Background: IGBP1/α4 interacts with microtubule-associated MID1 to regulate PP2A within the mTOR pathway.
Results: MID1 catalyzes the polyubiquitination and degradation of α4, demonstrating for the first time a mechanism for α4 regulation.
Conclusion: The tandem RING-Bbox domains are required for α4 polyubiquitination and degradation.
Significance: Ectopic overexpression of IGBP1/α4 transforms cells. Ubiquitination of α4 impacts the stability and activity level of PP2A.
Keywords: Cancer, Cell Signaling, Cellular Immune Response, E3 Ubiquitin Ligase, PP2A, Protein Degradation, Ubiquitination
Abstract
Alpha4 (α4) is a key regulator of protein phosphatase 2A (PP2A) and mTOR in steps essential for cell-cycle progression. α4 forms a complex with PP2A and MID1, a microtubule-associated ubiquitin E3 ligase that facilitates MID1-dependent regulation of PP2A and the dephosphorylation of MID1 by PP2A. Ectopic overexpression of α4 is associated with hepatocellular carcinomas, breast cancer, and invasive adenocarcinomas. Here, we provide data suggesting that α4 is regulated by ubiquitin-dependent degradation mediated by MID1. In cells stably expressing a dominant-negative form of MID1, significantly elevated levels of α4 were observed. Treatment of cells with the specific proteasome inhibitor, lactacystin, resulted in a 3-fold increase in α4 in control cells and a similar level in mutant cells. Using in vitro assays, individual MID1 E3 domains facilitated monoubiquitination of α4, whereas full-length MID1 as well as RING-Bbox1 and RING-Bbox1-Bbox2 constructs catalyzed its polyubiquitination. In a novel non-biased functional screen, we identified a leucine to glutamine substitution at position 146 within Bbox1 that abolished MID1-α4 interaction and the subsequent polyubiquitination of α4, indicating that direct binding to Bbox1 was necessary for the polyubiquitination of α4. The mutant had little impact on the RING E3 ligase functionality of MID1. Mass spectrometry data confirmed Western blot analysis that ubiquitination of α4 occurs only within the last 105 amino acids. These novel findings identify a new role for MID1 and a mechanism of regulation of α4 that is likely to impact the stability and activity level of PP2Ac.
Introduction
Alpha4 (α4) was first identified as an immunoglobulin binding protein (also known as IGBP1) involved in signal transduction through the B-cell antigen receptor in mammalian B and T lymphocytes (1–5). α4 functions as a novel regulatory subunit of protein phosphatase 2A (PP2A),2 PP4, and PP6 (6–10). With respect to PP2A, α4 binds the catalytic subunit (PP2Ac), at which time it is thought to displace the scaffolding (PP2Aa, PR65) and regulatory subunits (PP2Ab) that typically constitute the PP2A heterotrimeric complex (6, 11–13). Under growth-promoting conditions, α4 down-regulates PP2A activity within the mTOR signaling pathway, resulting in the downstream activation of eIF-4E and S6 kinase and the initiation of cell cycle progression (4, 14, 15).
Regulation of PP2A is directly linked to the strong interaction between α4 and MID1, a novel microtubule-associated E3 ligase belonging to the TRIM (tripartite motif) family of proteins (11, 14). MID1 possesses an N-terminal RING domain, two B-box domains, and a coiled-coil motif (16). The RING, Bbox1, and Bbox2 domains adopt ββα-RING folds, and each possesses ubiquitin E3 ligase activity (17–19). In their native tandem configuration, the activity of the RING domain is significantly enhanced by the B-boxes implying cooperativity in function (19). The interaction between α4 and MID1 is mediated by the C-terminal domain (residues 236–280) of α4 and the Bbox1 domain of MID1, whereas the N-terminal domain (residues 1–236) of α4 is involved in binding PP2Ac (15, 20, 21).
As part of the MID1-α4-PP2Ac complex, α4 is thought to modulate the functionality of both PP2A and MID1, facilitating either dephosphorylation of MID1 and its dissociation from the microtubules or the MID1-dependent ubiquitination of PP2Ac and thus turnover of the microtubule pool of PP2A (10, 20, 22). Loss of function of MID1 is associated with a range of birth defects, including cleft lip/palate, cardiac septal defects, and hypospadias (23, 24). An increase in PP2Ac levels and decreased serine/threonine phosphorylation of cytoskeletal proteins seen in patient fibroblasts supports the notion that proper regulation of the microtubule associated PP2A activity is critical for normal development (9, 10).
Recent data suggest that α4 can play both a protective and destructive role in regulating PP2A activity and that this functionality is dependent on its monoubiquitination by MID1 (25). Given the promiscuity of PP2A, the MID1-α4-PP2Ac complex likely participates in the regulation of many different cellular processes. For example, this complex has been shown to directly regulate mTORC1 signaling as well as associate with the translational apparatus to regulate translation of various mRNAs including those for MID1 itself (6, 8, 10, 16, 26). In each case, it has been concluded that this regulatory role is conferred by MID1-mediated ubiquitination of PP2Ac (6, 8, 10, 28).
Notably, α4 has been found significantly overexpressed in many cancers, including primary hepatocellular carcinomas, breast cancer, and invasive adenocarcinoma (29, 30). Furthermore, its ectopic overexpression in immunocompromised mice is sufficient to transform embryonic kidney cells (30). These studies suggest that it may be oncogenic. As a key regulator of mTOR activity, which is a major anti-cancer target, it is important to understand how α4 is regulated. Here we demonstrate for the first time that α4 is directly regulated both in vitro and in vivo by MID1-mediated polyubiquitination.
MATERIALS AND METHODS
Generation of Stable MDCK Cell Lines
MDCK cells were cultured at 37 °C in DMEM (Thermo Scientific) supplemented with 10% fetal bovine serum (Thermo Scientific) and 1× GlutaMAX (Invitrogen). To generate MID1delCTD-expressing cell lines, MDCK cells were transfected with either a pEGFP-C2-MID1delCTD or pEGFP-C2 control expression construct (35) using TransIT-LT1 transfection reagent (Mirus Bio, Madison, WI). The MID1delCTD mutant involved a construct in which the MID1 reading frame was truncated near the start of the SPRY/B30.2 domain (K491stop; Fig. 1A). Stable cell lines were selected in 0.6 μg/ml Geneticin (Invitrogen) for 10 days. Antibiotic-resistant clonal cells were obtained by diluting the cells into 96-well plates at a concentration of less than one cell per well, and the growth of clonal cells was monitored under a light microscope as well as a fluorescent microscope for GFP expression. To confirm the expression of both intact GFP and GFP-MID1delCTD in selected clonal lines, extracts from each were examined by Western blotting. Briefly, cells cultured in a 10-cm plate were lysed in radioimmune precipitation assay buffer (Thermo Scientific) with protease inhibitors (Protease Inhibitor Mixture, EDTA-free, Thermo Scientific) for 15 min at 4 °C. Lysates were collected with a cell scraper and clarified by centrifugation for 10 min at 10,000 × g. Proteins were separated by SDS-PAGE (8–16%, Precise Protein Gel, Thermo Scientific, IL) and semi-dry-transferred to PVDF membranes (Millipore). Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline with 0.05% Tween 20, probed with anti-GFP (ab290, Abcam), and developed using chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific).
