Background: Two CDK subunits of the Mediator complex play pivotal roles in transcription by a mechanism that has not yet been elucidated.
Results: The histone arginine methyltransferase PRMT5 is a Mediator CDK-interacting protein.
Conclusion: Mediator-associated PRMT5 symmetrically dimethylates histone H4 arginine 3, and this might cause transcriptional repression.
Significance: This work enables further exploration of Mediator functions in transcriptional repression.
Keywords: CDK (Cyclin-dependent Kinase), DNA Methyltransferase, Phorbol Esters, RNA Polymerase II, siRNA, Transcription Regulation
Abstract
The Mediator complex (Mediator) plays pivotal roles in activating transcription by RNA polymerase II, but relatively little is known about its roles in repression. Here, we identified the histone arginine methyltransferase PRMT5 and WD repeat protein 77/methylosome protein 50 (WDR77/MEP50) as Mediator cyclin-dependent kinase (CDK)-interacting proteins and studied the roles of PRMT5 in the transcriptional regulation of CCAAT enhancer-binding protein (C/EBP) β target genes. First, we purified CDK8- and CDK19-containing complexes from HeLa nuclear extracts and subjected these purified complexes to mass spectrometric analyses. These experiments revealed that two Mediator CDKs, CDK8 and CDK19, individually interact with PRMT5 and WDR77, and their interactions with PRMT5 cause transcriptional repression of C/EBPβ target genes by regulating symmetric dimethylation of histone H4 arginine 3 (H4R3me2s) in the promoter regions of those genes. Furthermore, the recruitment of the DNA methyltransferase DNMT3A correlated with H4R3 dimethylation potentially leading to DNA methylation at the promoter proximal region and tight inhibition of preinitiation complex formation. In vertebrates, C/EBPβ regulates many genes involved in immune responses and cell differentiation. These findings shed light on the molecular mechanisms of the repressive roles of Mediator CDKs in transcription of C/EBPβ target genes and might provide clues that enable future studies of the functional associations between Mediators and epigenetic regulation.
Introduction
In eukaryotes, the expression of protein-coding genes is elaborately regulated primarily at the level of transcription by RNA polymerase II (Pol II)2 in collaboration with chromatin regulation (1–3). Certain cofactors, including the Mediator complex (Mediator), are involved not only in direct regulatory roles during transcription but also in epigenetic regulatory events such as histone modifications and chromatin remodeling.
Mediator was first identified by genetics in the budding yeast Saccharomyces cerevisiae. Several yeast proteins, originally named SRBs (suppressors of RNA polymerase B), were subsequently found to be the Mediator subunits. Mutations in the SRB proteins suppress the defects caused by deletion of the C-terminal domain heptapeptide repeats of the largest subunit of Pol II (4). Subsequent studies revealed that Mediator relieves squelching (activator-induced inhibition of transcription) during in vitro transcription in crude nuclear extracts (5). Yeast Mediator was purified by Kornberg and colleagues (6). Mediator has also been identified in and purified from mammals, in which it connects nuclear hormone receptors with the transcription machinery (7–11).
A recently proposed unified nomenclature for all Mediator subunits includes 34 MED proteins (MED1–MED31, MED1L, MED12L, and MED13L) as well as two cyclin-dependent kinase (CDK) proteins (CDK8 and CDK19) and their common counterpart cyclin C (12, 13). CDK8 is a component of a CDK/cyclin submodule, functions as a serine/threonine kinase, and is required for various developmental events, but not for cell survival, in metazoa (14, 15). According to the current consensus, CDK8 generally functions as a negative regulatory component of Mediator (16, 17). However, recent studies have shown that CDK8 also plays a positive role in transcriptional regulation (18, 19). We observed that at least two Mediator subcomplexes contain human CDK8 yet exert opposite effects on transcriptional activation. Thus, it is clear that CDK8 plays multiple roles in transcriptional regulation (20–23). Another kinase subunit of the human Mediator complex, CDK19 (formerly CDK11), was recently identified using multidimensional protein identification technology (MudPIT) (24). Human CDK19 shares a high degree of amino acid sequence identity with CDK8. In previous studies, we demonstrated that CDK19 forms a CDK8-independent Mediator complex (25). Furthermore, we observed that CDK19 is expressed in a tissue-specific manner, whereas CDK8 is ubiquitously expressed (26). DNA microarray analysis of the target genes of each CDK complex revealed extensive overlap in their target gene preferences (26). Therefore, we decided to explore the idea that Mediators play a pivotal role in transcriptional regulation (19, 25).
To gain insight into the molecular mechanisms of transcriptional repression by CDK8 and/or CDK19, we treated HeLa cells with phorbol 12-myristate 13-acetate (PMA) and examined the effects on C/EBPβ target genes. The transcriptional activator C/EBPβ is a regulator of acute phase responses such as innate and adaptive immunity, senescence, and receptor tyrosine kinase/Ras-mediated tumorigenesis (27–35). The transcription activities of C/EBPβ are controlled both by protein-protein interactions with transcriptional cofactors and by post-transcriptional modifications (31, 32). Phosphorylation of C/EBPβ triggers a conformational change in this protein that correlates with Mediator subtype exchange (27). This finding indicates that there are differences between the Mediator subtypes with respect to transcriptional regulation; however, the functional role of each Mediator subtype during transcriptional regulation remains unclear.
In addition to performing functional studies, we attempted to isolate CDK8- and/or CDK19-interacting proteins from HeLa cells. We identified novel functional interactions between the two CDK subunits and the histone arginine methyltransferase PRMT5 and its functionally interacting partner, WD repeat protein 77 (WDR77, also called methylosome protein 50, MEP50). Both CDK-containing Mediator complexes contained PRMT5 and WDR77, and they exhibited histone H4-specific arginine methyltransferase activity in vitro. Our results demonstrate that CDK8 and CDK19 recruit PRMT5 to the C/EBPβ-inducible gene promoters and that PRMT5 represses their transcription through symmetric dimethylation of histone H4 arginine 3 (H4R3me2s). The accumulated evidence supports a model in which the Mediators themselves regulate chromatin structure (36–38). In addition, H4R3me2s and PRMT5 itself interact with DNA methyltransferase 3α (DNMT3A) to repress gene transcription (39). Finally, we show that DNMT3A is recruited to the C/EBPβ-inducible gene promoters in a manner that depends on the CDKs. Based on these results, we propose a model in which Mediators participate in epigenetic regulation in a process that is essential for transcriptional regulation.
EXPERIMENTAL PROCEDURES
Cell Lines and siRNA-mediated CDK Knockdown
Cell lines stably expressing HA/FLAG-tagged (HF) CDK8 or CDK19 stably expressing cell lines were established previously (25). Normal HeLa S3 and CDK-expressing cells were maintained in DMEM supplemented with 5% calf serum at 37 °C in 5% CO2. For PMA induction, we treated HeLa S3 cells with 25 ng/ml PMA for 2 h at 37 °C. To deplete endogenous CDK8 and/or CDK19, we used synthetic siRNA pools targeting each gene. Nontarget control siRNA (D-001810-10-05; Dharmacon) and siRNAs against CDK8 (L-003242-00-0005; Dharmacon) and CDK19 (L-004689-00-0005; Dharmacon) were transfected at a concentration of 5 nm by using the Lipofectamine RNAi MAX transfection reagent (Invitrogen). After transfection, the cells were cultured for 72 h and then subjected to ChIP or RT-qPCR analysis.
Antibodies
The antibodies used in this study were as follows: anti-human C/EBPβ rabbit polyclonal (sc-150; Santa Cruz Biotechnology), anti-human CDK8/19 goat polyclonal (sc-1521, Santa Cruz Biotechnology), anti-human CDK8 mouse monoclonal (552053; BD Biosciences), anti-human CDK19 rabbit polyclonal (HPA007053; Sigma-Aldrich), anti-phosphorylated human Pol II C-terminal domain mouse monoclonal (4H8 ab5408; Abcam), anti-HA mouse monoclonal (12CA5 1583816; Roche), anti-human MED17 rabbit polyclonal (H00009440-M02; Abnova), anti-human PRMT5 rabbit polyclonal (07-405; Millipore), anti-human WDR77 rabbit polyclonal (ab57722; Abcam), anti-S-tag mouse monoclonal (71549-3; Novagen), anti-H4R3me2s rabbit polyclonal (ab5823; Abcam), anti-human DNMT3A rabbit polyclonal (sc-20703; Santa Cruz Biotechnology), anti-human MED6 rabbit polyclonal (sc-9434,; Santa Cruz), anti-human MED12 rabbit polyclonal (ab70842; Abcam), anti-human MED15 rabbit polyclonal (11566-1-AP; Protein tech. Group), anti-H4 rabbit polyclonal (ab7311; Abcam), and anti-γ-tubulin mouse monoclonal (sc-17788; Santa Cruz Biotechnology).
Chromatin Immunoprecipitation and RT-qPCR
HeLa S3 cells grown in 15-cm dishes to 80–90% confluence were fixed in 1% formaldehyde for 10 min at room temperature. The cross-linking reaction was stopped by addition of 125 mm glycine, and the cells were incubated for 5 min at room temperature and washed with PBS. Fixed cells were collected in a 1.5-ml tube by scraping, and then 300 μl of lysis buffer (50 mm Tris·HCl, pH 8.1, 10 mm EDTA, 1% SDS) was added to each tube. The cells were completely disrupted by chromatin shearing using a Bioruptor sonication device (UCD-250; Cosmo Bio) at medium intensity for 30 cycles (30 s on, 90 s off). Cell lysates were centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was diluted 10-fold with dilution buffer (16.7 mm Tris·HCl, pH 8.1, 167 mm NaCl, 1.2 mm EDTA, 1.1% Triton X-100) and then incubated overnight at 4 °C with 2 μg of the indicated antibody. Fifty microliters of protein G Dynabeads were suspended in Dynabeads blocking buffer (10 mm Tris·HCl, pH 7.5, 1 mm EDTA, 1 mg/ml BSA, 0.4 mg/ml salmon sperm DNA) and incubated overnight at 4 °C. The next day the beads were washed with 1 ml of low salt buffer (20 mm Tris·HCl, pH 8.1, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.1% SDS), 1 ml of high salt buffer (20 mm Tris·HCl, pH 8.1, 500 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.1% SDS), 1 ml of LiCl buffer (10 mm Tris·HCl, pH 8.1, 250 mm LiCl, 1 mm EDTA, 1% Nonidet P-40, 1% sodium deoxycholate), and 1 ml of TEN buffer (16 mm Tris·HCl, pH 7.5, 1 mm EDTA, 0.5% Nonidet P-40). After washing, immunocomplexes were eluted by incubation with 100 μl of elution buffer (1% SDS, 100 mm NaHCO3) for 1 h at room temperature, and the eluates were collected. Cross-links were reversed by overnight incubation at 65 °C. Next, the eluate was treated with RNase A for 1 h at 37 °C, followed by treatment with proteinase K for 2 h at 37 °C. For qPCR, DNA templates were purified using the QIAquick PCR purification kit (Qiagen), and the purified DNA was quantified using SYBR® Premix Ex TaqTM II (TaKaRa) on an Mx3000P QPCR system (Stratagene). 350 ng of total RNA purified from siRNA-transfected cells using the Nucleo Spin RNA II kit (TaKaRa) was subjected to reverse transcription using PrimeScriptTM RT Master Mix (TaKaRa). Synthesized cDNA was measured using SYBR® Premix Ex TaqTM II (TaKaRa) on an Mx3000P QPCR system (Stratagene).
Primer Sets for qPCR
The following primer sets were used in ChIP analyses: IL-8 forward, 5′-AAGAAACCACCGGAAGGAAC-3′; IL-8 reverse, 5′-ACTGCACCTTCACACAGAGC-3′; TNFα forward, 5′-GGCAGTCAGATCATCTTCTCG-3′; TNFα reverse, 5′-CAGCTTGAGGGTTTGCTACA-3′; β-actin forward, 5′-TGGCACCCAGCACAATGAA-3′; and β-actin reverse, 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′. The following primer sets were used in qPCR analyses: IL-8 promoter forward, 5′-CCATCAGTTGCAAATCGTGGA-3′; IL-8 promoter reverse, 5′-GAAGCTTGTGTGCTCTGCTG-3′; IL-8 exon forward, 5′-GATACTCCCAGTCTTGTCATTGC-3′; IL-8 exon reverse, 5′-AAACAAGTTTCAACCAGCAAGAA-3′; TNFα promoter forward, 5′-TACCGCTTCCTCCAGATGAG-3′; TNFα promoter reverse, 5′-AATCATTCAACCAGCGGAAA-3′; TNFα exon forward, 5′-GACCACCACTTCGAAACCTG-3′; and TNFα exon reverse, 5′-GTGGTTGCCAGCACTTCAC-3′.
Purification of HF-CDK8- and HF-CDK19-containing Mediator Complexes
Nuclear extracts were prepared from each tagged CDK-expressing HeLa S3 cell line. Nuclear extracts were equilibrated with Buffer C(0.3) (BC(0.3)) (300 mm KCl, 20 mm HEPES·KOH, pH 7.9, 20% (v/v) glycerol, 0.5 mm EDTA, 50 mm PMSF, 2 μg/ml antipain, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 0.8 μg/ml pepstatin, 0.5 mm EGTA, 10 mm 2-mercaptoethanol) containing 0.05% (v/v) Nonidet P-40. The equilibrated nuclear extracts were fractionated on an HPLC TSK-gel G4000SW XL gel filtration column (7.8-mm inner diameter × 30 cm; Tosoh). Fractions eluted at a molecular mass of ∼2 MDa were CDK-containing Mediator and were then incubated with 40 μl of BC(0.3)-equilibrated anti-FLAG M2-agarose resin or 40 μl of IgG-agarose resin (Sigma-Aldrich) for 4 h at 4 °C. After four washes with 1 ml of BC(0.3) containing 0.05% Nonidet P-40, the bound proteins were eluted by incubation for 30 min at 4 °C with 100 μl of BC(0.3) containing 0.05% Nonidet P-40 and 0.3 mg/ml FLAG peptide. The purified Mediator fractions were used for Western blotting and in vitro histone methyltransferase assays.
Preparation of CDK8- and CDK19-containing Mediator-associating Proteins
Nuclear extracts prepared as described above were equilibrated with BC(0.3) containing 0.05% (v/v) Nonidet P-40. The equilibrated nuclear extracts (200 mg of protein) were incubated with 100 μl of BC(0.3) equilibrated IgG-Sepharose (GE Healthcare) for 1 h at 4 °C to remove nonspecific proteins. Each supernatant was incubated with 200 μl of anti-FLAG M2-agarose (Sigma-Aldrich) for 4 h at 4 °C. After washing five times with 10 ml of BC(0.3) containing 0.05% Nonidet P-40, bound proteins were eluted from beads by incubation for 30 min at 4 °C with 500 μl of BC(0.3) containing 0.05% Nonidet P-40 and 0.3 mg/ml FLAG peptide. The eluates were incubated for 30 min at 4 °C with 100 μl of protein G-agarose (Sigma-Aldrich) to adsorb the released IgG. The resulting supernatants were collected and used for LC-MS/MS analyses.
Affinity Pulldown Analyses
The human PRMT5 cDNA (FLJ90770AAAN) and human WDR77 cDNA (FLJ12798AAAN) were obtained from NBRC (NITE Biological Resource Center, Kisarazu, Japan). To generate plasmids compliant with the Gateway system, we converted the pTriEX-2 protein-expression plasmid (Millipore) into a Gateway-compliant plasmid using the Gateway® vector conversion system (Invitrogen). The cDNAs were subcloned into the converted pTriEX-2 vector using the Gateway system to yield constructs suitable for expression of S-tagged PRMT5 (S-PRMT5) or WDR77 (S-WDR77) in HeLa S3 cells and bacteria. Purified HF-CDK8, HF-CDK19, S-PRMT5, or S-WDR77 expression plasmid (3 μg) was transfected into HeLa S3 cells in a 10-cm dish using the LipofectamineTM LTX reagent (Invitrogen). After a 24-h incubation, the cells were harvested by scraping and then lysed in BC(0.3) containing 0.05% Nonidet P-40 by sonication (UR-21P; TOMY) for 10 s. Cell lysates were incubated with 20 μl of S protein-agarose (Novagen) for 6 h at 4 °C. After four washes with 1 ml of BC(0.3) containing 0.05% Nonidet P-40, precipitated proteins were subjected to Western blotting.
GST Pulldown Analyses
Bacterially expressed GST-tagged CDK8 or CDK19 (500 ng) was tethered on a glutathione-Sepharose resin (GE Healthcare) and incubated for 2 h at 4 °C with 200 ng of bacterially expressed S-PRMT5 or S-WDR77 in 500 μl of BC(0.1) (100 mm KCl, 20 mm HEPES·KOH, pH 7.9, 20% (v/v) glycerol, 0.5 mm EDTA, 50 mm PMSF, 2 μg/ml antipain, 2 μg/ml aprotinin, 1 μg/ml leupeptin, 0.8 μg/ml pepstatin, 0.5 mm EGTA, and 10 mm 2-mercaptoethanol) containing 0.05% Nonidet P-40. For S-PRMT5, precipitates were washed three times with 1 ml of BC(0.1) containing 0.05% Nonidet P-40, and for S-WDR77, precipitates were washed three times with 1 ml of BC(0.3) containing 0.05% Nonidet P-40. Finally, bound proteins were analyzed by Western blotting using an anti-S-tag antibody.
