Background: The RecA protein of Deinococcus radiodurans is essential for extreme radiation resistance.
Results: A nucleoprotein state of the DrRecA protein exists that is inactive with respect to ATP hydrolysis. SSB is essential for DrRecA-mediated DNA strand exchange.
Conclusion: There are at least two functional states of DrRecA filaments bound to DNA.
Significance: A potential new mode of regulation is revealed for DrRecA.
Keywords: ATPases, DNA, DNA Enzymes, DNA Recombination, DNA Repair, Electron Microscopy (EM), Deinococcus radiodurans, RecA, SSB
Abstract
The RecA protein of Deinococcus radiodurans (DrRecA) has a central role in genome reconstitution after exposure to extreme levels of ionizing radiation. When bound to DNA, filaments of DrRecA protein exhibit active and inactive states that are readily interconverted in response to several sets of stimuli and conditions. At 30 °C, the optimal growth temperature, and at physiological pH 7.5, DrRecA protein binds to double-stranded DNA (dsDNA) and forms extended helical filaments in the presence of ATP. However, the ATP is not hydrolyzed. ATP hydrolysis of the DrRecA-dsDNA filament is activated by addition of single-stranded DNA, with or without the single-stranded DNA-binding protein. The ATPase function of DrRecA nucleoprotein filaments thus exists in an inactive default state under some conditions. ATPase activity is thus not a reliable indicator of DNA binding for all bacterial RecA proteins. Activation is effected by situations in which the DNA substrates needed to initiate recombinational DNA repair are present. The inactive state can also be activated by decreasing the pH (protonation of multiple ionizable groups is required) or by addition of volume exclusion agents. Single-stranded DNA-binding protein plays a much more central role in DNA pairing and strand exchange catalyzed by DrRecA than is the case for the cognate proteins in Escherichia coli. The data suggest a mechanism to enhance the efficiency of recombinational DNA repair in the context of severe genomic degradation in D. radiodurans.
Introduction
The bacterium Deinococcus radiodurans has an extraordinary capacity to survive the effects of exposure to ionizing radiation, routinely surviving doses of 5 kilogray with no lethality (1). This phenotype is apparently due to an evolutionary adaptation to the effects of desiccation (2, 3). Many mechanisms have been proposed to explain this extreme resistance to radiation (1, 4–10), several only marginally related to DNA repair processes. Nevertheless, the accurate reconstitution of the Deinococcus genome from hundreds or thousands of fragments holds a special place in biology as an extraordinary feat of DNA repair. The central player in this process is the RecA protein. However, its mechanism for efficient DNA repair in the radioresistant D. radiodurans has not yet been carefully characterized.
Genome reconstitution after extreme irradiation occurs in two phases (4, 9, 11). The first, labeled extended synthesis-dependent strand annealing, serves to join fragments into longer chromosomal segments (4, 9). These are linked to form completed chromosomes in a phase 2 that relies on recombinational DNA repair. The entire process is completed in 3–4 h. The RecA protein of Deinococcus is required in phase 2, but also has an important role in phase 1 (9). Deinococcus strains that lack RecA function lose almost all of their radiation resistance (12, 13).
The prototypical RecA protein is that of Escherichia coli (EcRecA), studied in great detail over a period of 30 years (14–18). The EcRecA forms filaments on single-stranded DNA (ssDNA),2 aligns the bound DNA with a homologous duplex DNA (dsDNA), and promotes a reaction called DNA strand exchange. During this reaction, the duplex DNA strand that is complementary to the RecA-bound ssDNA is transferred to form a new duplex, and the identical strand of the duplex is displaced. In this process, EcRecA is a DNA-dependent ATPase. ATP is hydrolyzed throughout the strand exchange reaction (14–17, 19), conferring a number of important properties on the strand exchange process and rendering it efficient (14–17). ATP is always hydrolyzed when EcRecA is bound to ssDNA or dsDNA, and this property is widely used as an indirect but reliable measure of DNA binding (20–29). The rate of ATP hydrolysis by EcRecA nucleoprotein filaments on ssDNA is altered abruptly upon interaction with homologous dsDNA, as the filaments take on a new state amenable to DNA pairing and strand exchange (30, 31).
The ATPase activity of the EcRecA protein has been implicated in disassembly of EcRecA filaments and in the later stages of the DNA strand exchange reaction (14, 15, 23, 28, 32–37). ATP, but not its hydrolysis, is required for the formation of active EcRecA nucleoprotein filaments on ssDNA and for the homologous alignment of that ssDNA with a complementary duplex (35–39). Thus, a simplified view of EcRecA-mediated DNA strand exchange begins with the formation of active nucleoprotein filaments on ssDNA. The EcSSB protein facilitates filament extension in this phase and is displaced as the filaments form. The subsequent search for homology does not require ATP hydrolysis. Once homologous alignment is secured, ATP hydrolysis facilitates the formation of extensive tracts of the product heteroduplex DNA and also facilitates the eventual dissociation of the RecA filament.
The EcRecA protein is regulated at several levels. First, transcription of the recA gene is regulated as part of the SOS response (40, 41). Second, the C terminus of EcRecA is an autoregulatory flap; removal of 17 C-terminal residues enhances a number of EcRecA activities (15, 42–44). Third, and finally, certain activities of EcRecA, prominently filament assembly on and dissociation from DNA, are subject to regulation by an array of other proteins (16).
Based on changes in filament cooperativity, ATP hydrolysis rates, and other properties, there are at least four EcRecA filament states that can be identified at different stages of EcRecA-mediated DNA strand exchange (15, 30, 45). For example, when EcRecA nucleoprotein filaments are bound to ssDNA, ATP hydrolysis proceeds with a kcat of ∼30 min−1 at 37 °C. Addition of a homologous duplex DNA triggers a change in filament state that is manifested by an abrupt 30% decline in ATPase rate (31). Once EcRecA filaments are assembled on DNA, they are consistently active in the hydrolysis of ATP, although the rate is modulated by changes in filament state.
The Deinococcus RecA protein (DrRecA) appears to be quite similar to the EcRecA in many respects (46). However, at 37 °C, the DrRecA initiates DNA strand exchange primarily from the dsDNA instead of initiating from a bound ssDNA, inverse of other bacterial RecA proteins (47). Although the EcRecA can promote an inverse reaction under extraordinary reaction conditions (48), it is the preferred mode for the DrRecA under at least some conditions (47). The DrRecA protein forms filaments on dsDNA, nucleating much faster than EcRecA but extending slower (49). This combination of attributes would tend to create a larger number of shorter DrRecA filaments, an adaptation that may facilitate the repair of the large number of double strand breaks created by extreme irradiation events.
Deinococcus also encodes an SSB protein that is unusual. The DrSSB is approximately twice the size of a typical bacterial SSB (50–52), includes two OB folds instead of one, and functions as a dimer instead of a tetramer (50, 51, 53).
Genome reconstitution after heavy irradiation relies greatly on DNA double strand break repair, and the DrRecA and DrSSB proteins play key roles. The process of genome reconstitution is set up in its earliest steps, where DNA fragments are processed and DrRecA filaments are formed in potentially productive locations. This phase of “setting the stage” may involve quite a number of auxiliary proteins. However, we cannot understand how various proteins affect DrRecA until we better understand the properties of DrRecA itself. In this report, we further investigate one of the earliest stages of double strand DNA break repair, i.e. the formation of DrRecA protein filaments and filament dynamics. We also investigate the dependence of DrRecA function on DrSSB. The results reveal a highly constrained DrRecA protein that forms filaments on dsDNA, and ssDNA with SSB, but with no ATPase function at 30 °C and pH 7.5 or higher. Relative to EcRecA, changes in filament state are highly exaggerated in the DrRecA protein, highlighted by the ATPase inactive state that provides a potential new point of regulation. Activation of DrRecA is facilitated by a range of ligands and/or conditions, some of which reflect the situation likely to be present when double strand break repair is required. The results also show an unusually intimate functional relationship with DrSSB. We conclude our investigation with a working model that describes the potential dynamics of the RecA protein and its activities in D. radiodurans.
EXPERIMENTAL PROCEDURES
Protein Purification
The native DrRecA protein was purified as described (46). Its concentration was determined by absorbance at 280 nm with extinction coefficient 0.372 mg ml−1 (46). The wild type EcRecA (54), wild type EcSSB protein (54, 55), and wild type DrSSB proteins (51) were all purified in their native forms as described. Concentrations of these proteins were determined by absorbance at 280 nm using the extinction coefficients ϵ280 = 2.23 × 104 m−1 cm−1 (EcRecA), 2.38 × 104 m−1 cm−1 (EcSSB), and 4.10 × 104 m−1 cm−1 (DrSSB). All proteins were free of detectable nuclease contamination.
DNA Substrates
Circular single-stranded DNA and supercoiled DNA were derived from the bacteriophage M13mp18 and purified as described previously (30). Linear double-stranded DNA was created by digesting supercoiled M13mp18 DNA with PstI endonuclease using conditions recommended by the supplier. Circular single-stranded and double-stranded ΦX174 DNA was purchased from New England Biolabs. Double-stranded ΦX174 DNA was linearized by digestion with PstI to create the ΦX174 linear double-stranded DNA. Poly(dT)200 was purchased from Midland Certified Reagent Co. All DNA concentrations are reported in terms of total nucleotides.
ATP Hydrolysis Assays
A coupled spectrophotometric enzyme assay (56, 57) was used to measure the ATPase activities of the RecA proteins when binding to DNA. The assays were carried out in a Varian Cary 300 dual beam spectrophotometer equipped with a temperature controller and a 12-position cell changer. The cell path lengths were 1 cm, and band pass was 2 nm. The standard reaction condition was 25 mm Tris-OAc, pH 7.5 (90% cation), 5% w/v glycerol, 3 mm potassium glutamate, 10 mm magnesium acetate, 1 mm dithiothreitol (GoldBio), 2 mm NADH, and an ATP regeneration system (2 mm NADH, 10 units/ml pyruvate kinase, 10 units/ml lactate dehydrogenase, and 3 mm phosphoenolpyruvate; all purchased from Sigma) at 30 °C. Unless otherwise stated, reactions contained 3 μm DrRecA or EcRecA, 3 mm ATP, 5 μm single-stranded DNA, 10 μm linear double-stranded DNA, and 0.3 μm (monomer) SSB. The final pH of the reactions was 7.48. ATP hydrolysis by the RecA protein was indirectly measured by the decrease in NADH concentration monitored at 380 nm. The amount of ATP hydrolyzed was calculated using the extinction coefficient of NADH, 1.21 mm−1 cm−1. In some reactions, one or more reagents were added during the time course presented, resulting in a small abrupt change in absorbance due to the dilution effect. A correction has been included for this dilution effect for all data obtained subsequent to the reagent addition. All stated reagent concentrations represent those present after the final reagent addition.
ATPase pH-rate Profile
Reactions were carried out as described for the ATPase assays above, but with the ionization states of buffers varied as needed to adjust pH. The buffer MES was used for pH values between 5.53 and 7.26 (except pH 7.22), and Tris acetate was used in the pH range 7.22–8.7 (except for pH 7.26, which was MES). The pH values reported are final values taken at 30 °C in mock solutions containing all system components, but substituting storage buffers for proteins and DNA molecules. Reported values are taken from three to six separate measurements at each pH.
Strand Exchange Reactions
DNA strand exchange reactions were carried out at 30 °C for DrRecA and 37 °C for EcRecA. Standard reaction conditions included 25 mm Tris-OAc, pH 7.5 (90% cation), 3 mm potassium glutamate, 10 mm magnesium acetate, 5% w/v glycerol, 1 mm DTT, and an ATP regeneration system (3.3 mm phosphoenolpyruvate plus 10 units/ml pyruvate kinase). Reactions also included 3 μm DrRecA or EcRecA, 20 μm dsDNA, 10 μm ssDNA, and 1 μm (monomer) DrSSB or EcSSB, unless otherwise indicated. Aliquots of 10 μl were removed from each reaction at the indicated times and stopped by addition of 10 μl of a stopping solution (3:2 ratio of 18% w/v Ficoll and 20% SDS). The treated samples were run on a 0.8% agarose gel overnight at 20 mA. All products indicated on strand exchange gels were M13mp18 nicked circular products resulting from strand exchange between a circular ssDNA and linear dsDNA.
