Background: The in vivo mechanism of autophagy regulation remains unclear.
Results: The effect of insulin on mTORC1-autophagy is greater in muscle than in the liver, whereas that of amino acids is greater in liver than in muscle.
Conclusion: Autophagy is differentially regulated by insulin and amino acids in a tissue-dependent manner.
Significance: This information increases the understanding of protein homeostasis in vivo.
Keywords: Amino Acid, Autophagy, Insulin, mTOR Complex (mTORC), Protein Degradation
Abstract
Autophagy is a highly inducible intracellular degradation process. It is generally induced by nutrient starvation and suppressed by food intake. Mammalian (or mechanistic) target of rapamycin complex 1 (mTORC1) is considered to be the major regulator of autophagy, but the precise mechanism of in vivo regulation remains to be fully characterized. Here, we examined the autophagy-suppressive effect of glucose, insulin, and amino acids in the liver and muscle in mice starved for 1 day. Refeeding after starvation with a standard mouse chow rapidly suppressed autophagy in both tissues, and this suppression was inhibited by rapamycin administration almost completely in the liver and partially in muscle, confirming that mTORC1 is indeed a crucial regulator in vivo. As glucose administration showed no major suppressive effect on autophagy, we examined the role of insulin and amino acids using hyperinsulinemic-euglycemic clamp and intravenous amino acid infusion techniques. Insulin administration showed a clear effect on the mTORC1-autophagy pathway in muscle, but had only a very weak effect in the liver. By contrast, amino acids were able to regulate the mTORC1-autophagy pathway in the liver, but less effectively in muscle. These results suggest that autophagy is differentially regulated by insulin and amino acids in a tissue-dependent manner.
Introduction
Macroautophagy (referred to as autophagy hereafter) is one of the major degradation systems in the cell. A small region of the cytoplasm is first enclosed by the autophagosome, and then degraded when the autophagosome fuses with the lysosome (1, 2). Autophagy occurs constitutively at basal levels, which is important for turnover of cytoplasmic materials such as proteins and organelles both selectively and non-selectively (3–5). In addition, autophagy can be highly induced by nutrient starvation. The induced autophagy is important for providing amino acids and other factors, which can be used for synthesis of proteins and other metabolites, energy production, and gluconeogenesis under starvation conditions (4, 6). It is therefore essential to understand how autophagy is regulated, particularly in the whole body.
It has been shown that mammalian (or mechanistic) target of rapamycin (mTOR) complex 1 ((m)TORC1)2 plays a major role in autophagy regulation in eukaryotes (6–11). mTORC1 consists of mTOR, mLST8, raptor, PRAS40, and Deptor, and is activated by amino acid and insulin/growth factor signals (12–14). Whereas mTORC1 positively regulates protein translation, ribosome biogenesis, and cell size through phosphorylation of S6-kinase (S6K), 4E-BP1, and other substrates, it negatively regulates autophagy through phosphorylation of its putative substrates ULK1 and Atg13, which are part of the most upstream autophagy complex together with FIP200 and Atg101 (15, 16).
However, recent studies have shown that autophagy can also be regulated by a number of mTORC1-independent pathways or transcriptional factors (9, 17). For example, AMP-activated protein kinase not only inhibits the mTOR pathway, but also directly phosphorylates and activates ULK1 (18). Starvation-induced phosphorylation of Bcl-2 by JNK1 releases Beclin 1, which promotes autophagy (19). Starvation also activates transcription factor EB (TFEB), an important gene for lysosomal biogenesis, which induces various autophagy-related factors at the transcriptional level (20). Thus, it is uncertain whether mTORC1 is indeed a major regulator of autophagy under physiological conditions in vivo.
Furthermore, it is noteworthy that autophagy regulation can vary in different tissues/organs. The roles of individual amino acids and hormones in autophagy have been determined in great detail in the liver using an ex vivo perfusion system (21). However, such analyses are not entirely physiological, and have not been extensively performed in other tissues. In neonatal pigs, the differential effects of amino acids and insulin on protein synthesis were examined in vivo using euglycemic clamp methods; insulin has no effect on the rate of protein synthesis in the liver, whereas amino acids do in this organ (22), as well as in the pancreas and kidney (23). It has also been reported that amino acids alone can activate mTORC1 in the liver in vivo (22) and in ex vivo perfused liver (24). By contrast, insulin can stimulate protein synthesis in skeletal and cardiac muscles, skin, and spleen in neonatal pigs (23). However, whether autophagy is also regulated differently in different tissues by amino acids and insulin in vivo has not been determined. Thus, it is important to carefully assess the in vivo autophagy regulators.
In the present study, we investigated the relative contribution of glucose, insulin, and amino acids to regulation of mTORC1 and autophagy in the liver and muscle in vivo. To administer insulin and amino acids to starved mice, we utilized hyperinsulinemic-euglycemic clamp and intravenous amino acid administration methods, respectively. We found that glucose has no major role in autophagy regulation, and that the effect of insulin on the mTORC1-autophagy pathway is greater in muscle than in the liver, whereas amino acids regulate the mTORC1-autophagy pathway more effectively in the liver than in muscle.
EXPERIMENTAL PROCEDURES
Animals
C57BL/6J male mice were purchased from Sankyo Labo Service Corporation (Tokyo, Japan) and fed with a standard mouse chow (CRF-1, Charles River, Japan) and water. Most mice used in this study were between 10 and 12 weeks old.
For glucose stimulation, d-glucose (WAKO) was injected at a dose of 2.5 g/kg (intraperitoneal). For mTORC1 inhibition, rapamycin (LC Laboratories) was injected twice at a dose of 0.4 mg/kg (intraperitoneal) at 2 h and 16 h before sampling. A similar volume of vehicle was injected as a negative control.
For insulin and amino acid administration, mice underwent venous cannulation. On the day before the infusion, a polyethylene catheter was inserted into the right jugular vein under general anesthesia induced by 2,2,2,-tribromoethanol (Avertin®, Sigma). To prevent blood coagulation, the cannula was filled with saline containing heparin (Mochida Pharmaceutical) until the experiment. Insulin and amino acids were administered to conscious mice under non-stressed conditions. All animal experiments were approved by the Institutional Animal Care and Use Committee of Tokyo Medical and Dental University (No. 0130093A).
Hyperinsulinemic-euglycemic Clamp
Clamp studies were carried out as described previously (25, 26), with slight modifications. After starvation for 24 h, a primed continuous infusion of insulin (Humulin R, Lilly) was given (10 or 40 mU/kg/min). The blood glucose concentration was monitored every 5 min, and was maintained at 70–100 mg/dl by administration of 20% d-glucose for 1 h. Blood was sampled via the tail tip at 0, 30, and 60 min for measurement of the blood insulin concentration. For immunoblotting, fresh tissues were frozen in liquid nitrogen and stored at −80 °C until use.
Amino Acid Administration
After starvation for 24 h, a primed continuous infusion of an amino acid mixture (Amiparen®, Otsuka Pharmaceutical) was given at various concentrations (9.1, 18.2, and 36.4 mg/kg/min) for 1 h. Somatostatin (Sigma) was infused (7.14 μg/kg/min) simultaneously. Blood was sampled via the tail tip at 0, 30, and 60 min for measurement of plasma insulin and blood glucose levels. For immunoblotting and amino acid assay, fresh tissues were frozen in liquid nitrogen and stored at −80 °C until use.
Measurement of Plasma Insulin
Plasma insulin was determined using a commercially available enzyme-linked immunosorbent assay kit for mouse insulin (Shibayagi). The insulin level during a hyperinsulinemic-euglycemic clamp was measured using human insulin as a standard.
Measurement of Amino Acid Concentration
Mouse tissues were homogenized in 5 volumes of ice-cold 5% sulfosalicylic acid. After centrifugation at 10,000 × g for 15 min at 4 °C, free amino acids in the supernatant were measured using an L8500 amino acid analyzer (Hitachi, Ltd.).
Immunoblotting
Mouse tissues were homogenized in 8 volumes of ice-cold PBS supplemented with 1 mm PMSF, 0.4 mm Na orthovanadate, 0.4 mm EDTA, 10 mm NaF, and 10 mm Na pyrophosphate. Cells were lysed in a lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm phenylmethanesulfonyl fluoride (PMSF), 1 mm Na3VO4, and protease inhibitor mixture (complete EDTA-free protease inhibitor, Roche)). The homogenates and cell lysates were centrifuged at 500 × g at 4 °C for 10 min. After quantification of protein concentration by the Bradford method, protein extracts were subjected to SDS-PAGE and immunoblotting. Each lane represents a tissue extract from an independent mouse. The amount of protein was quantified using densitometric measurements for each lane with ImageJ software.
Antibodies against AKT pS473 (4051), AKT pT308 (9275), AKT (9272), PRAS40 pT246 (2997), S6K1 pT389 (9204), S6 pS235/236 (4856), S6 (2217), 4E-BP1 pS65 (9451), 4E-BP1 (9452), AMPKα pT172 (2535), AMPKα (2532), and ACC pS79 (3661) were purchased from Cell Signaling. Antibodies against α-tubulin (T9026) were purchased from Sigma-Aldrich. Rabbit anti-microtubule-associated protein 1 light chain 3 (LC3) antibody (NM1) against recombinant rat-LC3B was generated as previously described (27).
Statistical Analysis
All numerical data represent the mean ± S.E. Statistical comparisons between groups were made using 1-way analysis of variance (ANOVA).
RESULTS
Autophagy Is Suppressed by Feeding but Not by Glucose Administration in Muscle and Liver
To study the physiological modulation of autophagy in vivo, we used a starvation-refeeding system because autophagy is known to be rapidly suppressed by food intake (28, 29). C57BL/6J male mice were starved for 24 h, and then refed for 1 h. Feeding increased blood levels of glucose and insulin (Fig. 1A), accompanied by an increase in mTORC1 activity (phosphorylation of S6 kinase, S6, and 4E-BP1) in skeletal muscle and liver (Fig. 1B). The amount of LC3-II, which represents the number of autophagosomes (30), decreased in both skeletal muscle and liver during the 1-h refeeding period, confirming that autophagy is suppressed by feeding (Fig. 1B). Phosphorylation of AKT Ser-473 and PRAS40 Thr-246 was also enhanced by feeding in muscle, but not in the liver (Fig. 1B), suggesting that the upstream signaling and/or negative feedback loops are differently activated in muscle and liver.
FIGURE 1.
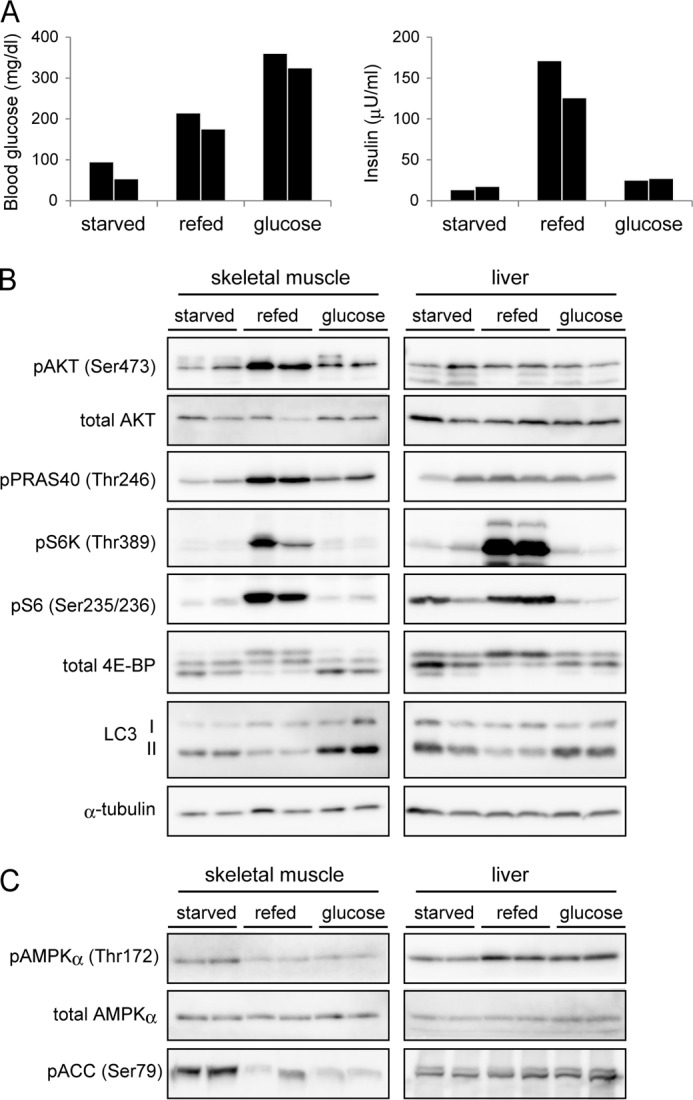
Autophagy is suppressed by refeeding but not by glucose administration in muscle and liver. A, mice were starved for 24 h, and then fed for 1 h (refed) or injected with glucose or saline (starved). Glucose was injected (2.5 g/kg intraperitoneal) at 1 h before sampling of blood and tissues. Plasma insulin and blood glucose concentrations were measured after feeding and glucose administration. Blood samples were collected from the tail vein. B, gastrocnemius muscle and liver samples were obtained from the 3 groups of mice described in A and subjected to immunoblot analysis of insulin-mTORC1 signaling and LC3 conversion. α-tubulin was used as a loading control. C, immunoblot analysis showing the effect of feeding and glucose administration on phosphorylation levels of AMPKα and its substrate acetyl-CoA carboxylase (ACC). Data from two independent mice are shown for each experimental group (A–C).
As feeding induced a clear elevation in the blood glucose level, we next determined the role of glucose in autophagy regulation in vivo. Intraperitoneal injection of glucose elevated the blood glucose concentration to a level higher than that of refed mice (Fig. 1A). Insulin secretion was only slightly induced by glucose injection, which may reflect that incretins (intestinal hormones) are important for feeding-induced insulin secretion (31, 32). After glucose injected in these mice, neither mTORC1 activation nor autophagy suppression was observed in muscle or liver (Fig. 1B). Glucose administration effectively suppressed the AMP-activated protein kinase (AMPK) signaling in muscle; phosphorylation of AMPK and one of its substrates acetyl-CoA carboxylase (ACC) was inhibited by glucose injection as well as by feeding (Fig. 1C). It suggests that glucose was in fact delivered to tissues in vivo. In the liver, both feeding and glucose administration paradoxically activated AMPK by an unknown reason, which may be through an AMP-independent pathway. These results suggest that glucose is not a major regulator of the feeding-induced suppression of autophagy in vivo.
mTORC1 Is Involved in Autophagy Suppression After Feeding
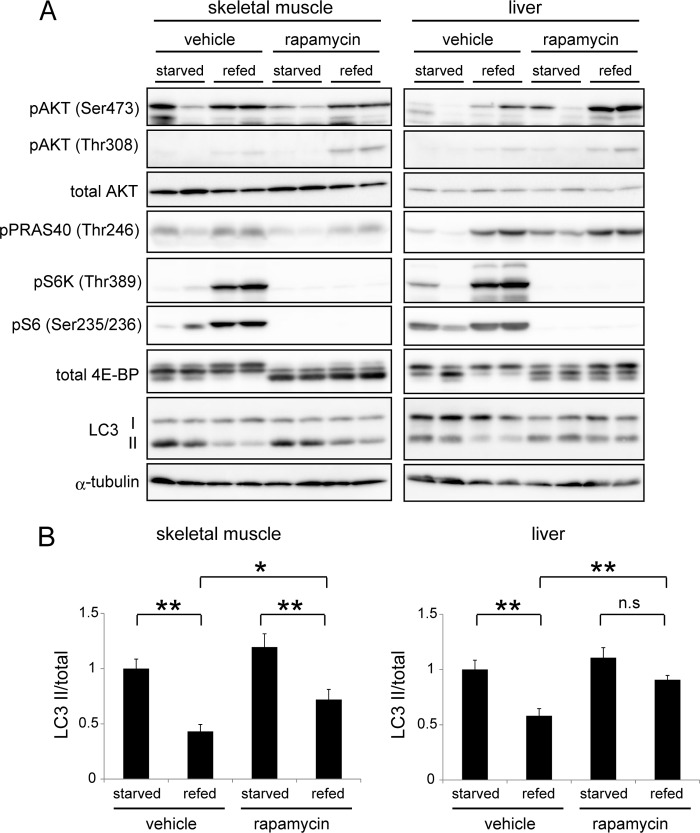
Although mTORC1 has been considered to be a master regulator of autophagy, recent studies suggest that there are mTOR-independent regulatory pathways (9, 17). To assess the importance of mTORC1 in autophagy regulation in vivo, we determined the effect of rapamycin, a specific inhibitor of mTOR, in refed mice. Pretreatment with rapamycin abolished feeding-induced phosphorylation of S6K, S6, and 4E-BP1 in both tissues (Fig. 2A). Accordingly, rapamycin treatment inhibited reduction of LC3-II almost completely in the liver (Fig. 2, A and B). Phosphorylation of AKT Ser-473 and Thr-308 were clearly observed in the presence of rapamycin, suggesting that mTOR-S6K-mediated feedback suppression is significant in the liver of refed mice. Rapamycin treatment also suppressed refed-induced LC3-II reduction in muscle of the same animals, but it was partial compared with the vehicle controls. These results confirm that mTOR is indeed a major regulator of autophagy in the liver, and at least partially in muscle.
FIGURE 2.
mTORC1 contributes to autophagy suppression after feeding. A, mice were starved for 24 h (starved) and then refed for 1 h (refed). Rapamycin (0.4 mg/kg), a specific inhibitor of mTORC1, or saline with 5% Tween 20 and 10% DMSO (vehicle) was intraperitoneally injected at 2 h and 16 h before tissue sampling. Muscle and liver samples were subjected to immunoblot analysis using the indicated antibodies. B, quantitative analysis of the immunoblot data in skeletal muscle and liver samples using densitometry scanning. The LC3-II/total LC3 (LC3-I+II) ratio were calculated. The anti-LC3 antibody used in this study reacts to both LC3-I and LC3-II with similar affinities (supplemental Fig. S2). Data are mean ± S.E. of 4 mice (two are shown in A and the other two are not shown). *, p < 0.05; **, p < 0.01; n.s., not significant.
The Effect of Insulin on the mTORC1-autophagy Pathway Is Greater in Muscle Than in Liver
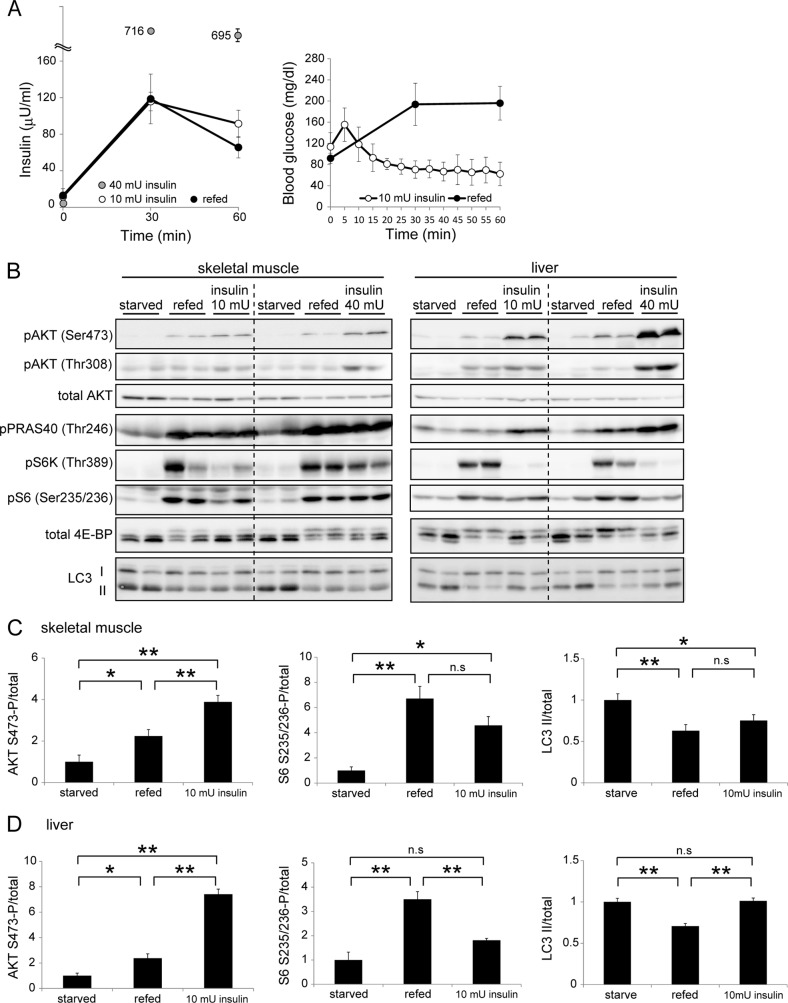
Because it is well known that mTORC1 is regulated by insulin and amino acids, next we determined the effect of insulin on autophagy in vivo. To avoid hypoglycemia and its secondary effects in starved mice, we performed hyperinsulinemic-euglycemic clamp experiments. Using this method, we could maintain constant blood glucose levels within a fasting range, even after insulin administration. When we infused insulin intravenously at 10 mU/kg/min to 24-h starved mice, the blood insulin level increased in a manner similar to that observed in fed mice (Fig. 3A). Infusion of insulin at 40 mU/kg/min increased plasma insulin concentration to a level that was much higher than that of fed mice.