In Vitro Histone Methyltransferase Assay
The HF-CDK8- or CDK19-containing Mediator fraction was incubated for 20 min at 30 °C with 25 ng each of bacterially expressed recombinant histones H2A, H2B, H3, and H4 (NEB) in histone methyltransferase reaction buffer (50 mm Tris·HCl, pH 8.5, 5 mm MgCl2, 4 mm DTT) in the presence of 1 mm S-(5′-adenosyl)-l-methionine (Sigma-Aldrich). Methylated histones were loaded onto an 18% acrylamide gel, electrophoresed, and subjected to Western blotting using anti-H4R3me2s and anti-histone H4 antibodies.
For fluorography analyses, equimolar amounts of purified complex and histones were incubated with 40 kBq of [3H]S-(5′-adenosyl)-l-methionine for 1 h at 30 °C. Methylated histones were separated by 18% polyacrylamide gel electrophoresis. Protein bands were stained with Coomassie Brilliant Blue, and the gels were equilibrated in water and then incubated with Enlight fluorographic enhancer (EnerGene) for 1 h at room temperature. After drying, the gels were exposed to x-ray film (Fuji film) for 2 weeks at −80 °C.
Microarray Analyses of Gene Expression Results after CDK8 and CDK19 Double Knockdown
HeLa S3 cells were seeded on 6-well plates at 4 × 104 cells/well in 2 ml of antibiotic-free DMEM 24 h before transfection. siRNA at a final concentration of 20 nm was transfected using Lipofectamine 2000 (Invitrogen). After culturing for 48 h, transfection medium was changed to fresh antibiotic-containing medium. Transfected cells were incubated for additional 12 h, and then RNA was prepared using RNeasy columns (Qiagen). Purified RNA (5 μg) was subjected to gene expression analysis using Human Genome U133 Plus 2.0 (Affymetrix) arrays followed by characterizations using the GeneSpring (Agilent) and Pathway Analysis (Ingenuity) software. In this analysis, we employed two individual siRNA against each CDK (CDK8 and CDK19) and three replicates of each condition. The entire set of microarray data has been deposited in the public database Gene Expression Omnibus, and the assigned accession number is GSE45210.
RESULTS
CDK8 and CDK19 Dissociate from C/EBPβ Target Gene Promoters upon Activation
Mediator complexes have been classified into at least two distinct subtypes, the large canonical holo-Mediator complex (L-Mediator; for simplicity, henceforth designated as Mediator) and the small core Mediator complex (S-Mediator). Mediator characteristically contains four submodules: head, middle, tail, and CDK/cyclin. S-Mediator uniquely possesses the additional subunit MED26 but lacks the CDK/cyclin submodule (40, 41). Previously, based on in vitro transcription studies, Mediator was thought to act as a negative transcriptional regulator, and S-Mediator was thought to act as a positive regulator (11). However, recent studies demonstrated positive transcriptional regulatory properties of Mediator, suggesting that it may require further classification into additional subtypes (3, 19, 23). The molecular mechanisms of Mediator's activities in transcriptional repression remain elusive.
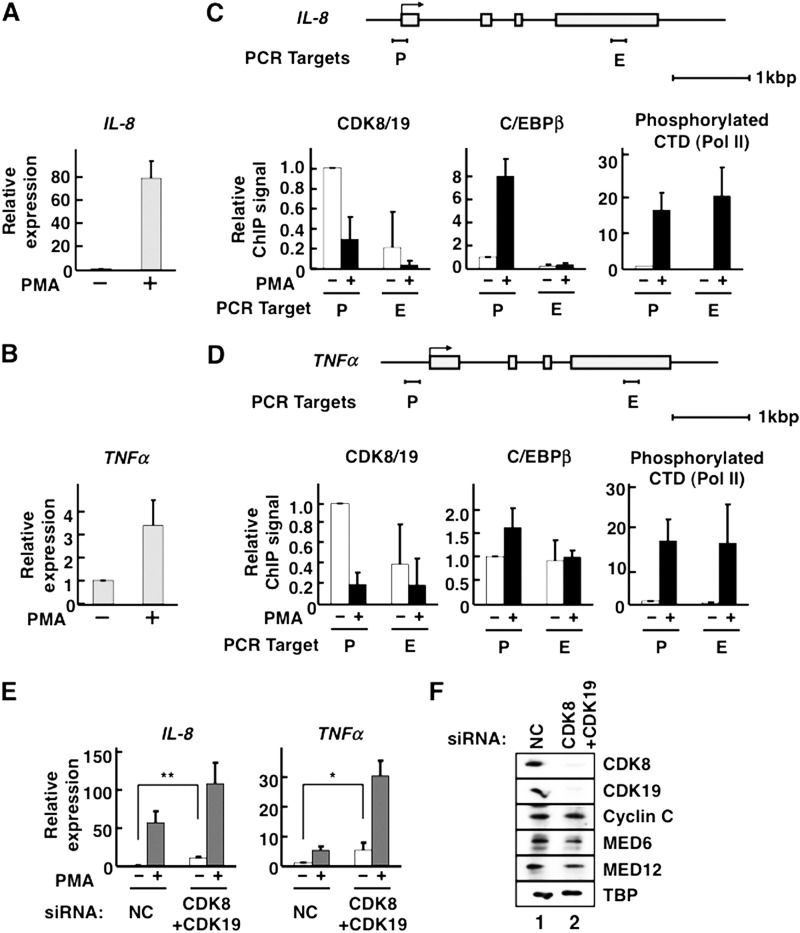
To study the mechanisms of transcriptional repression, we focused on Mediator's key enzymatic components, the CDK8 and CDK19 subunits. Previous reports demonstrated that CDK8 is released from C/EBPβ target gene promoter regions upon activation by C/EBPβ (27, 42), but the mechanism of CDK8 transcriptional repression is still unclear. To investigate the repressive functions of the two CDKs, we used C/EBPβ-inducible transcription in the HeLa S3 cell line as our model system and focused on the C/EBPβ target genes IL-8 and TNFα, as well as IL-1β and IL-12A (43, 44), as repression targets of CDK8 and CDK19. We activated C/EBPβ by adding the phorbol ester, PMA. PMA first activates PKC, which activates the MAPK signaling pathway; ultimately, activated MAPK phosphorylates C/EBPβ, and this phosphorylation leads to activation of C/EBPβ transcriptional activity in NIH3T3 and U937 cells (35). PMA treatment triggered transcription of the C/EBPβ target genes IL-8 and TNFα (Fig. 1, A and B). We also performed ChIP assays to analyze the localizations of C/EBPβ, CDK8, and CDK19 on the promoter regions of the target genes (Fig. 1, C and D). In these studies, we detected both CDK8 and CDK19 proteins with an anti-CDK8 antibody because this antibody cross-reacts with CDK19 in addition to CDK8 (26); therefore, the ChIP signals from this antibody are designated as CDK8/19 hereafter. Consistent with previous reports (35), PMA treatment facilitated C/EBPβ recruitment to the promoter regions of its target genes and triggered Pol II transcriptional activation (C-terminal domain phosphorylation) (Fig. 1, C and D). At each promoter, CDK8/19 signals were decreased by PMA treatment. This finding indicates that the dissociation of the Mediator CDKs (CDK8/19) from the activated promoters was caused by C/EBPβ-dependent transcriptional activation. CDK8 and CDK19 also dissociate from several other genes targeted by various transcriptional activators (27, 45, 46).
FIGURE 1.
Mediator CDKs repress C/EBPβ-inducible transcription. A–D, HeLa S3 cells were treated with 25 ng/ml of PMA for 2 h at 37 °C. A and B, qPCR for the measurement of IL-8 (A) or TNFα (B) mRNA (n = 2 in each case). C and D, occupancies of CDK8/19, C/EBPβ, and phosphorylated Pol II C-terminal domain (CTD) on IL-8 (C) and TNFα (D) genes. Schematic gene structures are displayed above the corresponding panels. Gray boxes indicate exons. P, promoter region; E, exonic region. The amount of each bound protein was measured by ChIP followed by qPCR. ChIP was individually carried out three times (n = 3). The data were normalized by defining the signal level without PMA treatment as 1. E, expression levels of IL-8 and TNFα genes were measured by qPCRs in CDK8/CDK19 double-knockdown cells treated with 25 ng/ml PMA for 2 h (n = 4). The data were normalized by defining the signal level without PMA treatment in the presence of nontarget control (NC) siRNA as 1. F, the effects of siRNA knockdown on protein expression of Mediator CDKs. After siRNA treatment, cell lysate proteins were separated by SDS-PAGE and transferred to PVDF membranes. Mediator subunits and the general transcription factor TBP (as a control) were detected by Western blotting using antibodies against CDK8, CDK19, cyclin C, MED6, MED12, and TBP, as indicated on the right side. In all panels, the error bars show S.D. *, p < 0.05; **, p < 0.01.
Correlation between Dissociation of CDKs and Transcriptional Activation
The observation of the repressor-like actions of CDK8 and CDK19 led us to hypothesize that these proteins repress C/EBPβ target gene transcription regardless of nuclear signaling (Fig. 1, C and D). To further investigate the relationships between transcriptional repression and CDK8/19 dissociation in HeLa S3 cells, we knocked down both CDKs in HeLa S3 cells using siRNAs (Fig. 1F) and measured IL-8 and TNFα mRNA levels by qPCR. The levels of these mRNAs were elevated by CDK8/19 double knockdown in the absence of PMA (Fig. 1E, white bars). Additionally, knockdown of Mediator CDKs increased the PMA-inducible transcription of those two genes (Fig. 1E, gray bars). Thus, dissociation of CDK8 and CDK19 from promoters is well correlated with transcriptional activation.
PRMT5 and WDR77 Interact with Both CDK8 and CDK19
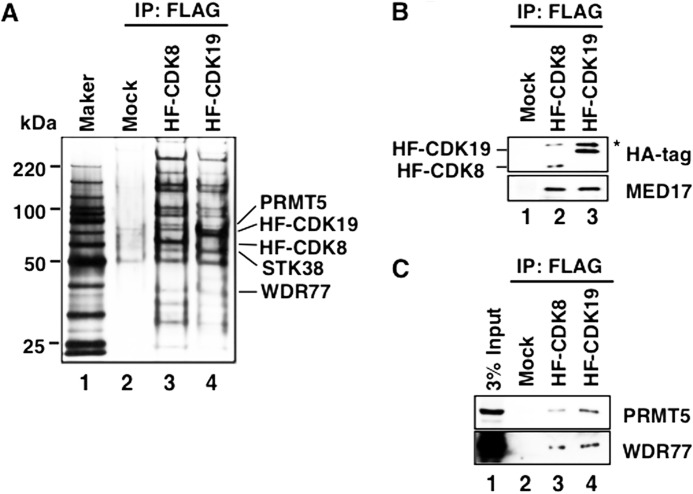
To identify CDK8- and/or CDK19-interacting proteins, we prepared large amounts of CDK8- and CDK19-containing Mediator complexes from HeLa nuclear extracts. These nuclear extracts were prepared from large cultures of previously prepared HA/FLAG-tagged CDK8 (HF-CDK8)- and CDK19 (HF-CDK19)-expressing stable cell lines previously established by our group (19, 25). Each CDK-containing Mediator complex was purified using anti-FLAG M2 agarose beads (Fig. 2, A and B). The silver-stained gels of each CDK8- or CDK19-containing Mediator fraction contained unknown bands that could not be considered Mediator subunits based on their molecular masses (Fig. 2A). We cut those bands from the gels and subjected them to LC-MS/MS analysis, resulting in the identification of the arginine methyltransferase PRMT5 and the WD repeat protein 77 (WDR77, also called MEP50) as candidates for Mediator CDK-interacting proteins. PRMT5 is a type II arginine methyltransferase that symmetrically catalyzes the addition of two methyl groups to two arginine guanidino-nitrogen atoms, i.e., the symmetric dimethylation of arginine residues (47). WDR77 is an important cofactor for PRMT5 catalytic activity (48). To validate the LC-MS/MS findings, we performed Western blotting on HF-CDK8- and HF-CDK19-containing Mediator complexes from individually prepared nuclear extracts, and we reproducibly detected these specific interactions in other purifications (Fig. 2C).
FIGURE 2.
The two CDK-containing Mediator complexes interact with PRMT5 and WDR77 in HeLa nuclear extracts. A, HF-CDK8- and HF-CDK19-containing Mediator complexes were purified from HeLa nuclear extracts expressing either HF-CDK8 or HF-CDK19, using an anti-FLAG M2 agarose column. Purified proteins were subjected to SDS-PAGE (4–15% gradient gel, Bio-Rad), and the gel was stained with silver. Lane 1, protein standard marker; lane 2, mock-purified fraction (Mock); lane 3, HF-CDK8-containing Mediator fraction (HF-CDK8); lane 4, HF-CDK19-containing Mediator fraction (HF-CDK19). B, Western blotting of affinity-purified HF-CDK8- and HF-CDK19-containing Mediator complexes. Each complex was detected using anti-HA and anti-MED17 antibodies. Lane 1, mock-purified fraction (Mock); lane 2, HF-CDK8-containing Mediator fraction (HF-CDK8); lane 3, HF-CDK19-containing Mediator fraction (HF-CDK19). C, the interactions of PRMT5 and WDR77 with HF-CDK8- and HF-CDK19-containing Mediator complexes were detected using anti-PRMT5 and anti-WDR77 antibodies, respectively. Lane 1, 3% of input HeLa nuclear extract; lane 2, mock-purified fraction (Mock); lane 3, HF-CDK8-containing Mediator fraction (HF-CDK8); lane 4, HF-CDK19-containing Mediator fraction (HF-CDK19). These complexes were purified from the nuclear extracts used in A. IP, immunoprecipitation.
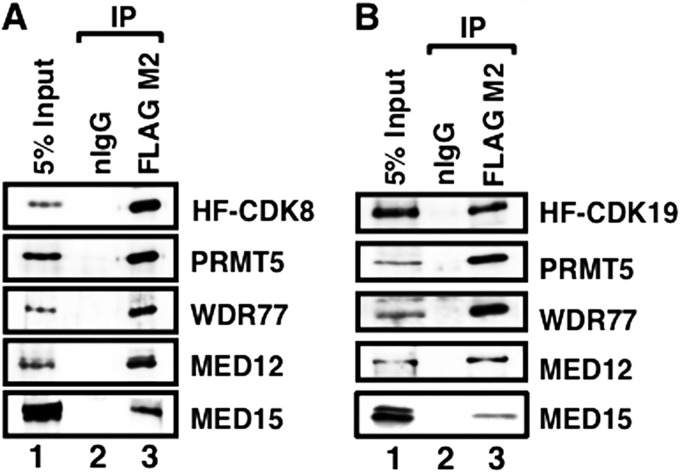
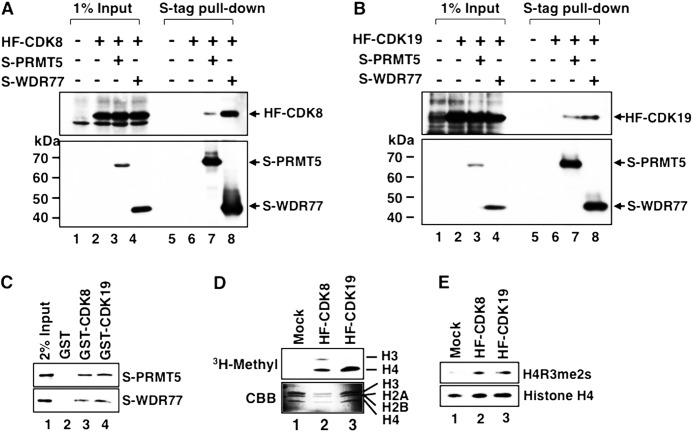
To further confirm the interactions between CDK8- or CDK19-containing intact Mediator and the two proteins we identified, we purified CDK8- and CDK19-containing Mediator complexes by HPLC gel filtration and anti-FLAG M2 agarose chromatography. In the eluate fractions of both CDK8- and CDK19-containing Mediators, we detected PRMT5 and WDR77 along with other Mediator subunits (MED12 and MED15) (Fig. 3, A and B), indicating that PRMT5 and WDR77 are associated with the Mediator complexes. Next, to determine whether PRMT5 and/or WDR77 bind directly to the two Mediator CDKs, we coexpressed S-tag-fused PRMT5 (S-PRMT5) or WDR77 (S-WDR77) with HF-CDK8 (Fig. 4A) or HF-CDK19 (Fig. 4B) in HeLa S3 cells. Both S-tagged proteins were precipitated with S-protein-agarose affinity beads. Ectopically expressed CDK8 and CDK19 were coprecipitated only when S-PRMT5 or S-WDR77 was simultaneously expressed. In addition, we showed that bacterially expressed S-PRMT5 or S-WDR77 bound to bacterially expressed GST-CDK8 and GST-CDK19 (Fig. 4C). These results indicate that PRMT5 and WDR77 bind independently and directly to both CDK8 and CDK19.
FIGURE 3.
PRMT5 and WDR77 interact with purified Mediator complexes. A and B, Western blotting of two-step purified HF-CDK8-containing (A) or HF-CDK19-containing (B) Mediator complex. Nuclear extracts from cells stably expressing HF-CDK8 were separated on an HPLC-TSK G4000SW XL gel filtration column. The 2-MDa fractions were further purified using anti-FLAG M2 agarose column or normal mouse IgG-conjugated agarose column. Lane 1, 5% input; lane 2, normal mouse IgG eluate (nIgG); lane 3, M2-agarose eluate (FLAG M2). Each protein indicated on the right side was detected with the corresponding antibody. IP, immunoprecipitation.
FIGURE 4.
Binding of PRMT5 and WDR77 to Mediator CDKs and detection of histone H4 arginine 3 symmetric dimethylation (H4R3me2s) activity in CDK-containing Mediator complexes. A and B, S-tagged PRMT5 (S-PRMT5) or WDR77 (S-WDR77) expression vector was cotransfected with the HF-CDK8 (A) or HF-CDK19 (B) expression vector into HeLa S3 cells. Expressed S-proteins were precipitated using anti-S-tag agarose and subjected to Western blotting. Lanes 1–4, 1% input of each cell lysate; lanes 5–8, S-tag pulldown fraction of each cell lysate. Arrowheads indicate each tagged protein. C, bacterially expressed GST-CDK8 or GST-CDK19 was immobilized on glutathione-Sepharose gel and incubated with each purified S-protein. After extensive washing, the retained proteins were detected by Western blotting. Lane 1, 2% input of S-PRMT5 or S-WDR77; lane 2, S-proteins bound to GST alone; lane 3, S-proteins bound to GST-CDK8; and lane 4, S-proteins bound to GST-CDK19. D, in vitro methyltransferase assay of anti-FLAG purified HF-CDK8 or HF-CDK19 complex. The purified complex was incubated with four purified bacterially expressed recombinant histone proteins in the presence of S-adenosyl-l-methyl-[3H]methionine. Fluorography (upper panel) and Coomassie Brilliant Blue (CBB)-stained histone proteins (lower panel) are shown. Lane 1, mock-purified fraction; lane 2, HF-CDK8-containing Mediator fraction; lane 3, HF-CDK19-containing Mediator fraction. E, in vitro methyltransferase assay using S-adenosylmethionine. Methylated histones were detected using anti-H4R3me2s and anti-histone H4 specific antibodies. Lanes 1–3 are as in D.
CDK8- and CDK19-containing Mediator Complexes Possess Histone Methyltransferase Activity
PRMT5 plays roles both in the nucleus and in the cytoplasm and has various substrates, including histones, cytoplasmic signal transducers, spliceosomal proteins, transcription factors, and PIWI proteins (49). In the context of transcriptional regulation, histones are particularly well known targets of PRMT5-directed methylation (39, 50, 51). The association of PRMT5 with both CDK8 and CDK19 indicates that this histone-modifying enzyme has a close relationship with Mediator (Fig. 4, A and B). To validate the functional role of PRMT5, we assayed the methyltransferase activities of purified HF-CDK8 and HF-CDK19-containing Mediator complexes and demonstrated that both complexes have histone H4-specific methyltransferase activity (Fig. 4D, lanes 2 and 3). The type II arginine methyltransferase PRMT5 and a group of type I arginine methyltransferases that catalyze the asymmetric dimethylation of arginine residues share common recognition sequences, including histone arginine residues. Symmetric dimethylation of H4R3 (H4R3me2s) is a known target of PRMT5 that has been linked to transcriptional repression, whereas asymmetric dimethylation of H4R3 (H4R3me2a), which is catalyzed by PRMT1 and PRMT6, is associated with transcriptional activation (52). To verify the specificity of the Mediator-associated methyltransferase activity for histone H4, we used an H4R3me2s-specific antibody (Abcam) to show that both CDK8- and CDK19-containing Mediator complexes could catalyze the H4R3me2s modification (Fig. 4E).
CDK8 and CDK19 Recruit PRMT5 to C/EBPβ Target Gene Promoters
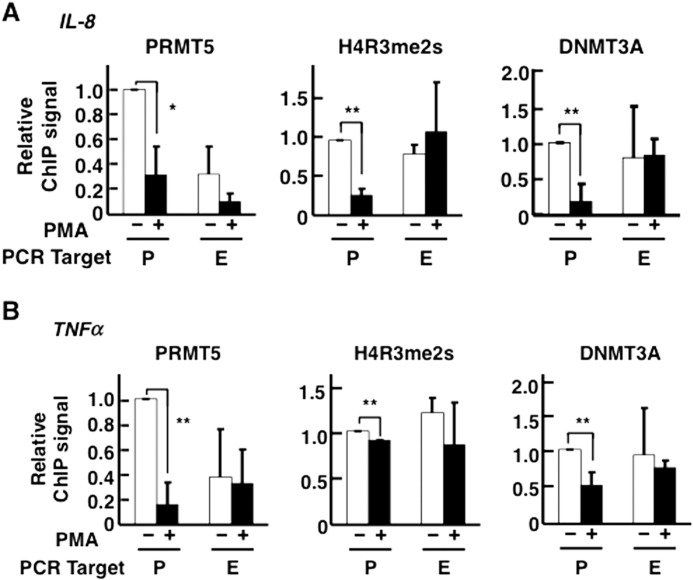
A recent genome-wide study demonstrated that H4R3me2s modification is enriched at transcriptionally repressed regions in the chromosomes of CD4+ T-cells (53). In addition, PRMT5 recruits DNA (cytosine-5)-methyltransferase 3α (DNMT3A), both directly and indirectly, to target regions where it represses expression of downstream genes (39). Because we suspected that the functional interactions between Mediator CDKs and histone modification enzymes, discussed in the previous section, are involved in Mediator CDK-dependent transcriptional repression, we analyzed the occupancy of C/EBPβ target gene promoters by PRMT5, using ChIP assays (Figs. 5 and 6). These assays revealed that PRMT5 was relatively enriched on C/EBPβ target gene promoter regions when these promoters were inactivated; furthermore, PMA treatment triggered PRMT5 dissociation from these promoter regions (Fig. 5, A and B, left panels). These dissociations seemed to correlate with the dissociation of CDK8/19 from these regions (Fig. 1, C and D). To study the consequences of PRMT5 dissociation, we determined the modification state and occupancies of these regions by antibodies against H4R3me2s and DNMT3A, respectively (Fig. 5, A and B, middle and right panels). The H4R3me2s modification signals were detected in both promoter and protein-coding regions (Fig. 5, A and B, middle panels). These signals, especially at the promoter regions, were decreased by PMA treatment. In addition, at the promoter regions, the DNMT3A signals were reduced by PMA treatment, correlating with the H4R3me2s signals at the promoters (Fig. 5, A and B, right panels). The DNMT3A signals were also detected in the protein-coding regions, again correlating with the H4R3me2s signals. We observed a significant correlation among the changes in the signal intensities of PRMT5, H4R3me2s, and DNMT3A on C/EBPβ target gene promoters after PMA treatment.
FIGURE 5.
Localization of PRMT5, H4R3me2s modification, and DNMT3A on C/EBPβ target genes. Occupancies on the promoter (P) and exon (E) regions of IL-8 (A) and TNFα (B) genes were measured by ChIP followed by qPCR. PMA treatment was performed at 25 ng/ml for 2 h. Immunoprecipitations were carried out individually three times (n = 3). The data were normalized by defining the signal level in the absence of PMA treatment as 1. The error bars show S.D. *, p < 0.05; **, p < 0.01.
FIGURE 6.
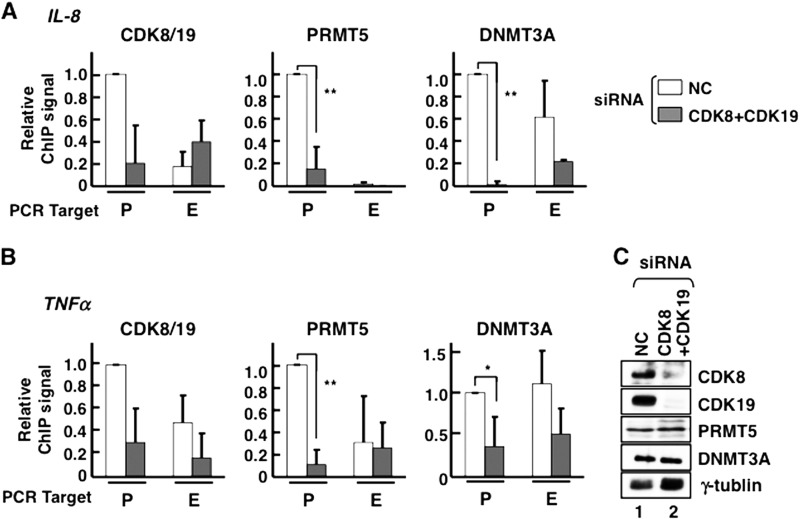
Double knockdown of CDK8 and CDK19 caused PRMT5 and DNMT3A dissociation from C/EBPβ target gene promoters. A and B, occupancies of CDK8/19, PRMT5, and DNMT3A on IL-8 (A) and TNFα (B) genes. To knock down protein expression of Mediator CDKs, an siRNA mixture targeting both CDK8 and CDK19 (CDK8+CDK19) or NC siRNA was transfected into HeLa S3 cells, and occupancies were measured by ChIP followed by qPCR. P, promoter regions; E, exonic regions. Immunoprecipitations were carried out individually three times (n = 3). The data were normalized by defining the signal level in the presence of NC siRNA as 1. The error bars show S.D. *, p < 0.05; **, p < 0.01. C, the efficiency of the CDK8 and CDK19 double knockdown was verified by Western blotting. Lane 1, nontarget control (NC); lane 2, siRNA mixture against both CDK8 and CDK19 (CDK8+CDK19). γ-Tubulin was used as a loading control.