Electron Microscopy
A modified Alcian method was used to visualize DrRecA filaments. Activated grids were prepared as described previously (43). Samples for electron microscopy analysis were prepared as follows. All incubations were carried out at 30 °C. An ATP regeneration system of 10 units/ml creatine phosphokinase and 12 mm phosphocreatine was included in every reaction. DrRecA (3 μm) was preincubated with 10 μm M13mp18 ldsDNA or 5 μm cssDNA in a common buffer containing the same reaction buffer indicated in the ATPase assays (25 mm Tris-OAc (90% cation), 5% (w/v) glycerol, 3 mm potassium glutamate, and 10 mm Mg(OAc)2) for 10 min. ATP was added to 3 mm, and the reaction was incubated for another 15 min, after which ATPγS was added to 3 mm to stabilize the filaments. The mixture was then incubated for a further 5 min. Whenever the experimental condition demanded SSB, DrSSB was added to 0.3 μm. The reaction mixture described above was diluted appropriately with 200 mm ammonium acetate, 10 mm HEPES, and 10% glycerol (final pH 7.5) to get the final concentration of DNA to 0.4 ng/μl. The sample was immediately adsorbed to the activated carbon grid for 3 min. The grid was then touched to a drop of the above buffer followed by floating on a drop of the same buffer for 1 min. The sample was then stained by touching to a drop of 5% uranyl acetate followed by floating on a fresh drop of the same solution for 30 s. Finally, the grid was washed by touching to a drop of double distilled water followed by immersion in two 10-ml beakers of double distilled water. After the sample was dried, it was rotary-shadowed with platinum. This protocol was designed for visualization of complete reaction mixtures, and no attempt was made to remove unreacted material. Although this approach should yield results that give a true insight into reaction components, it does lead to samples with a high background of unreacted proteins. To determine the proportion of the molecules observed that were either fully or partially coated by DrRecA protein, at least 10 separate regions of at least two grids were looked at, using an identical magnification for each sample.
A total of 11 DrRecA ldsDNA filament lengths were measured using MetaMorph analysis software to determine the extended length of DrRecA-ldsDNA. Each filament was measured three times, and the average length in nanometers, angstroms, and base pairs was calculated. We used the number of pixels in 500 nm as a standard and calculated the number of pixels along the length of a filament. The pixels were converted to nanometers and compared with the length in nanometers of a naked B-form M13mp18 ldsDNA (7249 bp times 0.34 nm per base). Imaging and photography were carried out with a TECNAI G2 12 Twin Electron Microscope (FEI Co.) equipped with a GATAN 890 CCD camera.
In experiments where types of DrRecA nucleoprotein filament species were quantified, at least 250 molecules were counted in two different grids analyzed per experiment. Every experiment and one repeat were counted, resulting in more than 500 filaments counted for each experimental condition. The qualitative sizes of each filament species (i.e. long, medium, short linears or big, medium, small circles, etc.) represented judgments constrained as described under “Results.” A subset of molecules in each category was directly measured, using the method described above. The numbers of molecules measured are provided in the relevant figures and figure legends. In these measurements, only the RecA-coated portion of the DNA was measured. The irregularity of SSB-coated regions precludes accurate length measurements.
RESULTS
This study focuses on characterizing the DNA binding and DNA recombination functions of the RecA protein from D. radiodurans. Previous characterization of DrRecA was done at 37 °C. However, the most common temperature for growing D. radiodurans cultures and for D. radiodurans in vivo studies is 30 °C (6, 58–62). To facilitate the accumulation of a database that is more physiologically relevant, we carried out the present studies at 30 °C. We used DrRecA-mediated ATP hydrolysis assays to monitor the ATPase activity of DrRecA and complemented them with direct visualization of the DrRecA nucleoprotein filaments with electron microscopy (EM).
Binding ssDNA in the Absence of SSB Is Limited by Secondary Structure
DrRecA hydrolyzes ATP when bound to single-stranded DNA (ssDNA). Kim et al. (46) previously reported that DrRecA preferentially binds to dsDNA at 37 °C even when ssDNA is present. Furthermore, the DrRecA protein promotes an inverse DNA strand exchange reaction, binding first to dsDNA and then carrying out a homology search on ssDNA (47). We initially carried out ATP hydrolysis assays as an indirect method for measuring DNA binding by DrRecA at 30 °C (Fig. 1A). We incubated DrRecA with circular single-stranded DNA (cssDNA), derived from M13mp18 bacteriophage, or poly(dT)200 lacking any secondary structure, for 10 min before adding ATP to each reaction to begin monitoring hydrolysis. At 37 °C, observed rates of ATP hydrolysis were comparable with those reported in earlier work (data not shown) (46). When bound to poly(dT)200 at 30 °C and pH 7.5, DrRecA hydrolyzed ATP with an apparent kcat of 6.80 ± 0.43 min−1 (assuming one DrRecA monomer bound for each three nucleotides of ssDNA) and a kcat of 4.81 ± 0.33 min−1 when bound to cssDNA alone. The calculated kcat estimate is based on an assumption that all potential binding sites on the DNA are occupied, and it may be an underestimate if the DrRecA protein is not coating the cssDNA uniformly.
FIGURE 1.
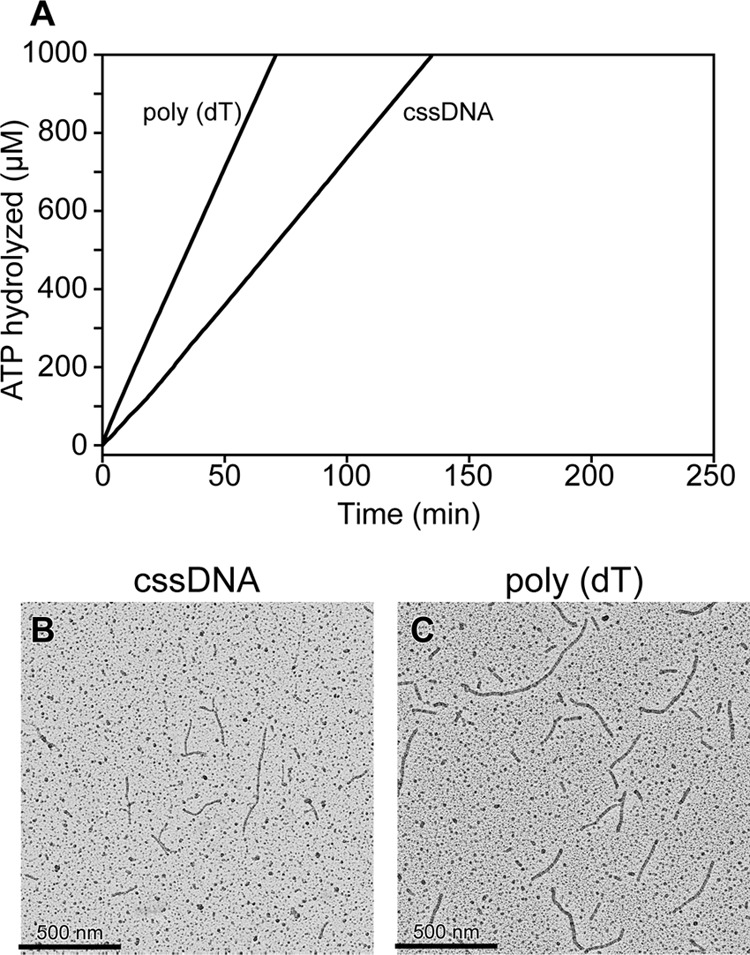
DrRecA ATPase activity on ssDNA. A, DrRecA (3 μm) was incubated with M13mp18 cssDNA, poly(dT)200 for 10 min at 30 °C before 3 mm ATP was added to start monitoring ATP hydrolysis. B, representative electron micrograph showing DrRecA bound to M13mp18 cssDNA for 15 min before the filaments were fixed with ATPγS and spread on an Alcian grid. C, representative micrograph showing DrRecA bound to poly(dT)200 for 15 min before the filaments were fixed with ATPγS and spread on an Alcian grid.
We used EM to directly visualize DrRecA on the M13mp18 cssDNA and poly(dT)200. On the M13mp18 cssDNA in the absence of the SSB, DrRecA formed irregular, short, and discontinuous filaments that most likely reflect its deficiency in melting out secondary structures within the circular ssDNA in the absence of the SSB (Fig. 1B). Because the poly(dT)200 DNA substrate lacks secondary structures, fully filamented DrRecA nucleoprotein filaments were observed (Fig. 1C). We note with interest that the DrRecA-poly(dT)200 filaments observed are much longer than what we would expect of DrRecA bound to 1 molecule of (dT)200 and are probably a result of multiple DrRecA-(dT)200 nucleoprotein filaments assembled end-to-end. Similar observations have been made with the EcRecA protein (63, 64).
SSB Suppresses DrRecA ATPase but Is Important for DrRecA Filament Formation on ssDNA
In vitro, the SSB protein is typically used with E. coli RecA to remove secondary structures in ssDNA to facilitate formation of contiguous EcRecA nucleoprotein filaments, resulting in a stimulation of ATP hydrolysis (65). We have observed that the SSB protein has the opposite effect on RecA-mediated ATP hydrolysis in the D. radiodurans system by inhibiting DrRecA ATP hydrolysis, although DrRecA is bound to M13mp18 cssDNA (Fig. 2A). Reaction 1 of Fig. 2A shows DrRecA ATPase while bound to M13mp18 cssDNA in the absence of DrSSB. Reaction 2 of Fig. 2A represents DrRecA ATPase activity if DrSSB was added to the reaction at the same time as ATP was added to begin the reaction (time 0 min). Reaction 3 of Fig. 2A represents DrRecA ATPase activity if DrSSB was added to the reaction 20 min after DrRecA had been hydrolyzing ATP while bound to cssDNA. We observed a strong ATPase inhibition mediated by the SSB protein in reactions 2 and 3. After the addition of SSB in reaction 3, ATP hydrolysis was initially accelerated but was almost completely abolished 30–45 min later. There was no drop in ATP hydrolysis in the reaction that did not contain DrSSB.
FIGURE 2.
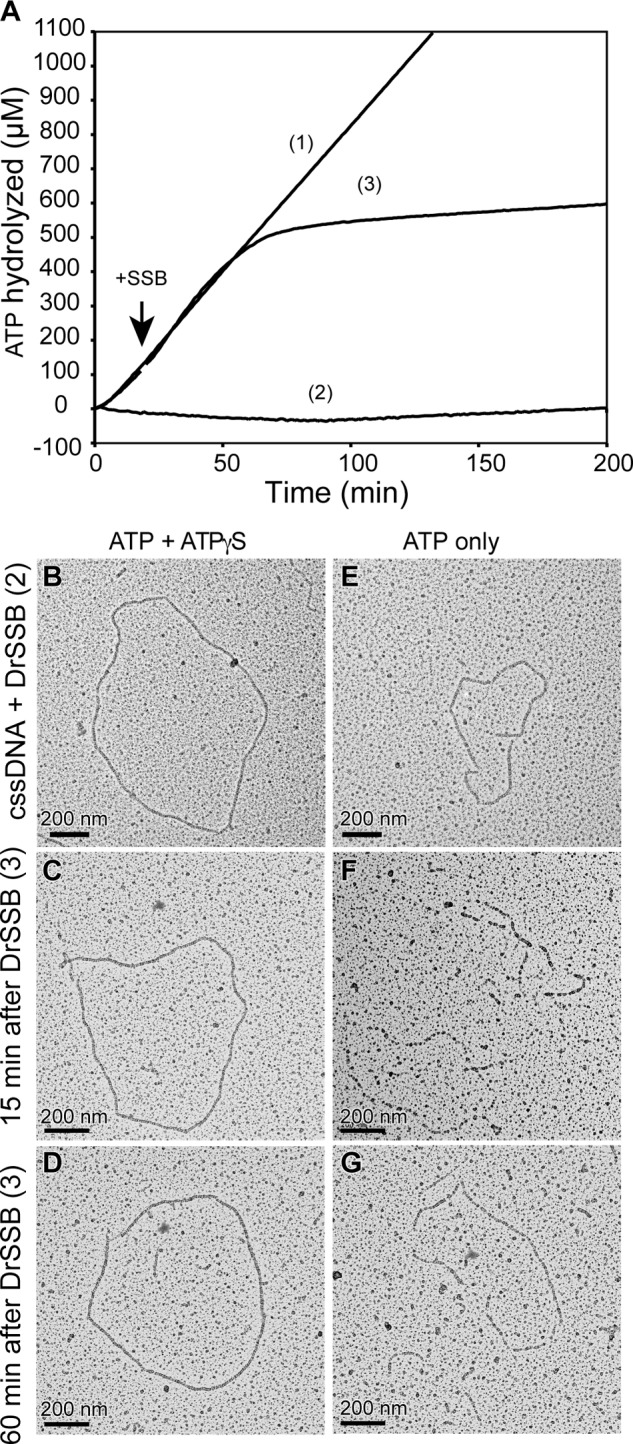
DrSSB inhibits DrRecA ATPase on cssDNA. A, DrRecA ATPase assay shows the following: reaction 1, DrRecA bound to cssDNA alone; reaction 2, DrRecA bound to cssDNA with DrSSB added at 0 min; and reaction 3, DrRecA bound to cssDNA with DrSSB added at 20 min. B, representative electron micrograph corresponding to reaction 2 at 15 min and fixed with ATPγS. C, representative electron micrograph corresponding to reaction 3 at 15 min after DrSSB addition and fixed with ATPγS. D, representative electron micrograph corresponding to reaction 3 at 60 min after DrSSB addition and fixed with ATPγS. E, same as B without ATPγS fixing. F, same as C without ATPγS fixing. G, same as D without ATPγS fixing.
Initially, the lack of ATP hydrolysis suggested that DrSSB was a much stronger competitor for ssDNA than DrRecA under these conditions and was displacing the RecA protein from the cssDNA. However, when we directly visualized reaction 2 containing DrSSB at 15 min by EM, we observed complete and fully extended DrRecA-cssDNA nucleoprotein filaments despite the absence of ATP hydrolysis (Fig. 2B). Reaction 3 was also visualized by EM at 15 min (Fig. 2C) and 60 min (Fig. 2D) after the addition of DrSSB, corresponding to the accelerated and suppressed stages of ATP hydrolysis, respectively. After 15 min, DrRecA nucleoprotein filaments were mostly complete with occasional gaps showing SSB-bound ssDNA. At the 60- min time point, DrRecA had fully coated and extended the cssDNA, despite the extremely low level of ATP hydrolysis. In all three spreads (Fig. 2, B–D) and their replicates, circular DrRecA nucleoprotein filaments were found in abundance, and little SSB-coated cssDNA was seen.
In these EM experiments, ATPγS was typically added just prior to spreading to stabilize the filaments, as done in many past studies (27, 32, 66–70). With ATP only, even slow hydrolysis of bound ATP will destabilize the filaments as ATP is removed from the surrounding solution in the final stages of spreading. To help ensure that our observations were not an artifact of the ATPγS addition, we also spread the samples immediately without fixing the filaments with ATPγS (Fig. 2, E–G). If DrRecA bound the cssDNA with very slow or no ATP hydrolysis, we should still be able to visualize the filaments without ATPγS fixation. Although the filaments were slightly broken and not as well formed without the ATPγS addition, they were still clearly detectable and found in abundance. The ill-formed DrRecA-cssDNA filaments were probably due to slight dissociation of DrRecA from the DNA as ATP is minimally hydrolyzed and not replaced during the spreading process. The DrRecA filaments are easily distinguished from complexes of DrSSB on ssDNA, which have a highly condensed and beaded appearance similar to what has been reported for EcSSB (53).
Suppression of the DrRecA ATPase activity was also observed when we used SSB from E. coli, but the EcRecA ATPase activity was not suppressed by SSB from either E. coli or D. radiodurans (Fig. 3). Either DrRecA or EcRecA was added to reactions containing cssDNA coated with DrSSB or EcSSB as indicated in the figure. With SSB added first, EcRecA binds to ssDNA slowly but eventually coats the DNA and hydrolyzes ATP. ATPase kinetics by EcRecA were similar whether DrSSB or EcSSB was used. In contrast, ATPase activity by DrRecA with either DrSSB- or EcSSB-coated cssDNA was very slow, although the reaction seen with EcSSB was somewhat faster than that seen with DrSSB.
FIGURE 3.
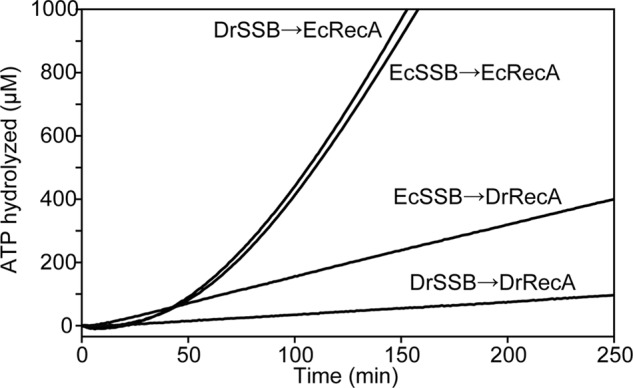
EcSSB suppresses DrRecA ATPase activity. M13mp18 cssDNA was coated with DrSSB or EcSSB for 10 min before addition of DrRecA or EcRecA to start ATPase monitoring. Both EcSSB and DrSSB inhibited DrRecA ATPase and did not affect EcRecA ATPase activity.
Under our conditions, the amount of DrSSB required to maximally suppress the ATPase activity of DrRecA activity on ssDNA is 300 nm monomers (150 nm dimers) or perhaps a bit less. In the experiments described, 5 μm nucleotides of cssDNA and 3 μm DrRecA were used. This equates to 1 DrSSB dimer for every 33 nucleotides of ssDNA or 1 dimer for every 20 DrRecA monomers. Interestingly, supersaturating concentrations of DrSSB, up to 1 μm monomer, relieved the inhibition of the DrRecA ATPase and even stimulated it (Fig. 4). Similar effects were seen with addition of higher concentrations of EcSSB (data not shown). At 400 or 500 nm, the effects of DrSSB exhibit a hybrid, biphasic effect. This likely reflects an apparent stimulation of the ATPase at early times due to the formation of contiguous filaments, followed by a relatively slow transition to the inactive form at these higher, less inhibitory concentrations of the DrSSB.
FIGURE 4.
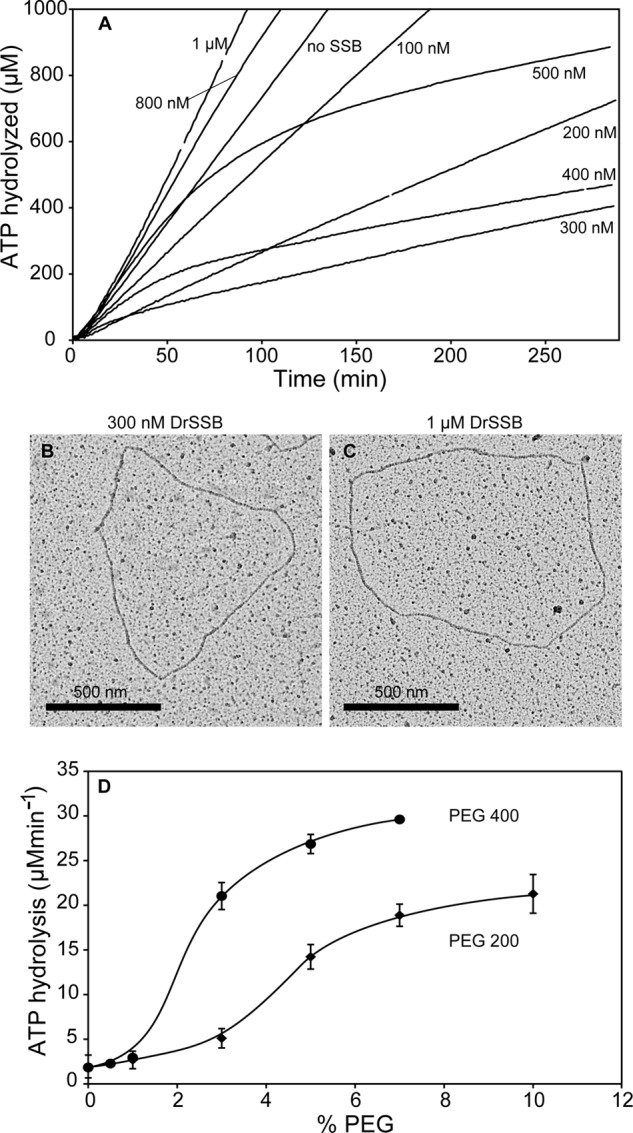
Maximal DrRecA ATPase inhibition is 300 nm DrSSB (monomer) in 5 μm ssDNA. A, ATPase assay with increasing DrSSB concentrations from 0 to 1 μm. B, DrRecA-cssDNA (3 μm DrRecA and 5 μm cssDNA) filament with 300 nm DrSSB. C, DrRecA-cssDNA (3 μm DrRecA and 5 μm cssDNA) filament with 1 μm DrSSB. D, effects of two different forms of polyethylene glycol on the ATP hydrolytic activity of DrRecA protein on ssDNA in the presence of DrSSB. Reactions contain 3 μm DrRecA, 5 μm cssDNA, and 0.3 μm DrSSB.
The effects of DrSSB and EcSSB on the DrRecA ATPase activity do not exhibit any tight stoichiometries or otherwise exhibit properties one might expect if the DrRecA protein formed a complex with either SSB. We have found no evidence for such a complex. Instead, we attribute these effects of the SSB proteins to a solute or excluded volume effect (71). We were able to mimic the activation effect with polyethylene glycol (Fig. 4D). The relief of the ATPase inhibition by the excess DrSSB provides additional evidence that the inhibitory effect of DrSSB does not reflect a displacement of the DrRecA from the DNA, as the higher levels of DrSSB clearly fail to do so. DrRecA filaments formed in the presence of either 300 nm or 1 μm DrSSB do not appear to be detectably different by EM (Fig. 4, B and C). We have also confirmed by analytical ultracentrifugation that the oligomeric state of DrSSB remains dimeric at both concentrations of DrSSB (data not shown).
The similarity of the DrRecA filaments at both DrSSB concentrations, despite the large difference in rates of ATP hydrolysis, suggests the presence of at least two activity states in the DrRecA filaments. Relatively subtle effects of DrSSB concentration and perhaps other factors govern the transition between states. That transition is not all or none, but is gradual, as seen with the effects of polyethylene glycol in Fig. 4D. However, the filament state transition in DrRecA brings about a greater change in ATPase activity than any comparable transition observed to date with the Escherichia coli RecA protein.
Double-stranded DNA Binding Inhibits DrRecA ATPase Activity
When DrRecA was incubated with the 7249-bp double-stranded DNA from M13mp18 bacteriophage, linearized with PstI restriction enzyme (Fig. 5A), no ATP hydrolysis was observed. A reaction with DrRecA and cssDNA is also included in Fig. 5A to allow for a direct comparison of the ATPase activity in the presence of the two DNA substrates, cssDNA and ldsDNA, both in the absence of SSB. We found the absence of ATP hydrolysis intriguing because ldsDNA is the preferred substrate at 37 °C (46). We utilized EM to directly visualize DrRecA in the presence of the ldsDNA and ATP at 30 °C, fixed with ATPγS (Fig. 5B). Although DrRecA did not hydrolyze ATP with ldsDNA, we observed an abundance of complete and fully coated DrRecA nucleoprotein filaments. ATP was required for this binding (data not shown), and samples that were spread directly and rapidly on a grid without addition of ATPγS still exhibited a large population of DrRecA filaments (Fig. 5C). As with the filaments on ssDNA, the DrRecA-dsDNA filaments spread without the addition of ATPγS generally contained some gaps. This was again most likely due to a sudden destabilization as the ATP was abruptly removed in the spreading protocol, and some bound ATP was hydrolyzed. Similar to observations described above with cssDNA in the presence of DrSSB, DrRecA-mediated ATPase activity was negligible even though DrRecA was bound to dsDNA and formed mostly complete filaments.
FIGURE 5.
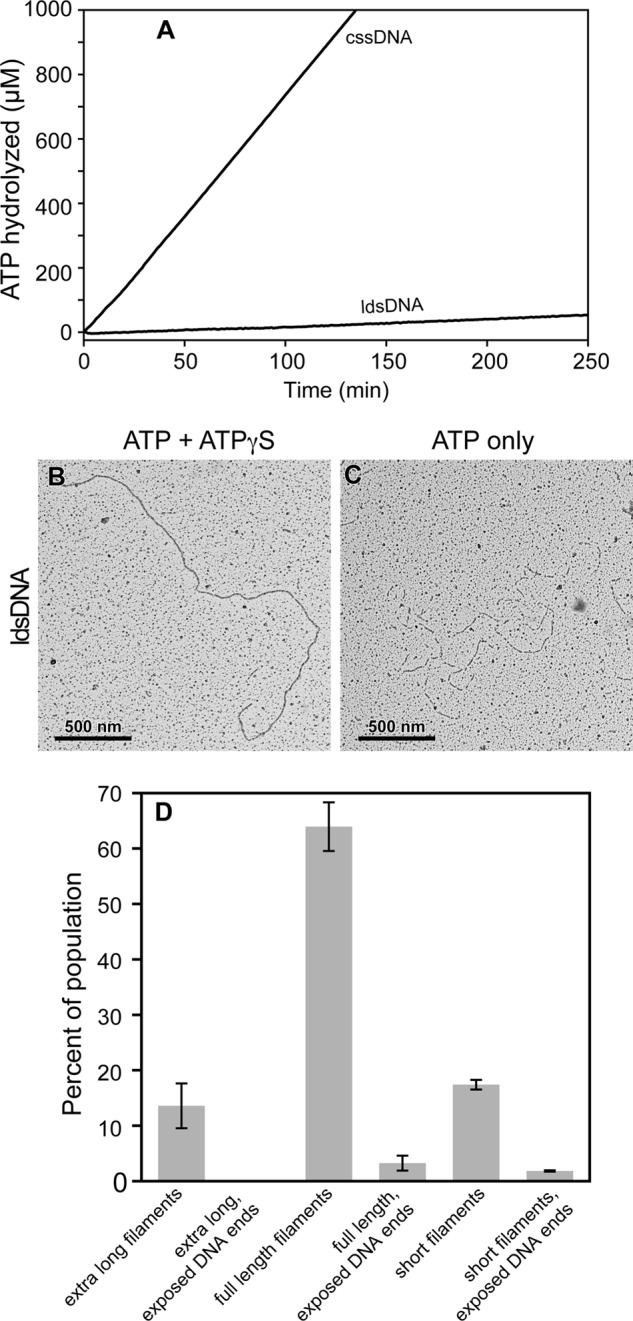
Lack of ATP hydrolysis when DrRecA is bound to dsDNA. A, DrRecA was incubated with M13mp18 cssDNA or ldsDNA for 10 min at 30 °C before ATP was added to begin ATPase monitoring. B, representative micrograph showing a full-length DrRecA-ldsDNA filament fixed with ATPγS. C, representative micrograph showing DrRecA-ldsDNA filaments not fixed with ATPγS. D, two independent EM experiments with DrRecA and ldsDNA were observed, and a total of 547 filaments was counted to make the total population. Precise length may vary within each class of filament. Full-length filaments were measured in detail to determine the fully extended DrRecA-dsDNA nucleoprotein filament length.