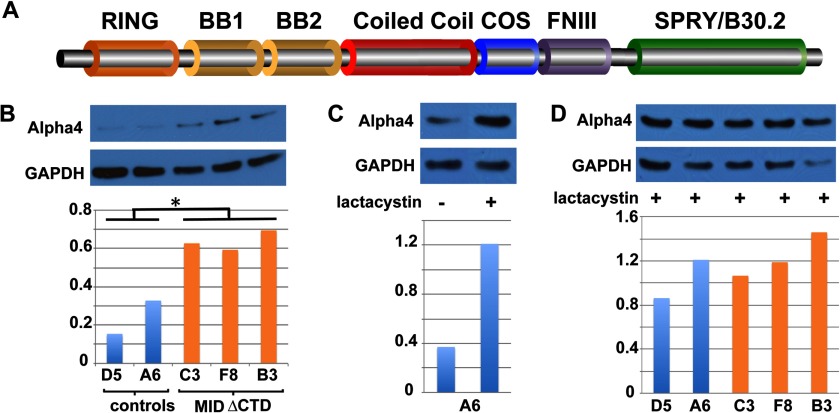
FIGURE 1.
MID1 facilitates the in vivo proteasome degradation of α4. A, shown is a schematic of MID1 depicting the different domains. The RING and Bbox domains are cysteine- and histidine-rich regions that coordinate two zinc ions. The coiled-coil domain is important for MID1 dimerization. The COS box is an extended region of the coiled-coil region that is required for microtubule association. The MID1 protein also contains a fibronectin type III (FNIII) motif and a C-terminal SPRY/B30.2 domain. The latter domain received its name based on homology to a protein domain encoded by exon B30–2 in human MHC-I and to regions found within the spla and the ryanodine receptor. B, levels of α4 are significantly elevated (p = 0.135) in permanent MDCK cells expressing EGFP-tagged MID1delCTD, a dominant negative MID1, when compared with control cells expressing EGFP only. Results from multiple independent cell lines (mutant: C3, F8, B3; control: D5, A6) are shown. The level of α4 was determined by Western blot and quantified as described under “Materials and Methods.” C, control MDCK cells (A6 line) were cultured in the presence (+) or absence (−) of lactacystin for 3 h, and the levels of α4 protein were compared. D, after the addition of lactacystin to control (EGFP) and EGFP-MID1delCTD-expressing MDCK cells, there was no significant difference in α4 levels (p = 0.3846). All experiments were performed at least twice with multiple cell lines.
Proteasome Inhibition Studies and Analysis of α4 Protein Levels
GFP control and MID1delCTD-expressing MDCK cell lines were cultured with or without 10 μm lactacystin (Millipore) in complete DMEM media for 3 h before cell lysates were prepared as described above. Western blot analysis was performed using a mouse anti-α4 antibody (clone 5F6, Millipore) and a goat anti-mouse peroxidase conjugated secondary antibody (catalog #A-4416, Sigma). The membranes were then stripped and probed with a rabbit anti-GAPDH antibody (catalog #IMG-5143A, IMGENEX, San Diego, CA) and a goat anti-rabbit peroxidase conjugated secondary antibody (catalog #A-0545, Sigma) as a loading control. A digital image of the Western blot bands was obtained by scanning non-saturating x-ray film exposures, and the band intensities were quantified with Image J. A two-tailed t test was used (p < 0.05) to determine the significance of differences in normalized protein levels in the control and mutant cells.
Functional Screening Using a Bacterial Two-hybrid System
Because of the much faster generation time of bacteria over yeast and the fact that plasmid DNA is more readily recovered from bacteria, we chose the BacterioMatch® II Two Hybrid system as a means to efficiently and functionally screen randomly generated mutations in MID1 B-box1 for loss of ability to bind α4. The ability of two proteins of interest to interact in this system is assessed by growth of co-transformants on selective medium containing 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of histidine synthesis, and confirmed by growth of transformants on dual-selective medium containing both 3-AT and streptomycin. Assessment of self-activation of fusion constructs and testing of the expected interaction pairs was performed essentially as described (BacterioMatch® II Two Hybrid System Vector kit manual, Stratagene).
To generate bait and prey constructs, full-length α4 was digested with BamHI and EcoRI, gel-purified, and cloned into similarly digested pTRG vector (BacterioMatch® II, Stratagene). The MID1 Bbox1 domain was amplified from full-length MID1 using the M1BB1-F (5′-GTG AAT TCT ACT AGT GCC GAG AAG GTC CTC TGC CAG TT-3′) and M1BB1-R (5′-GTG TCT CGA GTC AGT CCG GAA TTG GCT CAA TCA GAC-3′) primers, and the product was digested with EcoRI and XhoI and directionally ligated into similarly digested pBT to generate pBT-M1BB1. The resultant pTRG-αlpha4 and pBT-M1BB1 constructs were confirmed by sequencing. In this system, α4 and the MID1 B-box1 domain showed a robust and specific interaction.
To generate random mutations in the MID1 B-box1 domain, we employed an error-prone PCR mutagenesis protocol. Briefly, the same M1BB1-F and M1BB1-R primers, as utilized in the initial cloning of the wild-type MID1 B-box1 domain into pBT, were used with a custom-prepared non-proofreading DNA polymerase (CusTaq) in a reaction containing 300 nm MnCl2. The addition of MnCl2 at this concentration was predicted to generate an average of 1 mutation per ∼200 bp and, therefore, approximately a single mutation per 180-bp M1BB1 fragment. In addition, dNTP ratios were biased, with dCTP and dTTP concentrations each increased from 200 nm to 1 mm. Resultant M1BB1 PCR fragments were subsequently directionally cloned into pBT after EcoRI-XhoI double digestion. The mutagenized M1BB1-pBT ligation mix was then transformed into the bacterial host that already contained the pTRG-α4. 850 individual colonies were subsequently patched together with positive, negative, and self-activation control co-transformations onto selective and non-selective plates for screening.
Cell Transfection and Immunofluorescence Studies
Cos1 cells were maintained at 37 °C with 5% CO2 in DMEM (Invitrogen) supplemented with 10% fetal calf serum, 1% glutamine, 1% penicillin, and 1% streptomycin. Transfection of constructs was done using FuGENE transfection reagent (Promega) in accordance with the manufacturer's instructions. 18 h post-transfection, cells were washed in 1× PBS and fixed in 4% formaldehyde. Where antibody staining was required, cells were permeabilized in 0.2% Nonidet-P40 (ICN Biomedicals Inc.) and incubated with mouse anti-Myc 9E10 monoclonal antibody and/or rabbit anti-MID1 polyclonal antisera and detected with anti-mouse Texas Red and/or anti-rabbit Cy5 (Jackson ImmunoResearch) secondary antibodies, respectively. Antibody incubations were light-shielded at 37 °C for 2 h. Slides were mounted with Hoechst nuclear stain and antifade reagent. Staining was visualized using an Olympus AX70 light microscope with filters.
Plasmid Construction for Recombinant Protein Production
The His6-tagged Bbox1-Bbox2 (B1B2; His-71—Glu-214) was cloned into the pET151d vector with a TOPO directional kit (Invitrogen) using the forward primer 5′-CAC CAT GGA CGG GCT CAA GCG CAA CGT CAC CCT ACA G-3′ and the reverse primer 5′-TCA CTC ACT CAA AGC TGC CAC CTG ATG ATC-3′. The His6-tagged α4 C-terminal construct was made by amplifying the DNA encoding residues Glu-236—Gly-339 using the forward primer 5′-CCA AGG TTC ATA TGG AGA GGC GTC CAG TGA AAC CCT T-3′ and the reverse primer 5′-CCA AGG TTA AGC TTT CAG CCC ATG TTC TGT CGG TTC CC-3′. The PCR product was cloned into the pSKB3 vector, a derivative of pET28, for expression as an N-terminal His6-tag fusion. DNA sequencing confirmed the orientation and integrity of all vectors before they were transformed into BL21(DE3) cells. Changing leucine 146 to glutamine within the MID1 constructs was performed by a standard mutagenesis protocol.