FIGURE 3.
The effect of insulin on the mTORC1-autophagy pathway is greater in muscle than in liver. A, mice were starved for 24 h, and then fed for 1 h (refed) or insulin (10 or 40 mU/kg/min) was administered for 1 h using the euglycemic glucose clamp technique. Plasma insulin and blood glucose concentrations were measured during feeding and insulin administration. Data are mean ± S.E. of 4 mice for the starved and refed groups, and of 5 mice for the insulin-treated group. B, representative immunoblot analysis showing the effect of feeding and insulin administration (10 and 40 mU/kg/min) on phosphorylation levels of AKT, PRAS40, S6K1, S6, and 4E-BP1 and protein levels of LC3. C and D, quantitative analysis of the immunoblot data (starved, refed, and 10 mU/kg/min insulin) of skeletal muscle (C) and liver (D) samples using densitometry scanning. The phosphorylation levels of AKT Ser-473 and S6 Ser-235/236 were normalized relative to total protein content. Data are mean ± S.E. of 4 mice for the starved and refed groups, and of 5 mice for the insulin-treated group (two are shown in B and the others are not shown).*, p < 0.05; **, p < 0.01; n.s., not significant.
In the skeletal muscle of insulin-treated mice (both 10 and 40 mU/kg/min), phosphorylation of AKT, PRAS40, S6K, S6, and 4E-BP1, and reduction of LC3-II, were observed, similar to the changes seen in muscles of fed mice (Fig. 3, B and C). These results suggest that insulin-mTORC1 signaling is sufficient to suppress autophagy induced by food withdrawal.
Similarly, phosphorylation of AKT and PRAS40 was observed in the liver of insulin-treated mice. However, even though phosphorylation levels of these molecules were higher than those observed in refed mice, mTORC1 was hardly activated by insulin administration in the liver even at a high dose (40 mU/kg/min) (Fig. 3, B and D). Consistently, LC3-II generation was not significantly inhibited in the liver of these mice (Fig. 3, B and D). These data from the glucose clamp experiments suggest that sensitivity to insulin for autophagy suppression is greater in muscle than in the liver, and that insulin alone can account for feeding-induced autophagy suppression in muscle, but not in the liver.