As shown above, PRMT5 dissociation from the C/EBPβ target gene promoters was highly correlated with the eviction of CDK8/19 from the promoter regions upon PMA-induced transcriptional activation. These observations suggest that both CDK8 and CDK19 recruit PRMT5 to the C/EBPβ target gene promoter regions. We depleted CDK8 and CDK19 expression by transfecting HeLa S3 cells with an siRNA mixture against both CDK8 and CDK19 and then measured the occupancy of target gene promoters by each protein using ChIP (Fig. 6). Depletion of both CDK8 and CDK19 triggered a clear reduction of the CDK8/19 signal in C/EBPβ target gene promoter regions. CDK8 and CDK19 double depletion had no effect on PRMT5 and DNMT3A expression levels (Fig. 6C), but the PRMT5 and DNMT3A signals clearly decreased on C/EBPβ target gene promoters (Fig. 6, A and B).
DISCUSSION
How Mediators act both positively and negatively in transcriptional regulation has been a long standing question (3). In this study, we investigated the mechanisms of transcriptional repression by large Mediator complexes, which contain the CDK/cyclin submodule. Mediators play key roles in the regulation of Pol II transcriptional activation by controlling recruitment of the general transcription machinery to promoters (3). Compared with the functional relationships between Mediators and the general transcriptional machinery, the relationships between Mediators and the histone modification factors are less well understood. A previous study demonstrated that MED12, one of the largest Mediator subunits within the CDK/cyclin submodule, interacts with the repressive histone methyltransferase G9a and represses neuronal gene expression (54); this repressive mechanism is representative of the regulation of histone modification machinery by Mediators. We found in HeLa S3 cells that two CDKs, CDK8 and CDK19, each of which can serve as a subunit of canonical holo-Mediator, interact directly with the histone arginine methyltransferase PRMT5 and its cofactor WDR77 to repress transcription.
Dissociation of Mediator CDKs from C/EBPβ Target Genes Changes the Transcriptional Status from Repression to Activation
We used the phorbol ester PMA transcriptional induction system to study the pivotal role of Mediator CDKs in transcription. Upon PMA treatment of the HeLa S3 cell line, we observed C/EBPβ-dependent transcriptional activation (Fig. 1, A–D). Mediator CDKs, on the other hand, dissociated from the promoter regions (Fig. 1, C and D). When both CDKs were knocked down, transcriptional activation was further increased (Fig. 1E). Our observations clearly indicate that CDK8 and/or CDK19 act as repressors of the C/EBPβ target genes in the absence of MAPK signaling and that these CDKs play key roles in the transition from transcriptional repression to activation.
PRMT5 and WDR77 Are Mediator CDK-interacting Proteins
To determine which proteins cause Mediator CDK-dependent transcriptional repression, we tried to identify CDK8- and/or CDK19-interacting proteins from purified CDK-containing Mediator complexes (Fig. 2A). We identified two proteins, the histone arginine methyltransferase PRMT5 and the WD repeat protein WDR77, as both CDK8- and CDK19-containing Mediator-interacting proteins. We further characterized these interactions using two step-purified CDK-containing Mediator complexes and demonstrated that both PRMT5 and WDR77 stably associate with both types of CDK-containing Mediators (Fig. 3, A and B). These data indicate that PRMT5 and WDR77 bind to both CDK8- and CDK19-containing Mediators. Further biochemical analyses proved that both PRMT5 and WDR77 bound directly to CDK8 and CDK19 (Fig. 4, A–C). Taken together, these results clearly demonstrated that the two Mediator CDKs directly interact with PRMT5 as well as WDR77.
A Transcriptional Repression Model of C/EBPβ Target Genes by CDK-containing Mediators
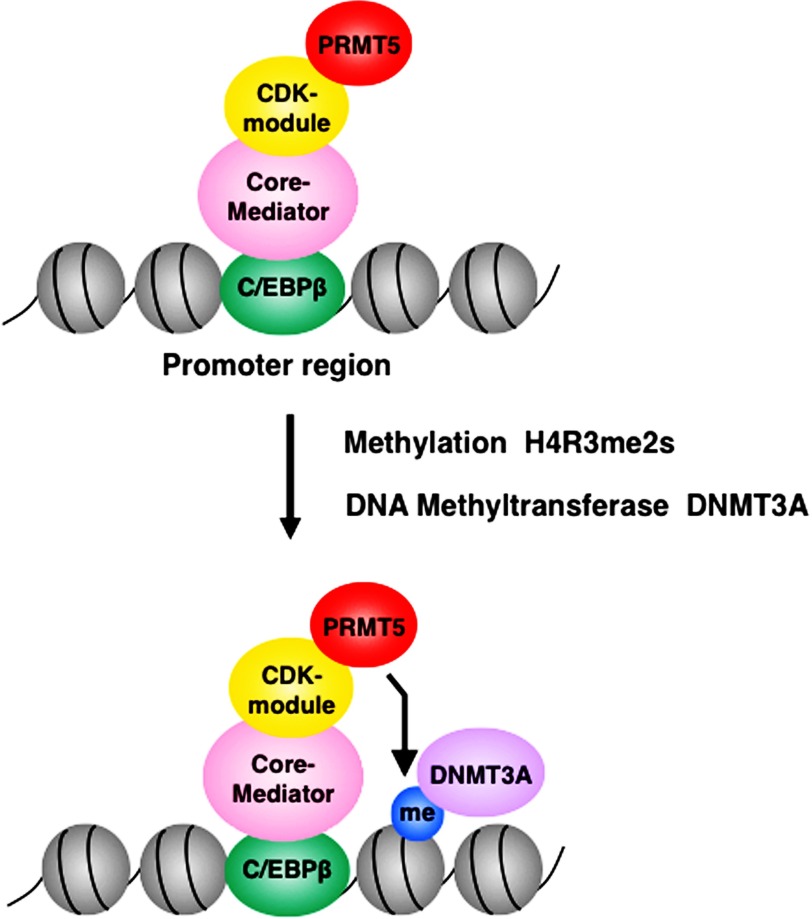
Both CDK8 and CDK19-containing Mediator fractions possessed histone H4 arginine 3 symmetric dimethylation (H4R3me2s) activity (Fig. 4, D and E). Based on the ChIP analyses, dissociation of CDK8 and CDK19 from promoters, dissociation of PRMT5 from promoters, and reduction of the H4R3me2s modification at promoters were mutually correlated (Figs. 5 and 6). These results clearly indicate that the H4R3me2s activity of the two CDK-containing Mediators is primarily due to their interactions with PRMT5. This specificity implies that the transcriptionally repressive functions of Mediator CDKs stem from histone modifications that repress transcription. In addition, these events were highly correlated with DNMT3A binding at the promoter regions of four C/EBPβ target genes (Figs. 5 and 6) (39). These results indicate that DNMT3A recruitment leads to the repression of target gene transcription. Taking all of these findings into consideration, we propose a novel model for transcriptional repression by Mediator CDKs (Fig. 7): (i) the large Mediator complex containing the CDK/cyclin submodule, which binds to C/EBPβ, associates with PRMT5 (and WDR77); (ii) PRMT5 symmetrically dimethylates histone H4R3 (H4R3me2s); (iii) the DNA methyltransferase DNMT3A and then targets this methylated H4R3 to bind and methylate CpG islands around the promoter region; and (iv) as a consequence, these three sequential events cause transcriptional repression of C/EBPβ target genes. In the near future, this model might help to solve several mechanistic questions regarding Mediator-based gene repression.
FIGURE 7.
Schematic model for PRMT5-dependent transcriptional repression of C/EBPβ target genes. CDK-containing Mediators are recruited to the TAD domain of C/EBPβ target genes, where they associate with PRMT5 and WDR77. PRMT5 then symmetrically dimethylates histone H4 arginine 3 (H4R3me2s), leading to recruitment of the DNA methyltransferase DNMT3A and ultimately to transcriptional repression of C/EBPβ target genes.
Regulation of H4R3me2s Modification Independent of PRMT5 Recruitment
We showed that the two Mediator CDKs physically interact with PRMT5 and WDR77 in HeLa S3 cells. The CDK8/19 occupancies of both gene promoters tested decreased to the background level (defined as exonic signals) after PMA treatment (Fig. 1, C and D). The reduced PRMT5 signal on promoters after PMA treatment was highly correlated with the CDK8/19 signals (Fig. 5). However, the H4R3me2s signal exhibited little correlation with the PRMT5 signal. In particular, at the TNFα gene promoter region, the PRMT5 signal was reduced 70–80% by PMA treatment, whereas H4R3me2s modification in this region was reduced only 20%. In exonic regions, there was a 20–40% change in the PRMT5 signal after PMA treatment, but H4R3me2s signals in exons were almost equal to those on the promoter. We suspect two underlying mechanisms caused this difference between PRMT5 occupancies and H4R3me2s modifications. First, the H4R3 residue can also be symmetrically methylated by the type II arginine methyltransferase PRMT7 (55); thus, in exonic regions, PRMT7 activity may compensate for loss of PRMT5. Second, we suspect the effects of competing regulation by opposing histone demethylases. A recent study showed that H4R3me2s is demethylated by JMJD6 (Jumonji domain-containing 6) (56). Based on the low occupancy of PRMT5 at exonic regions, we hypothesize that H4R3me2s modifications in exonic regions may be regulated by PRMT7 and/or another novel methyltransferase, rather than by PRMT5. Given that histone methylation is competitively or collaboratively regulated by multiple factors, we also suspect that H4R3me2s modifications in the TNFα promoter region are collaboratively regulated by other histone modification factors. In the future, there should be more detailed investigations into the underlying coordination of such factors.
Downstream Mechanism of CDK8- and CDK19-dependent DNMT3A Recruitment
PRMT5 is a transcriptionally repressive histone modification enzyme, and its modifications of H4R3me2s act repressively when recognized by DNMT3A (39). In addition, H4R3me2s modifications are localized to repressive regions of chromosomes (53). Our results demonstrate that histone demethylation and transcriptional activation of C/EBPβ target genes occur within less than 2 h. Although our results demonstrate a relationship, it seems likely that DNA methylation-independent transcriptional repression is also involved in the transcriptional repression of C/EBPβ target genes by Mediator CDKs. Indeed, DNMT3A interacts with the histone deacetylase HDAC1, as well as with the repressive histone methyltransferases SUV39H1 and SETDB1 (57, 58). These interactions with HDAC1 and/or histone methyltransferases might repress C/EBPβ target gene transcription. siRNA-mediated knockdown of CDK8 and CDK19 promoted PRMT5 dissociation from the promoter, similar to the effect of PMA treatment, clearly indicating that PRMT5 is recruited to C/EBPβ target gene promoter regions by Mediator CDKs and may repress target gene transcription via DNMT3A, as proposed in the model shown in Fig. 7.
Potential Functional Differences between CDK8 and CDK19
Even in light of the model we propose, questions remain regarding the functions of Mediator CDKs during transcriptional regulation. In the regulation of several types of immediate-early gene expression, CDK8 acts as a transcriptional activator (19, 21, 59). The functional bivalency of CDK8 is quite enigmatic, but structural studies may provide a clue. Mediators exhibit a structural change when they bind various types of transcriptional activators (60) and therefore exist as multiple structurally distinct subcomplexes. Some of these subcomplexes may bind tightly to PRMT5 and act as corepressors, as described in this paper, whereas others may lack PRMT5 and act as coactivators. On the other hand, the CDK/cyclin submodule functions in transcriptional activation of immediate-early genes in a Mediator-independent manner (59). We observed that PRMT5 bound holo-Mediator complexes (Figs. 2–4), pointing to the need for further studies into the functional differences between CDK8- and CDK19-containing Mediators, as well as the functions of the free forms of the two CDK/cyclin heterodimers. PRMT5 also symmetrically dimethylates histone H3 arginine 2 (H3R2me2s) (61). In contrast to H4R3me2s, H3R2me2s activates transcription by recruiting WDR5 and causing the methyltransferase complex MLL to trimethylate H3K4. Therefore, an additional regulatory mechanism probably underlies the specificity of PRMT5 histone modification.
Our study demonstrated that Mediator CDKs play regulatory roles in both histone modification and transcription of C/EBPβ target genes. In addition, we attempted to characterize the functions of Mediator CDKs at C/EBPβ target genes in HeLa S3 cells by performing microarray analysis (Table 1); the results showed that the repressive target genes of Mediator CDKs were highly correlated with the target genes of C/EBPβ. In future studies, we will use CDK knock-out mice to understand the roles of Mediators in important physiological processes.
TABLE 1.
Summary of genes transcriptionally repressed by CDK8 and CDK19 double knockdown
A total of 197 transcripts were identified as significantly changed relative to the negative control. These transcripts were analyzed using the Ingenuity Pathway Analysis software to categorize them according to their pathways (significance p < 0.05). The numbers of genes from the C/EBPβ and NF-κB target gene pathways are shown.
| Type of gene | Number of genes | p value |
|---|---|---|
| All genes repressed by double knockdown of CDK8 and CDK19 | 197 (100%) | |
| C/EBPβ target genes | 8 (4.1%) | 0.0379 |
| NF-κB target genes | 12 (6.1%) | 0.000294 |
Acknowledgments
We thank Dr. Hideaki Tagami for critical reading of this manuscript, Dr. Akiyoshi Fukamizu for the antibody against symmetrically dimethylated histone H4 arginine 3 (H4R3me2s), and Dr. Ichiro Takasaki and Dr. Yutaka Hirose for advice on the RT-qPCR and ChIP analyses. We also thank Dr. Hiroaki Sakurai and our colleagues at the University of Toyama for helpful discussions.
This work was supported in part by grants from the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (to Y. O.) and from the Japan Society for the Promotion of Science through “Research Fellowships for Young Scientists” (to T. T.).
- Pol II
- RNA polymerase II
- CDK
- cyclin-dependent kinase
- PMA
- phorbol 12-myristate 13-acetate
- DNMT
- DNA methyltransferase
- HF
- HA/FLAG-tagged
- qPCR
- quantitative PCR
- C/EBP
- CCAAT enhancer-binding protein.
REFERENCES
- 1. Lee T. I., Young R. A. (2000) Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34, 77–137 [DOI] [PubMed] [Google Scholar]
- 2. Li B., Carey M., Workman J. L. (2007) The role of chromatin during transcription. Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 3. Malik S., Roeder R. G. (2010) The metazoan Mediator coactivator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nonet M. L., Young R. A. (1989) Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123, 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D. (1990) A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61, 1209–1215 [DOI] [PubMed] [Google Scholar]
- 6. Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 [DOI] [PubMed] [Google Scholar]
- 7. Fondell J. D., Ge H., Roeder R. G. (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. U.S.A. 93, 8329–8333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rachez C., Suldan Z., Ward J., Chang C. P., Burakov D., Erdjument-Bromage H., Tempst P., Freedman L. P. (1998) A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12, 1787–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun X., Zhang Y., Cho H., Rickert P., Lees E., Lane W., Reinberg D. (1998) NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2, 213–222 [DOI] [PubMed] [Google Scholar]
- 10. Gu W., Malik S., Ito M., Yuan C. X., Fondell J. D., Zhang X., Martinez E., Qin J., Roeder R. G. (1999) A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 3, 97–108 [DOI] [PubMed] [Google Scholar]
- 11. Näär A. M., Lemon B. D., Tjian R. (2001) Transcriptional coactivator complexes. Annu. Rev. Biochem. 70, 475–501 [DOI] [PubMed] [Google Scholar]
- 12. Bourbon H. M., Aguilera A., Ansari A. Z., Asturias F. J., Berk A. J., Bjorklund S., Blackwell T. K., Borggrefe T., Carey M., Carlson M., Conaway J. W., Conaway R. C., Emmons S. W., Fondell J. D., Freedman L. P., Fukasawa T., Gustafsson C. M., Han M., He X., Herman P. K., Hinnebusch A. G., Holmberg S., Holstege F. C., Jaehning J. A., Kim Y. J., Kuras L., Leutz A., Lis J. T., Meisterernest M., Naar A. M., Nasmyth K., Parvin J. D., Ptashne M., Reinberg D., Ronne H., Sadowski I., Sakurai H., Sipiczki M., Sternberg P. W., Stillman D. J., Strich R., Struhl K., Svejstrup J. Q., Tuck S., Winston F., Roeder R. G., Kornberg R. D. (2004) A unified nomenclature for protein subunits of Mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14, 553–557 [DOI] [PubMed] [Google Scholar]
- 13. Conaway R. C., Sato S., Tomomori-Sato C., Yao T., Conaway J. W. (2005) The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30, 250–255 [DOI] [PubMed] [Google Scholar]