Expression and Purification of Recombinant Proteins
Cells expressing full-length α4 (His6-α4), the N-terminal domain (His6-α4N, residues 1–235), and the C-terminal domain (His6-α4C, residues 236–339) were grown in LB media at 37 °C to an A600 of ∼0.6 and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 18 h at 16 °C (15). Harvested cells were resuspended in buffer 50 mm Tris, 200 mm NaCl, 10% glycerol (pH 8.0) with protease inhibitors and lysed with lysozyme. The lysates were clarified by centrifugation at 4 °C for 30 min at 20,000 × g. The supernatant was purified with Ni2+-NTA resin (Qiagen) and ion-exchange chromatography by FPLC (Bio-Rad). The MID1 RING-Bbox1-Bbox2 domains (His6-RB1B2), MID1 RING-Bbox1 domains (His6-RB1), MID1 Bbox1-Bbox2 domains (His6-B1B2, residues 71–214), and His6-RING92 (MID1 residues 1–92 that includes the RING domain) were expressed and purified as previously described (19, 31).
In Vitro Ubiquitination Assay Using Purified Recombinant Proteins
Human Ub-activating enzyme E1, human Ub-conjugating enzymes (UbcH5c), and ubiquitin were purchased from R&D Systems Inc. (Minneapolis, MN). Ubiquitination assays of the MID1 E3 ligase domains in the presence of α4, α4C, and α4N were performed in a total volume of 30 μl. The mixture contained 1 unit of inorganic pyrophosphatase, 1 mm DTT, 5 mm ATP-Mg2+, 0.125 μm E1, 2.5 μm E2 (UbcH5c), and 30–50 μm Ub in 20 mm Tris-HCl (pH 7.5) and different amounts of purified recombinant proteins. The RB1B2-L146Q autoubiquitination assay was performed as previously described (32–34). The mixtures were incubated at 30 °C, and the reaction was terminated with 2× SDS-PAGE non-reducing sample loading buffer. The reaction products were subsequently analyzed by SDS-PAGE and chemiluminescent Western blot using the following antibodies: α4 N-terminal (catalog #α4 V-20) and C-terminal antibodies (catalog #α4 D-20, Santa Cruz Biotechnology). Control reactions were performed by omitting individual components from the assay.
Mass Spectrometry Studies of Ubiquitinated Products
The ubiquitinated protein bands were excised from the SDS-PAGE gels and treated with mass spectrometry grade trypsin. The cysteine residues were reduced and alkylated with methylmethanethiosulfonate. The peptides were extracted from each gel piece, dried by vacuum centrifugation, resuspended in 0.1% TFA and desalted using a C18 ZipTip column. The samples were then analyzed using a 4700 MALDI TOF-TOF mass spectrometer equipped with a Nd:YAG 200 Hz laser. Raw data files were subjected to a database search using Gly-Gly modifications of Lys and Cys modifications that represented ubiquitin stubs generated by trypsinolysis. Putative Gly-Gly modified peptides were reviewed for statistically significant Mascot and X!tandem peptide identification scores and for contiguous b-fragment and y-fragment ions.
RESULTS
MID1 Facilitates in Vivo Proteasomal Regulation of α4
To investigate the function of MID1 in epithelia, we attempted to generate permanent Madin-Darby canine kidney epithelial cells expressing an EGFP-tagged wild-type MID1. Unfortunately, repeated attempts to generate stable MDCK cells overexpressing full-length MID1 were unsuccessful as the cells exhibited cell division defects and increased cell death. We suspected this was due to reduced PP2A activity because of increased MID1-mediated polyubiquitination. Instead, we generated permanent MDCK epithelial cells expressing an EGFP-tagged mutant form of MID1 that is missing the C-terminal SPRY/B30.2 domain. Our previous studies as well as those of others had established that this mutant form of MID1, which is the most common type of mutation found in patients with X-linked Opitz syndrome, forms cytoplasmic aggregates (35) that in patient fibroblasts leads to reduced polyubiquitination and increased levels of PP2Ac (10). Importantly, this mutant MID1 retains the ability to dimerize with wild-type MID1 and thereby sequesters it away from its usual location, reducing overall endogenous MID1 activity (35, 36). Thus, the mutant MID1 (MID1delCTD) functions as a dominant negative (35, 37). In all cases cell lines expressing low levels of the GFP or GFP-MID1delCTD were maintained. For the MID1delCTD lines, this low level expression correlated with relatively few and smaller cytoplasmic aggregates of the mutant MID1 protein than normally seen in transient transfection experiments. This also correlated with a cell size similar to that of control cells, unlike cells that express high levels of the mutant.3 Multiple clonal cell lines were used for all experiments to serve as biological replicates.
MDCK cells expressing the dominant negative MID1delCTD showed significantly higher levels of α4 than the control GFP-expressing cells (p = 0.0135; Fig. 1B). To determine the basis for this increased level of α4, we treated the cells with the proteasome inhibitor lactacystin. We observed that inhibition of the proteasome resulted in increased α4 protein levels ∼3-fold in the control cells, which only expressed endogenous MID1 (Fig. 1C) supporting the notion that α4 is regulated by the proteasome. Furthermore, when α4 levels in lactacystin-treated control and MID1delCTD-expressing cells were compared, there was no significant difference in levels (p = 0.3846; Fig. 1D), supporting the notion that the elevated α4 levels in untreated MID1delCTD cells is due to disruption of MID1-dependent proteasome-mediated degradation.
MID1 Facilitates Polyubiquitination of α4 in Vitro
We recently characterized the interaction between the MID1 Bbox1 domain and α4 (6, 8, 21) using a 45-amino acid peptide derived from the C-terminal region of α4 (α445, residues 236–280) that was shown to bind strongly to MID1 (6). We observed that α445 interacted with the Bbox1 domain at a site that was close to Lys-153, the lysine that is autoubiquitinated (19, 21). α445 was observed to be monoubiquitinated at one of the three lysine residues (19, 21).
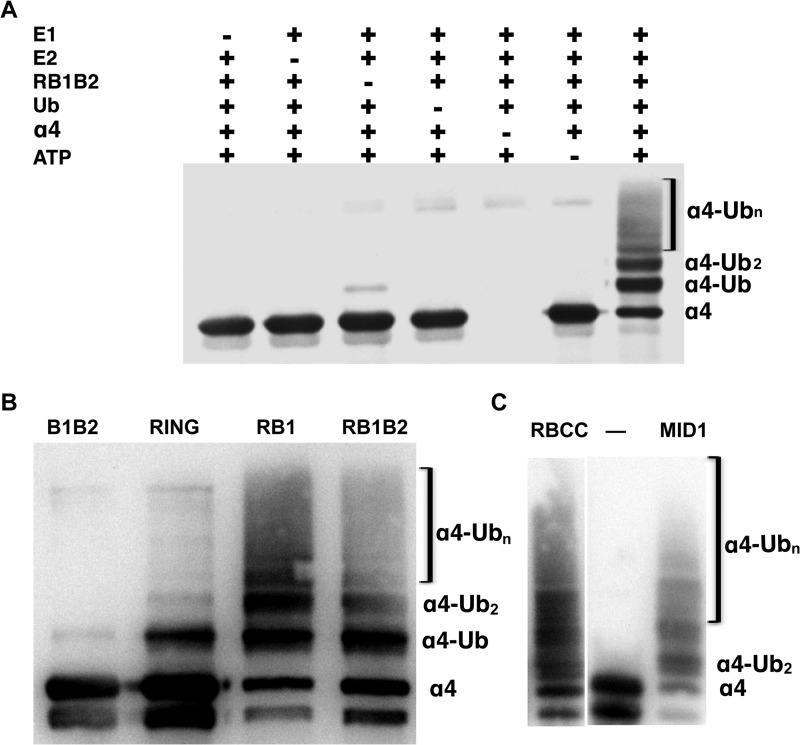
In light of the results from the MDCK cells, we sought to determine whether the full-length α4 is directly targeted for polyubiquitination by MID1. To test this using a well established in vitro ubiquitination assay, we used the first three domains (RING, Bbox1, Bbox2) of MID1 that we previously showed possessed E3 ligase activity. Using full-length α4 in the presence of a MID1 construct consisting of the RING-Bbox1-Bbox2 domains (RB1B2, residues 1–214), we observed polyubiquitinated α4 products, as indicated by the high molecular weight “smear pattern” bands (Fig. 2A). For the control experiments, in which individual components of the assay were omitted, no polyubiquitination of α4 was observed. A very low level of monoubiquitination of α4 was observed when no MID1 protein was included.