Amino Acids Regulate the mTORC1-autophagy Pathway Greater in the Liver than in Muscle
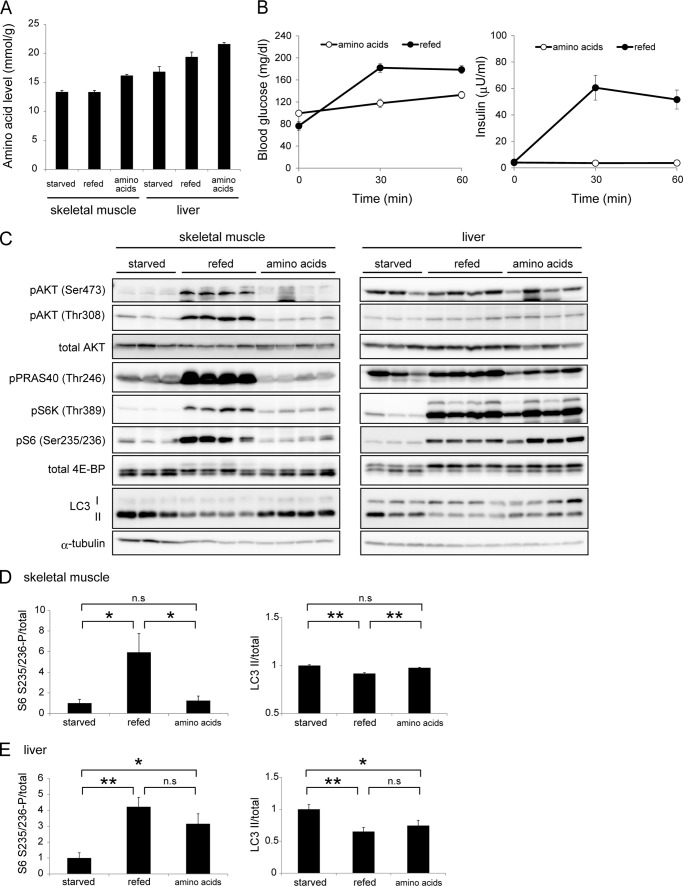
Finally, we determined the effect of amino acids on autophagy suppression in vivo. Intravenous infusion of an amino acid mixture (9.1, 18.2, and 36.4 mg/kg/min) for 1 h successfully elevated the total amino acid concentration both in the liver and muscle (supplemental Figs. S1A and 4A). After amino acid infusion, the level of blood glucose was not changed, but that of insulin was slightly increased (supplemental Fig. S1B). To prevent the endogenous secretion of insulin, somatostatin was simultaneously infused, which suppressed the increase in the blood insulin level during amino acid administration (Fig. 4B). Thus, the effect of these factors could be eliminated in this experiment.
FIGURE 4.
Amino acids induce mTORC1 activation greater in the liver than in muscle. A, mice were starved for 24 h, and then refed for 1 h or an amino acid mixture (18.2 mg/kg/min with somatostatin) was administered for 1 h. Free amino acid concentrations of a total of 20 amino acids in the liver and muscle were measured and expressed as mmol per mg wet weight. B, plasma insulin and blood glucose concentrations were measured during feeding and amino acid administration in the mice described in A. C, immunoblot analysis showing the effect of amino acid administration on phosphorylation levels of AKT, PRAS40, S6K1, S6, and 4E-BP1 and protein levels of LC3 in the gastrocnemius muscle and liver in the mice described in A. D and E, quantitative analysis of the immunoblot data of skeletal muscles (D) and liver (E) obtained from starved, refed, or amino acid-treated (18.2 mg/kg/min) mice. The phosphorylation levels of S6 Ser-235/236 were normalized relative to total protein contents. Data are mean ± S.E. of 3 mice for the starved group, and of 4 mice for the refed and amino acid-treated groups. *, p < 0.05; **, p < 0.01; n.s., not significant.
Although infusion of amino acids did not activate AKT, it led to activation of mTORC1 only in the liver, but not in muscle; S6K, S6, and 4E-BP1 were phosphorylated only in the liver (Fig. 4, C, D, and E). This effect was opposite to that of insulin. Accordingly, the amount of LC3-II was decreased in the liver but not in muscle (Fig. 4, C, D, and E). Overall, the effect of amino acid administration was similar to that of feeding in the liver. It should be noted that, without the somatostatin treatment, amino acids could partially stimulate mTORC1 even in skeletal muscle (supplemental Fig. S1C). Thus, a small amount of insulin secretion might be able to lead to mTORC1 activation in muscle. These results suggest that sensitivity to amino acids in terms of mTORC1 activation and autophagy suppression is greater in the liver than in muscle, and that amino acids could be the major autophagy regulator in the liver.
DISCUSSION
We have demonstrated that mTORC1 and autophagy are mainly regulated by amino acids in the liver in vivo, whereas they can be regulated by insulin in skeletal muscle. The glucose level does not seem to have a major impact on regulation of mTORC1 and autophagy. The differential roles of insulin and amino acids on mTORC1 and autophagy are generally consistent with those in protein synthesis reported in neonatal pigs; both amino acids and insulin stimulate protein synthesis in the muscle, but only amino acids do in the liver (22, 23). However, we cannot completely rule out any physiological significance of insulin on liver autophagy. In fact, autophagic activity is increased in the liver in streptozotocin-induced type 1 diabetic (insulin-deficient) rodent models (33, 34), suggesting that insulin could suppress autophagy in the liver. A possible explanation for this discrepancy is that even though insulin is not critical for acute suppression of autophagy, it could be important for long-term regulation of autophagy, possibly through transcriptional control of some key autophagy genes (34).
How can insulin alone (without amino acid administration) activate mTORC1 and suppress autophagy in the muscle? To be activated, mTORC1 needs to translocate to the lysosomal surface, which is promoted by amino acids and Rag GTPases (13, 14). One possible mechanism for the amino acid-independent mTORC1 activation in muscle could be that basal levels of intramuscular, but not intrahepatic, amino acids are sufficient for the translocation of mTORC1 to the lysosome even under starvation conditions. We have investigated this possibility by cell fractionation experiments, but did not observe a clear difference in lysosomal localization of the mTORC1 components between the liver and muscle. Another possibility is that insulin signaling may be more profoundly inhibited during starvation in the liver than in muscle, and supplementation of insulin alone may not be enough to activate mTORC1 in the liver. For example, glucagon, one of the major starvation hormones, was shown to reduce mTORC1 activity, which may be high in the liver (35, 36). Future studies will be required to explain the difference between these two organs with regard to mTORC1 regulation.