We directly measured the lengths of ldsDNA filaments fixed with ATPγS using the MetaMorph software (Fig. 5D). Three major classes of fully coated filaments exist in the population of the 547 filaments counted. The most common species, making up the majority of the population of filaments counted (63.9 ± 4.39%), appeared to coat the entire length of the duplex DNA, measuring 3627 ± 66.8 nm. Because the length of M13mp18 is 2464 nm having 7249 bp, these full-length DrRecA ldsDNA filaments were calculated to be 47.2 ± 2.7% longer than the naked B-form of M13mp18 ldsDNA. We interpreted these filaments to be complete unit length filaments, with an ATP-mediated extension consistent with that observed in EcRecA filaments (72, 73). The two less frequently observed filament length classes were “extra long filaments” that were apparent multiples of the unit length filament, formed by noncovalent association of filament ends (as seen previously with poly(dT)200), and “short filaments” that were of varying lengths, shorter than the unit length. We interpreted the latter filaments to be pieces of broken DNA bound by DrRecA or DNA unsaturated with DrRecA with naked DNA ends not clearly visible. Extra long fully filamented dsDNA made up 13.6 ± 4.03% and the short filaments 17.4 ± 0.87% of the EM population. A small subset of the full-length and short filaments had some visible naked dsDNA at the ends that was not coated with DrRecA. These partially coated DNAs minimally made up 3.25 ± 1.34% and 1.83 ± 0.12% of the population, respectively. No unbound DNA end segments were seen on extra long filaments.
To explore the general mechanism of ATPase suppression when DrRecA is bound to dsDNA, we examined the ATPase activity as a function of pH and temperature. The kcat of ATP hydrolysis was measured over a range of pH, from pH 5.5 to 9.0 at 30 °C, using linearized M13mp18 dsDNA as a binding substrate (Fig. 6A). We found that the ATPase activity of DrRecA peaked sharply at pH 7 and was negligible at pH 7.5 or above. A sharp decline was also observed below pH 7.0.
FIGURE 6.
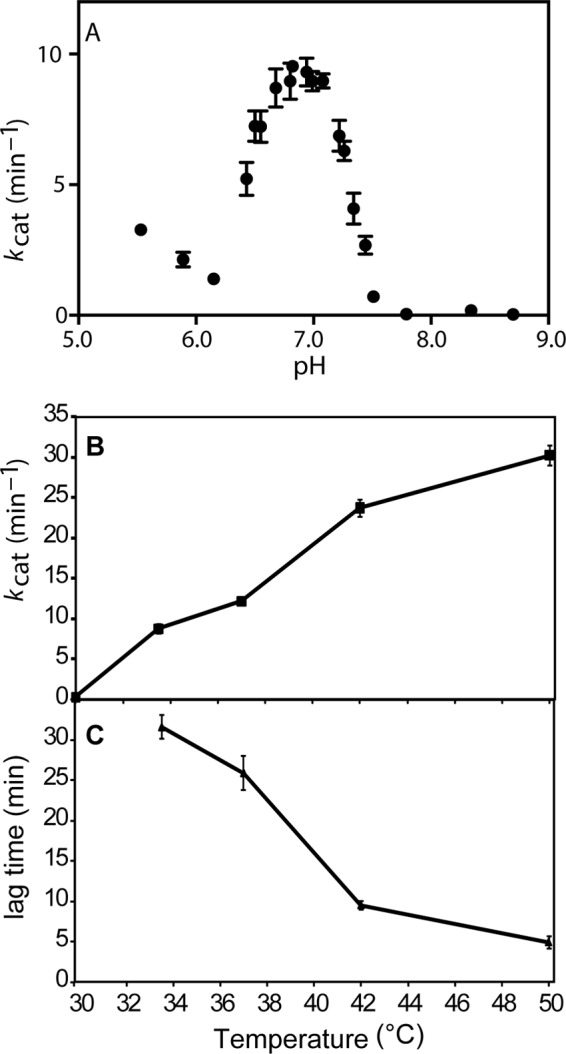
Characterizing DrRecA ATPase activity as a function of pH and temperature. A, kcat of ATP hydrolysis by DrRecA incubating with linear dsDNA at 30 °C varies with pH. ATP hydrolysis is highest at pH 7 and is suppressed at pH values higher than 7.5. B, ATPase activity by DrRecA bound to dsDNA at pH 7.5 increases with increasing temperature. C, lag time required to achieve steady state of ATP hydrolysis at temperatures between 30 and 50 °C decreases with increasing temperature.
The approximately bell-shaped pH-rate profile between pH 6.15 and 7.5 could not be fit by standard models (74, 75) built around a single ionizable group being responsible for the increase in kcat observed on each side of the pH 7 peak. The slope on each side of the curve was much too steep. Efforts to model the transition underlying the curve between pH 6.15 and 7.5 are ongoing. Fitting the curve requires either that there are multiple ionizations responsible for the slopes on either side of the curve or (more likely for a RecA filament) that the transition from inactive to active ATPase states in these filaments is highly cooperative.3 If the curve reflects a cooperative transition and a single ionization on either side of the peak, then there are at least two key amino acid residues with pKa values close to neutrality in either the active or inactive states. However, we note that there is only one His residue in the Deinococcus RecA protein. Thus, the results suggest that one or more of the amino acid residues giving rise to the activation seen in Fig. 6A have significantly perturbed pKa values. There are changes in the placement of charged amino acid residues in the DrRecA protein, relative to EcRecA, that may have a large impact on the electrostatic surface potential (76).
The kcat value of ATP hydrolysis and the lag time required to achieve steady state in the presence of dsDNA were also measured over a range of temperatures, from 30 to 50 °C, at pH 7.5. The rate of ATP hydrolysis was essentially zero at 30 °C but increased with increasing temperature (Fig. 6B). Where a measurable ATPase activity was observed, the measured lag time to steady state decreased with increasing temperatures (Fig. 6C).
ATPase activity as a function of temperature and pH is consistent with previous data reported by Kim et al. (46) that DrRecA hydrolyzed ATP at 37 °C. We note that the EcRecA ATPase also exhibits a dependence on temperature (77). The difference between EcRecA and DrRecA is that the ATPase activity by EcRecA is reliably coupled to DNA binding, whereas the ATPase activity by DrRecA is not.
The temperature effects seen with EcRecA are readily interpreted as a reflection of the activation barrier for ATP hydrolysis and the processes to which it is coupled (25). In contrast, the ATPase activity of the DrRecA protein exhibits a DNA-bound state that is inactive with respect to ATP hydrolysis. An activation observed with increasing temperature, changes in pH, or increases in the concentration of volume exclusion agents suggests a change in DrRecA filament state.
ATP Hydrolysis by DrRecA-dsDNA Filaments Is Triggered by Addition of ssDNA and SSB
The question now becomes the following. How does this transition from inactive to active filament forms affect the function of DrRecA in the cell and facilitate genome reconstitution after irradiation?
We sought to discover a condition that would stimulate ATPase activity when DrRecA was bound to dsDNA. DrRecA, at a concentration (3 μm) that is somewhat below that required to saturate the DNA, was incubated with ATP and 20 μm M13mp18 ldsDNA for 15 min at pH 7.5 and 30 °C while ATPase activity was monitored (Fig. 7A). There was no ATP hydrolysis. After 15 min, we added a variety of possible stimulants. In particular, cssDNA was added in a variety of forms and using several different protocols at 10 μm as follows: 1) heterologous ΦX174 cssDNA or 2) homologous M13mp18 cssDNA alone; 3) 1 μm DrSSB-coated ΦX174 or 4) 1 μm DrSSB-coated M13mp18 cssDNA; 5) ΦX174 cssDNA (heterologous) followed by 1 μm DrSSB 5 min later or 6) M13mp18 cssDNA (homologous) followed by 1 μm DrSSB 5 min later. In all cases, the addition of ssDNA stimulated ATP hydrolysis.
FIGURE 7.

Single-stranded DNA activates the ATPase of DrRecA filaments on dsDNA. Addition of homologous ssDNA and DrSSB initiate DNA strand exchange. A, DrRecA was incubated with excess M13mp18 ldsDNA and ATP to form DrRecA-dsDNA filaments. After 15 min, the following DNA substrates were added: reaction 1, heterologous (het) ΦX174 cssDNA; reaction 2, homologous (hom) M13mp18 cssDNA; reaction 3, DrSSB-bound heterologous cssDNA; reaction 4, DrSSB bound homologous cssDNA; reaction 5, heterologous cssDNA, or reaction 6, homologous cssDNA followed by DrSSB addition after 5 min. B, aliquot of reactions 1–6 were run on a 0.8% agarose gel. Only homologous reactions also containing DrSSB resulted in M13mp18 DNA recombination, indicated by the M13 product band, which is nicked circular dsDNA.
In the reactions where cssDNA alone was added, DrRecA began hydrolyzing the ATP immediately, reaching a maximal rate of 17.8 μm/min after several minutes (see protocol 1 (above), heterologous cssDNA). Both heterologous and homologous ssDNAs triggered the ATPase activation and did so fairly quickly. In the reactions where the added ssDNA was coated with DrSSB (see protocols 3 and 4), a stimulation of ATPase by DrRecA was again observed, albeit with a lag of more than 30 min. Addition of DrSSB 5 min after introducing ssDNA led to a much more rapid rate increase to 20.9 μm/min in the reaction with heterologous DNA (see protocol 5) and 17.7 μm/min in the reaction with homologous DNA (see protocol 6). The new rate in this case was sustained after the DrSSB was added. In general, the ATPase stimulation was clearly not dependent on homology in the ssDNA.
An aliquot of each reaction was removed at the end of the ATPase assay and run on a 0.8% agarose gel to determine whether DNA strand exchange had occurred (Fig. 7B). Interestingly, the only protocols that resulted in measurable DNA strand exchange were the ones with both homologous DNA and SSB (see protocols 4 and 6 above). No reaction was observed, not even the formation of detectable reaction intermediates, when homologous ssDNA was added without DrSSB (see protocol 2 above). Under these conditions, the overall results suggest that ATP hydrolysis by DrRecA protein does not occur until all DNA substrates needed for a potential DNA strand exchange are present. Whereas the ATP hydrolysis activity is triggered by any ssDNA, DNA strand exchange requires DNA homology and also occurs only when DrSSB is present.
DrRecA Nucleoprotein Filament Dynamics
We wanted to understand DrRecA filament dynamics reflecting the different ATPase activities shown in Fig. 7. Our first question was whether the stimulated ATPase occurred while DrRecA remained bound to ldsDNA, or instead whether it reflected a transfer to the introduced cssDNA. Because the rates of ATP hydrolysis in reactions 1 and 2 of Fig. 7 were more than 2-fold higher than that seen when DrRecA is bound to cssDNA alone, and SSB suppresses ATP hydrolysis by DrRecA bound to ssDNA (see Fig. 2), we suspected that the ATPase activity reflected DrRecA-ldsDNA filaments that had been activated by the cssDNA. Reactions 1 and 2 were directly visualized by electron microscopy using the same conditions as indicated in Fig. 7 with DrRecA at a slightly subsaturating concentration. After the addition of either heterologous ΦX174 cssDNA (Fig. 8A) or homologous M13mp18 cssDNA (Fig. 8B) to DrRecA-M13mp18 ldsDNA filaments, the reactions were incubated for 30 min before they were fixed with ATPγS for 5 min and spread on an Alcian grid. We deliberately used linear dsDNA and circular ssDNA (of different lengths) to help us determine the particular DNA substrate to which DrRecA was bound.
FIGURE 8.