FIGURE 2.
MID1 catalyzes the polyubiquitination of α4 in a RING- and B-box1-dependent manner. A, a Western blot shows in vitro ubiquitination of full-length α4. Lane 1 consists of all components of the ubiquitination assay except the E1-activating enzyme (E1). The second through fifth lanes represent a control experiment in which specific components of the assay were omitted. The high molecular weight-shifted bands or smear pattern represented polyubiquitinated α4. Antibody was specific for the C terminus. Ub, ubiquitin. B, a Western blot shows ubiquitination assays of α4 with the RING and Bbox domains of MID1. From left to right, the assay was performed separately with His6-Bbox1-Bbox2 (B1B2, residues 71–214), His6-RING (RING, residues1–92), His6-RING-Bbox1 (RB1, residues1–164), and His6-RING-Bbox1-Bbox2 (RB1B2, residues 1–214). Antibody for the Western blot was specific for the C-terminal domain of α4. The band slightly lower than the band labeled α4 indicates partial degraded product of α4. C, a Western blot shows the polyubiquitination of α4 by a RING-Bbox1-Bbox2-Coiled-Coil (RBCC) construct and full-length MID1. The first lane shows the control assay in which E3 ligase is omitted.
RING and Bbox1 Are Necessary for α4 Polyubiquitination
In our previous studies, we showed that the RING domain possessed greater E3 ligase activity than either of the Bbox domains or the Bbox domains in tandem based on the amount of polyubiquitinated products observed in in vitro ubiquitination assays (19). When the RING and Bbox domains are linked, as observed in native MID1, the activity of the RING domain is significantly enhanced compared with the RING domain alone (19). To decipher the minimal MID1 construct that could promote the ubiquitination of α4, assays were performed with RING, B1B2, RING-Bbox1 (RB1), and RB1B2 domains.
α4 has been shown to strongly interact with the Bbox1 domain (7, 10, 21). However, in the presence of Bbox1-Bbox2 (B1B2) domains only a trace amount of monoubiquitinated full-length α4 was observed (Fig. 2B). Interestingly, using the RING domain alone (residues 1–92), a strong monoubiquitinated band and very weak higher molecular weigh bands were observed (Fig. 2B), suggesting some level of polyubiquitination of α4. In contrast to the B1B2 and RING domain-only constructs, the RB1 and RB1B2 constructs resulted in polyubiquitination of α4 (Figs. 2, A and B). Testing of larger constructs of MID1 (e.g. RING-Bbox1-Bbox2-Coiled-Coil (RBCC) and full-length MID1) resulted in similar levels of polyubiquitination of α4 to those of the RB1 and RB1B2 constructs (Fig. 2C), consistent with the fact that domains C-terminal to the RING and Bbox domains have no impact on ubiquitination of α4. These results also validated our use of RB1 and RB1B2 constructs instead of full-length MID1 for the ubiquitination assay of α4.
Interaction of MID1 Bbox1 and α4 Is Essential for α4 Polyubiquitination
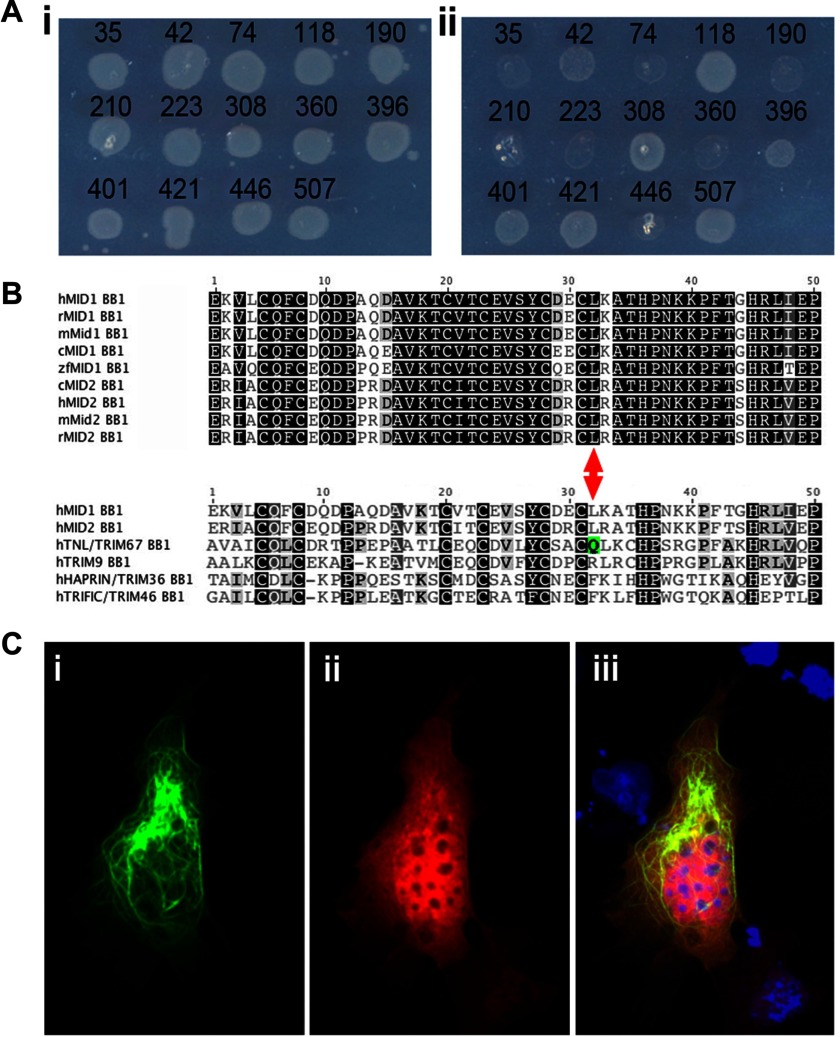
As stated above, the MID1 RING domain alone could monoubiquitinate α4. However, only when the Bbox1 domain was present was α4 polyubiquitinated. To investigate the requirement of direct binding of α4 to Bbox1 for polyubiquitination, we used a mutant MID1 that was identified in an unbiased functional screen for mutations that disrupt the strong interaction of α4 with Bbox1. Briefly, in this screen 850 randomly mutated clones were assessed using a bacterial two-hybrid system. Of the 850 clones, nine showed obvious reduced ability to grow on the selection plates (Fig. 3A), suggesting these clones carried mutations that interfered with α4 binding to Bbox1. Sequencing of these nine clones revealed a variety of mutations including a number of nonsense mutations as well as single base deletions. Three clones contained missense mutations, but two of these carried more than one mutation. The only clone with a single missense mutation encoded a Bbox1 domain with the amino acid substitution, leucine 146 to glutamine (L146Q). Sequence alignment of the Bbox1 domains of MID1 orthologs from human, rat, mouse, chick, and zebrafish together with the cognate MID2 orthologous sequences, which also bind α4, revealed that Leu-146 is highly conserved (Fig. 3B). When the sequence of MID1 and MID2 was aligned with the other TRIM proteins of the C-I subfamily, which do not bind α4, we observe variation at this position. Notably, TRIM67 has a glutamine in the equivalent position supporting the notion that the L146Q substitution maintains the overall Bbox1 topology and function and that the leucine residue is directly involved in α4 binding.
FIGURE 3.
Bbox1 domain is essential for MID1-α4 interaction. A, a random mutagenesis based bacterial two-hybrid screen was conducted to identify point mutations that abolished interaction between the MID1 Bbox1 and α4. 14 of 850 colonies from the original screen were re-spotted onto a master (i) and selection (ii) plates for validation. Those showing reduced or no growth on selection plates were sequenced to identify the underlying mutation. B, shown is sequence alignment of MID1 and MID2 Bbox1 domain sequences from human (h), rat (r), mouse (m), chicken (c), and zebrafish (z). Fully conserved residues are highlighted in black. Dark and light gray shading highlight slightly less conservation. In the bottom half of the alignment, the human MID1 and MID2 Bbox1 domains are compared with the Bbox1 domains of the other C-I TRIM subfamily proteins. Note the presence of a glutamine residue (Q, highlighted with green shading) in human TNL/TRIM67 in the equivalent place to Leu-146 in MID1 and MID2. C, shown is ectopic co-expression of EGFP-MID1-L146Q (i) and myc-tagged α4 in Cos1 cells (ii) and the merged images (iii). EGFP-MID1-L146Q expression was detected by GFP fluorescence (green). The α4 expression was visualized by immunocytochemistry using an anti-myc antibody (red). The L146Q mutant shows a microtubule distribution indistinguishable from wild-type MID1 (data not shown). However, in contrast to when α4 was co-expressed with wild-type MID1, α4 was not recruited to the microtubules when co-expressed with the L146Q mutant.
To validate whether this amino acid substitution truly abolished the interaction with α4, the mutation was introduced into full-length MID1 and expressed as a GFP fusion protein in Cos1 cells (Fig. 3C). In notable contrast to almost all Opitz syndrome-related MID1 mutations (35), the L146Q mutant did not appear to affect the ability of MID1 to associate with the microtubule network. However, when co-expressed with myc-tagged α4, α4 failed to associate with the microtubule cytoskeleton (Fig. 3C), as seen when expressed with wild-type MID1 (39). These data confirm the importance of leucine 146 for the interaction with α4.
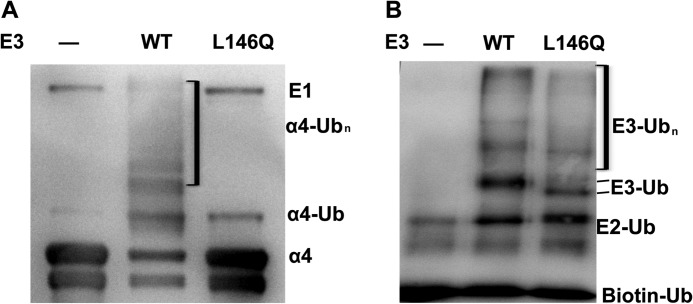
To test the effect of the L146Q mutation on the ability of MID1 to ubiquitinate α4, we expressed the RB1B2 fragment containing the mutation (RB1B2-L146Q) and used it in the same in vitro ubiquitination assays. The results showed that the mutant was unable to polyubiquitinate α4 (Fig. 4A). However, α4 did still appear to be monoubiquitinated, consistent with the result obtained with the RING domain alone. As noted, the RING domain has not previously been shown to physically interact with α4. Intriguingly, in the control experiments, RB1B2-L146Q had full auto-polyubiquitination activity (Fig. 4B) similar to wild-type RB1B2. Taken together with the lack of impact of the L146Q mutation on microtubule binding of full-length MID1, these data indicate that the structure and activity of RB1B2 is not affected by the specific mutation but that it only affects binding and polyubiquitination of α4.
FIGURE 4.
Leucine 146 of Bbox1 domain is essential for MID1-α4 interaction and α4 polyubiquitination. A, a Western blot assay shows the ubiquitination of α4 in the presence of wt RB1B2 (WT) and mutant RB1B2_L146Q in which Leu-146 was mutated to glutamine. The first lane represents a control assay in which MID1 was omitted. Antibody was specific for α4 C-terminal region. B, a Western blot shows the autoubiquitination of RB1B2_L146Q and wt RB1B2 (WT). Antibody was specific for the N-terminal domain of MID1.
Polyubiquitination of α4 Occurs within Its C-terminal End
To determine where on full-length α4 polyubiquitination occurs, we conducted reactions with either the N-terminal (residues 1–235) or C-terminal (residues 236–339) regions of α4. Polyubiquitination was observed within the C-terminal fragment (Fig. 5Ai), but no ubiquitin modification was seen within the N-terminal half (Fig. 5Aii). To identify the site of ubiquitin modification of α4, trypsin digestion and peptide mapping mass spectrometry analyses were performed on the ubiquitinated full-length form (Fig. 5B) as well as unmodified α4 and ubiquitin for comparison (Fig. 5B). The spectra for the unreacted and reacted α4 were the same with the exception being the shift of the peptide at 2060.11 in the unmodified α4 to a peptide with m/z at 2175.7 in the ubiquitin-modified α4. The data identified Lys-287 on α4 as the predominant site of ubiquitination. The mass spectrometry analyses were performed twice, and in both cases the sequence coverage was 25% for both unreacted and modified reacted α4. Although there may be the possibility of additional sites, the mass spectrometry data support our in vitro assay data confirming that the ubiquitin modification of α4 occurs only within the C-terminal domain of α4. In addition, we observed a predominantly monoubiquitinated α4 product using a mutant ubiquitin in which all of its lysine residues were mutated to arginine residues (data not shown).
FIGURE 5.
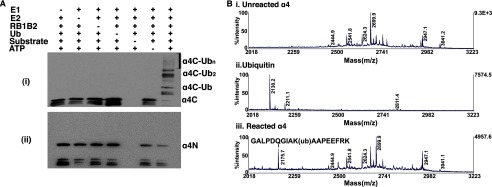
Identification of α4 ubiquitination site. Ai, a Western blot assay shows the ubiquitination of the C-terminal domain (α4C, residues 236–339) of α4. Antibody was C-terminal-specific. ii, a Western blot assay shows the ubiquitination of the N-terminal domain (α4N, residues 1–235) of α4. Antibody was specific for the N-terminal region of α4. Ub, ubiquitin. B, a portion of the tandem mass spectra shows fragmentations of unmodified α4 (i), ubiquitinated α4 (ii), and ubiquitin (iii). The peptide (GALPDQGIAKAAPEEFRK) was identified showing Lys-287 as the site of ubiquitination. Tandem mass spectrometry was performed in the linear ion trap, and y-ion and b-ion series were assigned by a database search assuming potential Gly-Gly residues covalently attached to any of the Lys residues on the protein.
DISCUSSION
α4 is a unique regulatory subunit of PP2Ac and forms a trimeric complex with the E3 ubiquitin ligase, MID1. The α4-PP2Ac-MID1 complex plays key roles in a variety of cellular processes, including regulating cell cycle progression, cell growth in response to nutrients, and lymphocyte migration (6, 11, 12, 30). Notably, overexpression of α4 has been associated with various tumors, including primary hepatocellular carcinomas, breast cancer, and invasive adenocarcinoma (29, 30), highlighting the importance of understanding the mechanisms that regulate its levels and function.
Using stable MDCK cells expressing a dominant negative form of MID1, MID1delCTD, in which the SPRY/B30.2 domain is missing, we observed a 3-fold increase in the amount of cellular α4 compared with the control cells. Although the truncated MID1delCTD protein can theoretically function as an E3 ligase, it is likely unable to facilitate the efficient ubiquitination of target proteins on the microtubules because it readily forms cytoplasmic aggregates, to which endogenous MID1 is also pulled. Notably, this difference in α4 levels between control and mutant cells was abolished after inhibition of the proteasome, indicating that MID1 targets at least a subset of α4 for ubiquitin-mediated degradation. To demonstrate that this effect on α4 is indeed direct, we have carried out in vitro polyubiquitination assays using various domain combinations of MID1 and performed mass spectrometry to identify the site of ubiquitination on α4.
Our data from experiments using different MID1 domain combinations suggest that the MID1 RING domain can facilitate monoubiquitination and a low level of polyubiquitination of α4. This result was surprising as there is no previous evidence that the RING domain interacts with α4 (7, 10). Although the mechanism of this action is unclear, the increased monoubiquitination and weak polyubiquitination of α4 may be due to weak interaction between the RING domain and the C-terminal domain of α4 or that the RING-E2 complex may enhance the E2 enzyme functionality and transfer of ubiquitin to α4.
In contrast, the Bbox1 domain in the absence of the RING domain is unable to facilitate ubiquitination of α4 even though it interacts strongly with α4 and has a similar fold as RING E3 ligase domains (7, 10, 18, 21). The weak monoubiquitinated band observed of α4 in the presence of B1B2 (Fig. 2B, first lane) suggest that the Bbox domains alone are not able to target α4 to the E2 enzyme. It is unclear why the Bbox domains are unable to catalyze the ubiquitination of α4. In our previous studies, the Bbox domains were shown to possess E3 ligase activity, albeit considerably less than the classic RING domain. On the other hand, the Bbox domains from TRIM16, which is technically not a TRIM protein because it lacks a RING domain but nevertheless has 28% identity to MID1 (40), were shown to facilitate polyubiquitination. We postulate that the binding of the C terminus of α4 to the Bbox1 domain alters the interface on Bbox1 that would typically be involved in E2-E3 interactions. Indeed, our NMR studies revealed that α445 binds on a surface of the Bbox1 domain that overlaps the surface that would be required for interaction with the E2 enzyme (21, 41, 42). Consistent with this, the presence of α445 markedly reduced the autoubiquitination activity of B1B2 (19).
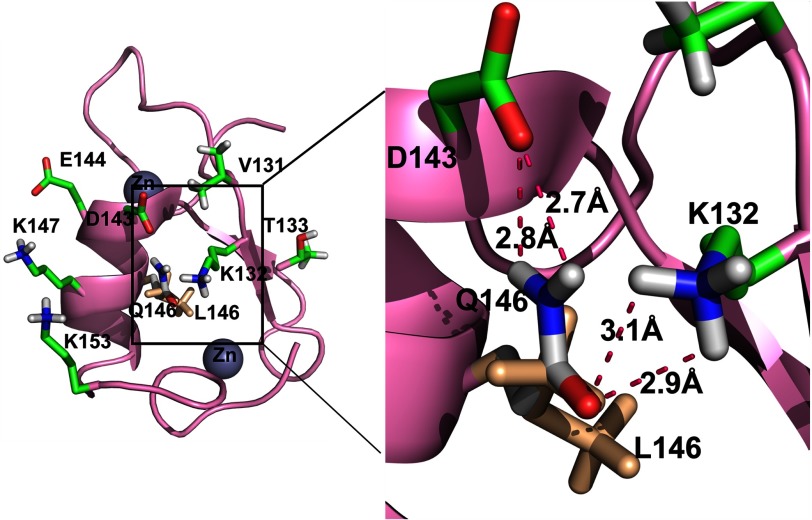
It is clear from the data that the MID1 Bbox1 domain functions to bind and recruit α4 and that this interaction is essential for its polyubiquitination. Specifically, we have shown that Leu-146 is critical for the MID1-α4 interaction, and its mutation to glutamine completely disrupted the polyubiquitination of α4. These data demonstrate that the ubiquitination of α4 is directly linked to its association with MID1. To understand how the L146Q mutation might affect the structure and function of MID1, we modeled a glutamine in place of Leu-146 and observed that the side chain carbonyl and amino group of Gln-146 extend past one of the methyl groups of the leucine. These groups are positioned in close proximity to form multiple hydrogen bonds with Asp-143 and Lys-132, both of which were previously shown to be involved in binding the C-terminal region of α4 (21). We postulate that these hydrogen bonds affect the α4 binding surface on Bbox1 but not its overall tertiary structure (Fig. 6). Although these results do not rule out other possibilities for the role of Bbox1, they do indicate that physical association of α4 with the MID1 Bbox1 is essential for regulating polyubiquitination of α4.
FIGURE 6.
Model of L146Q mutation of Bbox1. A ribbon representation of MID1 Bbox1 shows a proposed model of the effect of the L146Q mutation. Labeled are the residues on one surface of Bbox1 that are shown by NMR studies to be involved with binding the C terminus of α4 (21). The position of Leu-146 overlay with Gln-146 is shown in greater magnification to depict potential hydrogen bonds that can result between Gln-146 and Asp-143 and Lys-132.
Mechanistically the polyubiquitination of α4 requires both the RING and Bbox1 domains. In addition to binding and possibly orienting α4 for polyubiquitination, the Bbox1 domain may function to enhance the E3 ligase functionality of the RING domain (i.e. function like an E4 enzyme). This observation suggests that the RING domains of the large TRIM/Bbox1-Bbox2-Coiled-Coil (RBCC) family may all require the invariantly associated Bbox domains, located just C-terminal to the RING domain, to enhance their E3 ligase activity as well as determine substrate specificity. For instance, TRIM5α, which consists of the RING-Bbox2 domains, requires Bbox2 for anti-retroviral activity (43). Deletion or mutation that disrupts Bbox2 structure resulted in loss of TRIM5α antiviral activity even though the protein is expressed and localized properly (43). Similar functions have been observed with the RING domains of BARD1 and MdmX, which have been shown to enhance the E3 functionality of BRCA1 and MDM2, respectively (44–48). In all these cases, these E4 enzymes possess limited E3 ligase activities but are routinely observed to heterodimerize with a RING domain and function as ubiquitin chain elongation factors. As expected, E3 ligase functionality of the RING-Bbox domains is maintained completely within the TRIM motif (Fig. 2C) and does not require either the coiled coil domain of the TRIM motif or the C-terminal half of MID1.
Our results showing polyubiquitination of α4 are in contrast to a recent report by Watkins et al. (25), which showed only monoubiquitination of full-length α4 by MID1 when using UbcH5b (UBE2D2) and UbcH5c (UBE2D3) as the E2 enzymes (25). It was reported that monoubiquitination of α4 promotes its cleavage by calpain, and that this results in increased processing of PP2Ac (25). That said, close examination of the ubiquitination data of Watkins et al. (25) suggests the presence of diubiquitinated α4; higher molecular weight products were not shown with the Western blot. In addition, it was reported that a ubiquitin interacting motif within the N-terminal region of α4 was important for directing the level of ubiquitination of α4 and PP2A (38). However, in our data we observed that the proposed ubiquitin interacting motif had no effect on the level of ubiquitination of α4; in fact, ubiquitination occurred independently of the ubiquitin interacting motif, as observed with the polyubiquitination of the C terminus. Nonetheless, taken together these data indicate that the extent of ubiquitination of α4 regulates its function and level, perhaps via a combination of classic proteasome-mediate degradation and calpain-mediated turnover. Furthermore, we suggest that this is in part determined by the specific E2 enzymes available within the cell, thus providing an additional level of regulation of α4 and PP2A activity.
