Background: ADAMDEC1 is a putative ADAM-like metalloprotease with a short domain structure and a noncanonical active site.
Results: Recombinant ADAMDEC1 cleaves α2-macroglobulin and casein. The activity is significantly enhanced upon reconstituting the consensus zinc-binding site.
Conclusion: ADAMDEC1 is secreted as a proteolytic active, glycosylated metalloprotease.
Significance: The ADAMDEC1 has adapted a reduced catalytic activity, possibly compensating the loss of auxiliary specificity determining domains.
Keywords: ADAM ADAMTS, Metalloprotease, Post-translational Modification, Protein Evolution, Protein Expression, Metzincin
Abstract
ADAMDEC1 (Decysin-1) is a putative ADAM (a disintegrin and metalloprotease)-like metalloprotease with an unknown physiological role, selectively expressed in mature dendritic cells and macrophages. When compared with other members of the ADAM family, ADAMDEC1 displays some unusual features. It lacks the auxiliary cysteine-rich, EGF, and transmembrane domains, as well as the cytoplasmic tail. The active site of ADAMDEC1 is unique by being the only mammalian ADAM protease with a non-histidine zinc ligand, having an aspartic acid residue instead. Here we demonstrate that ADAMDEC1, despite these unique features, functions as an active metalloprotease. Thus, ADAMDEC1 is secreted as a mature, glycosylated, and proteolytically active metalloprotease, capable of cleaving macromolecular substrates. In the recombinant form, three of the four potential N-linked glycosylation sites are modified by carbohydrate attachment. Substitution of basic residues at the predicted proprotein convertase cleavage site blocks proprotein processing, revealing both specific ADAMDEC1-dependent and specific ADAMDEC1-independent cleavage of the prodomain. The pro-form of ADAMDEC1 does not have proteolytic activity, demonstrating that the prodomain of ADAMDEC1, like in other members of the ADAM family, confers catalytic latency. Interestingly, the proteolytic activity of mature ADAMDEC1 can be significantly enhanced when a canonical ADAM active site with three zinc-coordinating histidine residues is introduced.
Introduction
ADAMDEC1 (Decysin-1) mRNA was first identified in 1997 by Mueller et al. (1) and predicted to encode a novel ADAM2 (a disintegrin and metalloprotease)-like protease based on sequence homology with members of the ADAM family of metzincin metalloproteases. ADAMDEC1 bears closest resemblance to ADAM-28 and ADAM-7, with sequence identities of 47 and 36%, respectively, whose genes also cluster together with ADAMDEC1 on chromosome 8p12 in the human genome (2). Phylogenetic analysis suggests that the partial gene duplication event giving rise to ADAMDEC1 and ADAM-7 and -28 from a common ancestor followed the mammalian divergence from amphibians, indicating that ADAMDEC1 is of relative late descendant when compared with other ADAMs (2, 3).
The ADAM family constitutes type-1 transmembrane metzincin metalloproteases, which in addition to the metzincin/reprolysin-type metalloprotease domain have multiple C-terminal auxiliary domains including a disintegrin-like domain, a cysteine-rich domain, an EGF-like domain, a transmembrane domain, and a cytoplasmic tail (4, 5). The domains positioned C-terminal to the metalloprotease domain are involved in cell adhesion, integrin binding, signaling events, and substrate recognition (5–7). In metzincin superfamily members, the metalloprotease domain contains the active site consisting of an elongated zinc-binding motif (HEXXHXXG/NXXH/D) with three zinc-coordinating ligands (underlined) immediately followed by a family-specific residue (5, 8). For the catalytic active ADAM subfamily members, the consensus sequence conforms to HEXXHXXGXXHD with three histidine residues coordinating the zinc ion, which together with a glutamic acid residue polarizes the catalytic solvent molecule promoting the nucleophile attack on the substrate, a catalytic mechanism similar to that of thermolysin (9). The conserved glycine within the active site allows for a sharp turn positioning the third histidine for zinc binding (4). In addition, ADAMs contain a conserved methionine (placed in the so-called Met-turn) positioned beneath the three zinc-binding histidines acting as a hydrophobic base (4). Catalytically active ADAM proteases are synthesized as zymogens with a large N-terminal prodomain shown to maintain catalytic latency (5, 10).
Like other ADAMs, ADAMDEC1 mRNA encodes an N-terminal signal peptide for directing secretion and a relatively large (173-residue) prodomain, predicted to be cleaved off during maturation. Indeed, furin has been shown in vitro to be capable of processing the prodomain in the murine ortholog (11). The mature protein only comprises a metzincin metalloprotease domain and a short disintegrin-like domain. Consequently, ADAMDEC1 lacks most of the auxiliary domains otherwise present in the ADAMs and is predicted to be secreted as a soluble protein (1, 2). The metzincin metalloprotease domain harbors a putative zinc-binding active site consensus sequence (HEXXHXXGXXD). However, the third zinc-binding ligand in ADAMDEC1 is an aspartic acid residue, as found in some bacterial metzincins, instead of the histidine residue found in all other proteolytically active ADAMs. Due to the differences in primary structure, ADAMDEC1 is regarded as the first (and only) member of a novel subgroup of mammalian ADAMs (1).
Little is known about the physiological role of this putative metalloprotease, but ADAMDEC1 is expressed in monocytes at low levels and at increased levels during 1α,25-dihydroxyvitamin D3-induced differentiation into mature macrophages (12). Moreover, expression is strongly induced by spontaneous as well as by CD40- or LPS-stimulated maturation of immature dendritic cells (1, 12). ADAMDEC1 expression was found to be up-regulated in atherosclerotic plaques (13), pulmonary sarcoidosis (14), intestinal inflammation (15), intracranial tumor (craniopharyngioma) (16), and infection of lymphoblastoid cells by Epstein-Barr virus (17), whereas it is down-regulated in colorectal cancer (18). In addition, ADAMDEC1 is up-regulated during pregnancy (19). Collectively, the expression pattern of ADAMDEC1 suggests that it plays a role in the immune response (2). However, ADAMDEC1 may have additional physiological roles as well.
Given the lack of functional and structural characterization of this unusual ADAM-like protein, we investigated the putative proteolytic activity of human ADAMDEC1 expressed in a mammalian expression system. We demonstrate that ADAMDEC1 is a secreted, active enzyme capable of cleaving α2-macroglobulin (α2M) and casein. In addition, we show that the prodomain confers catalytic latency. The same prodomain undergoes specific ADAMDEC1-dependent cleavage, which can occur in trans. The proteolytic activity of mature ADAMDEC1 is enhanced by reconstructing the canonical ADAM active site consensus sequence with three histidine residues coordinating the zinc ion. Finally, we demonstrate that three of the four potential sites are modified by N-linked glycosylation.
EXPERIMENTAL PROCEDURES
Materials
The FreeStyle 293 expression system and all additives, SDS-PAGE, and Western blot materials (Novex/iBlot System) were from Invitrogen. Azocasein and (1,10)-phenanthroline were from Sigma-Aldrich and VWR International, respectively. PNGase F endoglycosidase was from New England Biolabs. Plasma-derived human α2M was a kind gift from Prof. Lars Sottrup-Jensen, Department of Molecular Biology and Genetics, Aarhus University, Denmark and Henrik Østergaard, Department of Hemophilia Biochemistry, Novo Nordisk A/S.
Plasmids
ADAMDEC1 sequence information was obtained from GenBankTM (accession number O15204). Human ADAMDEC1 cDNA, optimized for mammalian cell expression, was purchased from GeneArt (Invitrogen) and subcloned into the pCI-neo vector (Promega), resulting in pCI-ADAMDEC1. An ADAMDEC1 variant with an HPC4 affinity tag inserted 2 amino acids downstream of the proposed furin-like recognition site (RISR203) was created by overlap extension PCR. Subsequently, point mutations were introduced into the ADAMDEC1 cDNA, with or without the HPC4 tag, using the QuikChange XL site-directed mutagenesis kit (Agilent Technologies). All primers were purchased from MWG Eurofins (Ebersberg, Germany), and primer sequences are available in Table 1.
TABLE 1.
DNA primer list
| Sequence | |
|---|---|
| Subcloning | |
| Forward | ATACATATGCTCAGAGGCATCTCTCAGCTGCCTGCCGTG |
| Reverse | GTGCTCGAGCTCGGTGGTGTGGTTAGGGGCGTCGCCGCC |
| Insertion of N-terminal HPC4 tag | |
| Forward | TAGGCTAGCATGCTCAGAGGCAT |
| Internal forward | CAATCAGACGCGGATCCACCTGATCTTCCAGGGACCGGGAGATCC |
| Internal reverse | GGTGGATCCGCGTCTGATTGATGGCAAAAAGTCCCCTGAGAAGG |
| Reverse | AAAGGGAAGCGGCCGCCTACTA |
| Mutagenesisa | |
| E353A | GGAGTGATGTCCCACGCGCTGGGCCACGTGCTG |
| D362H | GTGCTGGGCATGCCTCACGTGCCTTTCAACACC |
| H362insert | CGTGCTGGGCATGCCTCATGACGTGCCTTTCAACACC |
| R200K/R2003A | GCAGGGCCCTATCAAGATCTCCGCGTCCCTGAAGTCCCC |
| H54A | GCTGCACATCCTGGCCAAGCGGGAGATCAAGAAC |
| K55A | GCTGCACATCCTGCACGCGCGGGAGATCAAG |
| R56A | GCACATCCTGCACAAGGCGGAGATCAAGAACAACC |
| E57A | GCACATCCTGCACAAGCGGGCGATCAAGAACAACCAGACCG |
| I58A | GCACAAGCGGGAGGCCAAGAACAACCAGACCG |
| K59A | GCACAAGCGGGAGATCGCGAACAACCAGACCG |
| K160A | CCAGAGATACCAGATCGCGCCTCTGAAGTCC |
| P161A | CCAGAGATACCAGATCAAGGCTCTGAAGTCCACCG |
| L162A | CCAGATCAAGCCTGCGAAGTCCACCGACGAGAAGG |
| K163A | CCAGATCAAGCCTCTGGCGTCCACCGACGAGAAGG |
| N61Q | GGGAGATCAAGAACCAACAGACCGAGAAGCACGGC |
| N184Q | GAGCAGGACCCTGCCCAACACACCTGCGGCGTG |
| N237Q | CAAGAACTACAACGAGCAACTGACCCTGATCCGG |
| N466Q | GCGGCGACGCCCCTCAACACACCACCGAGTG |
a Mutagenesis primers are stated as the forward primer sequence. For each forward primer, a reverse complementary primer was added to each mutagenesis reactions.
Protein Expression and Western Blot Analysis
HEK293-F or -6E cells were grown in FreeStyle 293 expression medium at 37 °C, 8% CO2, and 125 rpm; for HEK293-6E cells, the medium was supplemented with 25 μg/ml G-418 Geneticin and 0.1% (v/v) Pluronic F-68. Cells were transfected with 1 μg of DNA/106 cells using 293fectin and Opti-MEM according to the manufacturer's protocol. Media were harvested 3–4 days after transfection, sterile-filtered, and stored at −80 °C. For SDS-PAGE and Western blot analysis, proteins were denatured by boiling in LDS sample buffer. DTT was added to reduced samples to 50 mm prior to boiling. Samples were separated in 4–12% Bis-Tris gels using MES or MOPS running buffer. Proteins were transferred to nitrocellulose membranes using the iBlot dry blotting system and blocked in 5%(W/V) skim milk in wash buffer (20 mm Tris (pH 7.4), 140 mm NaCl, 5 mm CaCl2, 0.05%(v/v) Tween 20). 0.43 μg/ml murine anti-human ADAMDEC1 mAb (Abcam ab57224) and 1.3 μg/ml HRP-labeled rabbit anti-mouse IgG (Dako) were used for detection of ADAMDEC1 expression. N-Linked glycans were removed under denaturing conditions using PNGase F according to the manufacturer's protocol. For N-terminal sequencing, proteins were separated by SDS-PAGE and transferred to PVDF membranes. