Background: Helix 12 mutations of ERα reverse antagonists to agonists.
Results: Antagonist-induced homodimerization of mutant ERα LBD coincides with DNA binding activity and antagonist reversal activity.
Conclusions: Antagonist-dependent LBD homodimerization is an important step for antagonist reversal activity.
Significance: This mechanism may be associated with the partial agonist activity of selective estrogen receptor modulators.
Keywords: Estrogen, Estrogen Receptor, Mutant, Protein Domains, Transcription, Fulvestrant, SERMs, Tamoxifen, Antagonist Reversal, Dimerization
Abstract
A ligand-dependent nuclear transcription factor, ERα has two transactivating functional domains (AF), AF-1 and AF-2. AF-1 is localized in the N-terminal region, and AF-2 is distributed in the C-terminal ligand-binding domain (LBD) of the ERα protein. Helix 12 (H12) in the LBD is a component of the AF-2, and the configuration of H12 is ligand-inducible to an active or inactive form. We demonstrated previously that the ERα mutant (AF2ER) possessing L543A,L544A mutations in H12 disrupts AF-2 function and reverses antagonists such as fulvestrant/ICI182780 (ICI) or 4-hydoxytamoxifen (OHT) into agonists in the AF2ER knock-in mouse. Our previous in vitro studies suggested that the mode of AF2ER activation is similar to the partial agonist activity of OHT for WT-ERα. However, it is still unclear how antagonists activate ERα. To understand the molecular mechanism of antagonist reversal activity, we analyzed the correlation between the ICI-dependent estrogen-responsive element-mediated transcription activity of AF2ER and AF2ER-LBD dimerization activity. We report here that ICI-dependent AF2ER activation correlated with the activity of AF2ER-LBD homodimerization. Prevention of dimerization impaired the ICI-dependent ERE binding and transcription activity of AF2ER. The dislocation of H12 caused ICI-dependent LBD homodimerization involving the F-domain, the adjoining region of H12. Furthermore, F-domain truncation also strongly depressed the dimerization of WT-ERα-LBD with antagonists but not with E2. AF2ER activation levels with ICI, OHT, and raloxifene were parallel with the degree of AF2ER-LBD homodimerization, supporting a mechanism that antagonist-dependent LBD homodimerization involving the F-domain results in antagonist reversal activity of H12-mutated ERα.
Introduction
Estrogen regulates physiological responses in target cells by means of intracellular estrogen receptors (ERs)2. Major estrogenic activity appears through the nuclear ERα and ERβ that activate target genes related to biological tissue responses directly as ligand-dependent transcription factors (1, 2). ERs consist of homologous structural domains, designated A to F (Fig. 1), that are shared between the nuclear receptor superfamily (3, 4). ERα has two transactivating functional (AF) domains, AF-1 and AF-2. AF-1 is localized in the A/B domains, and AF-2 is distributed in the E-domain or ligand-binding domain (LBD) of the ERα protein. Helix 12 (H12) of ERα located in the LBD is a component of the AF-2 domain. The configuration of H12 is changed by agonist or antagonist binding to the active or inactive form of ERα protein, respectively (5). ERα makes a homodimer to bind the estrogen-responsive element (ERE) within the promoter region of target genes to regulate transcription. ERα contains two dimerization signals, a ligand-inducible major dimerization function in the LBD and a constitutive weak dimerization function associated with the DNA binding domain (DBD) or C-domain (6, 7). The mutation of L511R on helix 11 (H11) of mouse ERα has been reported to result in an E2 non-active mutant because of disruption of homodimerization (6). The results of crystallography support a structure that shows H11 making an interface of ERα LBD monomers to form the dimer (5, 8).
FIGURE 1.
Schematic diagrams of mouse ERα WT and mutants used in this study. A, Schematic of WT and mutated mouse ERα used for transcription assay. B, Schematic of Gal4-DBD-fused (light gray) or VP16-AD-fused (dark gray) WT and mutated ERα LBDs used for the mammalian two-hybrid assay.
We demonstrated previously the in vivo biological functionality of the ERα H12 mutant that possesses L543A,L544A mutations (AF2ER) (9, 10). These mutations disrupted the AF-2 function and resulted in a reversal of antagonists, such as fulvestrant/ICI182780 (ICI) and tamoxifen into agonists both in vitro in cell-based experiments and in vivo in the AF2ER knock-in (AF2ERKI) mouse. The AF2ERKI mouse expresses AF2ER mutant protein in the classical estrogen target tissues, but endogenous estradiol (E2) does not activate AF2ER. Thus, the phenotype of the AF2ERKI mouse is quite similar to the ERα knockout (αERKO) mouse that expresses no ERα protein (11), indicating that this mutant receptor is essentially inactive. However, unlike αERKO mice, ICI and TAM activate the AF2ER mutant ERα and mediate physiological responses in the AF2ERKI mice, such as uterotrophic effects in the female (9) and male reproductive tract functions (10). The mode of ICI-mediated AF2ER activation is similar to the partial agonist activity of 4-hydroxytamoxifen (OHT) on WT ERα. Namely, the overexpression of transcription coactivator, p300/cAMP-response element-binding protein (CREB)-binding protein enhanced both ICI-mediated AF2ER activation and OHT-mediated WT ERα activation through the A/B domains of ERα protein in a similar manner (9). This observation supported the previous findings, which suggest that the OHT-mediated partial agonistic activity for WT ERα can only be mediated by AF-1 (12). However, there is little information available about the mechanism of antagonist-mediated ERα activation. In this report, we analyze the correlation between ICI-dependent ERE-mediated transcription activity and LBD dimerization activity of AF2ER for a further understanding of the molecular mechanisms of antagonist-mediated ERα transcription activation.
We demonstrate here that the ICI-dependent ERE-mediated transcriptional activity of AF2ER is associated with the AF2ER-LBD dimerization activity. Furthermore, prevention of dimerization impaired the ICI-dependent AF2ER activation and ERE binding of AF2ER. Additionally, we suggest that the dislocation of H12 causes ICI-dependent LBD homodimerization involving the F-domain of ERα, which facilitates dimerization. Involvement of the F-domain in the antagonist-dependent LBD dimerization occurred not only with AF2ER but also with WT ERα. Understanding the mechanism of antagonist-mediated AF2ER activation will provide insights into the mechanism of partial agonist/antagonist activity of selective estrogen receptor modulators (SERMs).