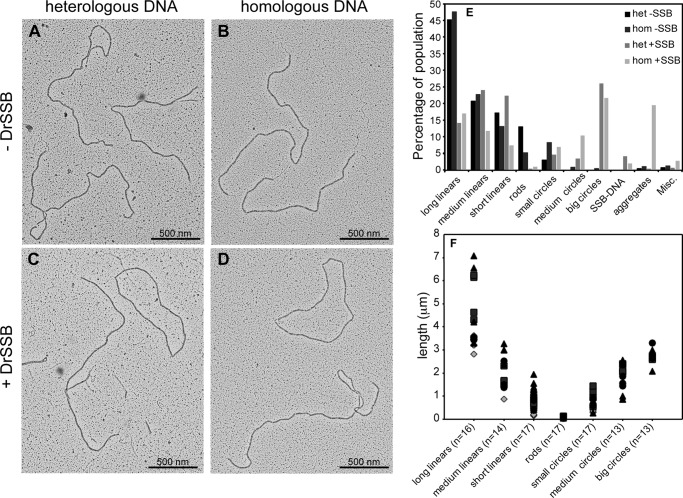
DrRecA-ldsDNA and DrRecA-cssDNA filaments are both observed primarily in the presence of SSB. A, representative electron micrograph showing M13mp18 ldsDNA and heterologous ΦX174 cssDNA in the absence of SSB (Het −SSB); B, M13mp18 ldsDNA and homologous cssDNA without SSB (Hom −SSB); C, M13mp18 ldsDNA and heterologous ΦX174 cssDNA with DrSSB (Het +SSB); and D, M13mp18 ldsDNA and homologous cssDNA with DrSSB (Hom +SSB). All components were added in the same order as indicated by corresponding reactions in Fig. 6 and visualized 50 min after the addition of ATP. Circular DrRecA filaments are primarily observed in reactions containing SSB. E, DrRecA-DNA filament types were counted and represented in a bar graph showing the decrease in long linear filaments and increase in big circular filaments with addition of SSB. F, examples of all types of RecA filaments described in E were directly measured as described under “Experimental Procedures.” The total numbers measured are provided along with the descriptions in the labels along the horizontal axis. At least four individual molecules were measured in each class of DrRecA filaments among the eight samples (duplicates of each condition). Diamonds represent molecules from heterologous DNA without SSB; squares represent molecules from the heterologous DNA with SSB; triangles represent molecules from homologous DNA without SSB, and circles represent molecules from homologous DNA with SSB. Long linear molecules range between 2.8 and 7.1 μm, medium linear between 0.9 and 3.3 μm, short linear range between 0.2 and 1.9 μm, and rods are very short molecules between 0.04 and 0.1 μm. Small circles range between 0.3 and 1.5 μm; medium circles are between 0.9 and 2.6 μm, and large circles typically range between 2.1 and 3.3 μm. No medium or big circles were observed in the heterologous DNA without SSB samples.
Careful counts of filaments seen on two grids revealed that the majority of filaments observed were linear DrRecA-dsDNA filaments (Fig. 8E). To support the judgments of DrRecA filament types observed, a subset of over a dozen molecules was directly measured, and the lengths are presented in Fig. 8F. Linear filaments, of various lengths, made up 83.1% of the 579 filaments counted in reaction 1, which contains heterologous DNA substrates in the absence of DrSSB (het-SSB). Similarly, linear filaments of various lengths made up 83.3% of all 521 filaments counted in reaction 2, which contains homologous DNA substrates in the absence of DrSSB (hom-SSB). Only 2.94 and 9.4% of the filaments were circles (indicative of ssDNA) in reactions 1 and 2, respectively. Although these small circular filaments could possibly be short linears on dsDNA joined at the ends, they appeared only in samples containing circular ssDNA. Very short DrRecA filaments, called “rods,” made up 13.0 and 5.18% of all the filaments in reactions 1 and 2, respectively. These rods could be DrRecA bound to cssDNA containing secondary structures as seen previously in Fig. 1B or oligomers of DrRecA in solution. Based on the majority of filaments being DrRecA-ldsDNA, our EM and ATPase results suggest that the stimulated ATPase activity observed was most likely to be ATP hydrolyzed by the DrRecA bound to ldsDNA, with cssDNA, heterologous or homologous, as the primary stimulant.
Our second question in understanding DrRecA filament dynamics was how DrSSB affected DrRecA-DNA filament formation. EM experiments were prepared with conditions corresponding to reactions 5 and 6 of Fig. 7, which contain DrSSB. The reactions were fixed with ATPγS after 30 min of incubation with DrSSB (Fig. 8, C and D). Interestingly, unlike reactions 1 and 2 without DrSSB, circular DrRecA-ssDNA nucleoproteins filaments were now observed in some abundance along with linear DrRecA-dsDNA filaments. These came in a variety of sizes, because the shorter DrRecA filaments on ssDNA tended to close up on themselves at the ends. The circular forms of all types again appear only in samples that included circular ssDNA. Because DrRecA was present in a subsaturating amount and harsh pipetting was avoided, it is unlikely that DrRecA-cssDNA filaments were formed by the DrRecA protein in solution or DrRecA protein displaced from dsDNA by vigorous mixing. Instead, this result suggested that DrSSB may trigger a transfer of DrRecA from the ldsDNA to the cssDNA. Filament quantification for reactions 5 and 6 revealed a decrease in long linear filaments and linear filaments in general and an increase in circular filaments (Fig. 8E). In the reaction using heterologous DNA with SSB added later (reaction 5), circular filaments increased over 10-fold, from 2.94 to 34.07%, and most of the filaments were large and fully coated circles. The size of the circles was consistent with the presence of DrRecA protein on the added ΦX174 ssDNA. In the reaction using homologous DNA with SSB added later (reaction 6), circular filaments increased from 9.4% (most of which were small circles) to 38.9% of mostly large and fully coated circles. The ∼4-fold increase was most likely an underestimate as the reaction with homologous DNA and SSB formed many large aggregates that we suspect to be intermediates of recombination. Thus, the addition of DrSSB facilitates a transfer of significant amounts of the DrRecA protein from dsDNA to ssDNA. Quantification of filaments from reactions 5 and 6 were calculated from counting 1183 and 666 total filaments, respectively.
We further investigated the DrRecA transfer facilitated by DrSSB by doing careful counts of DrRecA-DNA filaments at time points 0, 5, 10, and 20 min after DrSSB addition (Fig. 9). DrRecA was incubated with linear dsDNA and ATP for 15 min before circular ssDNA was added. DrSSB was added after 5 min to begin time point collection. All time points were fixed with ATPγS before EM spreading. When homologous DNA substrates were used, we observed large filament aggregates that could not be accurately resolved (data not shown). We suspect that these aggregates are joint molecules formed from homologous recombination of the DNA substrates by DrRecA. Therefore, the counts in Fig. 9 reflect the use of heterologous linear dsDNA and circular ssDNA to allow for clear detection of DrRecA filaments. Between 448 and 719 filaments were counted for each time point. The bar graph in Fig. 9A shows the decrease in linear DrRecA-dsDNA filaments and an increase in big circular DrRecA-ssDNA filaments over time. The representative EM images in Fig. 8 show additional evidence that there may be a transfer of DrRecA molecules from dsDNA to ssDNA, stimulated by DrSSB. As for Fig. 8, the lengths of a significant subset of molecules in each category are presented in Fig. 9B to support the judgments.
FIGURE 9.
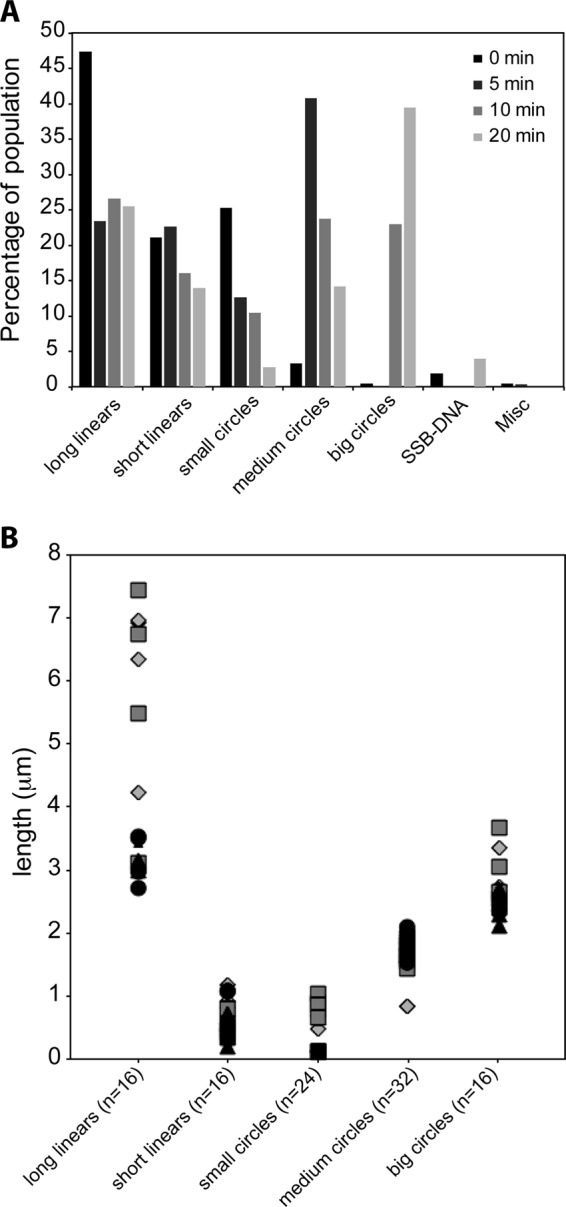
Abundance of DrRecA-cssDNA filaments increases with time in the presence of DrSSB. DrRecA was incubated with M13mp18 ldsDNA and ATP for 15 min before ΦX174 cssDNA was added for 5 min followed by DrSSB. The reaction was stopped at 0, 5, 10, and 20 min after the addition of DrSSB, fixed with ATPγS, and spread on Alcian grids. A, bar graph shows the decrease in long linear filaments with time, and increase in circular filaments, especially big circular filaments. B, at least four individual molecules were measured in each class of DrRecA filament in eight samples (duplicates of each time point). Diamonds represent molecules from 0-min samples, squares from 5-min samples, triangles from 10-min samples, and circles from 20-min samples. Long linear molecules range between 2.7 and 7.4 μm and short linear range between 0.2 and 1.1 μm. Small circles range between 0.1 and 1.3 μm; medium circles are between 0.8 and 2.1 μm, and large circles typically range between 2.1 and 3.7 μm. No small circles with small gaps were observed at the 10- and 20-min time point samples.
SSB Is Crucial for Strand Exchange by DrRecA
To investigate the role of DrSSB in strand exchange, we carried out in vitro three-strand exchange experiments with EcRecA or DrRecA, in each case comparing the reaction seen with the cognate SSB protein to a reaction without SSB (Fig. 10). The EcRecA protein reactions were carried out at 37 °C. As seen in past work (78, 79), EcSSB is important and stimulates the reaction. However, it is not essential to observe significant strand exchange, as seen in Fig. 10A. Substantial amounts of strand exchange intermediates are formed even without EcSSB, and some of these are converted to products. In contrast, the DrRecA reaction without DrSSB exhibited no detectable joint molecules (intermediates) or product formation (Fig. 10B). The reaction with DrSSB began forming product after 45 min and achieved ∼90% product formation after 3 h. The inability of DrRecA to catalyze recombination at any level without the help of DrSSB is similar to data reported for Streptococcus pneumoniae RecA and SSB and Saccharomyces cerevisiae Rad51 and the eukaryotic single-stranded DNA-binding protein, RPA (80–83).
FIGURE 10.
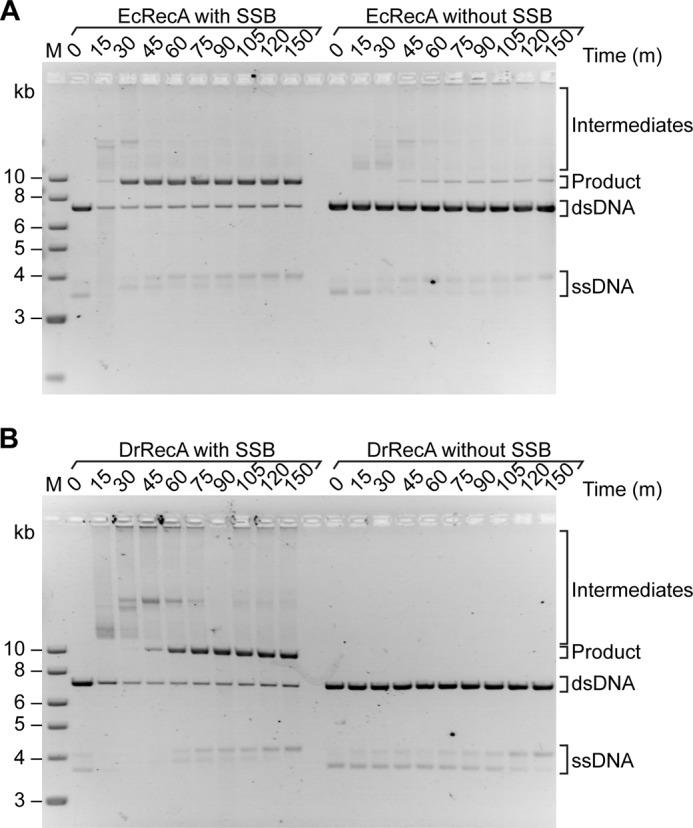
DrSSB is required for DNA pairing and strand exchange promoted by DrRecA protein. A, EcRecA; B, DrRecA was added to 10 μm M13mp18 cssDNA for 10 min (30 °C for DrRecA reaction and 37 °C for EcRecA reaction) followed by the addition of ATP for 10 min. M13mp18 ldsDNA (20 μm) and 1 μm of the corresponding cognate SSB protein were added to the two reactions on the left (with SSB, as indicated). Only the M13mp18 ldsDNA, and no SSB protein, was added to the reactions on the right (without SSB as indicated). Aliquots were removed at the indicated time points and run on a 0.8% agarose gel. Note that in these gels, the M13mp18 ssDNA tends to run as a doublet. The change in proportion of the two bands during some reactions does not necessarily reflect conversion of the circular species to duplex and build up of displaced linear ssDNA. As documented in earlier studies (31, 100–102), the secondary structure-rich M13-derived ssDNAs can take on at least two conformational arrangements that affect gel migration. Doublets routinely appear in gels even though more than 90–95% of the ssDNA is present as circles by EM examination (31, 98–100). The equilibrium between species can be affected by temperature and interaction with proteins such as DrRecA and DrSSB. The doublet is also evident in Fig. 12.
