Background: Fibronectin matrix assembly is mediated by cell traction at the RGD loop of 10FNIII, which is predicted to unravel β-strands A and B.
Results: Sequence SLLISWD from strand B initiates fibronectin multimerization.
Conclusion: SLLISWD mediates cell-mediated fibronectin fibril assembly.
Significance: This matrix motif provides insight into physiological fibronectin fibrillogenesis with utility in initiating matrix assembly for tissue repair.
Keywords: Computer Modeling, Extracellular Matrix, Peptides, Protein Cross-linking, Protein Self-assembly, RGD, Anastellin, Cryptic Sites, Mechanobiology
Abstract
Fibronectin (FN) assembly into extracellular matrix is tightly regulated and essential to embryogenesis and wound healing. FN fibrillogenesis is initiated by cytoskeleton-derived tensional forces transmitted across transmembrane integrins onto RGD binding sequences within the tenth FN type III (10FNIII) domains. These forces unfold 10FNIII to expose cryptic FN assembly sites; however, a specific sequence has not been identified in 10FNIII. Our past steered molecular dynamics simulations modeling 10FNIII unfolding by force at its RGD loop predicted a mechanical intermediate with a solvent-exposed N terminus spanning the A and B β-strands. Here, we experimentally confirm that the predicted 23-residue cryptic peptide 1 (CP1) initiates FN multimerization, which is mediated by interactions with 10FNIII that expose hydrophobic surfaces that support 8-anilino-1-napthalenesulfonic acid binding. Localization of multimerization activity to the C terminus led to the discovery of a minimal 7-amino acid “multimerization sequence” (SLLISWD), which induces polymerization of FN and the clotting protein fibrinogen in addition to enhancing FN fibrillogenesis in fibroblasts. A point mutation at Trp-6 that reduces exposure of hydrophobic sites for 8-anilino-1-napthalenesulfonic acid binding and β-structure formation inhibits FN multimerization and prevents physiological cell-based FN assembly in culture. We propose a model for cell-mediated fibrillogenesis whereby cell traction force initiates a cascade of intermolecular exchange starting with the unfolding of 10FNIII to expose the multimerization sequence, which interacts with strand B of another 10FNIII domain via a Trp-mediated β-strand exchange to stabilize a partially unfolded intermediate that propagates FN self-assembly.
Introduction
The extracellular matrix (ECM)2 regulates cell and tissue development by presenting solid-phase adhesion sites for growth factors and cells within its constituent macromolecules (1). Although some cell binding and recognition sites are present on solvent-exposed surfaces, other “cryptic sites” are buried within the native structure only to be activated upon solvent exposure resulting from structural changes in the molecule. For example, proteolytic cleavage of ECM proteins fibronectin (FN), collagen, perlecan, fibulin, and thrombospondin unmasks cryptic sites with important pro-angiogenic and anti-angiogenic functions (2–6). Additionally, FN contains mechanically regulated cryptic ECM assembly sites exposed by cell traction force that stretch the molecule (7). The assembly of FN into insoluble fibrils within the ECM is essential to many processes including embryogenesis (8), wound healing (9), atherosclerosis (10), angiogenesis (11), and cancer (12). However, the steps by which cell traction induces FN fibril formation remain unknown.
FN, a large dimeric glycoprotein structured by repeating units of domains FNI, FNII, and FNIII (Fig. 1A), supports the adhesion of many cell types via binding to cell-surface transmembrane integrin receptors (13). FN assembly is primarily mediated by the α5β1 integrin (14), which binds the RGD sequence in the tenth FN type III (10FNIII) domain (15). FN fibrillogenesis also requires cell-mediated traction forces generated by a contractile actin cytoskeleton that transmits force onto their FN adhesions (16). This mechanical force is a key regulator of FN fibrillogenesis that exposes buried cryptic assembly sites within FN. Physical stretching of immobilized FN by 30–35% exposes a cryptic FN binding site for the 70-kDa fragment, which contains the N-terminal assembly domain (1–5FNI) required for fibrillogenesis (7, 17). Cell traction force stretches FN leading to conformational changes in the molecule that include domain unfolding of FNIII repeats (18–20), which are not stabilized by internal disulfide bonds unlike the FNI and FNII repeats. Interestingly, cryptic FN assembly sites have been identified primarily within the FNIII domains (21), including the RGD-displaying 10FNIII domain.
FIGURE 1.
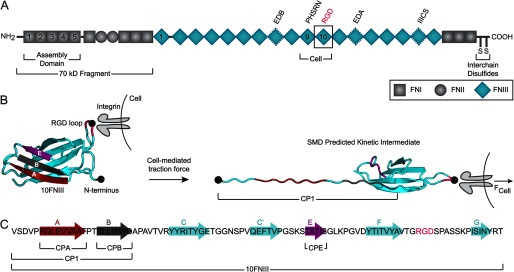
Structure of fibronectin and the cell-binding domain 10FNIII. A, the two arms of FN are linked by interchain disulfide bonds at the C termini. Each arm is composed of a combination of repeating FNI, FNII, and FNIII domains with insertions or deletions of alternatively spliced domains depending on the isoform. The cell-binding fragment of FN spans the 9–10FNIII repeats, which contain the RGD sequence in 10FNIII and the synergy site in neighboring 9FNIII. Anastellin, a C-terminal portion of the 1FNIII domain, has been shown to initiate multimerization of added FN. B, when cell traction imparts force (FCell) at the RGD loop (pink with central Gly depicted as a black circle) of 10FNIII (left) while anchored at the N terminus (black circle), the domain is predicted by SMD simulations to unfold to a kinetic intermediate with an unraveled N terminus exposing β-strands A (red) and B (gray) (labeled CP1) but not strand E (purple) (right) (26). C, the primary structure of 10FNIII highlights the residues that form β-strands in natively folded 10FNIII and sequences from which peptides were designed.
Although the RGD cell-binding sequence and neighboring synergy site are required for FN assembly by the α5β1 integrin (14, 22), how 10FNIII transduces force applied at its RGD loop to initiate fiber assembly remains unclear. Both single molecule atomic force microscopy experiments and steered molecular dynamics (SMD) simulations suggest that 10FNIII is one of the mechanically weakest FNIII domains (19, 23). Studies with thermally unfolded 10FNIII have identified cryptic assembly sites in the domain that support FN multimerization in vitro in a manner that does not require its C-terminal residues, and furthermore, this unfolded 10FNIII domain inhibits FN incorporation into fibroblast-deposited ECM (24). Additionally, point mutations P5A and P25A in the N terminus of 10FNIII partially destabilize the module to an intermediate structure prone to self-aggregation (25). Therefore, it is feasible that under physiological conditions, cell-generated mechanical forces applied at the RGD loop of FN could unfold 10FNIII to expose cryptic assembly sites that initiate fibrillogenesis.
We previously approached 10FNIII unfolding from a physiological perspective by using SMD simulations to look at its unfolding due to pulling at its RGD loop when anchored at the N terminus and found that the domain unfolds along a single pathway (26). The unfolding response due to pulling at the RGD loop differed from standard models of force application directed through the termini that resulted in multiple unfolding pathways for 10FNIII (26–28). Our simulations predicted 10FNIII to unfold to a partially unfolded kinetic intermediate with solvent-exposed N-terminal A and B β-strands in response to pulling at its physiological integrin-binding site. Here, we set out to test whether this predicted exposed region contributes cryptic assembly sites and to identify the minimal peptide sequence within the 10FNIII domain that is sufficient to induce FN self-assembly.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Purification
All peptides were synthesized by the Tufts University Core Facility (Boston, MA). Peptides were acetylated at the N terminus and amidated at the C terminus. A control peptide with the same sequence composition as cryptic peptide 1 (CP1) was generated as a scrambled sequence designated as CP1scr (DSALRSPVWIVTDSAEVPVLTLD). Peptide sequences CPA, CPE, and CPB(W6A) contain an additional N-terminal Tyr not found in the 10FNIII sequence to enable spectrophotometric concentration determination of the peptides in solution (Table 1). Peptide concentrations were determined from absorbance by using published calculated molar extinction coefficients (29): CP1, scrCP1, and CPB, ϵ280 = 5630 m−1 cm−1; and CPA, CPB(W6A), and CPE, ϵ278 = 1295 m−1 cm−1. Peptides were purified and analyzed by RP-HPLC on C18 columns (Agilent Technologies). Peptide molecular weight was confirmed by MALDI-TOF MS intact mass determination with a 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems).
TABLE 1.
Summary of characteristics for derived peptide mimics including peptide sequence, corresponding 10FNIII β-strand(s), and residue indices in fibronectin
| Sequence | β-Strand (s) | Residue indices | |
|---|---|---|---|
| CP1 | VSDVPRDLEVVAATPTSLLISWD | A, B | 1416–1438 |
| CPA | YRDLEVVAAT | A | 1421–1429 |
| CPB | SLLISWD | B | 1432–1438 |
| CPE | YTATIS | E | 1471–1475 |
Cloning and Purification of Recombinant 10FNIII
The recombinant 10FNIII domain was constructed with a C-terminal His6 affinity tag by modifying the pETCH-GST-8–11FNIII-His6 vector, which was a generous gift from Drs. Richard Clark and Xiang-Dong Ren (State University of New York at Stony Brook, Stony Brook, NY) (30). The 10FNIII domain in FN encoding amino acids 1416–1509 (Val-Ser-Asp-Val-…Asn-Tyr-Arg-Thr) was amplified by PCR from the pETCH-GST-8–11FNIII-His6 construct using two primers: 5′-CTTTAAGAAGGAGATATACATATGGTTTCTGATGTTCCGAGGGACCTG-3′ and 5′-GCTTAATGATGATGGTGGTGGTGTGTTCGGTAATTAATGGAAATTGGCTTGC-3′. The protein insert was confirmed by DNA sequencing. Protein expression was induced in the BL21(DE3) strain of Escherichia coli with 1 mm isopropyl β-d-1-thiogalactopyranoside at 37 °C for 6 h. Protein affinity purification was performed using HisPur cobalt resin (Thermo Scientific). Proteins were eluted in 50 mm sodium phosphate, 300 mm sodium chloride, and 150 mm imidazole (pH 7.4). The purified proteins were desalted on a Sephadex G-25 pre-packed PD-10 desalting column (GE Healthcare) equilibrated with Dulbecco's PBS containing calcium and magnesium (Invitrogen). Proteins were eluted with Dulbecco's PBS containing calcium and magnesium and concentrated with an Amicon 3-kDa filter (Millipore). The purity of protein was analyzed by SDS-PAGE and visualized with SYPRO Ruby protein gel stain (Lonza) photographed on a FluorChem M imager with a SYPRO Ruby filter (ProteinSimple). The 10FNIII protein concentration was determined from absorbance at 280 nm using published calculated molar extinction coefficients (29) (ϵ280 = 14,440 m−1 cm−1).
