOI] [PubMed] [Google Scholar]
- 5. Hale C. A., de Boer P. A. (1997) Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185 [DOI] [PubMed] [Google Scholar]
- 6. Pichoff S., Lutkenhaus J. (2005) Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55, 1722–1734 [DOI] [PubMed] [Google Scholar]
- 7. Jiménez M., Martos A., Vicente M., Rivas G. (2011) Reconstitution and organization of Escherichia coli proto-ring elements (FtsZ and FtsA) inside giant unilamellar vesicles obtained from bacterial inner membranes. J. Biol. Chem. 286, 11236–11241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rueda S., Vicente M., Mingorance J. (2003) Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 185, 3344–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pichoff S., Lutkenhaus J. (2002) Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21, 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aarsman M. E., Piette A., Fraipont C., Vinkenvleugel T. M., Nguyen-Distèche M., den Blaauwen T. (2005) Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55, 1631–1645 [DOI] [PubMed] [Google Scholar]
- 11. Rico A. I., García-Ovalle M., Palacios P., Casanova M., Vicente M. (2010) Role of Escherichia coli FtsN protein in the assembly and stability of the cell division ring. Mol. Microbiol. 76, 760–771 [DOI] [PubMed] [Google Scholar]
- 12. Bernhardt T. G., de Boer P. A. (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tonthat N. K., Arold S. T., Pickering B. F., Van Dyke M. W., Liang S., Lu Y., Beuria T. K., Margolin W., Schumacher M. A. (2011) Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 30, 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cho H., McManus H. R., Dove S. L., Bernhardt T. G. (2011) Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc. Natl. Acad. Sci. U.S.A. 108, 3773–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shiomi D., Margolin W. (2007) The C-terminal domain of MinC inhibits assembly of the Z ring in Escherichia coli. J. Bacteriol. 189, 236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dajkovic A., Lan G., Sun S. X., Wirtz D., Lutkenhaus J. (2008) MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr. Biol. 18, 235–244 [DOI] [PubMed] [Google Scholar]
- 17. Shen B., Lutkenhaus J. (2010) Examination of the interaction between FtsZ and MinCN in E. coli suggests how MinC disrupts Z rings. Mol. Microbiol. 75, 1285–1298 [DOI] [PubMed] [Google Scholar]
- 18. Bramkamp M., van Baarle S. (2009) Division site selection in rod-shaped bacteria. Curr. Opin. Microbiol. 12, 683–688 [DOI] [PubMed] [Google Scholar]
- 19. Shen B., Lutkenhaus J. (2011) Differences in MinC/MinD sensitivity between polar and internal Z rings in Escherichia coli. J. Bacteriol. 193, 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Juarez J. R., Margolin W. (2010) Changes in the Min oscillation pattern before and after cell birth. J. Bacteriol. 192, 4134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Männik J., Wu F., Hol F. J., Bisicchia P., Sherratt D. J., Keymer J. E., Dekker C. (2012) Robustness and accuracy of cell division in Escherichia coli in diverse cell shapes. Proc. Natl. Acad. Sci. U.S.A. 109, 6957–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li G., Young K. D. (2012) Isolation and identification of new inner membrane-associated proteins that localize to cell poles in Escherichia coli. Mol. Microbiol. 84, 276–295 [DOI] [PubMed] [Google Scholar]
- 23. Erickson H. P. (1995) FtsZ, a prokaryotic homologue of tubulin? Cell 80, 367–370 [DOI] [PubMed] [Google Scholar]
- 24. Erickson H. P. (1997) FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol. 7, 362–367 [DOI] [PubMed] [Google Scholar]
- 25. Löwe J., Amos L. A. (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206 [DOI] [PubMed] [Google Scholar]
- 26. Erickson H. P., Anderson D. E., Osawa M. (2010) FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mingorance J., Rivas G., Vélez M., Gómez-Puertas P., Vicente M. (2010) Strong FtsZ is with the force: mechanisms to constrict bacteria. Trends Microbiol. 18, 348–356 [DOI] [PubMed] [Google Scholar]
- 28. Mingorance J., Tadros M., Vicente M., González J. M., Rivas G., Vélez M. (2005) Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J. Biol. Chem. 280, 20909–20914 [DOI] [PubMed] [Google Scholar]
- 29. Mateos-Gil P., Márquez I., López-Navajas P., Jiménez M., Vicente M., Mingorance J., Rivas G., Vélez M. (2012) FtsZ polymers bound to lipid bilayers through ZipA form dynamic two dimensional networks. Biochim. Biophys. Acta 1818, 806–813 [DOI] [PubMed] [Google Scholar]