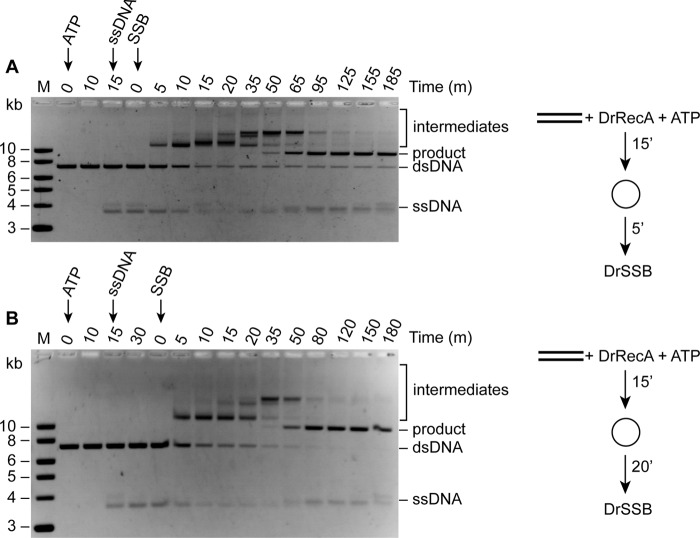
To further show that DrSSB is crucial for the initiation of strand exchange, we varied the time when DrSSB was added following the last component to the reaction (Fig. 11). DrRecA was added to M13mp18 ldsDNA first, followed by ATP for 10 min. Homologous cssDNA was then added to the reaction and incubated for either 5 or 20 min before DrSSB was added (Figs. 11, A and B, respectively). Intermediates were absent as long as the DrSSB was not present. Intermediates were observed almost immediately after the addition of DrSSB in all cases, and product formation began ∼35 min after the addition of DrSSB. These data again show that the presence of DrSSB is absolutely required for strand exchange catalyzed by DrRecA.
FIGURE 11.
DrSSB is required for DNA pairing and strand exchange promoted by DrRecA protein; time of addition experiments. DrRecA (2.4 μm) was incubated with 8 μm of M13mp18 ldsDNA for 5 min at 30 °C, followed by the addition of ATP as indicated in time point 0. The homologous cssDNA (4 μm) was added 15 min later. DrSSB (0.4 μm) was added either 5 min (A) or 20 min (B) later. The addition of DrSSB resets time points indicated in the gels above back to 0 to show that product formation began after 35 min in both reactions, although SSB was added 15 min later in the bottom reaction.
The concentration of DrSSB can also significantly influence the amount of recombined product generated by DrRecA (Fig. 12). In these strand exchange assays, DrRecA was again incubated with M13mp18 ldsDNA for 5 min before ATP was added. Homologous cssDNA was added 10 min later, followed by addition of DrSSB at varying concentrations, 0.1–2.0 μm. Aliquots were removed at time points 0–120 min after the addition of DrSSB and run on a 0.8% agarose gel. The rate of product formation catalyzed by DrRecA increased with increasing concentrations of DrSSB. In the presence of 0.1 μm DrSSB, minimal product began to appear after 90 min. At DrSSB concentrations of 1.0 μm or higher, product appeared in 45 min, and the reactions went nearly to completion. Overall, the evidence we have provided shows that not only is the SSB protein important for initiation of recombination in Deinococcus, it is also important for the kinetics of the recombination reaction promoted by the RecA protein. This could explain why higher concentrations of DrSSB may have a strong stimulatory effect on DrRecA ATPase activity and could play a role in stimulating DNA pairing and strand switching.
FIGURE 12.
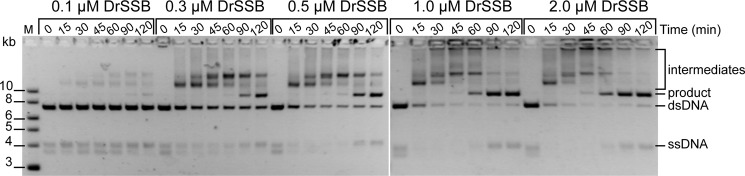
Product formation kinetics by DrRecA is dependent on DrSSB concentration. DrRecA was incubated with ldsDNA and ATP for 10 min before the addition of homologous cssDNA. The reaction was incubated for 5 min before 0.1, 0.3, 0.5, 1.0, or 2.0 μm DrSSB was added. Aliquots were removed at indicated time points and run on 0.8% agarose gel.
DISCUSSION
The work described in this study reveals a new regulatory mechanism at work with the DrRecA protein that has not previously been associated with any RecA protein. Relatively subtle changes in nucleoprotein filament state have been documented with EcRecA. However, the changes seen with the DrRecA nucleoprotein filaments are exaggerated and include an inactive state with respect to ATP hydrolysis that provides a potential new point of regulation. In particular, DrRecA filaments do not hydrolyze ATP when simply bound to dsDNA at pH 7.5 and 30 °C. Activation of the DrRecA is observed when ssDNA is added, a ligand that would be rendered abundant in the event of extreme desiccation or exposure to high levels of ionizing radiation. DrSSB is also absolutely required for DrRecA activity in DNA strand exchange, a tighter requirement than is seen for reactions promoted by the EcRecA protein.
The properties of these proteins are best considered in the context of the ecophysiology of the host organisms. E. coli spends most of its time in the lower intestine of warm-blooded organisms. The protein is normally unbound to DNA. Normally, EcRecA acts only when a stretch of single-stranded DNA is suddenly exposed by a stalled replication fork, the repair of which constitutes its primary biological function (84–87). The properties of DrRecA appear to reflect a much different approach to recombinational DNA repair. DrRecA is needed for genome reconstitution after desiccation (2, 3). DrRecA, along with several other proteins, are induced to high levels immediately after and probably during desiccation (the proteins are already present at high levels immediately after re-hydration) (58). The presence of an inactive form of DrRecA during desiccation, perhaps bound to dsDNA, could facilitate rapid response to improved growth conditions. Our working model for this re-activation is presented in Fig. 13.
FIGURE 13.
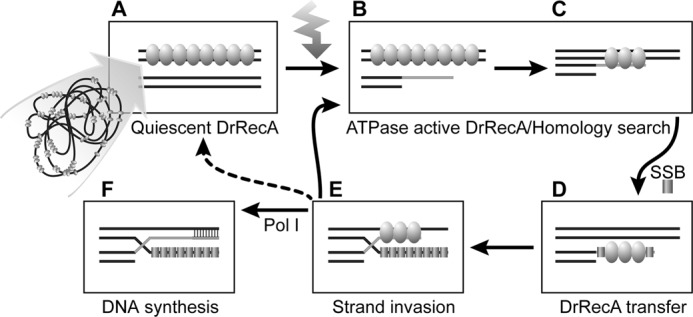
Proposed model of DrRecA mechanism in the cell. A, double-stranded genomic DNA is bound by DrRecA in a quiescent state. B, DNA damage, such as ionizing radiation, introduces ssDNA in the cell that activates the ATPase activity of DrRecA. C, ATP hydrolysis provides the energy for DNA homology search. D, presence of SSB triggers a transfer of DrRecA from dsDNA to ssDNA; E, initiates strand invasion and D-loop formation by DrRecA. F, DNA is synthesized by DNA polymerase I to reconstitute dsDNA. If additional ssDNA is present, the recombination cycle can start over again.
There are four main observations that contribute to this scheme. First, DrRecA can form nucleoprotein filaments at 30 °C on dsDNA, and ssDNA with DrSSB, while hydrolyzing little to no ATP. To our knowledge, this is the first report of an ATPase-inactive state of a bacterial recombinase bound to DNA. Second, the dsDNA-DrRecA filaments hydrolyze little or no ATP until they are activated by interaction with new ssDNA, with or without SSB. In effect, ATP is not hydrolyzed until all of the substrates needed for a possible DNA strand exchange are present. Third, DrSSB is a crucial component in the homologous recombination activity of DrRecA. Unlike the reactions characterized to date with other bacterial RecA proteins, except for RecA from Streptococcus (83), no detectable intermediates or products of DNA strand exchange are observed in the absence of SSB. Fourth and finally, SSB appears to trigger the transfer of at least some of the DrRecA protein from dsDNA to ssDNA. It is possible that DNA strand exchange is initiated from either dsDNA or ssDNA under these reaction conditions. We do not know if multiple transfers of DrRecA (perhaps back and forth between ssDNA and dsDNA) occur before strand exchange is initiated. In general, recombination carried out by DrRecA is tightly controlled. RecA functions in D. radiodurans can only be activated by the presence of specific ligands, such as ssDNA or the SSB protein. The lack of ATP hydrolysis by DrRecA until appropriate DNA substrates and cofactors are available may help to conserve resources in cells that have undergone extreme levels of oxidative damage (88). The more promiscuous hydrolysis of ATP seen with the EcRecA protein may be unsuitable for a bacterium that spends much of its time in a desiccated, metabolically compromised state. Eliminating DrRecA protein (and perhaps other ATP-requiring DNA repair proteins) when desiccation commences is also not a productive survival strategy, because these proteins are required when environmental conditions improve.
Activation of ATP hydrolysis can also be effected by certain changes in reaction conditions, including a decrease in pH or the addition of volume exclusion agents. This is likely to reflect a complex interplay of mobile amino acid residues and peptide segments that mediate the change in filament states, particularly at the subunit-subunit interface. The active site for ATP hydrolysis is at the interface between filament subunits, and amino acid residues from both flanking subunits cooperate in the hydrolytic reaction (26, 27). Mutational changes in key active site resides produce large effects on EcRecA activities (26, 27). We speculate that the transition between active and inactive states in DrRecA similarly reflects changes in the position and ionization state of ATPase active site residues, although the changes are exaggerated in the case of the DrRecA such that they can bring about an inactive state.
The sharp pH-dependent changes in ATPase activity documented in Fig. 6A suggest either multiple ionizable groups that can affect the activity of DrRecA or (more likely) a highly cooperative pH-dependent activation effect. Whereas the ionizations that affect activity exhibit apparent pKa values near neutrality, there is only one His residue in DrRecA protein. One or more ionizable amino acid side chains may thus be present in an environment where their pKa values are substantially perturbed. It is tempting to speculate that some of these groups are included in the more positively charged inner surface of the DrRecA filament that was documented by Rajan and Bell (76). At this surface, charge interactions with single-stranded DNA might bring about activating conformational changes at pH values above 7.5 that are brought about by group ionization below pH 7.5.
The changes in filament state are likely to be more complex than a simple change from one RecA protein conformation to another. RecA is part of a larger family of related ATPases that includes a range of helicases and the F1-ATPase (89, 90). Multiple distinct conformational changes are typically associated with the ATP hydrolytic cycle (91–99). In DrRecA, the filament undergoes a change from an inactive state to a state in which the ATP hydrolytic cycle is activated.