Multimerization Assay
Rhodamine-labeled bovine plasma FN (lyophilized powder; Cytoskeleton) was solubilized in ultrapure water (Milli-Q) at room temperature for 15 min prior to spectrophotometric concentration determination (ϵ565 = 70,000 m−1 cm−1). Peptide stock solutions were prepared in 50 mm Tris·HCl (pH 7.4) and various concentrations of CP1 (0–500 μm) or CPB (0–750 μm) were incubated with rhodamine-labeled FN (0.3 mg/ml) for 16 h at 37 °C. Following incubation, samples were analyzed by non-reducing SDS-PAGE analysis. Samples were prepared in NuPAGE lithium dodecyl sulfate non-reducing sample buffer and resolved on 3–8% Tris acetate SDS-PAGE gels (Invitrogen). Fluorescent gel images were scanned on a Typhoon FLA 9000 with a RITC filter (GE Healthcare), and densitometric analysis was performed using ImageJ (National Institutes of Health). Band intensities for each molecular weight species (reduced arms, disulfide-bonded dimer, and high molecular weight multimers) were assigned using reduced and non-reduced FN control samples. The percentage of multimers was calculated from the ratio of multimer intensity to the sum of intensities for the reduced arms, dimer, and multimers. Final sample preparations of rhodamine-labeled FN contain reduced arms (54 ± 2%) as the lyophilized stock contains β-mercaptoethanol leading to ∼2.5 mm β-mercaptoethanol in prepared reaction samples.
Turbidity Assay
The multimerization of human plasma FN (lyophilized powder; BD Biosciences) or plasminogen, von Willebrand factor, and FN-depleted human fibrinogen (FBG) (Enzyme Research Laboratories) by peptide was determined by measuring the optical density at 590 nm using an Agilent Cary 300 BioMelt UV-visible spectrophotometer (Agilent Technologies) in 10-mm path length quartz cuvettes (Starna Cells). Human plasma FN was solubilized according to the manufacturer's instructions. FN and FBG protein concentrations were determined by absorbance at 280 nm (FN, ϵ280 = 563,200 m−1 cm−1; FBG, ϵ280 = 498,300 m−1 cm−1). Peptides CP1 or CPB were prepared in 50 mm Tris·HCl (pH 7.4). After equilibrating peptides to 25 °C for 2 min, turbidity of each peptide sample was monitored for 15 min after which FN or FBG was added to each cuvette at a final concentration of 0.2 or 5 mg/ml, respectively. The change in optical density was continuously recorded for 2 h following the addition of FN or FBG. Absorbances are reported relative to the absorbance measured at the time of FN or FBG addition.
CD Spectroscopy
Peptide stock solutions were prepared in 10 mm sodium phosphate buffer (pH 7.4). All CD measurements were collected using a 1-mm path length quartz cuvette (Starna Cells) at 25 °C from 260 to 190 nm on a JASCO J-720 spectropolarimeter equipped with a JASCO PT-423S Peltier temperature controller. Data were collected in continuous scanning mode, and the spectra were averaged over 4 consecutive accumulations (scan speed 20 nm/min; data pitch 0.5 nm; response time 4 s; band width 1.0 nm; sensitivity 100 mdeg). A baseline measurement for buffer alone was subtracted from each spectrum. Data were expressed as mean residue ellipticity ([θ], deg cm2 dmol−1) as calculated using the relationship,
 |
where θobs is the measured signal in millidegrees, MRW is the mean residue weight (i.e. the molecular weight divided by the number of residues), c is the concentration of the molecule in mg/ml, and l is the optical path length of the sample cell in cm.
8-Anilino-1-napthalenesulfonic Acid (ANS) Binding Fluorescence Assay
Stock solutions of ANS (Sigma) were prepared in Dulbecco's PBS containing calcium and magnesium or 100 mm sodium phosphate buffer (pH 6.8) prior to spectrophotometric concentration determination (ϵ350 = 4,950 m−1 cm−1) (31). Anastellin (lyophilized powder; Sigma) was solubilized in Dulbecco's PBS containing calcium and magnesium at room temperature prior to spectrophotometric concentration determination ϵ280 = 16,960 m−1 cm−1) (29). Samples were prepared in Dulbecco's PBS containing calcium and magnesium with 50 μm ANS in the presence or absence of 50 μm anastellin or 50 μm 10FNIII with or without peptide. Emission spectra were collected at 25 °C at 5-nm intervals (1-s integration time) over a range from 380 to 650 nm with an excitation wavelength of 360 nm on a PTI QuantaMaster 40 spectrofluorometer with excitation and emission slit widths of 5 nm. A baseline measurement for buffer alone was subtracted from each spectrum. The maximum fluorescence of each emission spectrum was averaged across independent experiments. Relative fluorescence increase above background was calculated for each sample by subtracting the ANS fluorescence in the presence of buffer and normalizing to the fluorescence measured for ANS bound to anastellin.
Pelleting Assay
Human plasma FN (lyophilized powder; BD Biosciences) was solubilized according to the manufacturer's instructions, and peptides were prepared in 50 mm Tris·HCl (pH 7.4). Protein and peptide stocks were centrifuged (4,000 × g for 5 min) to sediment insoluble aggregates prior to spectrophotometric concentration determination. Samples were prepared in 50 mm Tris·HCl (pH 7.4) containing 5 mm EDTA and combinations of FN (0.3 mg/ml), anastellin (150 μm), CPB (500 μm), or CPB(W6A) (500 μm). After incubation for 16 h at 37 °C, samples were fractionated by centrifugation (20,000 × g for 10 min), pellets were rinsed with buffer, and insoluble material was collected by centrifugation (20,000 × g for 10 min). Insoluble material was resuspended in lithium dodecyl sulfate sample buffer containing NuPAGE reducing agent prior to analysis by SDS-PAGE (Invitrogen). Samples were resolved on 3–8% Tris acetate SDS-PAGE gels stained with SimplyBlue (Invitrogen) and photographed on a FluorChem M imager (ProteinSimple).
Thioflavin T (ThT) Binding Fluorescence Assay
A 1 mm stock solution of ThT (Sigma) was prepared in 50 mm Tris·HCl (pH 7.4) and filtered prior to dilution into ethanol for spectrophotometric concentration determination (ϵ416 = 26,620 m−1 cm−1) (32). Samples were prepared in 50 mm Tris·HCl (pH 7.4) with 20 μm ThT in the presence or absence of 10 μm 10FNIII or FBG and/or 500 μm CPB or CPB(W6A). Fluorescence emission at 482 nm was collected at 25 °C for 60 min (1 s integration time) with an excitation wavelength of 450 nm on a PTI QuantaMaster 40 spectrofluorometer with excitation and emission slit widths of 5 nm.
FN Matrix Incorporation Assay
Human lung fibroblasts (IMR90) were obtained from the American Type Culture Collection (ATCC) and maintained in Eagle's minimum essential medium (ATCC) supplemented with 10% fetal bovine serum (HyClone, Thermo Fisher Scientific), penicillin (100 units/ml), and streptomycin (100 μg/ml) (Invitrogen) at 37 °C under 5% CO2. The assembly of biotinylated FN into detergent-insoluble matrix was determined as previously described (33). IMR90 cultures were plated (2.5 × 104 cells/well) in complete medium in 96-well plates, cultured overnight, and then the medium was exchanged with complete medium supplemented with biotinylated FN (20 μg/ml) (Cytoskeleton) and 25 mm HEPES (pH 7.1) with or without anastellin, CPB, or CPB(W6A). Following incubation for 24 h, cells were rinsed three times in Dulbecco's PBS containing calcium and magnesium prior to scraping in deoxycholate buffer (2% deoxycholate, 2 mm phenylmethanesulfonyl fluoride, 2 mm EDTA, 2 mm N-ethylmaleimide, 2 mm iodoacetic acid, 20 mm Tris·HCl, pH 8.8). Cells were passed five times through a 23-gauge needle, and deoxycholate-insoluble material was fractionated by centrifugation (20,000 × g for 20 min at 4 °C). The pellet containing the deoxycholate-insoluble material was rinsed with deoxycholate buffer and collected by centrifugation (20,000 × g for 10 min at 4 °C). Insoluble material was recovered in Laemmli reducing sample buffer (Boston BioProducts) diluted in deoxycholate buffer and heated to 95 °C for 5 min. Samples were resolved on 4–20% Mini-Protean TGX gels (Bio-Rad) prior to electrotransfer onto 0.45-μm nitrocellulose membranes (Bio-Rad). After membranes were blocked (5% nonfat dry milk, 0.1% Tween 20, 150 mm NaCl, 50 mm Tris·HCl, pH 7.4), membranes were cut and probed either by horseradish peroxidase-conjugated streptavidin (1:200, Vector Laboratories) to quantify biotinylated FN incorporation or anti-vimentin monoclonal antibody (clone RV202, 1:100, Abcam) followed by horseradish peroxidase-conjugated horse anti-mouse IgG (1:1000, Vector Laboratories) to provide an internal protein loading control. Blots were developed by chemiluminescence with ECL prime Western blotting detection reagent (GE Healthcare) using HyBlot CL Autoradiography Film (Denville Scientific). The quantification of Western blots was performed by densitometric analysis of band intensities corresponding to biotinylated FN and vimentin bands using ImageJ (National Institutes of Health).
Statistical Analysis
Values are expressed as mean ± S.E. calculated across independent samples; p values were derived from Student's t test.
RESULTS
Unfolded N-terminal Sequence of Predicted 10FNIII Intermediate Initiates FN Multimerization
Our past SMD simulations modeling the unfolding pathway of 10FNIII under tension between its N terminus and the physiological integrin-binding RGD loop predicted an unfolded intermediate with an unraveled N terminus through the second β-strand (26) (Fig. 1B). Because this kinetic intermediate is sampled for extended simulation times while under tension, we proposed that such a partially unfolded structure might expose cryptic assembly sites that trigger FN assembly. We tested this hypothesis by evaluating the multimerization capacity of sequences representing the exposed 23 amino acids at the N terminus named CP1 and shorter regions encompassing β-strand sequences from the native 10FNIII structure (Table 1).
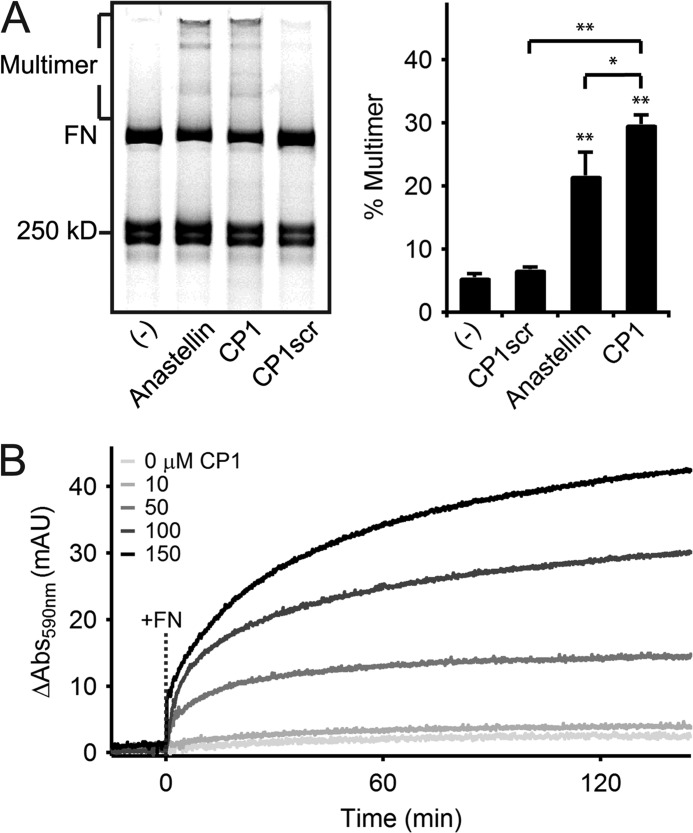
We used anastellin, a C-terminal fragment of 1FNIII that interacts with FN to induce the formation of disulfide-stabilized supramolecular FN known as superfibronectin (34), as a positive control. Non-reducing SDS-PAGE analysis shows that supramolecular FN multimers migrate as a distribution of discrete bands of molecular weights larger than that of disulfide-bonded molecular FN depending on the number of integrated molecules (Fig. 2A). Incubation of anastellin (150 μm) with rhodamine-labeled FN at a concentration similar to that of plasma FN in blood (35) (220:1 molar ratio of peptide to FN) led to 22 ± 4% of total FN (distributed as reduced FN arms, disulfide-bonded dimer, and multimers) forming high molecular weight multimers when analyzed by densitometry following separation by non-reducing SDS-PAGE (Fig. 2A). Interestingly, incubation of labeled FN with the CP1 peptide at the same concentration led to a 38 ± 24% increase in multimer formation over anastellin (Fig. 2A). Addition of reducing agent to the CP1-induced multimers prior to SDS-PAGE analysis collapsed the higher order bands suggesting multimer stabilization by disulfide bonds (data not shown). In contrast, the scrambled CP1 sequence (represented by peptide CP1scr) did not support FN multimerization when added at the same concentration (Fig. 2A). The CP1-induced multimerization effect was also observed with unlabeled FN, where the degree of aggregation was monitored in a turbidity assay (Fig. 2B). Although CP1 showed no multimerization in the absence of FN even at high peptide concentrations, turbidity increased only when FN was added to CP1 (molar ratio of CP1 to FN of 22:1 and 330:1 for CP1 concentrations of 10 and 150 μm, respectively) (Fig. 2B) demonstrating that FN multimerization kinetics depended on CP1 concentration (Fig. 2B).
FIGURE 2.
Polymerization of fibronectin by CP1, as compared with anastellin. A, non-reducing SDS-PAGE analysis of rhodamine-labeled FN (0.3 mg/ml) incubated with buffer (−), anastellin, CP1, or CP1scr at 150 μm for 16 h at 37 °C shows that both anastellin and CP1 induced the formation of a multimeric FN species (Multimer) with large discrete molecular weights relative to the disulfide-bonded molecular FN (FN) (left). Preparations of labeled FN contain reduced monomeric FN arms near the indicated 250-kDa standard (see “Experimental Procedures”). Densitometry of rhodamine fluorescence (right) reveals significant multimerization of FN by anastellin and CP1 (**, p < 0.01, two-tailed). CP1 shows significantly higher activity than anastellin (*, p < 0.05, one-tailed), whereas scrambling the CP1 sequence reduced its activity (**, p < 0.01, one-tailed) to background levels (p > 0.05, two-tailed). B, the development of turbidity at 590 nm at 25 °C by CP1 (10–150 μm) was monitored before and after the addition of unlabeled FN (0.2 mg/ml) at t = 0 min (dashed line marked +FN).
The Multimerization Effect of the CP1 Sequence in 10FNIII Is Cryptic
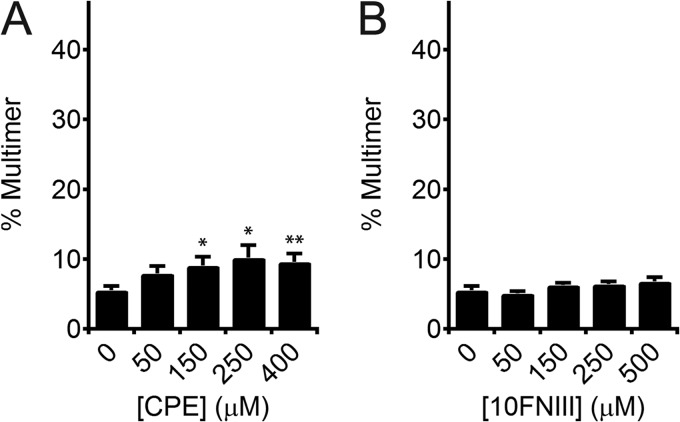
To test our predictions for the cryptic assembly sequence motivated by the SMD-calculated intermediate, we compared the multimerization capacity of sequence CP1 against that of β-strand E, which shares a face with CP1 in intact 10FNIII but is not solvent exposed in our proposed kinetic intermediate (Fig. 1B) (26). Addition of the CPE peptide (50 to 400 μm) to rhodamine-labeled FN induced minimal FN multimerization in the range of 8 ± 1 to 10 ± 1%, respectively (Fig. 3A). Although multimerization was statistically significant for CPE concentrations ≥150 μm (p < 0.05, two-tailed), the percentage of multimers initiated by CPE is significantly less (approximately one-third) than that initiated by 150 μm CP1 (p < 0.01, one-tailed). Moreover, we found that the multimerization activity of CP1 is cryptic within 10FNIII as addition of natively folded 10FNIII, which contains the CP1 sequence in its N terminus, at concentrations between 50 and 500 μm to rhodamine-labeled FN showed background levels of multimer formation (p > 0.05, two-tailed) (Fig. 3B).
FIGURE 3.
Effect of CPE and 10FNIII on fibronectin multimerization. A, densitometry of rhodamine fluorescence of labeled FN analyzed by SDS-PAGE shows low-level multimerization of FN induced by CPE (50–400 μm). Multimers were induced significantly above background by high concentrations of CPE (150 and 250 μm: *, p < 0.05, two-tailed; 400 μm: **, p < 0.01, two-tailed). B, densitometry of rhodamine fluorescence reveals that recombinant 10FNIII (50–500 μm) does not induce significant FN multimerization above background (p > 0.05, two-tailed).
CP1 Interacts with 10FNIII to Expose Buried Hydrophobic Surfaces
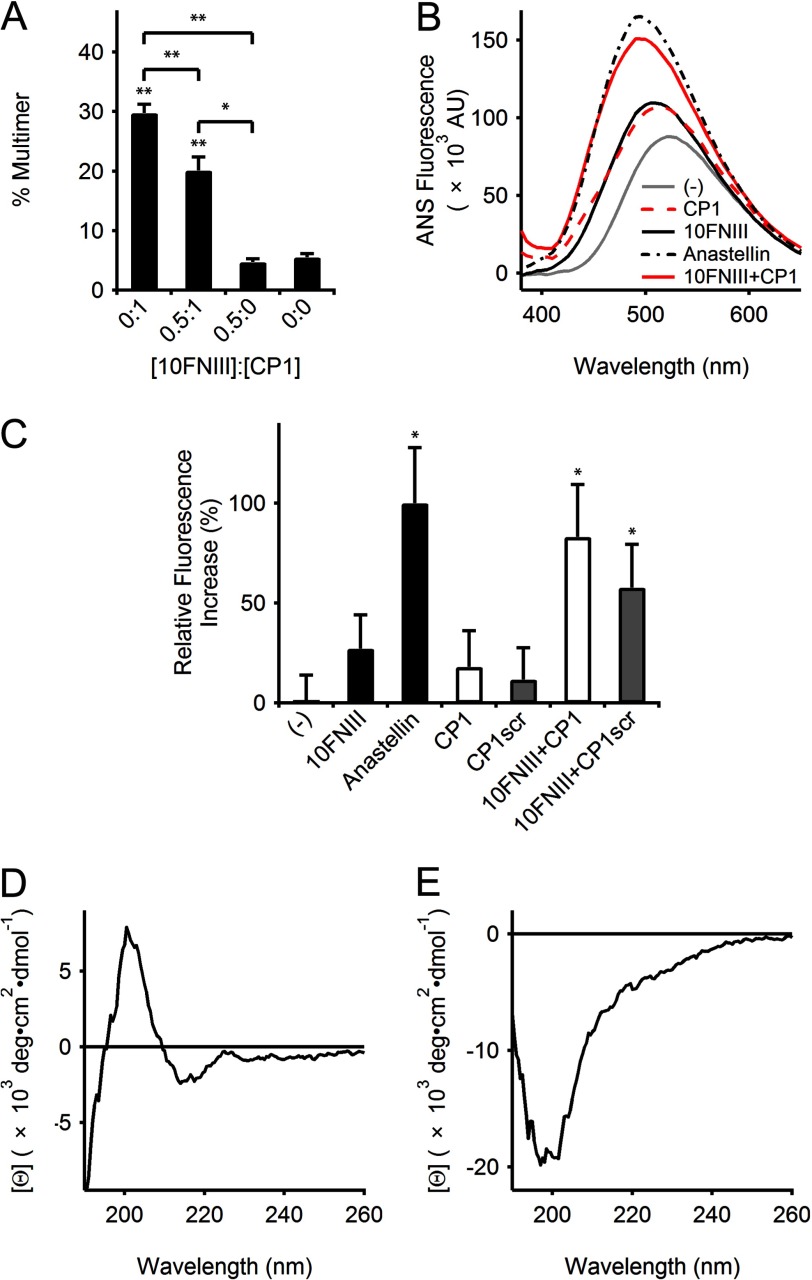
Because the CP1 sequence corresponds to 24% of the 10FNIII sequence at the N terminus, we investigated whether CP1-initiated multimerization of FN involved interactions with 10FNIII. The presence of 10FNIII in mixtures of CP1 (both at large molar excess to rhodamine labeled FN) reduced the percentage of observed multimers detected by SDS-PAGE (Fig. 4A). For a fixed concentration of CP1 (150 μm), adding 10FNIII at half the molar concentration of CP1 (yielding a molar ratio of 110:1 of 10FNIII to FN) attenuated, but did not abolish, multimer formation by nearly one-third (Fig. 4A). The control sample of 10FNIII (75 μm) in the absence of CP1 was not significantly different from background (p > 0.05, two-tailed). Increasing the molar ratio of 10FNIII to CP1 to 1:1 did not lead to further reduction in multimer formation (p > 0.05, two-tailed). These results suggest that CP1 interacts with 10FNIII.
FIGURE 4.
CP1 interacts with 10FNIII. A, densitometry of rhodamine-labeled FN multimers separated by SDS-PAGE showed that multimerization by 150 μm CP1 is significantly different from samples containing 75 μm 10FNIII (**, p < 0.01, two-tailed) or background (**, p < 0.01, two-tailed). However, FN multimer formation by CP1 was reduced in the presence of 75 μm 10FNIII (**, p < 0.01, one-tailed). The reduced multimerization in mixtures of CP1 and 10FNIII is significantly different from the samples for 10FNIII alone (*, p < 0.05, two-tailed) or background (**, p < 0.01, two-tailed). B, comparison of representative ANS (50 μm) emission intensities from a single experiment in the presence of buffer (gray line), CP1 (150 μm, red dash line), 10FNIII (50 μm, black solid line), anastellin (50 μm, black dash dot line), or a mixture of 10FNIII (50 μm) and CP1 (150 μm) (red solid line). C, quantification of ANS-dependent emission spectra maxima above background as a percentage of the maximal ANS emission in the presence of anastellin (50 μm) for samples containing 10FNIII (50 μm) with or without CP1 or CP1scr (150 μm). Data are pooled from independent experiments. ANS fluorescence due to anastellin and mixtures of 10FNIII with CP1 or CP1scr are significantly higher than ANS background (*, p < 0.05, one-tailed). D, CD spectrum of 10 μm 10FNIII. E, CD spectrum of 30 μm CP1.
We further investigated the interactions of CP1 with 10FNIII. Because 10FNIII is stabilized by a highly conserved buried hydrophobic core, peptide-induced disruptions of the FNIII-fold may yield solvent-exposed hydrophobic surfaces. ANS is a fluorescent dye that probes exposed hydrophobic surfaces whereby formation of noncovalent interactions between ANS and hydrophobic cavities leads to enhanced and blue-shifted fluorescence (36). Anastellin is known to present hydrophobic binding pockets for ANS (37), and thus, as expected, we observed an increase in fluorescence when ANS was combined with anastellin (Fig. 4B). Although ANS fluorescence was previously reported to increase 10-fold when bound to anastellin (37), we only observed about a 2-fold increase (Fig. 4B); however, this increase was found to be statistically significant (p < 0.05, one-tailed) (Fig. 4C). Moreover, using a Job plot to quantify the stoichiometry of ANS binding to anastellin, we find similar results to those previously reported by others (37). Addition of ANS to 10FNIII produced an enhanced and blue-shifted fluorescence above background as well (Fig. 4, B and C), suggesting that ANS is binding to hydrophobic pockets within 10FNIII. CD spectroscopy studies confirmed that 10FNIII is natively folded (Fig. 4D), as indicated by the presence of a negative signal at 190 nm and positive signals at 203 and 225 nm characteristic of β-sheets and turns (38). The offset in the CD spectra shifting the peak at 225 toward slightly negative values is within the noise of the measurement. We further confirmed 10FNIII stability with a thermal melting curve by tracking the increase in fluorescence of the single buried tryptophan (ratio of 350/320 nm emission intensity following excitation at 280 nm) as a function of temperature to confirm a transition near its published melting temperature of 88 °C (data not shown). Our observed ANS results for natively folded 10FNIII are consistent with published data describing conformational flexibility in 10FNIII in solution (39). We find that ANS does not significantly interact with CP1 (p > 0.05, one-tailed) (Fig. 4C) despite the CP1 sequence being unstructured, as shown by CD spectroscopy (Fig. 4E). However, ANS showed a strong interaction when CP1 (150 μm) was added to 10FNIII at a molar ratio of 3:1 (Fig. 4B), as demonstrated by the enhanced blue-shifted fluorescence peak relative to the spectrum observed for either 10FNIII or CP1 (Fig. 4B). Moreover, we observe weaker enhancement in ANS fluorescence by CP1scr in the presence of 10FNIII (Fig. 4C). These collective data show that CP1 interacts with 10FNIII in a manner that induces the formation of solvent-exposed hydrophobic cavities that are accessible for ANS binding.
A 7-Amino Acid Sequence in CP1 Is Sufficient to Initiate FN Multimerization
We explored whether β-strands A and B contribute to the multimerization activity of CP1 by testing short peptides encompassing these sequences, CPA and CPB, respectively, against CP1 at a high concentration (500 μm). SDS-PAGE analysis revealed that multimers were induced by high concentrations of CP1 as well as CPB, but not by CPA (Fig. 5A). Co-administration of CPA and CPB did not significantly enhance the degree of multimerization due to CPB alone (p = 0.05, two-tailed) (Fig. 5A). CPB also initiated the multimerization of unlabeled FN when analyzed using a centrifugation-based pelleting assay: insoluble multimers sedimented in mixtures with CPB (500 μm) or anastellin (150 μm), but not with untreated FN (Fig. 5B). The degree of multimerization by CP1 is concentration-dependent in the range of 10–500 μm with maximal activity at 150 μm (30 ± 1%, Fig. 5C). CPB, which is 30% the length of CP1, required a higher concentration (500 μm) to achieve similar multimerization as 150 μm CP1 (Fig. 5C). These results clearly show that the 7-amino acid sequence of CPB - SLLISWD - found at the C terminus of CP1 is sufficient to initiate FN multimerization.
FIGURE 5.
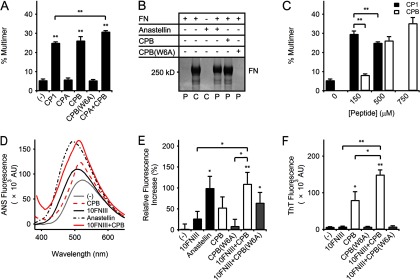
Effect of CPB on fibronectin and 10FNIII. A, densitometry of rhodamine-labeled FN multimers separated by SDS-PAGE showed significant FN multimerization by 500 μm CP1, CPB, or equimolar mixtures of CPA and CPB (**, p < 0.01, two-tailed). Combination of CPA and CPB minimally enhanced CPB activity (p = 0.05, two-tailed) but is significantly higher than that for equimolar CP1 (**, p < 0.01, two-tailed). A W6A point mutation in CPB abolished multimerization activity. B, SDS-PAGE analysis of unlabeled FN multimers sedimented by centrifugation compares the pellets (P) collected from mixtures of unlabeled FN (0.3 mg/ml) with anastellin (150 μm), CPB (500 μm), or CPB(W6A) (500 μm) incubated for 16 h at 37 °C. Control samples (C) corresponding to samples containing FN or anastellin alone (lanes 2 and 3, respectively) were not processed by centrifugation. C, densitometry analysis of CP1 or CPB (150–750 μm) mixtures with rhodamine-labeled FN separated by SDS-PAGE showed significant FN multimerization (p < 0.01, two-tailed). CP1 induced more multimers at 150 μm than at 500 μm (**, p < 0.01, one-tailed) and was significantly different from CPB at 150 μm (**, p < 0.01, two-tailed) but similar to 500–750 μm CPB (p > 0.05, two-tailed). D, comparison of representative ANS (50 μm) emission intensities from a single experiment in the presence of buffer (gray line), CPB (500 μm, red dash line), 10FNIII (50 μm, black solid line), anastellin (50 μm, black dash dot line), or a mixture of 10FNIII (50 μm) and CPB (500 μm) (red solid line). E, quantification of ANS-dependent emission spectra maxima above background as a percentage of the maximal ANS emission in the presence of anastellin (50 μm) for samples containing 10FNIII (50 μm) with or without 500 μm CPB or CPB(W6A). The graph shown represents pooled data from independent experiments. A significant increase in fluorescence was measured for anastellin (*, p < 0.05, one-tailed) and 10FNIII with CPB (**, p < 0.01, one-tailed) or CPB(W6A) (*, p < 0.05, one-tailed). Addition of CPB, but not CPB(W6A), to 10FNIII significantly increased ANS fluorescence above that for 10FNIII (*, p < 0.05, one-tailed) or CPB(W6A) (*, p < 0.05, one-tailed). F, comparison of ThT (20 μm) fluorescence at 482 nm after a 60-min incubation at 25 °C for 10FNIII (10 μm) in the presence or absence of 500 μm CPB or CPB(W6A). Significant ThT fluorescence was observed for CPB (*, p < 0.05, two-tailed) and a mixture of 10FNIII and CPB (**, p < 0.01, two-tailed). Addition of CPB to 10FNIII led to a significant increase in fluorescence above that for CPB (*, p < 0.05, one-tailed) or 10FNIII (**, p < 0.01, one-tailed). Addition of CPB(W6A) to 10FNIII did not lead to a significant change in fluorescence (p > 0.05, two-tailed).
SLLISWD Multimerization Sequence Interacts with 10FNIII to Expose Hydrophobic Pockets and Form β Structure
We evaluated whether CPB interacts with 10FNIII to induce exposure of hydrophobic surfaces in a similar manner to CP1. Addition of CPB at 10-fold molar excess to 10FNIII (500 and 50 μm, respectively) enhanced ANS fluorescence above that exhibited by CPB and significantly above that for 10FNIII alone (p < 0.05, one-tailed) (Fig. 5, D and E). The CD spectrum of CPB also confirmed its random coil-like behavior in solution (data not shown). These results demonstrate that the multimerization sequence is also capable of interacting with 10FNIII to form solvent-exposed hydrophobic surfaces that support ANS binding.
The formation of superfibronectin and 9FNIII multimers has been proposed to follow a polymerization mechanism of intermolecular β-strand exchange where unfolded intermediates present exposed β-sheet elements for multimerization (38, 40). Such a mechanism may be relevant to CPB-induced FN multimerization as the SLLISWD sequence has potential for β-sheet interactions with the parent domain given that its sequence is derived from the second β-strand of 10FNIII. ThT is a benzothiazole dye that undergoes a spectral shift leading to enhanced fluorescence when bound to β-sheet-rich structures and has been used to detect self-assembling β-rich fibrils formed by amyloidogenic precursors, immunoglobulin light chain, and 9FNIII (32, 38, 41). ThT fluorescence is similar to background in the presence of 10FNIII (p > 0.05, two-tailed) (Fig. 5F) but significant in the presence of CPB (p < 0.05, two-tailed). However, addition of CPB to 10FNIII (50:1 molar concentration of peptide to 10FNIII) led to an 87 ± 55% increase in fluorescence over that measured for CPB alone (p < 0.05, one-tailed) and a 19-fold increase in fluorescence as compared with 10FNIII alone (p < 0.01, one-tailed) (Fig. 5F). Furthermore, the ThT fluorescence profile showed a time-dependent increase following 10FNIII addition to CPB suggesting that CPB interactions with 10FNIII resulted in the formation of β-contacts.
A Key Trp Residue Is Required for Multimerization Sequence Activity
Residue Trp-22 in the second N-terminal β-strand of 10FNIII is a conserved residue found in all FNIII domains that is critical to the stabilization of their hydrophobic cores (23, 42, 43). This residue corresponds to the largest hydrophobic side chain within the multimerization sequence at position 6. An alanine point mutation at Trp-6 in the multimerization sequence (yielding peptide CPB(W6A)) abolished the ability of the sequence to multimerize labeled FN (p > 0.05, two-tailed) and unlabeled FN as detected by SDS-PAGE (Fig. 5, A and B, respectively), thus emphasizing the key role that Trp-6 plays in FN multimerization. Additionally the W6A mutation reduced the ability of the sequence in mixtures with 10FNIII to support ANS binding (Fig. 5E) and eliminated ThT-dependent fluorescence (Fig. 5F).
CPB, but Not CPB(W6A), Enhances FN Matrix Assembly by Fibroblasts
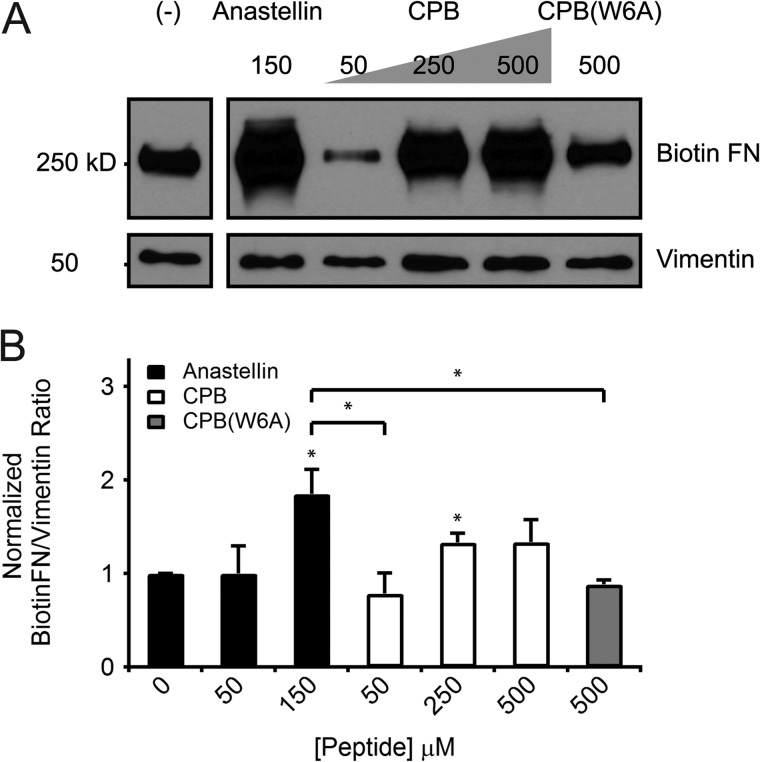
Localization of the multimerization sequence to a partially unfolded subsequence within the kinetic intermediate predicted by FN unfolding simulations that apply force at the integrin-binding site suggests that the multimerization sequence may play a role in physiological cell-mediated FN fibrillogenesis. This multimerization sequence has been shown thus far to initiate assembly of pure FN in vitro. To test the relevance of this sequence to physiological FN assembly, we monitored the incorporation of biotinylated FN into deoxycholate-extracted matrix by cell monolayers treated with peptide. Previous studies show that cell monolayers treated with anastellin increase FN deposition into matrix (34). Similarly, we found that treatment of IMR90 cells with anastellin at the same concentration (150 μm) at which FN multimerization was observed (Fig. 2A) enhances the incorporation of FN into matrix (p < 0.05, two-tailed) (Fig. 6, A and B). Likewise, the addition of high concentrations CPB (250 and 500 μm) to IMR90 cell monolayers also enhanced FN fibrillogenesis similar to 150 μm anastellin (p > 0.05, two-tailed), whereas treatment with mutant peptide CPB(W6A) (500 μm) had no effect (Fig. 6, A and B).
FIGURE 6.
CPB-mediated increases in fibronectin matrix assembly by cultured fibroblasts. A, a representative blot from a single experiment shows levels of biotinylated FN (top) incorporated by fibroblast monolayers into deoxycholate-insoluble matrix for samples normalized to vimentin (bottom). Cell layers were treated for 24 h with complete medium supplemented with biotinylated FN (20 μg/ml) containing 25 mm HEPES (pH 7.1) with or without anastellin (150 μm), CPB (50, 250, and 500 μm), or CPB(W6A) (500 μm) prior to extraction in 2% deoxycholate. B, biotinylated FN matrix incorporation assessed by quantifying the ratio of band densities for biotinylated FN and vimentin within each sample. Data are pooled from three independent experiments. Addition of 150 μm anastellin or 250 μm CPB to IMR90 cells significantly increased assembly of biotinylated FN into the matrix as compared with untreated cells (*, p < 0.05, two-tailed). Cells treated with high concentrations CPB (250 and 500 μm) showed similar levels of FN matrix incorporation as 150 μm anastellin (p > 0.05, two-tailed), unlike cells treated with 50 μm CPB or 500 μm CPB(W6A) (*, p < 0.05, two-tailed).
CPB Also Initiates FBG Multimerization
We also studied the effects of CPB on the plasma protein fibrinogen (FBG), which is an ECM protein that is involved in blood clot formation. Past work has shown that physical integration of FBG into ECM fibrils also requires FN matrix assembly (44) and that anastellin induces FBG polymerization, as well as FN assembly (2). Similar to anastellin, addition of CPB to FBG (5 mg/ml) led to an increase in spectrophotometric absorbance over time indicative of FBG polymerization in a peptide concentration-dependent manner (Fig. 7A). CPB-induced FBG multimerization was observed using molar concentrations of peptide to FBG at 33:1 and 50:1 (CPB at 500 and 750 μm, respectively). In addition, a time-dependent increase in ThT fluorescence is observed in mixtures of CPB and FBG (50:1 molar concentrations of peptide to FBG) (Fig. 7B), but not for mixtures of CPB(W6A) and FBG (Fig. 7C) or isolated samples of CPB, CPB(W6A), or FBG alone (Fig. 7, B and C) indicating that the CPB-induced FBG assemblies form β-containing structures.
FIGURE 7.
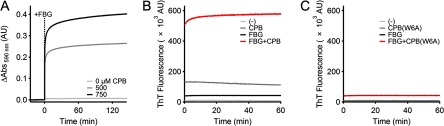
Effect of CPB on fibrinogen polymerization. A, the development of turbidity at 590 nm at 25 °C by CPB (500–750 μm) was monitored before and after the addition of FBG (5 mg/ml) at t = 0 min (dashed line marked +FBG). B, time traces of ThT-dependent (20 μm) fluorescence at 482 nm at 25 °C for buffer (light gray line), CPB (500 μm, dark gray line), FBG (10 μm, black line), or a mixture of FBG (10 μm) and CPB (500 μm) (red line). C, ThT-dependent fluorescence for buffer (light gray line), CPB(W6A) (500 μm, dark gray line), FBG (10 μm, black line), or a mixture of FBG (10 μm) and CPB(W6A) (500 μm) (red line) over time.
DISCUSSION
We have identified a 7-amino acid “multimerization sequence,” SLLISWD, within the cell-binding domain of FN that is sufficient to initiate polymerization of added FN or FBG. This discovery was motivated by mechanistic insight into cell-driven FN fibrillogenesis provided by SMD simulations that predicted N-terminal unraveling of 10FNIII to a partially unfolded intermediate state when force is applied at its RGD loop (26). A 23-amino acid peptide mimic designed from this unfolded terminus is shown to be a cryptic sequence in 10FNIII that initiates FN multimerization as its scrambled sequence, the unexposed β-strand E, and natively folded 10FNIII are unable to initiate similar multimerization results. We show that the multimerization process may involve interactions of this 23-amino acid sequence with 10FNIII, which leads to solvent exposure of hydrophobic surfaces. Further analysis of the CP1 sequence led us to discover a 7-amino acid multimerization sequence with a key Trp-6 residue localized to its C terminus that is sufficient to initiate FN multimerization. We now show using two different preparations of FN that both CP1 and CPB initiate FN multimerization using multiple different analytic techniques (i.e. non-reducing SDS-PAGE multimerization, turbidity, and pelleting assays). It is also important to note that the rhodamine-labeled FN used in our multimerization studies contains ∼2.5 mm β-mercaptoethanol, which can interfere with protein multimerization; thus, the degree of multimerization presented here may be an underestimate of the full potency of these peptides. Although previous studies have identified cryptic FN assembly sites within thermally denatured 10FNIII (24) and have shown that mutation of conserved proline residues at the N terminus (P5A or P25A) enhances self-aggregation (25), this is the first study that identified a specific sequence in 10FNIII that induces FN multimerization. Moreover, we confirm that this sequence is physiologically relevant as the addition of the multimerization sequence to cultured fibroblasts monolayers enhances FN fibrillogenesis in vitro.
The peptides presented in this study possess similar multimerization capabilities as anastellin, a 75-amino acid fragment of 1FNIII (2, 34). Comparison at a peptide to FN molar ratio of 220:1, CP1 remarkably induces a higher percentage of FN multimers than anastellin as detected by SDS-PAGE despite being less than one-third the length of anastellin. The shorter 7-amino acid multimerization sequence within CP1 requires a 3-fold higher molar ratio of peptide to FN to induce a similar degree of multimerization as the 23-amino acid CP1 sequence (733:1 versus 220:1, respectively). These observations suggest a significantly stronger FN binding affinity for CP1, which may provide additional favorable contacts to position the multimerization sequence to interact with FN. Like anastellin, we show the multimerization sequence also initiates FBG assembly. It should be noted, however, that the peptides described in this study do differ from anastellin in a few significant ways. First, comparison of the SLLISWD and anastellin sequences shows a 10-fold difference in length. Such short peptides are easily synthesized and readily amenable to chemical modification enabling the incorporation of bioorthogonal handles. This flexibility is beneficial to the development of multifunctional “smart” probes with ECM-forming activity relevant for applications addressing diseases of ECM remodeling, such as abnormal wound healing, fibrosis, or cancer, for example. Second, the multimerization sequence is derived from the mechanically weak 10FNIII domain that is recognized by integrins at the RGD loop. As 10FNIII provides the direct point of contact on FN for cell surface integrin receptors, this sequence may play a potential role in the mechanical process of cell-mediated FN fibrillogenesis. Third, CP1 and anastellin are complementary structural counterparts in homologous FNIII structures. Anastellin is a N-terminal deletion fragment of 1FNIII lacking the N terminus that includes strands A and B (45), which is the region spanning CP1 in 10FNIII. Despite differences in β strand coverage of the FNIII structure, both systems display hydrophobic surfaces to solvent. The multimerization sequence is unstructured and exposes hydrophobic binding surfaces upon interaction with 10FNIII, whereas anastellin possesses exposed hydrophobic binding pockets for ANS in its residual structure (37, 45).
FN multimerization has been identified as a hydrophobic process where cells convert FN into deoxycholate-insoluble fibers stabilized by noncovalent interactions (46). Stretching FN fibers induces conformational changes including exposure of hydrophobic surfaces and unraveling of secondary structure (20, 47). Long stretches of hydrophobic residues can form β-sheet fibril assemblies as demonstrated by the use of hydrophobic sequences derived from the amyloidogenic β-Alzheimer peptide to engineer fibril assembly into a 14-amino acid peptide (48). CP1 encompasses the most hydrophobic region of 10FNIII (26) including β-strands A and B, which possess 56 and 57% hydrophobic character, respectively. The identified multimerization sequence corresponds to one of the most hydrophobic β-strands in 10FNIII. It is clear that sequence specificity is important to the assembly process as strand A shows similar hydrophobic character but lacks the ability to initiate FN multimerization. Moreover, a point mutation of the largest hydrophobic residue (W6A) in the multimerization sequence reduced its ability to expose hydrophobic surfaces and form β-structure when mixed with 10FNIII and more importantly annihilated its ability to initiate FN multimerization and enhance physiological FN assembly. This Trp-6 residue corresponds to the single conserved Trp residue in strand B found in all FNIII domains that is important for stabilization of the hydrophobic core of the fold (23, 42, 43). In 10FNIII, a randomized mutation of this conserved Trp-22 residue in a yeast two-hybrid system severely destabilized complementation of a bisected 10FNIII to form a stable 10FNIII structure (49). It is possible that this Trp-6 residue in the multimerization sequence may play a role as a substitute for Trp-22 to stabilize an exposed hydrophobic core of a partially unfolded 10FNIII intermediate that subsequently propagates FN self-assembly.
We show with cell-based experiments that the multimerization sequence is relevant to physiological FN fibrillogenesis as the addition of the CPB peptide to cell monolayers enhances FN deposition into the ECM. FN assembly, in the context of superfibronectin and 9FNIII multimer formation, has been hypothesized to follow a process of intermolecular β-strand exchange, where β-strands from one partially unfolded domain complements a similarly unfolded intermediate of another domain to form a stable polymeric structure (38, 40, 45). FN assembly initiated by other peptide sequences in addition to anastellin, including peptides anginex, CLT1, and BBK32, has also been described to follow a β-structure-mediated process (50–52). Although these inducing sequences are not native to FN, they may share a common mechanism for assembly involving hydrophobic interactions and β-sheet formation. BBK32, derived from a surface-expressed lipoprotein, shows weak sequence homology to regions present in anastellin, specifically to the C/C′ loop and the F strand in 1FNIII (52). Anginex is a 33-amino acid peptide designed to mimic β-sheets (50), and CLT1 is a 10-mer identified by phage display that possesses a hydrophobic N terminus and hydrophilic C terminus proposed to form β-sheets similar to amyloidogenic fibrils (51). Because the multimerization sequence possesses similar properties to anastellin and these other peptides capable of initiating FN assembly, we suggest β-strand exchange as a potential mechanism for FN assembly initiated by the multimerization sequence.
We propose one model of β-strand exchange that is consistent with the results reported here whereby the multimerization sequence stabilizes a partially unfolded intermediate of 10FNIII by displacing the B strand of 10FNIII (and subsequently disrupting the N-terminal residues including the A strand) by exchange of its Trp-6 residue with the conserved Trp-22 in the hydrophobic core. The subsequent partially unfolded 10FNIII structure can thus propagate assembly by forming complementary intermolecular cross-links with additional domains via the multimerization sequence contained within its exposed N terminus (Fig. 8). Stabilization of a partially unfolded 10FNIII structure by exchange of Trp-6 in the multimerization sequence with the 10FNIII core during a β-strand exchange with strand B would yield exposed hydrophobic binding surfaces and a net gain of β-structure as we have observed. This proposed process would rely on the chemical instability of 10FNIII. Experiments reveal that recombinant 10FNIII possesses a high melting temperature of ∼88 °C (53, 54) and is resistant to denaturation in 6 m urea (20). These measures reflect global conformational stability of the domain as a whole but do not reveal local fluctuations within the structure. NMR experiments measuring the conformational exchange parameter show a high degree of conformational flexibility on the microsecond to millisecond time scales for strands A and B within 10FNIII (39). Although this past study focused on the time scale of fluctuations, local transient opening of the N terminus was revealed. We are not suggesting that strands A and B are completely exposed by spontaneous unfolding as this would lead to spontaneous FN multimerization. Instead, our hypothesis proposes that fluctuations that exist in the N terminus of 10FNIII may open up the domain to enable a portion of the multimerization sequence to interact with the 10FNIII core, which would feed back to promote additional (but still partial) unfolding of adjacent domains at the N terminus to then drive fibrillogenesis as a result of strand swapping with strand B. Our observations show that solutions of 10FNIII and the multimerization sequence exhibit exposed hydrophobic binding surfaces and β-structure formation following mixing without extended incubation times. Although the largest enhancement in ThT binding is shown for mixtures of the multimerization sequence with 10FNIII, it is noted that the multimerization sequence alone shows significant ThT fluorescence above background. However, the CD spectrum of CPB reflects a random coil signature suggesting that the peptide at low concentration is predominantly unstructured, and turbidity measurements of peptide at high concentration indicates minimal change in turbidity. The polymerization of FBG, a protein primarily composed of coiled-coils, by the multimerization sequence may also follow a β-strand swapping model as the FBG end-domain involved in polymerization includes a β-strand capable of forming intermolecular β-sheet contacts for assembly when exposed in a partially unfolded intermediate (55). Our ThT binding data supports this model whereby the multimerization sequence is shown to interact with FBG to form β-rich assemblies over time.
FIGURE 8.