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids used for reporter assay are as follows: pcDNA3-mERa; the pcDNA3 plasmid contains full-length mouse ERα (mERα1–599), pcDNA3–121-ERa; the pcDNA3 plasmid contains N-terminal 120-amino acid truncated mouse ERα (mERα121–599), pcDNA3-AF2ER; the pcDNA3 plasmid contains L543A,L544A mutated full-length mouse ERα (mERα1–599, L543A,L544A), and pcDNA3–121-AF2ER; the pcDNA3 plasmid contains L543A,L544A mutated N-terminal 120-amino acid truncated mouse ERα (mERα121–599, L543A,L544A) (9). The plasmid 3xERE-TATA-Luc (pGL3–3xERE-TATA-Int-Luc) was used for the ERE reporter gene. The plasmid pRL-TK Renilla luciferase expression plasmid (Promega) was used for internal control. To generate plasmids pcDNA3-mERaΔF and pcDNA3-AF2ERΔF, the cDNAs of mouse ERα WT and AF2ER were amplified by PCR using the following primer set: mE/F-5′, 5′-GGA TCC AGC ACA CTA AGA AGA ATA GCC CTG CCT T-3′; mERaΔF-3′ (XhoI), 5′-CAC TCT CGA GCT ACT GGC TGG GGC ATG AAG-3′. The amplified fragment was cloned into pCR2.1 (Invitrogen) by the TA cloning kit (Invitrogen) and sequenced (National Institute of Environmental Health Sciences sequencing laboratory). The fragments were excised from the plasmids pCR2.1-LBD(ERaΔF)_XhoI and pCR2.1-LBD(AF2ΔF)_XhoI by XhoI and then subcloned into the XhoI sites of the pcDNA3-mERa plasmid. The direction of the inserted fragment was determined by SmaI digestion. The plasmids pcDNA3-mERaΔH12, pcDNA3-mERa-L511R, and pcDNA3-AF2ER-L511R were generated by PCR-based, site-directed mutagenesis, and the following oligo DNAs were used for the mutagenesis: ΔH12_S, 5′-CCT CTA TGA TGC CCA CCG CCT TCA TGC CCC AGC CAG-3′; ΔH12_AS, 5′-GCG GTG GGC ATC ATA GAG GGG CAC AAC GTT CTT GCA TTT C-3′; L511R_S, 5′-GCC GCC TAG CTC AGC GCC TTC TCA TTC TTT CC-3′; and L511R_AS, 5′-GGA AAG AAT GAG AAG GCG CTG AGC TAG GCG GC-3′. PCR was performed using the Pfu Turbo DNA polymerase (Agilent Technologies), a pair of sense (S) and antisense (AS) oligo DNAs, and the plasmids pBluescript-mERa_XhoI or pBluescript-AF2ER_XhoI (the XhoI fragment from pcDNA3-mERa or pcDNA3-AF2ER was subcloned into the XhoI site of pBluescript) as a template, following the instructions of the manufacturer (Agilent Technologies). Mutated clones were confirmed by sequencing and then subcloned into the XhoI sites of pcDNA3-mERa. The direction of the inserted fragment was determined by NotI digestion. Plasmids used for mammalian two-hybrid assay are as follows: the plasmid pACT (Promega) was used for the prey, the plasmid pBIND (Promega) was used for the bait (this plasmid includes the Renilla luciferase expression cassette for internal control), and the plasmid pG5-Luc (Promega) was used for the Gal4 binding element reporter gene. The cDNAs of mouse ERα WT and the AF2ER LBD regions were amplified by PCR using the following primer set: mE/F-5′, 5′-GGA TCC AGC ACA CTA AGA AGA ATA GCC CTG CCT T-3′ and mE/F-3′, 5′-GGT ACC TGG GAG CTC TCA GAT CGT GTT GGG-3′. The amplified fragment was cloned into pCR2.1 by TA cloning kit and sequenced. To generate the plasmids pACT-LBD/WT, pACT-LBD/AF2ER, pBIND-LBD/WT, and pBIND-LBD/AF2ER, LBD fragments were excised from the plasmids pCR2.1-mE/F(WT) and pCR2.1-mE/F(AF2) by BamHI and KpnI and then subcloned into the BamHI and KpnI sites of pACT or pBIND. The plasmids pACT-LBD/WTΔF, pACT-LBD/AF2ERΔF, pBIND-LBD/WTΔF, and pBIND-LBD/AF2ERΔF were generated as follows. The cDNAs of mouse ERα WT and the AF2ER LBD regions were amplified by PCR using the following primer set: mE/F-5′, 5′-GGA TCC AGC ACA CTA AGA AGA ATA GCC CTG CCT T-3′ and mERaΔF-3′(KpnI), 5′-GGT ACC TAG CGA CTG GCT GGG GCA TGA-3′. The amplified fragments were cloned into pCR2.1 by TA cloning kit and sequenced. The fragments were excised from the plasmids pCR2.1-LBD(WTΔF)_KpnI and pCR2.1-LBD(AF2ΔF)_KpnI by BamHI and KpnI and then subcloned into the BamHI and KpnI sites of pACT or pBIND. The plasmids pACT-LBD/ΔH12, pBIND-LBD/ΔH12, pACT-LBD/WT-L511R, pBIND-LBD/WT-L511R, pACT-LBD/AF2ER-L511R, and pBIND-LBD/AF2ER-L511R were generated by PCR-based, site-directed mutagenesis. The same sets of oligo DNAs as described (ΔH12_S, ΔH12_AS, L511R_S, and L511R_AS) were used for the mutagenesis. The PCR was performed using the Pfu Turbo DNA polymerase, a pair of S and AS oligo DNAs, and the plasmids pCR2.1-mE/F(WT) or pCR2.1-mE/F(AF2) as a template. Mutated clones were confirmed by sequencing and then subcloned into the pACT or pBIND plasmids. Plasmids used for protein degradation assay are as follows. The plasmid pTet-off (Clontech) was used for the tetracycline-dependent suppression of the Tet-responsive element (TRE) containing plasmids-derived gene expression, and the plasmid pCMV-SPORT-β-gal was used for internal control. The plasmids pcTRE-mERa, pcTRE-AF2ER, pcTRE-ERa(L511R), and pcTRE-AF2ER(L511R) were generated as follows. The ScaI-EcoRI fragment containing the TRE with the minimal CMV promoter was excised from the pTRE plasmid (Clontech) and the excised fragment was subcloned into the ScaI and EcoRI sites of pcDNA3-mERa, pcDNA3-AF2ER, pcDNA3-ERa(L511R), and pcDNA3-AF2ER(L511R).
Cell Culture and Transfection Conditions for the Luciferase Assay
HepG2 (human hepatocellular carcinoma) cells were cultured in phenol red-free α-minimal essential medium supplemented with 10% FBS and 1% penicillin-streptomycin (Sigma-Aldrich). For transient transfections, the cells were cultured in phenol red-free medium supplemented with 10% charcoal-stripped FBS (Gemini-Bio) and seeded in 48-well plates at a density of 1.2 × 105 cells/well. The cells were transfected with the following DNA mixture for 6 h using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. For the reporter assay, a DNA mixture containing 50 ng of expression plasmids for WT or mutated ERα (Fig. 1A), 100 ng of reporter plasmid for 3xERE-TATA-luc, and 100 ng of Renilla luciferase expression plasmid pRL-TK (Promega) was transfected in each well. For the mammalian two-hybrid assay, a DNA mixture containing 50 ng of expression plasmids for Gal4-DBD fusion proteins (pBIND), 50 ng of expression plasmids for VP16 activation domain (AD) fusion proteins (pACT) (Fig. 1B), and 100 ng of pG5-Luc reporter plasmid was transfected in each well. The pBIND plasmid contains a Renilla luciferase expression unit for transfection normalization. To analyze the coactivator motif interaction with the LBD, a DNA mixture containing 50 ng of expression plasmid for Gal4-DBD-fused SRC1 NR-box (amino acids 621–765 of human SRC1a) (pM-SRC1-NR) (13), 50 ng of expression plasmids for VP16-AD fusion proteins (pACT), 100 ng of pG5-Luc reporter plasmid, and 100 ng of pRL-TK was transfected in each well.
Luciferase Assay
The cells were cultured in fresh medium supplemented with E2 (Sigma-Aldrich), ICI (Tocris Bioscience), OHT (Sigma-Aldrich), or raloxifene (Ral, Tocris Bioscience) 6 h after transfections. Luciferase and Renilla luciferase activities were assayed 18 h after treatments. Luciferase activity was normalized for transfection efficiency using Renilla luciferase as an internal control. All results are representative of at least three independent experiments and represent the mean ± S.D. of triplicate samples.
Western Blot Analysis
Cell lysates for the protein degradation assay and DNA binding assay were prepared by the following extraction method. Cells were lysed in an extraction buffer containing 50 mm Tris/HCl (pH8.0), 5 mm EDTA, 1% Nonidet P-40, 0.2% sarkosyl, 0.4 m NaCl, 1× Halt protease inhibitor mixture (Pierce), and 1 mm dithiothreitol by vortexing, followed by a 15-min incubation on ice. After centrifugation for 5 min (21,000 × g at 4 °C), the protein amount in the supernatant was determined by BCA assay (Pierce). Proteins were resolved by SDS-PAGE and subsequently transferred to nitrocellulose membranes. Blots were incubated overnight in 4 °C with primary antibody for ERα (1:650, catalog no. MC-20, Santa Cruz Biotechnology; 1:350, catalog no. H-184, Santa Cruz Biotechnology; or 1:200, catalog no. TE111.5D11, Thermo), β-actin (1:1500, catalog no. AC-74, Sigma), or β-galactosidase (1:1000, catalog no. ab616, Abcam). We found that the F-domain of mERα is the antigenic site of MC-20. Therefore, we used H-184 and TE111.5D11 to determine the ΔF protein expression. The blots were washed and then incubated with IRDye infrared dye-conjugated anti-rabbit antibody (LI-COR Biosciences) for ERα (MC-20 and H-184) and β-galactosidase or with IRDye infrared dye-conjugated anti-mouse antibody (LI-COR Biosciences) for ERα (TE111.5D11) and β-actin. The signals were visualized with an Odyssey infrared imaging system (LI-COR Biosciences).
Protein Degradation Assay
HeLa cells were cultured in phenol red-free DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. For transient transfections, the cells were cultured in phenol red-free medium supplemented with 10% charcoal-stripped FBS and seeded in 60-mm dishes at a density of 1.0 × 106 cells/dish. The cells were transfected with the following DNA mixture for 12 h using Lipofectamine 2000. Cells were transfected with DNA mixture containing 0.5 μg pcTRE-mERa, pcTRE-ERa(L511R), pcTRE-AF2ER, or pcTRE-AF2ER(L511R) plasmid, 1.5 μg of pTet-off plasmid, and 0.5 μg of β-galactosidase expression plasmid pCMV-SPORT-β-gal. After 12 h, cells were treated with 1 μg/ml doxycycline (Clontech) to suppress the synthesis of WT and mutant ERα from the pcTRE plasmids. Cells were cultured with or without 100 nm ICI and harvested 2, 4, and 6 h after doxycycline treatment.
DNA Binding Assay
The DNA binding assay was performed using the NoShift transcription factor assay kit (Novagen) with the following modifications to the instructions of the manufacturer. The cell lysates that expressed WT and mutant ERα proteins were incubated on ice for 90 min in the 24-μl reaction mixture containing 1xNoShift binding buffer (Novagen), 0.005 units/μl poly(dI-dC), 25 ng/μl salmon sperm DNA, and 0.5 pmol/μl biotinylated 1xERE (S, 5′-GTC CAA AGT CAG GTC ACA GTG ACC TGA TCA AAG TT-3′, and AS, 5′-AAC TTT GAT CAG GTC ACT GTG ACC TGA CTT TGG AC-3′) with or without ligands (1 μm E2 or 1 μm ICI). The DNA-protein complexes were put into streptavidin-coated 8-well strips (Novagen). The samples were incubated for 1 h at 37 °C. The DNA-protein complex-bound strips were washed by 1xNoShift washing buffer (Novagen), and then primary antibody for ERα (1:300, catalog no. MC-20, Santa Cruz Biotechnology) suspended in 1xNoShift antibody dilution buffer (Novagen) was added. After 1 h of incubation at 37 °C, the DNA-protein-antibody complex-bound strips were washed, and HRP-conjugated anti-rabbit antibody (1:1000, Cell Signaling Technology) was added. After 30 min of incubation at 37 °C, the strips were washed, and 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate solution (Thermo) was added. The samples were incubated at room temperature, and 1 n HCl was added to measure the absorbance at 450 nm with a plate reader.
Statistical Analysis
Statistical analysis was performed with two-way ANOVA with Bonferroni's multiple comparison test by GraphPad Prism (GraphPad software), and p < 0.001 was considered statistically significant.
RESULTS
The Pure ERα Antagonist Activates ERE-mediated Transcription through AF2ER
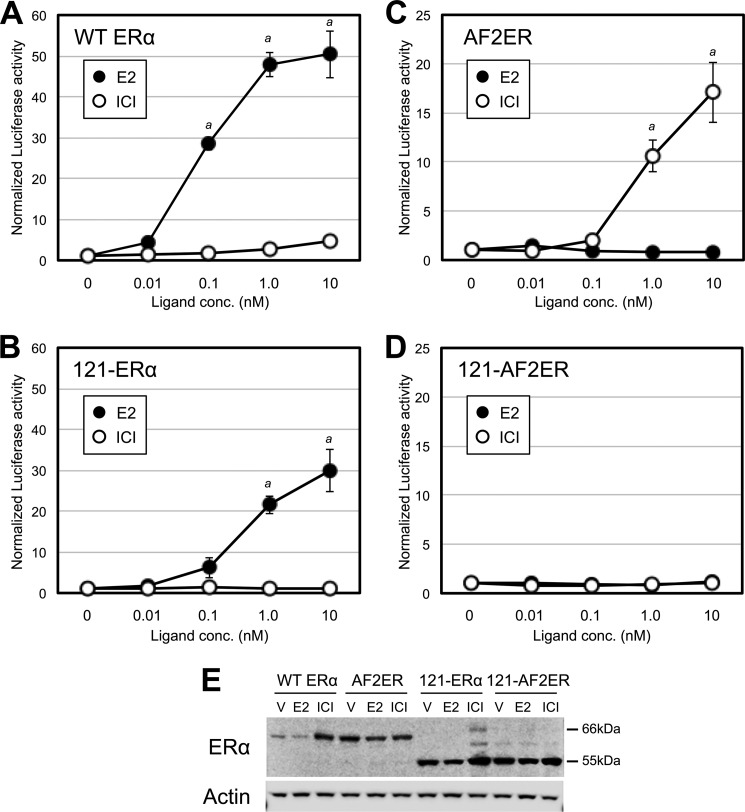
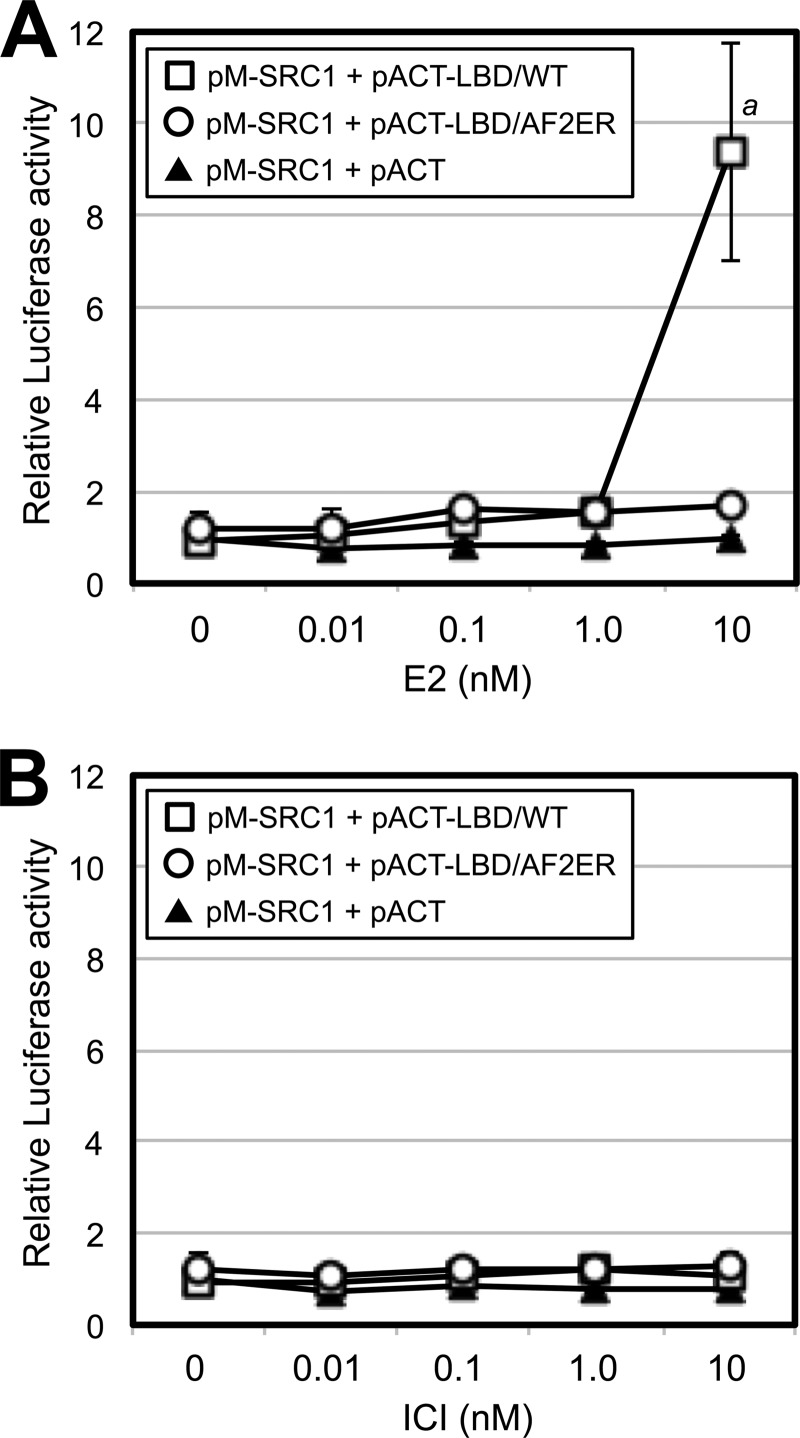
We demonstrated the antagonist reversal activity of AF2ER for ERE-mediated transcription with an in vitro transient transfection assay using ERα-negative HepG2 cells. As shown in Fig. 2, 0.1 nm and higher concentrations of E2 produced activation of the ERE reporter with WT ERα (Fig. 2A, ●) and a reduced activation with the N-terminal truncated ERα (121-ERα) because of the loss of AF-1 function (B, ●). The activities of WT ERα and 121-ERα were not induced by the pure ERα-antagonist ICI (Fig. 2, A and B, ○). In contrast, 1.0 and 10 nm ICI activated the ERE-mediated transcription of AF2ER (Fig. 2C, ○) but not with E2 (●). As shown in Fig. 2D, ICI-mediated AF2ER activation was diminished by the N-terminal truncation of AF2ER (121-AF2ER). Furthermore, we analyzed ligand-dependent p160 coactivator (SRC1) recruitment to the WT ERα and AF2ER LBDs using a mammalian two-hybrid assay. HepG2 cells were cotransfected with a Gal4-responsive reporter (pG5-Luc) and vector for the Gal4-DBD fused to the SRC1 NR-box that contains the three LXXLL motifs (13) in the presence of the vectors for VP16-AD fused to the amino termini of either the WT or AF2ER LBD. Cells were treated with a series of concentrations of E2 or ICI (0.01–10 nm). The SRC1 NR-box bound to the WT LBD with 10 nm E2 (Fig. 3A, □) but not ICI (B, □). On the other hand, the SRC1-NR-box did not bind to AF2ER-LBD, neither with E2 nor ICI (Fig. 3, A and B, ○). These results suggest that the AF2ER mutation has completely inactivated the AF-2 function and that AF-1 is necessary for ICI-mediated AF2ER activation.