- 30. Mukherjee A., Lutkenhaus J. (1999) Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J. Bacteriol. 181, 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mendieta J., Rico A. I., López-Viñas E., Vicente M., Mingorance J., Gómez-Puertas P. (2009) Structural and functional model for ionic (K+/Na+) and pH dependence of GTPase activity and polymerization of FtsZ, the prokaryotic ortholog of tubulin. J. Mol. Biol. 390, 17–25 [DOI] [PubMed] [Google Scholar]
- 32. Gueiros-Filho F. J., Losick R. (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16, 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Popp D., Iwasa M., Narita A., Erickson H. P., Maéda Y. (2009) FtsZ condensates: an in vitro electron microscopy study. Biopolymers. 91, 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gündoğdu M. E., Kawai Y., Pavlendova N., Ogasawara N., Errington J., Scheffers D. J., Hamoen L. W. (2011) Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 30, 617–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Milam S. L., Osawa M., Erickson H. P. (2012) Negative-stain electron microscopy of inside-out FtsZ rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys J. 103, 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stricker J., Maddox P., Salmon E. D., Erickson H. P. (2002) Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. U.S.A. 99, 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biteen J. S., Goley E. D., Shapiro L., Moerner W. E. (2012) Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. ChemPhysChem 13, 1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Z., Trimble M. J., Brun Y. V., Jensen G. J. (2007) The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fu G., Huang T., Buss J., Coltharp C., Hensel Z., Xiao J. (2010) In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS ONE 5, e12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Durand-Heredia J., Rivkin E., Fan G., Morales J., Janakiraman A. (2012) Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194, 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hale C. A., Shiomi D., Liu B., Bernhardt T. G., Margolin W., Niki H., de Boer P. A. (2011) Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 193, 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Durand-Heredia J. M., Yu H. H., De Carlo S., Lesser C. F., Janakiraman A. (2011) Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193, 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ebersbach G., Galli E., Møller-Jensen J., Löwe J., Gerdes K. (2008) Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol. Microbiol. 68, 720–735 [DOI] [PubMed] [Google Scholar]
- 44. Galli E., Gerdes K. (2010) Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol. Microbiol. 76, 1514–1526 [DOI] [PubMed] [Google Scholar]
- 45. Low H. H., Moncrieffe M. C., Löwe J. (2004) The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 341, 839–852 [DOI] [PubMed] [Google Scholar]
- 46. Dajkovic A., Pichoff S., Lutkenhaus J., Wirtz D. (2010) Cross-linking FtsZ polymers into coherent Z rings. Mol. Microbiol. 78, 651–668 [DOI] [PubMed] [Google Scholar]
- 47. Monahan L. G., Robinson A., Harry E. J. (2009) Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol. Microbiol. 74, 1004–1017 [DOI] [PubMed] [Google Scholar]
- 48. Mohammadi T., Ploeger G. E., Verheul J., Comvalius A. D., Martos A., Alfonso C., van Marle J., Rivas G., den Blaauwen T. (2009) The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry. 48, 11056–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shen B., Lutkenhaus J. (2009) The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinCC/MinD. Mol. Microbiol. 72, 410–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haney S. A., Glasfeld E., Hale C., Keeney D., He Z., de Boer P. (2001) Genetic analysis of the E. coli FtsZ·ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276, 11980–11987 [DOI] [PubMed] [Google Scholar]
- 51. Camberg J. L., Hoskins J. R., Wickner S. (2009) ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc. Natl. Acad. Sci. U.S.A. 106, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scheffers D. J. (2008) The effect of MinC on FtsZ polymerization is pH dependent and can be counteracted by ZapA. FEBS Lett. 582, 2601–2608 [DOI] [PubMed] [Google Scholar]
- 53. Pazos M., Natale P., Vicente M. (2013) A specific role for the ZipA protein in cell division: stabilization of the FtsZ protein. J. Biol. Chem. 288, 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buske P. J., Levin P. A. (2012) Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287, 10945–10957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van den Ent F., Löwe J. (2000) Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19, 5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yim L., Vandenbussche G., Mingorance J., Rueda S., Casanova M., Ruysschaert J. M., Vicente M. (2000) Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J. Bacteriol. 182, 6366–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rico A. I., García-Ovalle M., Mingorance J., Vicente M. (2004) Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol. Microbiol. 53, 1359–1371 [DOI] [PubMed] [Google Scholar]
- 58. Lara B., Rico A. I., Petruzzelli S., Santona A., Dumas J., Biton J., Vicente M., Mingorance J., Massidda O. (2005) Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol. Microbiol. 55, 699–711 [DOI] [PubMed] [Google Scholar]
- 59. Krupka M., Rivas G., Rico A. I., Vicente M. (2012) Key role of two terminal domains in the bidirectional polymerization of FtsA protein. J. Biol. Chem. 287, 7756–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Szwedziak P., Wang Q., Freund S. M., Löwe J. (2012) FtsA forms actin-like protofilaments. EMBO J. 31, 2249–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martos A., Monterroso B., Zorrilla S., Reija B., Alfonso C., Mingorance J., Rivas G., Jiménez M. (2012) Isolation, characterization and lipid-binding properties of the recalcitrant FtsA division protein from Escherichia coli. PLoS ONE 7, e39829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shiomi D., Margolin W. (2007) Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol. Microbiol. 66, 1396–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Corbin B. D., Geissler B., Sadasivam M., Margolin W. (2004) Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J. Bacteriol. 186, 7736–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Busiek K. K., Eraso J. M., Wang Y., Margolin W. (2012) The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J. Bacteriol. 194, 1989–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pichoff S., Shen B., Sullivan B., Lutkenhaus J. (2012) FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol. Microbiol. 83, 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Skoog K., Daley D. O. (2012) The Escherichia coli cell division protein ZipA forms homodimers prior to association with FtsZ. Biochemistry 51, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 67. Mosyak L., Zhang Y., Glasfeld E., Haney S., Stahl M., Seehra J., Somers W. S. (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19, 3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Potluri L. P., Kannan S., Young K. D. (2012) ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J. Bacteriol. 194, 5334–5342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Geissler B., Elraheb D., Margolin W. (2003) A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 100, 4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Helmstetter C., Cooper S., Pierucci O., Revelas E. (1968) On the bacterial life sequence. Cold Spring Harb. Symp. Quant. Biol. 33, 809–822 [DOI] [PubMed] [Google Scholar]
- 71. López-Montero I., Mateos-Gil P., Sferrazza M., Navajas P. L., Rivas G., Vélez M., Monroy F. (2012) Active membrane viscoelasticity by the bacterial FtsZ-division protein. Langmuir 28, 4744–4753 [DOI] [PubMed] [Google Scholar]
- 72. López-Montero I., López-Navajas P., Mingorance J., Vélez M., Vicente M., Monroy F. (2013) Membrane reconstitution of FtsZ-ZipA complex inside giant spherical vesicles made of E. coli lipids: large membrane dilation and analysis of membrane plasticity. Biochimica et biophysica acta. 1828, 687–698 [DOI] [PubMed] [Google Scholar]
- 73. López-Montero I., López-Navajas P., Mingorance J., Rivas G., Vélez M., Vicente M., Monroy F. (2013) Intrinsic disorder of the bacterial cell division protein ZipA: coil-to-brush conformational transition. FASEB J. 10.1096/fj.12-224337 [DOI] [PubMed] [Google Scholar]
- 74. Mingorance J., Rico A. I., Gómez-Puertas P. (2004) Bacterial morphogenes. in Molecules in Time and Space: Bacterial Shape, Division and Phylogeny (Vicente M., Tamames J., Valencia A., Mingorance J., eds) pp. 173–194, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 75. Goehring N. W., Gonzalez M. D., Beckwith J. (2006) Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 61, 33–45 [DOI] [PubMed] [Google Scholar]
- 76. Dubarry N., Barre F. X. (2010) Fully efficient chromosome dimer resolution in Escherichia coli cells lacking the integral membrane domain of FtsK. EMBO J. 29, 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dubarry N., Possoz C., Barre F. X. (2010) Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol. Microbiol. 78, 1088–1100 [DOI] [PubMed] [Google Scholar]