Importantly, the entire C-terminal MID1 binding domain (residues 236–339), whether as a domain by itself or as part of the full-length α4 protein, is necessary for polyubiquitination. The site of polyubiquitination (lysine-287) is outside the range of the residues used in our initial studies using α445 (residues 236–280) (19), which was only observed to be monoubiquitinated. This implies that the full C terminus helps to position this key lysine residue in close proximity to the active site of the activated E2 enzyme. The N-terminal region of α4 neither interacts with MID1 nor is it a target for MID1-mediated ubiquitination.
To summarize, our data support a new role for MID1, namely the regulation of α4 via various levels of ubiquitination that could alter α4 function and/or target it for proteasome degradation. In either case the prospect that α4 plays such a key role in cellular transformation and is linked to different types of cancer suggests that MID1 may also play an important role in tumor suppression. Further studies are needed to resolve the differential contribution of proteasome- and calpain-dependent regulation of PP2A activity in determining specific cellular responses.
Acknowledgments
We are grateful to Dr. Yetrib Hathout (Children's National Medical Center, Washington D. C.) for help with the acquisition and analysis of the mass spectrometry data and to Drs. Blair Hopwood and Leila Belle and to Terri King for technical assistance. The mass spectrometry studies were supported in part by NICHD, National Institutes of Health Core Grant 5P30HD040677-10.
This work was supported by National Science Foundation Awards 1102696 and 1052520 (to M. A. M.) and the Laurel Foundation Endowment for Craniofacial Research (to T. C. C.).
H. Du, Y. Huang, M. Zaghlula, E. Walters, T. C. Cox, and M. A. Massiah, unpublished observations.
- PP2A
- protein phosphatase 2A
- Ub
- ubiquitin
- MDCK
- Madin-Darby canine kidney
- EGFP
- enhanced GFP
- B1B2
- Bbox1-Bbox2
- RB1
- RING-Bbox1.
REFERENCES
- 1. Chuang E., Fisher T. S., Morgan R. W., Robbins M. D., Duerr J. M., Vander Heiden M. G., Gardner J. P., Hambor J. E., Neveu M. J., Thompson C. B. (2000) The CD28 and CTLA-4 receptors associate with the serine/threonine phosphatase PP2A. Immunity 13, 313–322 [DOI] [PubMed] [Google Scholar]
- 2. Inui S., Kuwahara K., Mizutani J., Maeda K., Kawai T., Nakayasu H., Sakaguchi N. (1995) Molecular cloning of a cDNA clone encoding a phosphoprotein component related to the Ig receptor-mediated signal transduction. J. Immunol. 154, 2714–2723 [PubMed] [Google Scholar]
- 3. Murata K., Wu J., Brautigan D. L. (1997) B cell receptor-associated protein α4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc. Natl. Acad. Sci. U.S.A. 94, 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Inui S., Sanjo H., Maeda K., Yamamoto H., Miyamoto E., Sakaguchi N. (1998) Ig receptor binding protein 1 (α4) is associated with a rapamycin-sensitive signal transduction in lymphocytes through direct binding to the catalytic subunit of protein phosphatase 2A. Blood 92, 539–546 [PubMed] [Google Scholar]
- 5. Onda M., Inui S., Maeda K., Suzuki M., Takahashi E., Sakaguchi N. (1997) Expression and chromosomal localization of the human α4/IGBP1 gene, the structure of which is closely related to the yeast TAP42 protein of the rapamycin-sensitive signal transduction pathway. Genomics 46, 373–378 [DOI] [PubMed] [Google Scholar]
- 6. Aranda-Orgillés B., Aigner J., Kunath M., Lurz R., Schneider R., Schweiger S. (2008) Active transport of the ubiquitin ligase MID1 along the microtubules is regulated by protein phosphatase 2A. PLoS ONE 3, e3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aranda-Orgillés B., Trockenbacher A., Winter J., Aigner J., Köhler A., Jastrzebska E., Stahl J., Müller E. C., Otto A., Wanker E. E., Schneider R., Schweiger S. (2008) The Opitz syndrome gene product MID1 assembles a microtubule-associated ribonucleoprotein complex. Hum. Genet. 123, 163–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schweiger S., Schneider R. (2003) The MID1/PP2A complex A key to the pathogenesis of Opitz BBB/G syndrome. Bioessays 25, 356–366 [DOI] [PubMed] [Google Scholar]
- 9. Short K. M., Hopwood B., Yi Z., Cox T. C. (2002) MID1 and MID2 homo- and heterodimerise to tether the rapamycin-sensitive PP2A regulatory subunit, α4, to microtubules. Implications for the clinical variability of X-linked Opitz GBBB syndrome and other developmental disorders. BMC Cell Biol. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trockenbacher A., Suckow V., Foerster J., Winter J., Krauss S., Ropers H. H., Schneider R., Schweiger S. (2001) MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat. Genet. 29, 287–294 [DOI] [PubMed] [Google Scholar]
- 11. Everett A. D., Brautigan D. L. (2002) Developmental expression of α4 protein phosphatase regulatory subunit in tissues affected by Opitz syndrome. Dev. Dyn. 224, 461–464 [DOI] [PubMed] [Google Scholar]
- 12. Kong M., Bui T. V., Ditsworth D., Gruber J. J., Goncharov D., Krymskaya V. P., Lindsten T., Thompson C. B. (2007) The PP2A-associated protein α4 plays a critical role in the regulation of cell spreading and migration. J. Biol. Chem. 282, 29712–29720 [DOI] [PubMed] [Google Scholar]
- 13. Kong M., Ditsworth D., Lindsten T., Thompson C. B. (2009) α4 is an essential regulator of PP2A phosphatase activity. Mol. Cell 36, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu E., Knutzen C. A., Krauss S., Schweiger S., Chiang G. G. (2011) Control of mTORC1 signaling by the Opitz syndrome protein MID1. Proc. Natl. Acad. Sci. U.S.A. 108, 8680–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smetana J. H., Oliveira C. L., Jablonka W., Aguiar Pertinhez T., Carneiro F. R., Montero-Lomeli M., Torriani I., Zanchin N. I. (2006) Low resolution structure of the human α4 protein (IgBP1) and studies on the stability of α4 and of its yeast ortholog Tap42. Biochim. Biophys. Acta 1764, 724–734 [DOI] [PubMed] [Google Scholar]
- 16. Short K. M., Cox T. C. (2006) Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 281, 8970–8980 [DOI] [PubMed] [Google Scholar]
- 17. Massiah M. A., Matts J. A., Short K. M., Simmons B. N., Singireddy S., Yi Z., Cox T. C. (2007) Solution structure of the MID1 B-box2 CHC(D/C)C(2)H(2) zinc-binding domain. Insights into an evolutionarily conserved RING fold. J. Mol. Biol. 369, 1–10 [DOI] [PubMed] [Google Scholar]
- 18. Massiah M. A., Simmons B. N., Short K. M., Cox T. C. (2006) Solution structure of the RBCC/TRIM B-box1 domain of human MID1. B-box with a RING. J. Mol. Biol. 358, 532–545 [DOI] [PubMed] [Google Scholar]
- 19. Han X., Du H., Massiah M. A. (2011) Detection and characterization of the in vitro e3 ligase activity of the human MID1 protein. J. Mol. Biol. 407, 505–520 [DOI] [PubMed] [Google Scholar]