Our working model to describe how the RecA protein may play a role in the radiation and desiccation resistance of D. radiodurans is outlined in Fig. 13. In the cell, DrRecA would be bound to the double-stranded genomic DNA or DNA fragments in an ATPase inactive state, or quiescent state, symbolized by the oval molecules (Fig. 13A). In this state, DrRecA might be stored directly on the DNA without hydrolyzing ATP and depleting resources. Some of the DrRecA might also be stored in some undetermined form outside the nucleoid, occasionally binding to genomic dsDNA. Upon re-hydration, the appearance of some ATP in the environment would render DrRecA function possible. The presence of significant amounts of ssDNA would signal to the cell that genomic DNA damage had occurred and DrRecA ATPase activity would be activated (Fig. 13, B and C). The presence of ssDNA would also lead to binding of the ssDNA by DrSSB. This in turn would lead to transfer of DrRecA from random cellular locations to regions of ssDNA where repair would be required (Fig. 13D). DrSSB protein expression is induced upon recovery from irradiation (53). It is unclear whether or not the transfer is a step required for strand exchange, although both RecA transfer and strand exchange required the presence of SSB. Interestingly, transfer of RecA to SSB-bound ssDNA might allow RecA strand exchange to initiate on the ssDNA and proceed in the direction considered normal for other related recombinases, or a subsequent transfer back to dsDNA might allow initiation from that DNA substrate. Additionally, SSB acts as a signal to initiate strand exchange and push the reaction to strand invasion and D-loop formation (Fig. 13E). This in turn would prime DNA synthesis by DNA polymerase to reconstitute the missing genomic information (Fig. 13F). When bound to dsDNA, RecA will remain ATPase-inactive. The presence of additional ssDNA in the cell would stimulate ATP hydrolysis by RecA, and the cycle can continue. In the cell, this “DNA-hopping” mechanism could play an important role in localizing the DrRecA protein specifically to sites where genomic DNA insults have occurred and require repair. In addition, having multiple levels of regulation to permit proper activation of RecA, such as requiring the presence of both ssDNA and SSB, indicates a more constrained RecA protein function in D. radiodurans. In vivo, strict regulation of DrRecA may be important to prevent unwanted mutagenesis and illegitimate recombination that would compromise genomic stability. It is possible that DrRecA may be adapted for an environment in which cells are recovering from desiccation or heavy doses of ionizing radiation, where energy-yielding metabolism may not yet be fully restored. ATPase-inactive RecA that is quickly localized to potential sites of damage when required would provide the most effective use of the recombinase.
DrSSB plays a pivotal role in the initiation and completion of the strand exchange reaction catalyzed by DrRecA. In E. coli, SSB added after RecA stabilizes the strand exchange reaction by removing secondary structures on the initial ssDNA for RecA to make a continuous filament and also by binding to the displaced ssDNA after strand switching. Although SSB is important, it is not essential for homologous recombination to go to completion in E. coli. In D. radiodurans, RecA is completely unable to initiate strand exchange in the absence of DrSSB. The inability of DrRecA to catalyze recombination without DrSSB is similar to properties reported for S. pneumoniae RecA and SSB and S. cerevisiae Rad51 and the eukaryotic single-stranded DNA-binding protein RPA (80, 82, 83). We speculate that the sequestration of the displaced ssDNA may be a critical function of DrSSB, with displacement energetically constrained when DNA strand exchange is attempted by DrRecA alone. In this scenario, detectable strand exchange will be driven forward only when the DrSSB is present to bind and sequester the displaced ssDNA strand. The coupling of DNA strand exchange and ssDNA sequestration could help ensure that genetic recombination occurs only when sufficient DrSSB is available to bind the displaced ssDNA, and it provides another mechanism of regulation by modulation of DrSSB availability. This scheme may also help preserve essential genomic resources, ensuring protection of displaced single strands by DrSSB as they are created.
Acknowledgments
We acknowledge helpful discussions with John R. Battista that contributed to the interpretation of the results in this paper. We also acknowledge with gratitude the assistance of D. B. Knowles and M. Thomas Record in the analysis and interpretation of the pH-rate profile data in Fig. 6A.
This work was supported, in whole or in part, by National Institutes of Health Grant GM32335 from the NIGMS (to M. M. C.). This work was also supported by an Advanced Opportunity Fellowship from the University of Wisconsin College of Agricultural and Life Sciences and a Genentech predoctoral fellowship (both to K. V. N.). Michael M. Cox is a board member and shareholder of Recombitech, Inc., which is applying recombinational DNA repair to problems in medical diagnostics. His relationship to the company is managed by the University of Wisconsin-Madison in accordance with its conflict of interest policies.
D. B. Knowles, personal communication.
- ssDNA
- single-stranded DNA
- ATPγS
- adenosine 5-O-(3-thiotriphosphate)
- SSB
- single-stranded DNA-binding protein
- cssDNA
- circular single-stranded DNA
- dsDNA
- double-stranded DNA
- ldsDNA
- linear double-stranded DNA.
REFERENCES
- 1. Cox M. M., Battista J. R. (2005) Deinococcus radiodurans–The consummate survivor. Nat. Rev. Microbiol. 3, 882–892 [DOI] [PubMed] [Google Scholar]
- 2. Mattimore V., Battista J. R. (1996) Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178, 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rainey F. A., Ray K., Ferreira M., Gatz B. Z., Nobre M. F., Bagaley D., Rash B. A., Park M. J., Earl A. M., Shank N. C., Small A. M., Henk M. C., Battista J. R., Kämpfer P., da Costa M. S. (2005) Extensive diversity of ionizing radiation-resistant bacteria recovered from Sonoran desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl. Environ. Microbiol. 71, 5225–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zahradka K., Slade D., Bailone A., Sommer S., Averbeck D., Petranovic M., Lindner A. B., Radman M. (2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443, 569–573 [DOI] [PubMed] [Google Scholar]
- 5. Minsky A. (2003) Structural aspects of DNA repair: the role of restricted diffusion. Mol. Microbiol. 50, 367–376 [DOI] [PubMed] [Google Scholar]
- 6. Daly M. J., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Zhai M., Leapman R. D., Lai B., Ravel B., Li S. M., Kemner K. M., Fredrickson J. K. (2007) Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 5, e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fredrickson J. K., Li S. M., Gaidamakova E. K., Matrosova V. Y., Zhai M., Sulloway H. M., Scholten J. C., Brown M. G., Balkwill D. L., Daly M. J. (2008) Protein oxidation: key to bacterial desiccation resistance? ISME J. 2, 393–403 [DOI] [PubMed] [Google Scholar]
- 8. Frenkiel-Krispin D., Minsky A. (2006) Nucleoid organization and the maintenance of DNA integrity in E. coli, B. subtilis, and D. radiodurans. J. Struct. Biol. 156, 311–319 [DOI] [PubMed] [Google Scholar]
- 9. Slade D., Lindner A. B., Paul G., Radman M. (2009) Recombination and replication in DNA repair of heavily irradiated Deinococcus radiodurans. Cell 136, 1044–1055 [DOI] [PubMed] [Google Scholar]
- 10. Levin-Zaidman S., Englander J., Shimoni E., Sharma A. K., Minton K. W., Minsky A. (2003) Ringlike structure of the Deinococcus radiodurans genome: A key to radioresistance? Science 299, 254–256 [DOI] [PubMed] [Google Scholar]
- 11. Daly M. J., Minton K. W. (1996) An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 178, 4461–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minton K. W. (1996) Repair of ionizing radiation damage in the radiation-resistant bacterium Deinococcus radiodurans. Mutat. Res. 363, 1–7 [DOI] [PubMed] [Google Scholar]
- 13. Earl A. M., Mohundro M. M., Mian I. S., Battista J. R. (2002) The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J. Bacteriol. 184, 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox M. M. (2004) in The Bacterial Chromosome (Higgins N. P., ed) pp 369–388, American Society of Microbiology, Washington, D. C [Google Scholar]
- 15. Cox M. M. (2007) in Topics in Current Genetics: Molecular Genetics of Recombination (Rothstein R., Aguilera A., eds) pp. 53–94, Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 16. Cox M. M. (2007) Regulation of bacterial RecA function. Crit. Rev. Biochem. Mol. Biol. 42, 41–63 [DOI] [PubMed] [Google Scholar]
- 17. Lusetti S. L., Cox M. M. (2002) The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71, 71–100 [DOI] [PubMed] [Google Scholar]
- 18. McGrew D. A., Knight K. L. (2003) Molecular design and functional organization of the RecA protein. Crit. Rev. Biochem. Mol. Biol. 38, 385–432 [DOI] [PubMed] [Google Scholar]
- 19. Cox M. M. (2007) Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 8, 127–138 [DOI] [PubMed] [Google Scholar]
- 20. Brenner S. L., Mitchell R. S., Morrical S. W., Neuendorf S. K., Schutte B. C., Cox M. M. (1987) RecA protein-promoted ATP hydrolysis occurs throughout RecA nucleoprotein filaments. J. Biol. Chem. 262, 4011–4016 [PubMed] [Google Scholar]
- 21. Kowalczykowski S. C., Clow J., Krupp R. A. (1987) Properties of the duplex DNA-dependent ATPase activity of Escherichia coli RecA protein and its role in branch migration. Proc. Natl. Acad. Sci. U.S.A. 84, 3127–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morimatsu K., Kowalczykowski S. C. (2003) RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol. Cell 11, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 23. Arenson T. A., Tsodikov O. V., Cox M. M. (1999) Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J. Mol. Biol. 288, 391–401 [DOI] [PubMed] [Google Scholar]
- 24. Baitin D. M., Bakhlanova I. V., Chervyakova D. V., Kil Y. V., Lanzov V. A., Cox M. M. (2008) Two RecA protein types that mediate different modes of hyperrecombination. J. Bacteriol. 190, 3036–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bedale W. A., Cox M. (1996) Evidence for the coupling of ATP hydrolysis to the final (extension) phase of RecA protein-mediated DNA strand exchange. J. Biol. Chem. 271, 5725–5732 [DOI] [PubMed] [Google Scholar]
- 26. Cox J. M., Abbott S. N., Chitteni-Pattu S., Inman R. B., Cox M. M. (2006) Complementation of one RecA protein point mutation by another–Evidence for trans catalysis of ATP hydrolysis. J. Biol. Chem. 281, 12968–12975 [DOI] [PubMed] [Google Scholar]
- 27. Cox J. M., Li H., Wood E. A., Chitteni-Pattu S., Inman R. B., Cox M. M. (2008) Defective dissociation of a “slow” RecA mutant protein imparts an Escherichia coli growth defect. J. Biol. Chem. 283, 24909–24921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cox J. M., Tsodikov O. V., Cox M. M. (2005) Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 3, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sakai A., Cox M. M. (2009) RecFOR and RecOR as distinct RecA loading pathways. J. Biol. Chem. 284, 3264–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haruta N., Yu X., Yang S., Egelman E. H., Cox M. M. (2003) A DNA pairing-enhanced conformation of bacterial RecA proteins. J. Biol. Chem. 278, 52710–52723 [DOI] [PubMed] [Google Scholar]
- 31. Schutte B. C., Cox M. M. (1987) Homology-dependent changes in adenosine 5′-triphosphate hydrolysis during RecA protein promoted DNA strand exchange: evidence for long paranemic complexes. Biochemistry 26, 5616–5625 [DOI] [PubMed] [Google Scholar]
- 32. Bork J. M., Cox M. M., Inman R. B. (2001) RecA protein filaments disassemble in the 5′ to 3′ direction on single-stranded DNA. J. Biol. Chem. 276, 45740–45743 [DOI] [PubMed] [Google Scholar]
- 33. Lindsley J. E., Cox M. M. (1990) Assembly and disassembly of RecA protein filaments occurs at opposite filament ends: relationship to DNA strand exchange. J. Biol. Chem. 265, 9043–9054 [PubMed] [Google Scholar]
- 34. Shan Q., Cox M. M. (1997) RecA filament dynamics during DNA strand exchange reactions. J. Biol. Chem. 272, 11063–11073 [DOI] [PubMed] [Google Scholar]
- 35. Kowalczykowski S. C., Krupp R. A. (1995) DNA-strand exchange promoted by RecA protein in the absence of ATP: implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc. Natl. Acad. Sci. U.S.A. 92, 3478–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Menetski J. P., Bear D. G., Kowalczykowski S. C. (1990) Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 87, 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rehrauer W. M., Kowalczykowski S. C. (1993) Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli RecA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 268, 1292–1297 [PubMed] [Google Scholar]
- 38. Cox M. M., Lehman I. R. (1981) RecA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc. Natl. Acad. Sci. U.S.A. 78, 3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shan Q., Cox M. M., Inman R. B. (1996) DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J. Biol. Chem. 271, 5712–5724 [DOI] [PubMed] [Google Scholar]