FIGURE 2.
ICI activates AF2ER-dependent, ERE-mediated transcription. HepG2 cells were cotransfected with the reporter gene (3xERE-TATA-luc), reference gene (pRL-TK), and expression vector for WT ERα (A); N-terminal truncated ERα (121-ERα) (B); AF2ER (C); or N-terminal truncated AF2ER (121-AF2ER) (D) and treated with either vehicle (0 nm), E2 (0.01–10 nm, ●), or ICI182780 (0.01–10 nm, ○). The luciferase activities are represented as fold change over vehicle (0 nm). Luciferase activity is represented as mean ± S.D. a, p < 0.001 against vehicle (0 nm) in each panel. E, Whole cell lysates extracted from vehicle-treated (V), E2-treated (1 nm), and ICI-treated (10 nm) transfected HepG2 cells were analyzed by immunoblotting with the anti-ERα antibody (MC-20, ERα). β-actin (Actin) was used as a loading control. A representative Western blot analysis is shown.
FIGURE 3.
AF2ER-LBD did not recruit the SRC1 NR-box. HepG2 cells were cotransfected with pG5-luc, pRL-TK, and the expression vector for the Gal4-DBD-fused SRC1 NR-box (pM-SRC1-NR) in the presence of expression vectors for VP16-AD (pACT, ▴), VP16-AD-fused ERα WT-LBD (pACT-LBD/WT, □) or VP16-AD-fused AF2ER-LBD (pACT-LBD/AF2ER, ○). A, cells were treated with vehicle (0 nm) or E2 (0.01–10 nm). B, cells were treated with vehicle (0 nm) or ICI (0.01–10 nm). The luciferase activities are represented as fold change over vehicle (0 nm) in the pACT and pM-SRC1-NR co-transfected cells (▴). Luciferase activity is represented as the mean ± S.D. a, p < 0.001 against vehicle (0 nm).
ICI-dependent AF2ER LBD Dimerization Correlates with the Transcription Activity of AF2ER
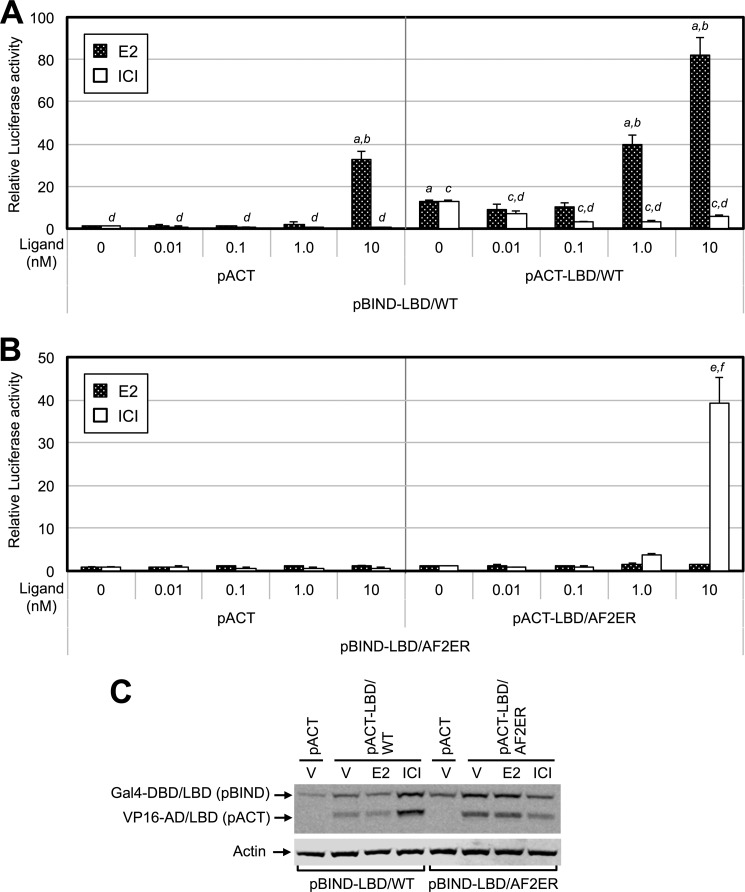
Because the dimerization of ERα is an important step for transcription activation, we evaluated the correlation between ligand-dependent LBD dimerization and transcription activities. To examine the activity of LBD homodimerization, we performed a mammalian two-hybrid assay. HepG2 cells were cotransfected with pG5-Luc and vectors for the Gal4-DBD fused to the amino termini of either WT or mutated ERα LBD (pBIND-LBD) in the presence of the vectors for VP16-AD alone (pACT) or VP16-AD fused to the amino termini of either WT or mutated ERα LBD (pACT-LBD). Cells were treated with a series of concentrations of E2 or ICI (0.01–10 nm). At first, we demonstrated ligand-dependent WT-LBD homodimerization. As shown in Fig. 4A, we observed a relatively high basal level of luciferase activity in the pBIND-LBD/WT and pACT-LBD/WT co-transfected cells without ligand (0 nm), and that activity was increased with 1 nm E2 (Fig. 4A, right panel, black column), suggesting that the WT-LBD makes a homodimer without ligand and that the level is increased by E2. Treatment with ICI decreased the level of the WT-LBD homodimer (Fig. 4A, right panel, white column). The combination of pBIND-LBD/WT with pACT induced luciferase activity at 10 nm E2 (Fig. 4A, left panel, black column), suggesting that pBIND-LBD/WT itself induces the transcription activity at a higher E2 concentration. Thus, using this method, we could not define whether the dimerization of WT-LBD was induced at higher E2 concentrations. On the other hand, the homodimer of AF2ER-LBD was not detected at any E2 concentrations (Fig. 4B, black column). The level of the AF2ER-LBD homodimer was increased by 10 nm ICI (Fig. 4B, right panel, white column), suggesting that the ICI-dependent AF2ER-LBD homodimerization coincides with the transcription activity of AF2ER. For the following two-hybrid and reporter assays, we used concentrations of E2 from 0–1 nm for WT ERα and concentrations of ICI from 0–10 nm for AF2ER.
FIGURE 4.
ICI-dependent LBD dimerization activity correlates with AF2ER activation. A, HepG2 cells were cotransfected with pG5-luc and expression vector for Gal4-DBD-fused WT-LBD (pBIND-LBD/WT) in the presence of expression vector for VP16-AD (pACT, left panel) or VP16-AD-fused WT-LBD (pACT-LBD/WT, right panel). The luciferase activity is represented as fold change over vehicle (0 nm) in the pACT and pBIND-LBD/WT co-transfected cells. B, HepG2 cells were cotransfected with pG5-luc and expression vector for Gal4-DBD-fused AF2ER-LBD (pBIND-LBD/AF2ER) in the presence of expression vector for VP16-AD (pACT, left panel) or VP16-AD-fused AF2ER-LBD (pACT-LBD/AF2ER, right panel). The luciferase activity is represented as fold change over vehicle (0 nm) in the pACT and pBIND-LBD/AF2ER co-transfected cells. Cells were treated with either vehicle (0 nm), E2 (0.01–10 nm, black column), or ICI (0.01–10 nm, white column). Luciferase activity is represented as mean ± S.D. a and c, p < 0.001 in each treatment against vehicle (0 nm) in the pACT and pBIND-LBD/WT co-transfected cells; b and d, p < 0.001 in each treatment against vehicle (0 nm) in the pACT-LBD/WT and pBIND-LBD/WT co-transfected cells; e, p < 0.001 in ICI treatment against vehicle (0 nm) in the pACT and pBIND-LBD/AF2ER co-transfected cells; e and f, p < 0.001 in ICI treatment against vehicle (0 nm) in the pACT-LBD/AF2ER and pBIND-LBD/AF2ER co-transfected cells. C, whole cell lysate was prepared from vehicle (V), E2 (1 nm), and ICI-treated (10 nm) transfected HepG2 cells and analyzed by immunoblotting with anti-ERα antibody (MC-20) to indicate the expression levels of Gal4-DBD/LBD and VP16-AD/LBD. β-actin (Actin) was used as a loading control. A representative Western blot analysis is shown.
Homodimerization Is Necessary for the Antagonist Reversal Activity of AF2ER
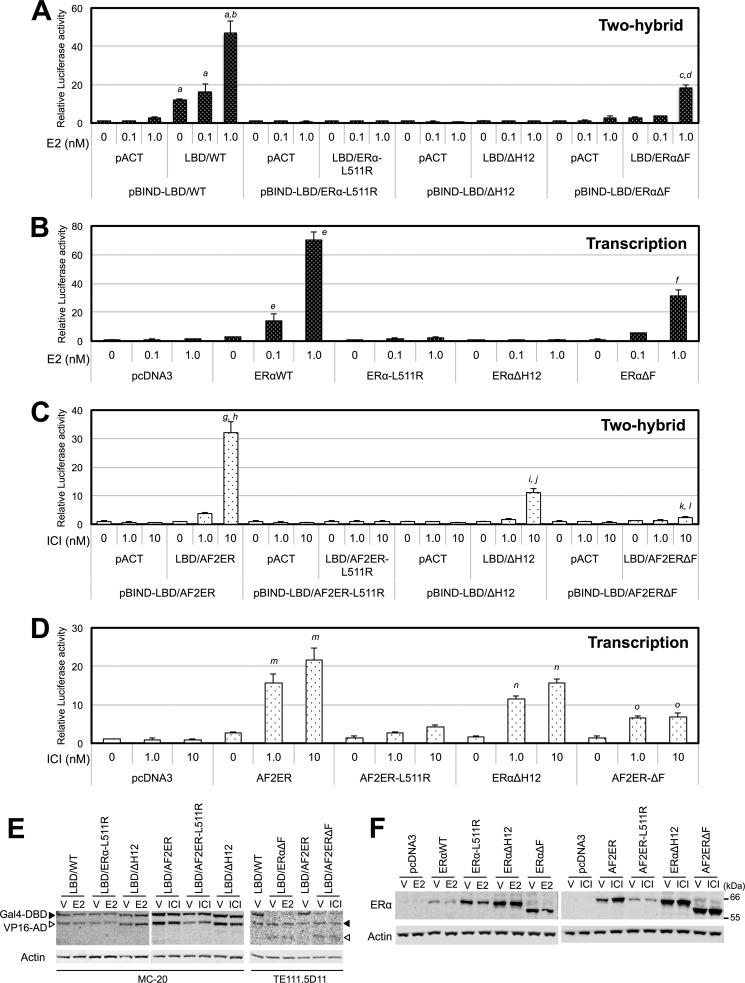
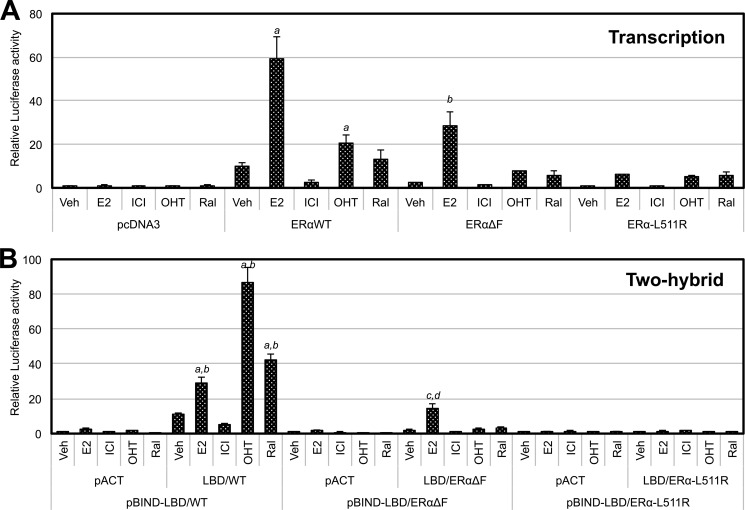
Mutation of leucine 511 to arginine (L511R) has been shown previously to inhibit the E2-dependent dimerization of mouse ERα (6). Thus, we generated the L511R-mutated ERα (ERα-L511R), AF2ER (AF2ER-L511R), and two-hybrid constructs of the LBD (LBD/ERα-L511R, LBD/AF2ER-L511R) to evaluate the significance of LBD dimer formation on the antagonist reversal activity of AF2ER. We found no luciferase activities from the pBIND-LBD/ERα-L511R with pACT-LBD/ERα-L511R- or pBIND-LBD/AF2ER-L511R with pACT-LBD/AF2ER-L511R-transfected cells treated with E2 or ICI, respectively (Fig. 5, A and C), suggesting that the L511R-mutated LBDs did not induce ligand-dependent homodimerization. The ligand-dependent ERE-mediated transcription activations of ERα-L511R and AF2ER-L511R were attenuated (Fig. 5, B and D), suggesting that homodimerization is required for the ligand-dependent ERα activation and the antagonist reversal activity of AF2ER.
FIGURE 5.
LBD dimer formation and transcription activities. A, HepG2 cells were cotransfected with pG5-luc and expression vector for Gal4-DBD fused WT or mutated ERα LBDs (pBIND-LBD/WT, pBIND-LBD/ERα-L511R, pBIND-LBD/ΔH12, or pBIND-LBD/ERαΔF) in the presence of the expression vector for VP16-AD (pACT) or VP16-AD-fused WT or mutated ERα LBDs (pACT-LBD/WT, pACT-LBD/ERα-L511R, pACT-LBD/ΔH12, or pACT-LBD/ERαΔF). Cells were treated with either vehicle (0 nm) or E2 (0.1 and 1 nm). B, HepG2 cells were cotransfected with 3xERE-TATA-luc, pRL-TK, and expression vector for WT ERα, ERα-L511R, ERαΔH12, or ERαΔF and treated with either vehicle (0 nm) or E2 (0.1 and 1 nm). C, HepG2 cells were cotransfected with pG5-luc and the expression vector for Gal4-DBD-fused AF2ER or mutated ERα LBDs (pBIND-LBD/AF2ER, pBIND-LBD/AF2ER-L511R, pBIND-LBD/ΔH12, or pBIND-LBD/AF2ERΔF) in the presence of the expression vector for VP16-AD (pACT), VP16-AD-fused AF2ER, or mutated ERα LBDs (pACT-LBD/AF2ER, pACT-LBD/AF2ER-L511R, pACT-LBD/ΔH12, or pACT-LBD/AF2ERΔF). Cells were treated with either vehicle (0 nm) or ICI (1 and 10 nm). D, HepG2 cells were cotransfected with 3xERE-TATA-luc, pRL-TK, and expression vector for AF2ER, AF2ER-L511R, ERαΔH12, or AF2ERΔF and treated with either vehicle (0 nm) or ICI (1 and 10 nm). The luciferase activities in A and C are represented as fold change over vehicle in each pACT and pBIND-LBD co-transfected sample. The luciferase activities in B and D are represented as fold change over vehicle in the empty expression vector-transfected (pcDNA3) cells. Luciferase activities are represented as mean ± S.D. a, c, g, i, and k, p < 0.001 against vehicle in each pACT and pBIND-LBD co-transfected sample; b, d, h, j, and l, p < 0.001 against vehicle in each pACT-LBD and pBIND-LBD co-transfected sample; e, f, m, n, and o, p < 0.001 against the vehicle level of each receptor. E, whole cell lysate was prepared from vehicle-treated (V), E2-treated (1 nm), and ICI-treated (10 nm) transfected HepG2 cells and analyzed by immunoblotting with anti-ERα antibody (MC-20 or TE111.5D11) to indicate the expression levels of Gal4-DBD/LBD (closed arrowhead) and VP16-AD/LBD (open arrowhead). β-actin (Actin) was used as a loading control. A representative Western blot analysis is shown. F, whole cell lysates extracted from vehicle-treated (V), E2-treated (1 nm), and ICI-treated (10 nm) transfected HepG2 cells were analyzed by immunoblotting with anti-ERα antibody (H-184, ERα) to indicate the expression levels of ERα WT and mutants. β-actin (Actin) was used as a loading control. A representative Western blot analysis is shown.
Disruption of Helix 12 Causes the Antagonist Reversal Activity
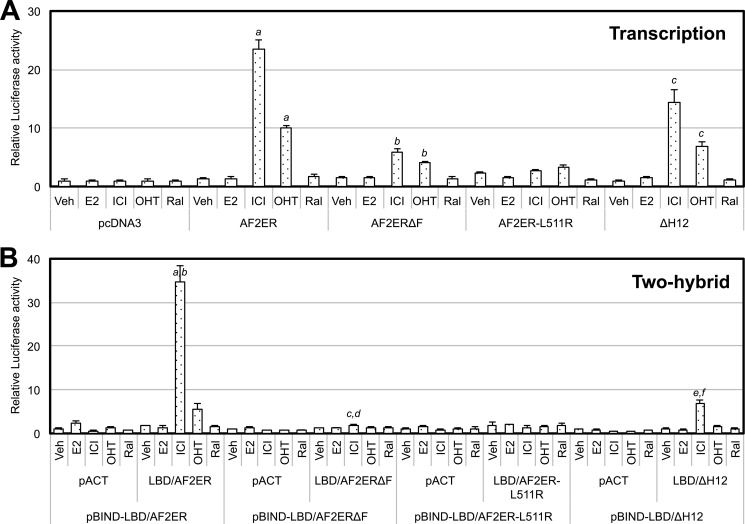
To evaluate the impact of the disruption of H12 on antagonist reversal activity, the entire H12 (seven amino acids, DLLLEML) was deleted from ERα (ΔH12), and ERαΔH12 and LBD/ΔH12 were generated. Surprisingly, we found that 10 nm ICI induced the luciferase activity in the pBIND-LBD/ΔH12 and pACT-LBD/ΔH12 co-transfected cells (Fig. 5C) but not E2 (A). The E2-dependent transcription activity of ERαΔH12 was diminished (Fig. 5B). In contrast, ERαΔH12 was activated by ICI, and that level was lower than AF2ER (Fig. 5D), correlating with the level of ICI-dependent LBD/ΔH12 homodimerization. These results may suggest that the H12 is disoriented in the AF2ER mutant, causing the antagonist reversal activity of AF2ER.
The F-domain Is Important for Antagonist Reversal Activity of AF2ER
The results from the ΔH12 mutation evoked the question of whether the disposition of H12 affects the function of the adjoining F-domain. We generated an F-domain-truncated ERα (ERαΔF), AF2ER (AF2ERΔF), and two-hybrid constructs of LBD (LBD/ERαΔF, LBD/AF2ERΔF) to evaluate the involvement of the F-domain in the antagonist reversal activity. In the mammalian two-hybrid assay, the basal level of luciferase activity in the LBD/ERαΔF-transfected cells was lower than the level of LBD/WT-transfected cells without ligand (0 nm), but it was increased with 1 nm E2 (Fig. 5A). The E2-dependent transcription activities of ERαΔF and WT were increased in parallel with the levels of LBD/ERαΔF and LBD/WT homodimerization, respectively (Fig. 5B). On the other hand, the level of luciferase activity in the ICI-treated pBIND-LBD/AF2ERΔF with pACT-LBD/AF2ERΔF-transfected cells was impaired but was detectable at 10 nm ICI (Fig. 5C). ICI-dependent AF2ERΔF transcription was attenuated and not induced in a dose-dependent manner (Fig. 5D). These results suggest that the F-domain plays a role in the antagonist reversal activity of AF2ER.
Homodimerization Affects the Ligand-dependent DNA Binding Activity
To evaluate the effect of homodimerization on DNA binding activity, we analyzed the ligand-dependent DNA binding activities of WT ERα, AF2ER, and L511R mutants (ERα-L511R and AF2ER-L511R). As shown in Fig. 6, the DNA binding level of WT was increased significantly by treatment with E2 or ICI compared with no-ligand (-Ligand). The DNA binding level of AF2ER was increased significantly by ICI treatment but not E2 compared with no ligand. These responses were eliminated in the ERα-L511R or AF2ER-L511R mutants. These results suggest that ICI-dependent AF2ER homodimerization influences ERE binding activity.
FIGURE 6.
Disruption of dimerization inhibits the ligand-dependent DNA binding of AF2ER. To detect the ligand-dependent ERE binding activities of WT and mutated ERα, a biotinylated ERE probe was incubated with either vehicle (-Ligand), 1 μm E2 (+E2), or ICI (+ICI) and HeLa cell extracts that were transfected with pcDNA3 (empty), WT ERα, ERα-L511R, AF2ER, or AF2ER-L511R expression plasmids. Detection was performed as described under “Experimental Procedures.” Relative DNA binding activity was represented as fold change over the pcDNA3-transfected cell level. Results are represented as the mean ± S.D. of three independent experiments. a and b, p < 0.001 against vehicle sample (-Ligand) of each receptor. A representative Western blot analysis shows the levels of ERα WT and mutants and endogenous β-actin in each extract used for the DNA binding assay (inset).
Dimerization Does Not Affect ICI-mediated ERα Protein Degradation
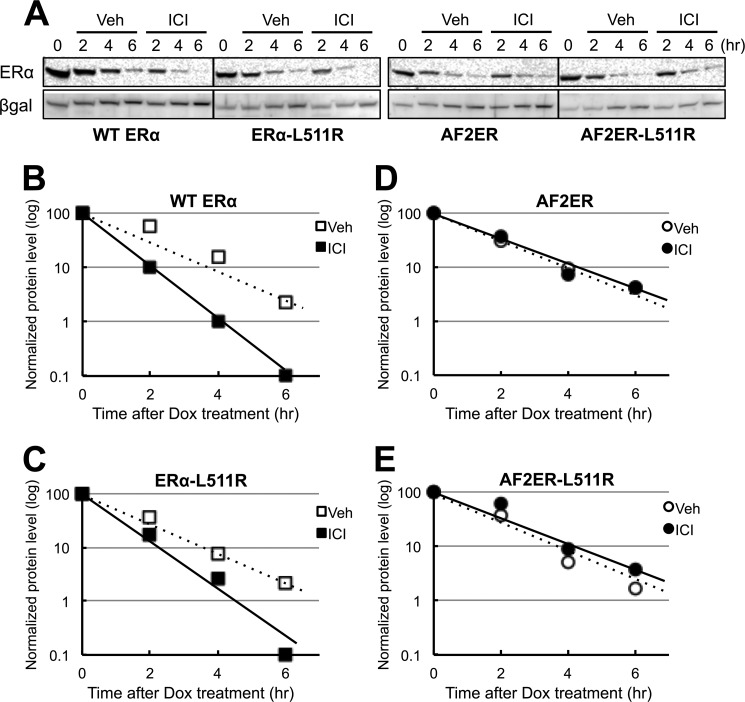
We analyzed the effect of ICI on AF2ER protein stability because it is well known that ICI induces the degradation of ERα protein (14, 15). To exclude the possibility of any chemical effect on the expression level of transfected cDNAs, we employed the tetracycline-dependent transcription repression method (Tet-Off system) for determining the protein degradation. As shown in Fig. 7B, ICI induced the degradation of WT protein. On the contrary, ICI did not induce AF2ER protein degradation (Fig. 7D). To evaluate the effect of dimerization on ERα protein degradation, we analyzed the effect of ICI on ERα-L511R and AF2ER-L511R mutants. The profiles of ERα-L511R and AF2ER-L511R mutants were identical to WT and AF2ER, respectively (Fig. 7, C and E), suggesting that dimerization does not affect ICI-dependent ERα protein degradation.
FIGURE 7.
AF2ER mutation prevents ICI-mediated proteolysis. A, WT ERα-, ERα-L511R-, AF2ER-, and AF2ER-L511R-expressing cells were treated with doxycycline (Dox,1 μg/ml) and were incubated with (ICI) or without (Veh) 100 nm ICI as described under “Experimental Procedures.” Cells were harvested at 2, 4, and 6 h after Dox treatment and then lysed and analyzed by immunoblotting with anti-ERα antibody (ERα, MC-20) and anti-β-galactosidase antibody (βgal). A representative Western blot analysis is shown. B–E, the level of ERα was normalized by βgal and plotted in a logarithmic scale. □ and ○ indicate vehicle (Veh) treatment. ■ and ● indicate ICI treatment. Representative results of three independent experiments are shown.
The Effect of LBD Homodimerization Correlates to Antagonist Reversal Activity of AF2ER
We analyzed the effect of other types of antagonists, OHT and Ral, on the transcription and LBD homodimerization activities of AF2ER, AF2ERΔF, AF2ER-L511R, ERαΔH12 (Fig. 8), WT ERα, ERαΔF, andERα-L511R (Fig. 9) using an ERE reporter assay and a mammalian two-hybrid assay, respectively. AF2ER and ERαΔH12-mediated transcription were activated by OHT to a lower level than ICI but were not activated by Ral (Fig. 8A). The levels of ligand-dependent LBD/AF2ER and LBD/ΔH12 homodimerization were parallel to the transcription activities of AF2ER and ERαΔH12, respectively (Fig. 8B). On the other hand, OHT and Ral induced WT-LBD homodimerization more than E2 (Fig. 9B). However, the transcription activity of WT ERα was in opposition to the level of WT-LBD homodimerization (Fig. 9A). Furthermore, F-domain truncation from WT-LBD (LBD/ERαΔF) dramatically depressed the OHT and Ral-dependent homodimerization of LBD/ERαΔF (Fig. 9B).
FIGURE 8.
The effect of SERMs on AF2ER, AF2ERΔF, AF2ER-L511R, and ERαΔH12 activities. A, HepG2 cells were cotransfected with 3xERE-TATA-luc, pRL-TK, and the expression vector for AF2ER, AF2ERΔF, AF2ER-L511R, ΔH12, or pcDNA3 and then treated with either vehicle (Veh), 10 nm E2, ICI, OHT, or Ral. The luciferase activity is represented as fold change against the empty expression vector (pcDNA3) in the presence of each ligand. Luciferase activity is represented as mean ± S.D. a, b, and c, p < 0.001 against the vehicle level of each receptor. B, HepG2 cells were cotransfected with pG5-luc and the following combinations: pBIND-LBD/AF2ER in the presence of pACT or pACT-LBD/AF2ER, pBIND-LBD/AF2ERΔF in the presence of pACT or pACT-LBD/AF2ERΔF, pBIND-LBD/AF2ER-L511R in the presence of pACT or pACT-LBD/AF2ER-L511R, and pBIND-LBD/ΔH12 in the presence of pACT or pACT-LBD/ΔH12. Cells were treated with either vehicle, 10 nm E2, ICI, OHT, or Ral. The luciferase activity is represented as a fold change over vehicle in each pACT and pBIND-LBD co-transfected sample. Luciferase activity is represented as mean ± S.D. a, c, and e, p < 0.001 against vehicle in each pACT and pBIND-LBD co-transfected sample; b, d, and f, p < 0.001 against vehicle in each pACT-LBD and pBIND-LBD co-transfected sample.
FIGURE 9.
The effects of SERMs on WT ERα, ERαΔF, and ERα-L511R activities. A, HepG2 cells were cotransfected with 3xERE-TATA-luc, pRL-TK, and the expression vector for WT ERα, ERαΔF, ERα-L511R, or pcDNA3 and then treated with either vehicle (Veh), 1 nm E2, ICI, OHT, or Ral. The luciferase activity is represented as fold change against the pcDNA3 in the presence of each ligand. Luciferase activity is represented as mean ± S.D. a and b, p < 0.001 against the vehicle level of each receptor. B, HepG2 cells were cotransfected with pG5-luc and the following combinations: pBIND-LBD/WT in the presence of pACT or pACT-LBD/WT, pBIND-LBD/ERαΔF in the presence of pACT or pACT-LBD/ERαΔF, and pBIND-LBD/ERα-L511R in the presence of pACT or pACT-LBD/ERα-L511R. Cells were treated with either vehicle, 1 nm E2, ICI, OHT, or Ral. The luciferase activity is represented as a fold change over vehicle in each pACT and pBIND-LBD co-transfected sample. Luciferase activity is represented as mean ± S.D. a and c, p < 0.001 against vehicle in each pACT and pBIND-LBD co-transfected sample; b and d, p < 0.001 against vehicle in each pACT-LBD and pBIND-LBD co-transfected sample.
DISCUSSION
ICI is a pure antagonist for WT ERα and blocks transcription activity (16). A portion of the antagonist effect of ICI results from the stimulation of ERα proteolysis occurring in vitro (Fig. 7B and Ref. 14) and in vivo (15). We found that the AF2ER mutations prevent ICI-mediated ERα protein degradation (Fig. 7D). This may be a partial explanation for ICI-mediated AF2ER activation. Meanwhile, ICI reduces WT-LBD dimerization and induces AF2ER-LBD dimerization (Fig. 4). However, dimerization did not have any impact on ICI-mediated ERα proteolysis (Fig. 7, C and E). Tamrazi et al. (17) reported that ERα ligands, including ICI, stabilize the WT-LBD homodimer compared with no ligand using an in vitro FRET-based assay. The differences between their in vitro results and our findings in the mammalian two-hybrid assay suggest that the ICI-bound WT ERα may introduce cellular factors to prevent homodimerization independently of proteolysis. It has been reported that the inhibition of ERα dimerization reduces the binding activity to the consensus ERE in an EMSA (6, 18). However, the ligand dependence of ERE binding has not been clearly defined using the EMSA method. We employed a colorimetric immunoassay in which the complex of biotinylated ERE and ERα on a streptavidin-coated plate is detected by an ERα-specific antibody. These data suggest that the ERE-binding activity of AF2ER is increased by ICI but not E2 and that activity was prevented by the disruption of dimer formation with the L511R mutation (Fig. 6). The DNA binding activity of WT was increased by E2 and also ICI, and that activity was diminished by the L511R mutation the same as with the AF2ER (Fig. 6). This result would support that ICI stabilizes the homodimerization of WT ERα in vitro, as Tamrazi et al. reported (17), and it may cause the induction of DNA binding activity with ICI in our in vitro DNA binding assay. Recently, Hilmi et al. (19) reported that ICI induces SUMOylation of human ERα protein in the cells and that the mutations of H12 (L539A or L540A, which correspond to mouse L543A and L544A) strongly reduced SUMOylation in the presence of ICI. We observed that ICI reduced the level of ERα WT-LBD homodimerization in the two-hybrid assay (Fig. 4). Therefore, ICI-mediated, posttranslational modification of WT ERα may cause the prevention of ERα dimer formation in vivo. Taken together, these results suggest that the prevention of ICI-dependent proteolysis maintains the dimer formation of AF2ER and that this increases ERE binding and transcription activation.
Previous studies have also reported that the H12 mutation in the ERα LBD changes antagonists into agonists (20–22). The L543A/L544A and M547A/L548A mouse ERα mutants exhibit reduced basal transcription activity and have lost the ability to respond to E2, but these mutants are activated by anti-estrogens, ICI164384, and OHT (20). These hydrophobic amino acids (Leu-543, Leu-544, Met-547, and Leu-548) are localized on the same surface of H12 (23). The Leu-544 of mouse ERα correlates to Leu-540 in the human ERα. Montano et al. (21) have reported that the characteristics of the L540Q human ERα mutant are quite similar to the L543A,L544A mouse ERα (AF2ER) mutant. Recently, H12 mutations (M543V and M543A/L544A) in human breast cancers were reported, and both mutants were activated by ICI and OHT more than by E2 (24). The position of Met-543 and Leu-544 of human ERα correlates with the Met-547 and Leu-548 of mouse ERα that were reported by Mahfoudi et al. (20). These results suggest that the disruption of the nonpolar surface of H12 provokes antagonist reversal activity, and the possibility of tamoxifen resistance in breast cancer might arise from such a mechanism.
Here we suggest that ICI-mediated antagonist reversal activity can also be induced by the removal of the entire H12 and that the subsequent F-domain is involved in this activity (Fig. 5). Interestingly, OHT and Ral induced the dimer formation of WT-LBD more potently than E2, and F-domain truncation strongly depressed the ability of OHT/Ral-mediated WT-LBD dimerization (Fig. 9B). These results would indicate that the F-domain is closely associated with the antagonist-dependent ERα LBD dimerization. Because the F-domain is directly adjacent to H12, the location of the F-domain would be influenced by H12. Crystallographic analyses have shown that SERMs generate a differential orientation of H12, resulting in partial agonist/antagonist activity of SERMs (25, 26). Our current findings provide further consideration for the orientation of the F-domain toward understanding the precise activity of SERMs.
The crystallographic analysis indicated that the possible H12 position would not allow for a secure structure with ICI bound to the ER LBD (27). Because the H12-mutated ERα (AF2ER) is activated by ICI, we questioned whether the AF2ER-LBD could recruit p160 coactivators in a similar manner as the WT-LBD. However, the AF2ER-LBD did not recruit the SRC1-NR-box with ICI, nor with E2 (Fig. 3). This result is consistent with the fact that the N-terminal-truncated AF2ER (121-AF2ER) does not have any transcriptional activity with ligands (Fig. 2). Our earlier results suggest that the SRC-1 accelerates the p300/CBP-dependent AF2ER activation through the N-terminal transcriptional activation function (9). Our current view is that the AF2ER-LBD alone does not possess functional transcription activity and that the N-terminal (AB domains)-derived transcription activity is mediated by ligand-dependent AF2ER activation. Although Métivier et al. (28) indicated that a physical interaction between the A-domain and the LBD of ERα causes the repression of unliganded ERα transcription activity, it is still controversial how AF-1 activity is controlled in the unliganded or antagonist-bound, non-active WT ERα. Further examination will be necessary with the possibility that antagonist-induced dimerization of AF2ER-LBD may release the A-domain from the LBD to activate AF2ER transcriptional activity.
Acknowledgments
We thank John Otstot and Jason Malphurs of the NIEHS Sequencing Laboratory Group for their DNA sequencing and confirmations of the constructed vectors used in the experiments. We also thank Drs. Tatsuya Sueyoshi and Yin Li for critical reading of the manuscript.
This work was supported by Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences Grant Z01ES70065 (to K. S. K.).
- ER
- estrogen receptor
- AF
- transactivating function
- LBD
- ligand-binding domain
- H12
- helix 12
- ERE
- estrogen-responsive element
- DBD
- DNA-binding domain
- ICI
- fulvestrant/ICI182780
- E2
- estradiol
- OHT
- 4-hydroxytamoxifen
- SERM
- selective estrogen receptor modulator
- S
- sense
- AS
- antisense
- TRE
- Tet-responsive element
- AD
- activation domain
- Ral
- raloxifene
- SUMO
- small ubiquitin-like modifier.
REFERENCES
- 1. Hall J. M., Couse J. F., Korach K. S. (2001) The multifaceted mechanisms of estradiol and estrogen receptor signaling. J. Biol. Chem. 276, 36869–36872 [DOI] [PubMed] [Google Scholar]
- 2. Heldring N., Pike A., Andersson S., Matthews J., Cheng G., Hartman J., Tujague M., Ström A., Treuter E., Warner M., Gustafsson J. A. (2007) Estrogen receptors. How do they signal and what are their targets. Physiol. Rev. 87, 905–931 [DOI] [PubMed] [Google Scholar]
- 3. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) The nuclear receptor superfamily. The second decade. Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Germain P., Staels B., Dacquet C., Spedding M., Laudet V. (2006) Overview of nomenclature of nuclear receptors. Pharmacol. Rev. 58, 685–704 [DOI] [PubMed] [Google Scholar]
- 5. Brzozowski A. M., Pike A. C., Dauter Z., Hubbard R. E., Bonn T., Engström O., Ohman L., Greene G. L., Gustafsson J. A., Carlquist M. (1997) Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389, 753–758 [DOI] [PubMed] [Google Scholar]
- 6. Fawell S. E., Lees J. A., White R., Parker M. G. (1990) Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell 60, 953–962 [DOI] [PubMed] [Google Scholar]
- 7. Schwabe J. W., Chapman L., Finch J. T., Rhodes D. (1993) The crystal structure of the estrogen receptor DNA-binding domain bound to DNA. How receptors discriminate between their response elements. Cell 75, 567–578 [DOI] [PubMed] [Google Scholar]
- 8. Tanenbaum D. M., Wang Y., Williams S. P., Sigler P. B. (1998) Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5998–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arao Y., Hamilton K. J., Ray M. K., Scott G., Mishina Y., Korach K. S. (2011) Estrogen receptor α AF-2 mutation results in antagonist reversal and reveals tissue selective function of estrogen receptor modulators. Proc. Natl. Acad. Sci. U.S.A. 108, 14986–14991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arao Y., Hamilton K. J., Goulding E. H., Janardhan K. S., Eddy E. M., Korach K. S. (2012) Transactivating function (AF) 2-mediated AF-1 activity of estrogen receptor α is crucial to maintain male reproductive tract function. Proc. Natl. Acad. Sci. U.S.A. 109, 21140–21145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hewitt S. C., Kissling G. E., Fieselman K. E., Jayes F. L., Gerrish K. E., Korach K. S. (2010) Biological and biochemical consequences of global deletion of exon 3 from the ERα gene. FASEB J. 24, 4660–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berry M., Metzger D., Chambon P. (1990) Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 9, 2811–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang C. Y., Norris J. D., Grøn H., Paige L. A., Hamilton P. T., Kenan D. J., Fowlkes D., McDonnell D. P. (1999) Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries. Discovery of peptide antagonists of estrogen receptors α and β. Mol. Cell Biol. 19, 8226–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dauvois S., Danielian P. S., White R., Parker M. G. (1992) Antiestrogen ICI 164,384 reduces cellular estrogen receptor content by increasing its turnover. Proc. Natl. Acad. Sci. U.S.A. 89, 4037–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gibson M. K., Nemmers L. A., Beckman W. C., Jr., Davis V. L., Curtis S. W., Korach K. S. (1991) The mechanism of ICI 164,384 antiestrogenicity involves rapid loss of estrogen receptor in uterine tissue. Endocrinology 129, 2000–2010 [DOI] [PubMed] [Google Scholar]
- 16. Howell A. (2006) Pure oestrogen antagonists for the treatment of advanced breast cancer. Endocr.-Relat. Cancer 13, 689–706 [DOI] [PubMed] [Google Scholar]
- 17. Tamrazi A., Carlson K. E., Daniels J. R., Hurth K. M., Katzenellenbogen J. A. (2002) Estrogen receptor dimerization. Ligand binding regulates dimer affinity and dimer dissociation rate. Mol. Endocrinol. 16, 2706–2719 [DOI] [PubMed] [Google Scholar]
- 18. Lees J. A., Fawell S. E., White R., Parker M. G. (1990) A 22-amino-acid peptide restores DNA-binding activity to dimerization-defective mutants of the estrogen receptor. Mol. Cell Biol. 10, 5529–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hilmi K., Hussein N., Mendoza-Sanchez R., El-Ezzy M., Ismail H., Durette C., Bail M., Rozendaal M. J., Bouvier M., Thibault P., Gleason J. L., Mader S. (2012) Role of SUMOylation in full antiestrogenicity. Mol. Cell Biol. 32, 3823–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mahfoudi A., Roulet E., Dauvois S., Parker M. G., Wahli W. (1995) Specific mutations in the estrogen receptor change the properties of antiestrogens to full agonists. Proc. Natl. Acad. Sci. U.S.A. 92, 4206–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montano M. M., Ekena K., Krueger K. D., Keller A. L., Katzenellenbogen B. S. (1996) Human estrogen receptor ligand activity inversion mutants. Receptors that interpret antiestrogens as estrogens and estrogens as antiestrogens and discriminate among different antiestrogens. Mol. Endocrinol. 10, 230–242 [DOI] [PubMed] [Google Scholar]
- 22. Lupien M., Jeyakumar M., Hebert E., Hilmi K., Cotnoir-White D., Loch C., Auger A., Dayan G., Pinard G. A., Wurtz J. M., Moras D., Katzenellenbogen J. A., Mader S. (2007) Raloxifene and ICI182,780 increase estrogen receptor association with a nuclear compartment via overlapping sets of hydrophobic amino acids in activation function 2 helix 12. Mol. Endocrinol. 21, 797–816 [DOI] [PubMed] [Google Scholar]
- 23. Schwartz J. A., Brooks S. C. (1997) Neutral mutations to three acidic AF2 residues in the mouse estrogen receptor confer agonist activity to A-ring isomers of estradiol. J. Steroid Biochem. Mol. Biol. 62, 173–184 [DOI] [PubMed] [Google Scholar]
- 24. Nichols M., Cheng P., Liu Y., Kanterewicz B., Hershberger P. A., McCarty K. S. (2010) Breast cancer-derived M543V mutation in helix 12 of estrogen receptor α inverts response to estrogen and SERMs. Breast Cancer Res. Treat. 120, 761–768 [DOI] [PubMed] [Google Scholar]
- 25. Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 26. Wu Y. L., Yang X., Ren Z., McDonnell D. P., Norris J. D., Willson T. M., Greene G. L. (2005) Structural basis for an unexpected mode of SERM-mediated ER antagonism. Mol. Cell 18, 413–424 [DOI] [PubMed] [Google Scholar]
- 27. Pike A. C., Brzozowski A. M., Walton J., Hubbard R. E., Thorsell A. G., Li Y. L., Gustafsson J. A., Carlquist M. (2001) Structural insights into the mode of action of a pure antiestrogen. Structure 9, 145–153 [DOI] [PubMed] [Google Scholar]
- 28. Métivier R., Stark A., Flouriot G., Hübner M. R., Brand H., Penot G., Manu D., Denger S., Reid G., Kos M., Russell R. B., Kah O., Pakdel F., Gannon F. (2002) A dynamic structural model for estrogen receptor-α activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol. Cell 10, 1019–1032 [DOI] [PubMed] [Google Scholar]