- 20. Aranda-Orgillés B., Rutschow D., Zeller R., Karagiannidis A. I., Köhler A., Chen C., Wilson T., Krause S., Roepcke S., Lilley D., Schneider R., Schweiger S. (2011) Protein phosphatase 2A (PP2A)-specific ubiquitin ligase MID1 is a sequence-dependent regulator of translation efficiency controlling 3-phosphoinositide-dependent protein kinase-1 (PDPK-1). J. Biol. Chem. 286, 39945–39957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du H., Massiah M. A. (2011) NMR studies of the C-terminus of α4 reveal possible mechanism of its interaction with MID1 and protein phosphatase 2A. PLoS ONE 6, e28877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J., Prickett T. D., Elliott E., Meroni G., Brautigan D. L. (2001) Phosphorylation and microtubule association of the Opitz syndrome protein mid-1 is regulated by protein phosphatase 2A via binding to the regulatory subunit α4. Proc. Natl. Acad. Sci. U.S.A. 98, 6650–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Falco F., Cainarca S., Andolfi G., Ferrentino R., Berti C., Rodriguez Criado G., Rittinger O., Dennis N., Odent S., Rastogi A., Liebelt J., Chitayat D., Winter R., Jawanda H., Ballabio A., Franco B., Meroni G. (2003) X-linked Opitz syndrome. Novel mutations in the MID1 gene and redefinition of the clinical spectrum. Am. J. Med. Genet. A 120A, 222–228 [DOI] [PubMed] [Google Scholar]
- 24. Graham J. M., Jr., Wheeler P., Tackels-Horne D., Lin A. E., Hall B. D., May M., Short K. M., Schwartz C. E., Cox T. C. (2003) A new X-linked syndrome with agenesis of the corpus callosum, mental retardation, coloboma, micrognathia, and a mutation in the α4 gene at Xq13. Am. J. Med. Genet. A 123A, 37–44 [DOI] [PubMed] [Google Scholar]
- 25. Watkins G. R., Wang N., Mazalouskas M. D., Gomez R. J., Guthrie C. R., Kraemer B. C., Schweiger S., Spiller B. W., Wadzinski B. E. (2012) Monoubiquitination promotes calpain cleavage of the protein phosphatase 2A (PP2A) regulatory subunit α4, altering PP2A stability and microtubule-associated protein phosphorylation. J. Biol. Chem., 287, 24207–24215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berti C., Fontanella B., Ferrentino R., Meroni G. (2004) Mig12, a novel Opitz syndrome gene product partner, is expressed in the embryonic ventral midline and co-operates with Mid1 to bundle and stabilize microtubules. BMC Cell Biol. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delete in proof
- 28. Yang J., Roe S. M., Prickett T. D., Brautigan D. L., Barford D. (2007) The structure of Tap42/α4 reveals a tetratricopeptide repeat-like fold and provides insights into PP2A regulation. Biochemistry 46, 8807–8815 [DOI] [PubMed] [Google Scholar]
- 29. Sakashita S., Li D., Nashima N., Minami Y., Furuya S., Morishita Y., Tachibana K., Sato Y., Noguchi M. (2011) Overexpression of immunoglobulin (CD79a) binding protein1 (IGBP-1) in small lung adenocarcinomas and its clinicopathological significance. Pathol. Int. 61, 130–137 [DOI] [PubMed] [Google Scholar]
- 30. Chen L. P., Lai Y. D., Li D. C., Zhu X. N., Yang P., Li W. X., Zhu W., Zhao J., Li X. D., Xiao Y. M., Zhang Y., Xing X. M., Wang Q., Zhang B., Lin Y. C., Zeng J. L., Zhang S. X., Liu C. X., Li Z. F., Zeng X. W., Lin Z. N., Zhuang Z. X., Chen W. (2011) α4 is highly expressed in carcinogen-transformed human cells and primary human cancers. Oncogene 30, 2943–2953 [DOI] [PubMed] [Google Scholar]
- 31. Tao H., Simmons B. N., Singireddy S., Jakkidi M., Short K. M., Cox T. C., Massiah M. A. (2008) Structure of the MID1 tandem B-boxes reveals an interaction reminiscent of intermolecular ring heterodimers. Biochemistry 47, 2450–2457 [DOI] [PubMed] [Google Scholar]
- 32. Berleth E. S., Pickart C. M. (1990) Several mammalian ubiquitin carrier proteins, but not E2(20K), are related to the 20-kDa yeast E2, RAD6. Biochem. Biophys. Res. Commun. 171, 705–710 [DOI] [PubMed] [Google Scholar]
- 33. Mastrandrea L. D., You J., Niles E. G., Pickart C. M. (1999) E2/E3-mediated assembly of lysine 29-linked polyubiquitin chains. J. Biol. Chem. 274, 27299–27306 [DOI] [PubMed] [Google Scholar]
- 34. Pickart C. M. (2000) Ubiquitin biology. An old dog learns an old trick. Nat. Cell. Biol. 2, E139–E141 [DOI] [PubMed] [Google Scholar]
- 35. Cox T. C., Allen L. R., Cox L. L., Hopwood B., Goodwin B., Haan E., Suthers G. K. (2000) New mutations in MID1 provide support for loss of function as the cause of X-linked Opitz syndrome. Hum. Mol. Genet. 9, 2553–2562 [DOI] [PubMed] [Google Scholar]
- 36. Buchner G., Montini E., Andolfi G., Quaderi N., Cainarca S., Messali S., Bassi M. T., Ballabio A., Meroni G., Franco B. (1999) MID2, a homologue of the Opitz syndrome gene MID1. Similarities in subcellular localization and differences in expression during development. Hum. Mol. Genet. 8, 1397–1407 [DOI] [PubMed] [Google Scholar]
- 37. Granata A., Quaderi N. A. (2003) The Opitz syndrome gene MID1 is essential for establishing asymmetric gene expression in Hensen's node. Dev. Biol. 258, 397–405 [DOI] [PubMed] [Google Scholar]
- 38. LeNoue-Newton M., Watkins G. R., Zou P., Germane K. L., McCorvey L. R., Wadzinski B. E., Spiller B. W. (2011) The E3 ubiquitin ligase- and protein phosphatase 2A (PP2A)-binding domains of the α4 protein are both required for α4 to inhibit PP2A degradation. J. Biol. Chem. 286, 17665–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Short K. M., Hopwood B., Yi Z., Cox T. C. (2002) MID1 and MID2 homo- and heterodimerise to tether the rapamycin-sensitive PP2A regulatory subunit, α4, to microtubules. Implications for the clinical variability of X-linked Opitz GBBB syndrome and other developmental disorders. BMC Cell Biology 3, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bell J. L., Malyukova A., Holien J. K., Koach J., Parker M. W., Kavallaris M., Marshall G. M., Cheung B. B. (2012) TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PLoS ONE 7, e37470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dou H., Buetow L., Sibbet G. J., Cameron K., Huang D. T. (2012) BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Structure of a c-Cbl-UbcH7 complex. RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 43. Javanbakht H., Diaz-Griffero F., Stremlau M., Si Z., Sodroski J. (2005) The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5α. J. Biol. Chem. 280, 26933–26940 [DOI] [PubMed] [Google Scholar]
- 44. Brzovic P. S., Rajagopal P., Hoyt D. W., King M. C., Klevit R. E. (2001) Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol. 8, 833–837 [DOI] [PubMed] [Google Scholar]
- 45. Mallery D. L., Vandenberg C. J., Hiom K. (2002) Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 21, 6755–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xia Y., Pao G. M., Chen H. W., Verma I. M., Hunter T. (2003) Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 278, 5255–5263 [DOI] [PubMed] [Google Scholar]
- 47. Okamoto K., Taya Y., Nakagama H. (2009) Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 583, 2710–2714 [DOI] [PubMed] [Google Scholar]
- 48. Wang X., Jiang X. (2012) Mdm2 and MdmX partner to regulate p53. FEBS Lett. 586, 1390–1396 [DOI] [PubMed] [Google Scholar]