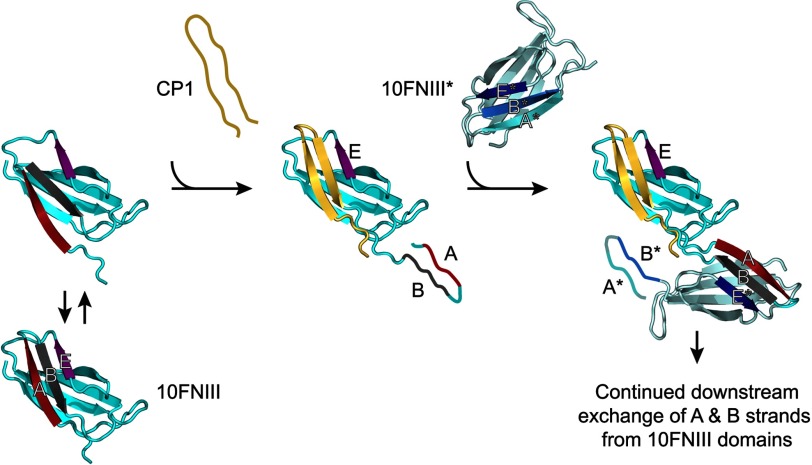
The proposed β-strand swapping mechanism for initiating fibronectin assembly. A conformationally flexible 10FNIII (left) whose B β-strand (and subsequently residues N-terminal to it) is displaced by the multimerization sequence from the C terminus of CP1 yields a partially unfolded intermediate stabilized by the Trp-6 residue in the multimerization sequence. The newly released N terminus in the partially unfolded 10FNIII (center) presents its own cryptic multimerization motif from β-strand B that participates in additional domain swapping interactions with a second FNIII domain (e.g. 10FNIII*) to form β-strand-mediated intermolecular cross-links (right). Repeated downstream domain swapping with additional FN molecules thus leads to FN assembly.
The similarity of the FN multimers initiated by the multimerization sequence in solution, a thermodynamic process, to the initiation of physiologic FN fibrils formed by mechanically driven cell traction force remains to be determined given that the relative chemical and mechanical stabilities of the FNIII domains are dissimilar (19). A recent study with mutant FN containing a single cysteine point mutation within the E strand in 10FNIII at position Ala-1472 showed that integration into growing fibrils by cell-mediated stretching led to minimal labeling by a fluorescein-conjugated maleimide in solvent (20). These experimental observations do not exclude our current hypothesis given that this A1472C mutation in the E strand does not probe unfolding within our proposed region of interest at the N terminus. The results reported here agree that the sequence of strand E does not efficiently initiate FN assembly. Our model requires further testing to determine whether the N terminus of 10FNIII is exposed during physiological FN fibrillogenesis.
Our proposed model for cell-mediated FN fibrillogenesis stipulates that the N terminus of 10FNIII unfolds. However, it is known that cell adhesion to RGD on FN is enhanced by a synergy site in the N-terminal neighboring domain 9FNIII for integrins α5β1 and αIIbβ3, but not αVβ3 (56–58). In particular, FN assembly by α5β1, but not by αVβ3, requires the synergy site (59). Because α5β1 is the main receptor that mediates FN assembly (14) and increasing the spacing between the PHSRN and RGD sites significantly reduces cell adhesion (60, 61), one would suspect that N-terminal unfolding of 10FNIII would prohibit enhanced adhesion and subsequent fibrillogenesis. However, it has been shown that activation of the α5β1 integrin overcomes the requirement of the synergy site for improved adhesion to the RGD sequence and FN assembly (58, 59). Therefore, our proposed model of unfolding of the N terminus in 10FNIII, and thus separating the synergy site and the RGD loop, is feasible to describe cell-mediated FN fibrillogenesis once the α5β1 integrin is bound to FN and activated.
The results described in this study showed that the structure proposed by SMD simulations modeling of the unfolding of 10FNIII along a physiological pulling geometry revealed new insights into specific regions of the domain that are important to FN assembly. Thus, these data demonstrate the advantage of using SMD simulations to gain atomistic insight into physiologically relevant intermediate states that are not easily observed with experimental approaches alone and illustrate the synergy in combining computational models with biochemical and biophysical assays to identify biologically relevant motifs within a force-responsive protein. This model-driven approach is in contrast to the discovery of anastellin, whose superfibronectin properties were identified prior to simulations that predicted an anastellin-like unfolded intermediate of 1FNIII with exposed N-terminal A and B β-strands (34, 62). We have identified a minimal multimerization sequence from the 10FNIII domain in FN that initiates assembly of FN and FBG, proteins that are important in promoting tissue repair. Although there have been other identified peptide sequences capable of inducing FN assembly (50–52), this is the first sequence that is identified in 10FNIII, the mechanically weak domain that contains the initial integrin binding site that transmits tensional force onto FN. Our identification of a cryptic multimerization sequence in 10FNIII provides new insight into the initial steps in the physiological process of cell-mediated FN fibrillogenesis as well as a new motif for initiating FN assembly to stimulate ECM formation.
Acknowledgments
We gratefully acknowledge the generous gift of the pETCH-GST-8–11FNIII-His6 vector from Drs. Richard Clark and Xiang-Dong Ren.
This work was supported, in whole or in part, by National Institutes of Health Grant PO1 CA045548, by United States Department of Defense Breast Cancer Innovator Award BC074986, and a grant from the Wyss Institute for Biologically Inspired Engineering at Harvard University (to D. E. I.).
- ECM
- extracellular matrix
- FN
- fibronectin
- 10FNIII
- tenth FN type III
- SMD
- steered molecular dynamics
- CP1
- cryptic peptide 1
- CP1scr
- scrambled cryptic peptide 1
- FBG
- fibrinogen
- ANS
- 8-anilino-1-napthalenesulfonic acid
- ThT
- thioflavin T.
REFERENCES
- 1. Hynes R. O. (2009) The extracellular matrix. Not just pretty fibrils. Science 326, 1216–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yi M., Ruoslahti E. (2001) A fibronectin fragment inhibits tumor growth, angiogenesis, and metastasis. Proc. Natl. Acad. Sci. U.S.A. 98, 620–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Reilly M. S., Boehm T., Shing Y., Fukai N., Vasios G., Lane W. S., Flynn E., Birkhead J. R., Olsen B. R., Folkman J. (1997) Endostatin. An endogenous inhibitor of angiogenesis and tumor growth. Cell 88, 277–285 [DOI] [PubMed] [Google Scholar]
- 4. Mongiat M., Sweeney S. M., San Antonio J. D., Fu J., Iozzo R. V. (2003) Endorepellin, a novel inhibitor of angiogenesis derived from the C terminus of perlecan. J. Biol. Chem. 278, 4238–4249 [DOI] [PubMed] [Google Scholar]
- 5. Xie L., Palmsten K., MacDonald B., Kieran M. W., Potenta S., Vong S., Kalluri R. (2008) Basement membrane derived fibulin-1 and fibulin-5 function as angiogenesis inhibitors and suppress tumor growth. Exp. Biol. Med. (Maywood) 233, 155–162 [DOI] [PubMed] [Google Scholar]
- 6. Sid B., Sartelet H., Bellon G., El Btaouri H., Rath G., Delorme N., Haye B., Martiny L. (2004) Thrombospondin 1. A multifunctional protein implicated in the regulation of tumor growth. Crit. Rev. Oncol. Hematol. 49, 245–258 [DOI] [PubMed] [Google Scholar]
- 7. Zhong C., Chrzanowska-Wodnicka M., Brown J., Shaub A., Belkin A. M., Burridge K. (1998) Rho-mediated contractility exposes a cryptic site in fibronectin and induces fibronectin matrix assembly. J. Cell Biol. 141, 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marsden M., DeSimone D. W. (2001) Regulation of cell polarity, radial intercalation and epiboly in Xenopus. Novel roles for integrin and fibronectin. Development 128, 3635–3647 [DOI] [PubMed] [Google Scholar]
- 9. Mosher D. F. (1975) Cross-linking of cold-insoluble globulin by fibrin-stabilizing factor. J. Biol. Chem. 250, 6614–6621 [PubMed] [Google Scholar]
- 10. Feaver R. E., Gelfand B. D., Wang C., Schwartz M. A., Blackman B. R. (2010) Atheroprone hemodynamics regulate fibronectin deposition to create positive feedback that sustains endothelial inflammation. Circ. Res. 106, 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou X., Rowe R. G., Hiraoka N., George J. P., Wirtz D., Mosher D. F., Virtanen I., Chernousov M. A., Weiss S. J. (2008) Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes Dev. 22, 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hynes R. O. (1973) Alteration of cell-surface proteins by viral transformation and by proteolysis. Proc. Natl. Acad. Sci. U.S.A. 70, 3170–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johansson S., Svineng G., Wennerberg K., Armulik A., Lohikangas L. (1997) Fibronectin-integrin interactions. Front. Biosci. 2, d126–146 [DOI] [PubMed] [Google Scholar]
- 14. Fogerty F. J., Akiyama S. K., Yamada K. M., Mosher D. F. (1990) Inhibition of binding of fibronectin to matrix assembly sites by anti-integrin (α5β1) antibodies. J. Cell Biol. 111, 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pierschbacher M. D., Ruoslahti E. (1984) Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 309, 30–33 [DOI] [PubMed] [Google Scholar]
- 16. Wu C., Keivens V. M., O'Toole T. E., McDonald J. A., Ginsberg M. H. (1995) Integrin activation and cytoskeletal interaction are essential for the assembly of a fibronectin matrix. Cell 83, 715–724 [DOI] [PubMed] [Google Scholar]
- 17. McDonald J. A., Quade B. J., Broekelmann T. J., LaChance R., Forsman K., Hasegawa E., Akiyama S. (1987) Fibronectin's cell-adhesive domain and an amino-terminal matrix assembly domain participate in its assembly into fibroblast pericellular matrix. J. Biol. Chem. 262, 2957–2967 [PubMed] [Google Scholar]
- 18. Smith M. L., Gourdon D., Little W. C., Kubow K. E., Eguiluz R. A., Luna-Morris S., Vogel V. (2007) Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 5, e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oberhauser A. F., Badilla-Fernandez C., Carrion-Vazquez M., Fernandez J. M. (2002) The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 319, 433–447 [DOI] [PubMed] [Google Scholar]