- 40. Michel B. (2005) After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 3, e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fonville N. C., Bates D., Hastings P. J., Hanawalt P. C., Rosenberg S. M. (2010) Role of RecA and the SOS response in thymineless death in Escherichia coli. PLoS Genet. 6, e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eggler A. L., Lusetti S. L., Cox M. M. (2003) The C terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J. Biol. Chem. 278, 16389–16396 [DOI] [PubMed] [Google Scholar]
- 43. Lusetti S. L., Wood E. A., Fleming C. D., Modica M. J., Korth J., Abbott L., Dwyer D. W., Roca A. I., Inman R. B., Cox M. M. (2003) C-terminal deletions of the Escherichia coli RecA protein–Characterization of in vivo and in vitro effects. J. Biol. Chem. 278, 16372–16380 [DOI] [PubMed] [Google Scholar]
- 44. Lusetti S. L., Shaw J. J., Cox M. M. (2003) Magnesium ion-dependent activation of the RecA protein involves the C terminus. J. Biol. Chem. 278, 16381–16388 [DOI] [PubMed] [Google Scholar]
- 45. Shan Q., Cox M. M. (1996) RecA protein dynamics in the interior of RecA nucleoprotein filaments. J. Mol. Biol. 257, 756–774 [DOI] [PubMed] [Google Scholar]
- 46. Kim J. I., Sharma A. K., Abbott S. N., Wood E. A., Dwyer D. W., Jambura A., Minton K. W., Inman R. B., Daly M. J., Cox M. M. (2002) RecA protein from the extremely radioresistant bacterium Deinococcus radiodurans: Expression, purification, and characterization. J. Bacteriol. 184, 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim J.-I., Cox M. M. (2002) The RecA proteins of Deinococcus radiodurans and Escherichia coli promote DNA strand exchange via inverse pathways. Proc. Natl. Acad. Sci. U.S.A. 99, 7917–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zaitsev E. N., Kowalczykowski S. C. (2000) A novel pairing process promoted by Escherichia coli RecA protein: inverse DNA and RNA strand exchange. Genes Dev. 14, 740–749 [PMC free article] [PubMed] [Google Scholar]
- 49. Hsu H.-F., Ngo K. V., Chitteni-Pattu S., Cox M. M., Li H.-W. (2011) Investigating Deinococcus radiodurans RecA protein filament formation on double-stranded DNA by a real-time single-molecule approach. Biochemistry 50, 8270–8280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bernstein D. A., Eggington J. M., Killoran M. P., Misic A. M., Cox M. M., Keck J. L. (2004) Crystal structure of the D. radiodurans single-stranded DNA-binding protein suggests a novel mechanism for coping with DNA damage. Proc. Natl. Acad. Sci. U.S.A. 101, 8575–8580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eggington J. M., Haruta N., Wood E. A., Cox M. M. (2004) The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kozlov A. G., Eggington J. M., Cox M. M., Lohman T. M. (2010) Binding of the Dimeric Deinococcus radiodurans single-stranded DNA binding protein to single-stranded DNA. Biochemistry 49, 8266–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. George N. P., Ngo K. V., Chitteni-Pattu S., Norais C. A., Battista J. R., Cox M. M., Keck J. L. (2012) Structure and cellular dynamics of Deinococcus radiodurans single-stranded DNA (ssDNA)-binding protein (SSB)-DNA complexes. J. Biol. Chem. 287, 22123–22132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Petrova V., Chitteni-Pattu S., Drees J. C., Inman R. B., Cox M. M. (2009) An SOS inhibitor that binds to free RecA protein: The PsiB protein. Mol. Cell 36, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lohman T. M., Green J. M., Beyer R. S. (1986) Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λ PL control. Biochemistry 25, 21–25 [DOI] [PubMed] [Google Scholar]
- 56. Lindsley J. E., Cox M. M. (1989) Dissociation pathway for RecA nucleoprotein filaments formed on linear duplex DNA. J. Mol. Biol. 205, 695–711 [DOI] [PubMed] [Google Scholar]
- 57. Morrical S. W., Lee J., Cox M. M. (1986) Continuous association of Escherichia coli single-stranded DNA-binding protein with stable complexes of RecA protein and single-stranded DNA. Biochemistry 25, 1482–1494 [DOI] [PubMed] [Google Scholar]
- 58. Tanaka M., Earl A. M., Howell H. A., Park M. J., Eisen J. A., Peterson S. N., Battista J. R. (2004) Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics 168, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Harris D. R., Tanaka M., Saveliev S. V., Jolivet E., Earl A. M., Cox M. M., Battista J. R. (2004) Preserving genome integrity: The DdrA protein of Deinococcus radiodurans R1. PLoS Biol. 2, e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Daly M. J., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Zhai M., Venkateswaran A., Hess M., Omelchenko M. V., Kostandarithes H. M., Makarova K. S., Wackett L. P., Fredrickson J. K., Ghosal D. (2004) Accumulation of Mn(II) in Deinococcus radiodurans facilitates γ-radiation resistance. Science 306, 1025–1028 [DOI] [PubMed] [Google Scholar]
- 61. Liu Y., Zhou J., Omelchenko M. V., Beliaev A. S., Venkateswaran A., Stair J., Wu L., Thompson D. K., Xu D., Rogozin I. B., Gaidamakova E. K., Zhai M., Makarova K. S., Koonin E. V., Daly M. J. (2003) Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 100, 4191–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Carroll J. D., Daly M. J., Minton K. W. (1996) Expression of recA in Deinococcus radiodurans. J. Bacteriol. 178, 130–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nordén B., Elvingson C., Kubista M., Sjöberg B., Ryberg H., Ryberg M., Mortensen K., Takahashi M. (1992) Structure of RecA-DNA complexes studied by combination of linear dichroism and small-angle neutron scattering measurements on flow-oriented samples. J. Mol. Biol. 226, 1175–1191 [DOI] [PubMed] [Google Scholar]
- 64. Register J. C., 3rd, Griffith J. (1986) RecA protein filaments can juxtapose DNA ends: an activity that may reflect a function in DNA repair. Proc. Natl. Acad. Sci. U.S.A. 83, 624–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kowalczykowski S. C., Krupp R. A. (1987) Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J. Mol. Biol. 193, 97–113 [DOI] [PubMed] [Google Scholar]
- 66. Shan Q., Bork J. M., Webb B. L., Inman R. B., Cox M. M. (1997) RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 265, 519–540 [DOI] [PubMed] [Google Scholar]
- 67. Egelman E. H., Stasiak A. (1986) Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-γ-S or ATP. J. Mol. Biol. 191, 677–697 [DOI] [PubMed] [Google Scholar]
- 68. Egelman E. H., Stasiak A. (1993) Electron microscopy of RecA-DNA complexes: two different states, their functional significance and relation to the solved crystal structure. Micron 24, 309–324 [Google Scholar]
- 69. Drees J. C., Lusetti S. L., Cox M. M. (2004) Inhibition of RecA protein by the Escherichia coli RecX protein–modulation by the RecA C terminus and filament functional state. J. Biol. Chem. 279, 52991–52997 [DOI] [PubMed] [Google Scholar]
- 70. Gruenig M. C., Renzette N., Long E., Chitteni-Pattu S., Inman R. B., Cox M. M., Sandler S. J. (2008) RecA-mediated SOS induction requires an extended filament conformation but no ATP hydrolysis. Mol. Microbiol. 69, 1165–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Knowles D. B., LaCroix A. S., Deines N. F., Shkel I., Record M. T. (2011) Separation of preferential interaction and excluded volume effects on DNA duplex and hairpin stability. Proc. Natl. Acad. Sci. U.S.A. 108, 12699–12704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Di Capua E., Engel A., Stasiak A., Koller T. (1982) Characterization of complexes between RecA protein and duplex DNA by electron microscopy. J. Mol. Biol. 157, 87–103 [DOI] [PubMed] [Google Scholar]
- 73. Sattin B. D., Goh M. C. (2004) Direct observation of the assembly of RecA/DNA complexes by atomic force microscopy. Biophys. J. 87, 3430–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bevilacqua P. C. (2003) Mechanistic considerations for general acid-base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry 42, 2259–2265 [DOI] [PubMed] [Google Scholar]
- 75. Segel I. H. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, pp. 884–925, John Wiley & Sons Inc., Hoboken, NJ [Google Scholar]
- 76. Rajan R., Bell C. E. (2004) Crystal structure of RecA from Deinococcus radiodurans: Insights into the structural basis of extreme radioresistance. J. Mol. Biol. 344, 951–963 [DOI] [PubMed] [Google Scholar]
- 77. Pugh B. F., Cox M. M. (1988) General mechanism for RecA protein binding to duplex DNA. J. Mol. Biol. 203, 479–493 [DOI] [PubMed] [Google Scholar]
- 78. Lavery P. E., Kowalczykowski S. C. (1992) A postsynaptic role for single-stranded DNA-binding protein in recA protein-promoted DNA strand exchange. J. Biol. Chem. 267, 9315–9320 [PubMed] [Google Scholar]
- 79. Kowalczykowski S. C., Dixon D. A., Eggleston A. K., Lauder S. D., Rehrauer W. M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58, 401–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Soustelle C., Vedel M., Kolodner R., Nicolas A. (2002) Replication protein A is required for meiotic recombination in Saccharomyces cerevisiae. Genetics 161, 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Eggler A. L., Inman R. B., Cox M. M. (2002) The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J. Biol. Chem. 277, 39280–39288 [DOI] [PubMed] [Google Scholar]
- 82. Sugiyama T., Zaitseva E. M., Kowalczykowski S. C. (1997) A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 272, 7940–7945 [DOI] [PubMed] [Google Scholar]
- 83. Steffen S. E., Katz F. S., Bryant F. R. (2002) Complete inhibition of Streptococcus pneumoniae RecA protein-catalyzed ATP hydrolysis by SSB protein: Implications for the mechanism of SSB protein-stimulated DNA strand exchange. J. Biol. Chem. 277, 14493–14500 [DOI] [PubMed] [Google Scholar]
- 84. Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., Marians K. J. (2000) The importance of repairing stalled replication forks. Nature 404, 37–41 [DOI] [PubMed] [Google Scholar]
- 85. Kowalczykowski S. C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25, 156–165 [DOI] [PubMed] [Google Scholar]
- 86. Kuzminov A. (2001) DNA replication meets genetic exchange: Chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8461–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cox M. M. (2001) Historical overview: Searching for replication help in all of the rec places. Proc. Natl. Acad. Sci. U.S.A. 98, 8173–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Daly M. J., Gaidamakova E. K., Matrosova V. Y., Kiang J. G., Fukumoto R., Lee D. Y., Wehr N. B., Viteri G. A., Berlett B. S., Levine R. L. (2010) Small molecule antioxidant proteome–Shields in Deinococcus radiodurans. PLoS One 9, e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Iyer L. M., Leipe D. D., Koonin E. V., Aravind L. (2004) Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31 [DOI] [PubMed] [Google Scholar]
- 90. Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 [DOI] [PubMed] [Google Scholar]
- 91. Gai D., Zhao R., Li D., Finkielstein C. V., Chen X. S. (2004) Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, 47–60 [DOI] [PubMed] [Google Scholar]
- 92. Moore K. J., Lohman T. M. (1994) Kinetic mechanism of adenine nucleotide binding to and hydrolysis by the Escherichia coli Rep monomer. 1. Use of fluorescent nucleotide analogues. Biochemistry 33, 14550–14564 [DOI] [PubMed] [Google Scholar]
- 93. Dittrich M., Yu J., Schulten K. (2007) in Atomistic Approaches in Modern Biology: From Quantum Chemistry to Molecular Simulations (Reiher M., ed) pp. 319–347, Springer-Verlag, Berlin and Heidelberg [Google Scholar]
- 94. del Toro Duany Y., Klostermeier D. (2011) Nucleotide-driven conformational changes in the reverse gyrase helicase-like domain couple the nucleotide cycle to DNA processing. Phys. Chem. Chem. Phys. 13, 10009–10019 [DOI] [PubMed] [Google Scholar]
- 95. Gu M., Rice C. M. (2010) Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. U.S.A. 107, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hsieh J., Moore K. J., Lohman T. M. (1999) A two-site kinetic mechanism for ATP binding and hydrolysis by E. coli Rep helicase dimer bound to a single-stranded oligodeoxynucleotide. J. Mol. Biol. 288, 255–274 [DOI] [PubMed] [Google Scholar]
- 97. Lewis R., Dürr H., Hopfner K. P., Michaelis J. (2008) Conformational changes of a Swi2/Snf2 ATPase during its mechano-chemical cycle. Nucleic Acids Res. 36, 1881–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jezewska M. J., Bujalowski W. (1996) Global conformational transitions in Escherichia coli primary replicative helicase DnaB protein induced by ATP, ADP, and single-stranded DNA binding–Multiple conformational states of the helicase hexamer. J. Biol. Chem. 271, 4261–4265 [DOI] [PubMed] [Google Scholar]
- 99. Rajendran S., Jezewska M. J., Bujalowski W. (2000) Multiple-step kinetic mechanism of DNA-independent ATP binding and hydrolysis by Escherichia coli replicative helicase DnaB protein: Quantitative analysis using the rapid quench-flow method. J. Mol. Biol. 303, 773–795 [DOI] [PubMed] [Google Scholar]
- 100. Iype L. E., Wood E. A., Inman R. B., Cox M. M. (1994) RuvA and RuvB proteins facilitate the bypass of heterologous DNA insertions during RecA protein-mediated DNA strand exchange. J. Biol. Chem. 269, 24967–24978 [PubMed] [Google Scholar]
- 101. Iype L. E., Inman R. B., Cox M. M. (1995) Blocked RecA protein-mediated DNA strand exchange reactions are reversed by the RuvA and RuvB proteins. J. Biol. Chem. 270, 19473–19480 [DOI] [PubMed] [Google Scholar]
- 102. Schutte B. C., Cox M. M. (1988) Homology-dependent underwinding of duplex DNA in RecA protein generated paranemic complexes. Biochemistry 27, 7886–7894 [DOI] [PubMed] [Google Scholar]