- 14. Loncle N., Boube M., Joulia L., Boschiero C., Werner M., Cribbs D. L., Bourbon H. M. (2007) Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 26, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Westerling T., Kuuluvainen E., Mäkelä T. P. (2007) Cdk8 is essential for preimplantation mouse development. Mol. Cell. Biol. 27, 6177–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hengartner C. J., Myer V. E., Liao S. M., Wilson C. J., Koh S. S., Young R. A. (1998) Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2, 43–53 [DOI] [PubMed] [Google Scholar]
- 17. Akoulitchev S., Chuikov S., Reinberg D. (2000) TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407, 102–106 [DOI] [PubMed] [Google Scholar]
- 18. Liu Y., Kung C., Fishburn J., Ansari A. Z., Shokat K. M., Hahn S. (2004) Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 24, 1721–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Furumoto T., Tanaka A., Ito M., Malik S., Hirose Y., Hanaoka F., Ohkuma Y. (2007) A kinase subunit of the human mediator complex, CDK8, positively regulates transcriptional activation. Genes Cells 12, 119–132 [DOI] [PubMed] [Google Scholar]
- 20. Andrau J. C., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M. G., van de Peppel J., Werner M., Holstege F. C. (2006) Genome-wide location of the coactivator mediator. Binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22, 179–192 [DOI] [PubMed] [Google Scholar]
- 21. Donner A. J., Szostek S., Hoover J. M., Espinosa J. M. (2007) CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell 27, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knuesel M. T., Meyer K. D., Bernecky C., Taatjes D. J. (2009) The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Conaway R. C., Conaway J. W. (2011) Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 21, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sato S., Tomomori-Sato C., Parmely T. J., Florens L., Zybailov B., Swanson S. K., Banks C. A., Jin J., Cai Y., Washburn M. P., Conaway J. W., Conaway R. C. (2004) A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 [DOI] [PubMed] [Google Scholar]
- 25. Tsutsui T., Umemura H., Tanaka A., Mizuki F., Hirose Y., Ohkuma Y. (2008) Human mediator kinase subunit CDK11 plays a negative role in viral activator VP16-dependent transcriptional regulation. Genes Cells 13, 817–826 [DOI] [PubMed] [Google Scholar]
- 26. Tsutsui T., Fukasawa R., Tanaka A., Hirose Y., Ohkuma Y. (2011) Identification of target genes for the CDK subunits of the Mediator complex. Genes Cells 16, 1208–1218 [DOI] [PubMed] [Google Scholar]
- 27. Mo X., Kowenz-Leutz E., Xu H., Leutz A. (2004) Ras induces Mediator complex exchange on C/EBPβ. Mol. Cell 13, 241–250 [DOI] [PubMed] [Google Scholar]
- 28. Roesler W. J. (2001) The role of C/EBP in nutrient and hormonal regulation of gene expression. Annu. Rev. Nutr. 21, 141–165 [DOI] [PubMed] [Google Scholar]
- 29. Farmer S. R. (2006) Transcriptional control of adipocyte formation. Cell Metab. 4, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sebastian T., Johnson P. F. (2006) Anti-proliferative and mitogenic functions of the transcription factor C/EBPβ. Cell Cycle 5, 953–957 [DOI] [PubMed] [Google Scholar]
- 31. Nerlov C. (2007) The C/EBP family of transcription factors. A paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 17, 318–324 [DOI] [PubMed] [Google Scholar]
- 32. Zahnow C. A. (2009) CCAAT/enhancer-binding protein β. Its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev. Mol. Med. 11, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Gade P., Nallar S. C., Raha A., Roy S. K., Karra S., Reddy J. K., Reddy S. P., Kalvakolanu D. V. (2008) The Med1 subunit of transcriptional mediator plays a central role in regulating CCAAT/enhancer-binding protein-β-driven transcription in response to interferon-gamma. J. Biol. Chem. 283, 13077–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakajima T., Kinoshita S., Sasagawa T., Sasaki K., Naruto M., Kishimoto T., Akira S. (1993) Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc. Natl. Acad. Sci. U.S.A. 90, 2207–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kowenz-Leutz E., Twamley G., Ansieau S., Leutz A. (1994) Novel mechanism of C/EBPβ (NF-M) transcriptional control. Activation through derepression. Genes Dev. 8, 2781–2791 [DOI] [PubMed] [Google Scholar]
- 36. Meyer K. D., Donner A. J., Knuesel M. T., York A. G., Espinosa J. M., Taatjes D. J. (2008) Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 27, 1447–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S., Taatjes D. J., Dekker J., Young R. A. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin J. J., Lehmann L. W., Bonora G., Sridharan R., Vashisht A. A., Tran N., Plath K., Wohlschlegel J. A., Carey M. (2011) Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev. 25, 2198–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., Jane S. M. (2009) PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malik S., Roeder R. G. (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25, 277–283 [DOI] [PubMed] [Google Scholar]
- 41. Takahashi H., Parmely T. J., Sato S., Tomomori-Sato C., Banks C. A., Kong S. E., Szutorisz H., Swanson S. K., Martin-Brown S., Washburn M. P., Florens L., Seidel C. W., Lin C., Smith E. R., Shilatifard A., Conaway R. C., Conaway J. W. (2011) Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kowenz-Leutz E., Pless O., Dittmar G., Knoblich M., Leutz A. (2010) Crosstalk between C/EBPβ phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 29, 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matsusaka T., Fujikawa K., Nishio Y., Mukaida N., Matsushima K., Kishimoto T., Akira S. (1993) Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc. Natl. Acad. Sci. U.S.A. 90, 10193–10197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uematsu S., Kaisho T., Tanaka T., Matsumoto M., Yamakami M., Omori H., Yamamoto M., Yoshimori T., Akira S. (2007) The C/EBPβ isoform 34-kDa LAP is responsible for NF-IL-6-mediated gene induction in activated macrophages, but is not essential for intracellular bacteria killing. J. Immunol. 179, 5378–5386 [DOI] [PubMed] [Google Scholar]
- 45. Pavri R., Lewis B., Kim T. K., Dilworth F. J., Erdjument-Bromage H., Tempst P., de Murcia G., Evans R., Chambon P., Reinberg D. (2005) PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell 18, 83–96 [DOI] [PubMed] [Google Scholar]
- 46. Kim Y. K., Bourgeois C. F., Pearson R., Tyagi M., West M. J., Wong J., Wu S. Y., Chiang C. M., Karn J. (2006) Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 25, 3596–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Branscombe T. L., Frankel A., Lee J. H., Cook J. R., Yang Z., Pestka S., Clarke S. (2001) PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276, 32971–32976 [DOI] [PubMed] [Google Scholar]
- 48. Friesen W. J., Wyce A., Paushkin S., Abel L., Rappsilber J., Mann M., Dreyfuss G. (2002) A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277, 8243–8247 [DOI] [PubMed] [Google Scholar]
- 49. Karkhanis V., Hu Y. J., Baiocchi R. A., Imbalzano A. N., Sif S. (2011) Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 36, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aggarwal P., Vaites L. P., Kim J. K., Mellert H., Gurung B., Nakagawa H., Herlyn M., Hua X., Rustgi A. K., McMahon S. B., Diehl J. A. (2010) Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tae S., Karkhanis V., Velasco K., Yaneva M., Erdjument-Bromage H., Tempst P., Sif S. (2011) Bromodomain protein 7 interacts with PRMT5 and PRC2, and is involved in transcriptional repression of their target genes. Nucleic Acids Res. 39, 5424–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Di Lorenzo A., Bedford M. T. (2011) Histone arginine methylation. FEBS Lett. 585, 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu X., Hoang S., Mayo M. W., Bekiranov S. (2010) Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinfomatics 11, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ding N., Zhou H., Esteve P.-O., Chin H. G., Kim S., Xu X., Joseph S. M., Friez M. J., Schwartz C. E., Pradhan S., Boyer T. G. (2008) Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol. Cell 31, 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jelinic P., Stehle J. C., Shaw P. (2006) The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 4, e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chang B., Chen Y., Zhao Y., Bruick R. K. (2007) JMJD6 is a histone arginine demethylase. Science 318, 444–447 [DOI] [PubMed] [Google Scholar]
- 57. Fuks F., Burgers W. A., Godin N., Kasai M., Kouzarides T. (2001) Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20, 2536–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fuks F., Hurd P. J., Deplus R., Kouzarides T. (2003) The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31, 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Donner A. J., Ebmeier C. C., Taatjes D. J., Espinosa J. M. (2010) CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17, 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Meyer K. D., Lin S. C., Bernecky C., Gao Y., Taatjes D. J. (2010) p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 17, 753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Migliori V., Müller J., Phalke S., Low D., Bezzi M., Mok W. C., Sahu S. K., Gunaratne J., Capasso P., Bassi C., Cecatiello V., De Marco A., Blackstock W., Kuznetsov V., Amati B., Mapelli M., Guccione E. (2012) Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 19, 136–144 [DOI] [PubMed] [Google Scholar]