- 78. Corbin B. D., Wang Y., Beuria T. K., Margolin W. (2007) Interaction between cell division proteins FtsE and FtsZ. J. Bacteriol. 189, 3026–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang D. C., Peters N. T., Parzych K. R., Uehara T., Markovski M., Bernhardt T. G. (2011) An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl. Acad. Sci. U.S.A. 108, E1052–E1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gonzalez M. D., Akbay E. A., Boyd D., Beckwith J. (2010) Multiple interaction domains in FtsL, a protein component of the widely conserved bacterial FtsLBQ cell division complex. J. Bacteriol. 192, 2757–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gonzalez M. D., Beckwith J. (2009) Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. J. Bacteriol. 191, 2815–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. van den Ent F., Vinkenvleugel T. M., Ind A., West P., Veprintsev D., Nanninga N., den Blaauwen T., Löwe J. (2008) Structural and mutational analysis of the cell division protein FtsQ. Mol. Microbiol. 68, 110–123 [DOI] [PubMed] [Google Scholar]
- 83. Mohammadi T., van Dam V., Sijbrandi R., Vernet T., Zapun A., Bouhss A., Diepeveen-de Bruin M., Nguyen-Distèche M., de Kruijff B., Breukink E. (2011) Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30, 1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fraipont C., Alexeeva S., Wolf B., van der Ploeg R., Schloesser M., den Blaauwen T., Nguyen-Distèche M. (2011) The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–259 [DOI] [PubMed] [Google Scholar]
- 85. Bertsche U., Kast T., Wolf B., Fraipont C., Aarsman M. E., Kannenberg K., von Rechenberg M., Nguyen-Distèche M., den Blaauwen T., Höltje J. V., Vollmer W. (2006) Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 61, 675–690 [DOI] [PubMed] [Google Scholar]
- 86. Derouaux A., Wolf B., Fraipont C., Breukink E., Nguyen-Distèche M., Terrak M. (2008) The monofunctional glycosyltransferase of Escherichia coli localizes to the cell division site and interacts with penicillin-binding protein 3, FtsW, and FtsN. J. Bacteriol. 190, 1831–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gerding M. A., Ogata Y., Pecora N. D., Niki H., de Boer P. A. (2007) The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 63, 1008–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bernard C. S., Sadasivam M., Shiomi D., Margolin W. (2007) An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 64, 1289–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Peters N. T., Dinh T., Bernhardt T. G. (2011) A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J. Bacteriol. 193, 4973–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Osawa M., Anderson D. E., Erickson H. P. (2008) Reconstitution of contractile FtsZ rings in liposomes. Science 320, 792–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lu C., Reedy M., Erickson H. P. (2000) Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J. Bacteriol. 182, 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Díaz J. F., Kralicek A., Mingorance J., Palacios J. M., Vicente M., Andreu J. M. (2001) Activation of cell division protein FtsZ. Control of switch loop T3 conformation by the nucleotide γ-phosphate. J. Biol. Chem. 276, 17307–17315 [DOI] [PubMed] [Google Scholar]
- 93. Lan G., Daniels B. R., Dobrowsky T. M., Wirtz D., Sun S. X. (2009) Condensation of FtsZ filaments can drive bacterial cell division. Proc. Natl. Acad. Sci. U.S.A. 106, 121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ghosh B., Sain A. (2008) Origin of contractile force during cell division of bacteria. Phys. Rev. Lett. 101, 178101. [DOI] [PubMed] [Google Scholar]
- 95. Ghosh B., Sain A. (2011) Force generation in bacteria without nucleotide-dependent bending of cytoskeletal filaments. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 83, 051924 [DOI] [PubMed] [Google Scholar]
- 96. Small E., Marrington R., Rodger A., Scott D. J., Sloan K., Roper D., Dafforn T. R., Addinall S. G. (2007) FtsZ polymer-bundling by the Escherichia coli ZapA orthologue, YgfE, involves a conformational change in bound GTP. J. Mol. Biol. 369, 210–221 [DOI] [PubMed] [Google Scholar]
- 97. Coltharp C., Xiao J. (2012) Superresolution microscopy for microbiology. Cell. Microbiol. 14, 1808–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Martos A., Jiménez M., Rivas G., Schwille P. (2012) Towards a bottom-up reconstitution of bacterial cell division. Trends Cell Biol. 22, 634–643 [DOI] [PubMed] [Google Scholar]