- 20. Lemmon C. A., Ohashi T., Erickson H. P. (2011) Probing the folded state of fibronectin type III domains in stretched fibrils by measuring buried cysteine accessibility. J. Biol. Chem. 286, 26375–26382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Geiger B., Bershadsky A., Pankov R., Yamada K. M. (2001) Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2, 793–805 [DOI] [PubMed] [Google Scholar]
- 22. Nagai T., Yamakawa N., Aota S., Yamada S. S., Akiyama S. K., Olden K., Yamada K. M. (1991) Monoclonal antibody characterization of two distant sites required for function of the central cell-binding domain of fibronectin in cell adhesion, cell migration, and matrix assembly. J. Cell Biol. 114, 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Craig D., Gao M., Schulten K., Vogel V. (2004) Tuning the mechanical stability of fibronectin type III modules through sequence variations. Structure 12, 21–30 [DOI] [PubMed] [Google Scholar]
- 24. Hocking D. C., Smith R. K., McKeown-Longo P. J. (1996) A novel role for the integrin-binding III-10 module in fibronectin matrix assembly. J. Cell Biol. 133, 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steward A., Adhya S., Clarke J. (2002) Sequence conservation in Ig-like domains. The role of highly conserved proline residues in the fibronectin type III superfamily. J. Mol. Biol. 318, 935–940 [DOI] [PubMed] [Google Scholar]
- 26. Gee E. P., Ingber D. E., Stultz C. M. (2008) Fibronectin unfolding revisited. Modeling cell traction-mediated unfolding of the tenth type-III repeat. PLoS One 3, e2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao M., Craig D., Vogel V., Schulten K. (2002) Identifying unfolding intermediates of FN-III(10) by steered molecular dynamics. J. Mol. Biol. 323, 939–950 [DOI] [PubMed] [Google Scholar]
- 28. Paci E., Karplus M. (1999) Forced unfolding of fibronectin type 3 modules. An analysis by biased molecular dynamics simulations. J. Mol. Biol. 288, 441–459 [DOI] [PubMed] [Google Scholar]
- 29. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang R., Clark R. A., Mosher D. F., Ren X. D. (2005) Fibronectin's central cell-binding domain supports focal adhesion formation and Rho signal transduction. J. Biol. Chem. 280, 28803–28810 [DOI] [PubMed] [Google Scholar]
- 31. Azzi A. (1974) The use of fluorescent probes for the study of membranes. Methods Enzymol. 32, 234–246 [DOI] [PubMed] [Google Scholar]
- 32. Wall J., Murphy C. L., Solomon A. (1999) In vitro immunoglobulin light chain fibrillogenesis. Methods Enzymol. 309, 204–217 [DOI] [PubMed] [Google Scholar]
- 33. McKeown-Longo P. J., Mosher D. F. (1983) Binding of plasma fibronectin to cell layers of human skin fibroblasts. J. Cell Biol. 97, 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morla A., Zhang Z., Ruoslahti E. (1994) Superfibronectin is a functionally distinct form of fibronectin. Nature 367, 193–196 [DOI] [PubMed] [Google Scholar]
- 35. Yamada K. M., Kennedy D. W., Kimata K., Pratt R. M. (1980) Characterization of fibronectin interactions with glycosaminoglycans and identification of active proteolytic fragments. J. Biol. Chem. 255, 6055–6063 [PubMed] [Google Scholar]
- 36. Stryer L. (1965) The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non-polar binding sites. J. Mol. Biol. 13, 482–495 [DOI] [PubMed] [Google Scholar]
- 37. Ohashi T., Augustus A. M., Erickson H. P. (2009) Transient opening of fibronectin type III (FNIII) domains. The interaction of the third FNIII domain of FN with anastellin. Biochemistry 48, 4189–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Litvinovich S. V., Brew S. A., Aota S., Akiyama S. K., Haudenschild C., Ingham K. C. (1998) Formation of amyloid-like fibrils by self-association of a partially unfolded fibronectin type III module. J. Mol. Biol. 280, 245–258 [DOI] [PubMed] [Google Scholar]
- 39. Carr P. A., Erickson H. P., Palmer A. G., 3rd (1997) Backbone dynamics of homologous fibronectin type III cell adhesion domains from fibronectin and tenascin. Structure 5, 949–959 [DOI] [PubMed] [Google Scholar]
- 40. Ohashi T., Erickson H. P. (2005) Domain unfolding plays a role in superfibronectin formation. J. Biol. Chem. 280, 39143–39151 [DOI] [PubMed] [Google Scholar]
- 41. Biancalana M., Makabe K., Koide A., Koide S. (2009) Molecular mechanism of thioflavin-T binding to the surface of β-rich peptide self-assemblies. J. Mol. Biol. 385, 1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Halaby D. M., Poupon A., Mornon J. (1999) The immunoglobulin fold family. Sequence analysis and 3D structure comparisons. Protein Eng. 12, 563–571 [DOI] [PubMed] [Google Scholar]
- 43. Cota E., Hamill S. J., Fowler S. B., Clarke J. (2000) Two proteins with the same structure respond very differently to mutation. The role of plasticity in protein stability. J. Mol. Biol. 302, 713–725 [DOI] [PubMed] [Google Scholar]
- 44. Pereira M., Rybarczyk B. J., Odrljin T. M., Hocking D. C., Sottile J., Simpson-Haidaris P. J. (2002) The incorporation of fibrinogen into extracellular matrix is dependent on active assembly of a fibronectin matrix. J. Cell Sci. 115, 609–617 [DOI] [PubMed] [Google Scholar]
- 45. Briknarová K., Akerman M. E., Hoyt D. W., Ruoslahti E., Ely K. R. (2003) Anastellin, an FN3 fragment with fibronectin polymerization activity, resembles amyloid fibril precursors. J. Mol. Biol. 332, 205–215 [DOI] [PubMed] [Google Scholar]
- 46. Chen H., Mosher D. F. (1996) Formation of sodium dodecyl sulfate-stable fibronectin multimers. Failure to detect products of thiol-disulfide exchange in cyanogen bromide or limited acid digests of stabilized matrix fibronectin. J. Biol. Chem. 271, 9084–9089 [DOI] [PubMed] [Google Scholar]
- 47. Little W. C., Schwartlander R., Smith M. L., Gourdon D., Vogel V. (2009) Stretched extracellular matrix proteins turn fouling and are functionally rescued by the chaperones albumin and casein. Nano Lett. 9, 4158–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Otzen D. E., Kristensen O., Oliveberg M. (2000) Designed protein tetramer zipped together with a hydrophobic Alzheimer homology. A structural clue to amyloid assembly. Proc. Natl. Acad. Sci. U.S.A. 97, 9907–9912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dutta S., Koide A., Koide S. (2008) High-throughput analysis of the protein sequence-stability landscape using a quantitative yeast surface two-hybrid system and fragment reconstitution. J. Mol. Biol. 382, 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Akerman M. E., Pilch J., Peters D., Ruoslahti E. (2005) Angiostatic peptides use plasma fibronectin to home to angiogenic vasculature. Proc. Natl. Acad. Sci. U.S.A. 102, 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Knowles L. M., Malik G., Hood B. L., Conrads T. P., Pilch J. (2012) CLT1 targets angiogenic endothelium through CLIC1 and fibronectin. Angiogenesis 15, 115–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Prabhakaran S., Liang X., Skare J. T., Potts J. R., Höök M. (2009) A novel fibronectin binding motif in MSCRAMMs targets F3 modules. PLoS One 4, e5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Litvinovich S. V., Ingham K. C. (1995) Interactions between type III domains in the 110-kDa cell-binding fragment of fibronectin. J. Mol. Biol. 248, 611–626 [DOI] [PubMed] [Google Scholar]
- 54. Cota E., Clarke J. (2000) Folding of β-sandwich proteins. Three-state transition of a fibronectin type III module. Protein Sci. 9, 112–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yakovlev S., Litvinovich S., Loukinov D., Medved L. (2000) Role of the β-strand insert in the central domain of the fibrinogen γ-module. Biochemistry 39, 15721–15729 [DOI] [PubMed] [Google Scholar]
- 56. Bowditch R. D., Hariharan M., Tominna E. F., Smith J. W., Yamada K. M., Getzoff E. D., Ginsberg M. H. (1994) Identification of a novel integrin binding site in fibronectin. Differential utilization by β3 integrins. J. Biol. Chem. 269, 10856–10863 [PubMed] [Google Scholar]
- 57. Aota S., Nomizu M., Yamada K. M. (1994) The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J. Biol. Chem. 269, 24756–24761 [PubMed] [Google Scholar]
- 58. Danen E. H., Aota S., van Kraats A. A., Yamada K. M., Ruiter D. J., van Muijen G. N. (1995) Requirement for the synergy site for cell adhesion to fibronectin depends on the activation state of integrin α5β1. J. Biol. Chem. 270, 21612–21618 [DOI] [PubMed] [Google Scholar]
- 59. Sechler J. L., Corbett S. A., Schwarzbauer J. E. (1997) Modulatory roles for integrin activation and the synergy site of fibronectin during matrix assembly. Mol. Biol. Cell 8, 2563–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Grant R. P., Spitzfaden C., Altroff H., Campbell I. D., Mardon H. J. (1997) Structural requirements for biological activity of the ninth and tenth FIII domains of human fibronectin. J. Biol. Chem. 272, 6159–6166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Spitzfaden C., Grant R. P., Mardon H. J., Campbell I. D. (1997) Module-module interactions in the cell binding region of fibronectin. Stability, flexibility and specificity. J. Mol. Biol. 265, 565–579 [DOI] [PubMed] [Google Scholar]
- 62. Gao M., Craig D., Lequin O., Campbell I. D., Vogel V., Schulten K. (2003) Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates. Proc. Natl. Acad. Sci. U.S.A. 100, 14784–14789 [DOI] [PMC free article] [PubMed] [Google Scholar]