The difference between the liver and muscle in the regulation of the mTORC1-autophagy pathway would be physiologically relevant. In contrast to a highly dynamic change in the blood level of insulin, change in the amino acid level in peripheral blood is small under starvation conditions (37). However, the liver is exceptional, because it receives its blood supply directly from the intestine through the portal vein. The change in the amino acid concentration in plasma from the portal vein is larger than in plasma from the arteries after protein feeding (38). Thus, it would be reasonable to assume that the mTORC1-autophagy pathway is regulated mainly by insulin rather than amino acids in muscle, in which amino acid availability is relatively stable, and by amino acids in the liver, in which amino acid availability is highly changeable.
Dependence of autophagy regulation on mTORC1 appears to differ between muscle and liver. Rapamycin completely suppresses the effect of refeeding on autophagy in the liver but not in muscle, even though phosphorylation of mTORC1 substrates was effectively inhibited (Fig. 2), suggesting that an mTORC1-independent pathway plays a significant role in regulation of muscle autophagy. Considering that insulin is the major regulator of muscle autophagy, some signaling pathways other than the mTORC1 pathway may also be important for autophagy regulation downstream of the insulin receptor in muscle. The FoxO3 pathways, which are known to be important for autophagy regulation in muscle (39, 40), may also contribute to the acute control of autophagy.
Acknowledgments
We thank Drs. Masato Iwabu and Takashi Kadowaki (the University of Tokyo) for help with hyperinsulinemic-euglycemic clamp experiments, and Yuka Hiraoka, Dr. Tsutomu Fujimura, and Dr. Takashi Ueno (the Laboratory of Proteomics and Biomolecular Science, BioMedical Research Center, Juntendo University Graduate School of Medicine) for support with amino acid analysis.
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, the Funding Program for Next Generation World-Leading Researchers, and the Takeda Science Foundation (to N. M.).

This article contains supplemental Figs. S1 and S2.
- mTORC1
- mammalian (or mechanistic) target of rapamycin complex 1
- LC3
- microtubule-associated protein light chain 3
- AKT (PKB)
- protein kinase B
- S6K1
- S6 kinase 1
- 4E-BP1
- eIF4E-binding protein 1
- PRAS40
- proline-rich AKT substrate of 40 kDa.
REFERENCES
- 1. Tooze S. A., Yoshimori T. (2010) The origin of the autophagosomal membrane. Nat. Cell Biol. 12, 831–835 [DOI] [PubMed] [Google Scholar]
- 2. Mizushima N., Yoshimori T., Ohsumi Y. (2011) The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 3. Levine B., Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizushima N., Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 [DOI] [PubMed] [Google Scholar]
- 5. Reggiori F., Komatsu M., Finley K., Simonsen A. (2012) Autophagy: more than a nonselective pathway. Int. J. Cell Biol. 2012, 219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuma A., Mizushima N. (2010) Physiological role of autophagy as an intracellular recycling system: with an emphasis on nutrient metabolism. Semin. Cell Dev. Biol. 21, 683–690 [DOI] [PubMed] [Google Scholar]
- 7. Meijer A. J., Codogno P. (2004) Regulation and role of autophagy in mammalian cells. Int. J. Biochem. Cell Biol. 36, 2445–2462 [DOI] [PubMed] [Google Scholar]
- 8. Neufeld T. P. (2010) TOR-dependent control of autophagy: biting the hand that feeds. Curr. Opin. Cell Biol. 22, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kroemer G., Mariño G., Levine B. (2010) Autophagy and the integrated stress response. Mol. Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang D., Contu R., Latronico M. V., Zhang J., Rizzi R., Catalucci D., Miyamoto S., Huang K., Ceci M., Gu Y., Dalton N. D., Peterson K. L., Guan K. L., Brown J. H., Chen J., Sonenberg N., Condorelli G. (2010) MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J. Clin. Invest. 120, 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Efeyan A., Zoncu R., Chang S., Gumper I., Snitkin H., Wolfson R. L., Kirak O., Sabatini D. D., Sabatini D. M. (2013) Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dazert E., Hall M. N. (2011) mTOR signaling in disease. Curr. Opin. Cell Biol. 23, 744–755 [DOI] [PubMed] [Google Scholar]
- 13. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laplante M., Sabatini D. M. (2012) mTOR Signaling in Growth Control and Disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mizushima N. (2010) The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139 [DOI] [PubMed] [Google Scholar]
- 16. Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H. (2010) mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mehrpour M., Esclatine A., Beau I., Codogno P. (2010) Overview of macroautophagy regulation in mammalian cells. Cell Res. 20, 748–762 [DOI] [PubMed] [Google Scholar]
- 18. Mihaylova M. M., Shaw R. J. (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 13, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. (2008) JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 30, 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M. G., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A. (2011) TFEB Links Autophagy to Lysosomal Biogenesis. Science 332, 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mortimore G. E., Pösö A. R. (1987) Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 7, 539–564 [DOI] [PubMed] [Google Scholar]
- 22. O'Connor P. M., Kimball S. R., Suryawan A., Bush J. A., Nguyen H. V., Jefferson L. S., Davis T. A. (2004) Regulation of neonatal liver protein synthesis by insulin and amino acids in pigs. Am. J. Physiol. Endocrinol. Metab. 286, E994–E1003 [DOI] [PubMed] [Google Scholar]
- 23. Davis T. A., Fiorotto M. L., Burrin D. G., Reeds P. J., Nguyen H. V., Beckett P. R., Vann R. C., O'Connor P. M. (2002) Stimulation of protein synthesis by both insulin and amino acids is unique to skeletal muscle in neonatal pigs. Am. J. Physiol. Endocrinol. Metab. 282, E880–E890 [DOI] [PubMed] [Google Scholar]
- 24. Dennis M. D., Baum J. I., Kimball S. R., Jefferson L. S. (2011) Mechanisms involved in the coordinate regulation of mTORC1 by insulin and amino acids. J. Biol. Chem. 286, 8287–8296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kamei N., Tobe K., Suzuki R., Ohsugi M., Watanabe T., Kubota N., Ohtsuka-Kowatari N., Kumagai K., Sakamoto K., Kobayashi M., Yamauchi T., Ueki K., Oishi Y., Nishimura S., Manabe I., Hashimoto H., Ohnishi Y., Ogata H., Tokuyama K., Tsunoda M., Ide T., Murakami K., Nagai R., Kadowaki T. (2006) Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J. Biol. Chem. 281, 26602–26614 [DOI] [PubMed] [Google Scholar]
- 26. Kubota N., Terauchi Y., Kubota T., Kumagai H., Itoh S., Satoh H., Yano W., Ogata H., Tokuyama K., Takamoto I., Mineyama T., Ishikawa M., Moroi M., Sugi K., Yamauchi T., Ueki K., Tobe K., Noda T., Nagai R., Kadowaki T. (2006) Pioglitazone ameliorates insulin resistance and diabetes by both adiponectin-dependent and -independent pathways. J. Biol. Chem. 281, 8748–8755 [DOI] [PubMed] [Google Scholar]
- 27. Quy P. N., Kuma A., Pierre P., Mizushima N. (2013) Proteasome-dependent activation of mammalian target of rapamycin complex 1 (mTORC1) is essential for autophagy suppression and muscle remodeling following denervation. J. Biol. Chem. 288, 1125–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfeifer U., Strauss P. (1981) Autophagic vacuoles in heart muscle and liver. A comparative morphometric study including circadian variations in meal-fed rats. J. Mol. Cell Cardiol. 13, 37–49 [DOI] [PubMed] [Google Scholar]
- 29. Pfeifer U. (1987) in Lysosomes: Their role in protein breakdown (Glaumann H., Ballard F. J. eds), pp. 3–59, Academic Press, London [Google Scholar]
- 30. Mizushima N., Yoshimori T., Levine B. (2010) Methods in mammalian autophagy research. Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nauck M., Stöckmann F., Ebert R., Creutzfeldt W. (1986) Reduced incretin effect in type 2 (non-insulin-dependent) diabetes. Diabetologia 29, 46–52 [DOI] [PubMed] [Google Scholar]
- 32. Holst J. J., Gromada J. (2004) Role of incretin hormones in the regulation of insulin secretion in diabetic and nondiabetic humans. Am. J. Physiol. Endocrinol. Metab. 287, E199–E206 [DOI] [PubMed] [Google Scholar]
- 33. Lenk S. E., Bhat D., Blakeney W., Dunn W. A., Jr. (1992) Effects of streptozotocin-induced diabetes on rough endoplasmic reticulum and lysosomes of rat liver. Am. J. Physiol. 263, E856–E862 [DOI] [PubMed] [Google Scholar]
- 34. Liu H. Y., Han J., Cao S. Y., Hong T., Zhuo D., Shi J., Liu Z., Cao W. (2009) Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J. Biol. Chem. 284, 31484–31492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kimball S. R., Siegfried B. A., Jefferson L. S. (2004) Glucagon represses signaling through the mammalian target of rapamycin in rat liver by activating AMP-activated protein kinase. J. Biol. Chem. 279, 54103–54109 [DOI] [PubMed] [Google Scholar]
- 36. Baum J. I., Kimball S. R., Jefferson L. S. (2009) Glucagon acts in a dominant manner to repress insulin-induced mammalian target of rapamycin complex 1 signaling in perfused rat liver. Am. J. Physiol. Endocrinol. Metab. 297, E410–E415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palou A., Remesar X., Arola L., Herrera E., Alemany M. (1981) Metabolic effects of short term food deprivation in the rat. Horm. Metab. Res. 13, 326–330 [DOI] [PubMed] [Google Scholar]
- 38. Fafournoux P., Remesy C., Demigne C. (1990) Fluxes and membrane transport of amino acids in rat liver under different protein diets. Am. J. Physiol. 259, E614–E625 [DOI] [PubMed] [Google Scholar]
- 39. Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., Goldberg A. L., Schiaffino S., Sandri M. (2007) FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- 40. Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., Goldberg A. L. (2007) FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]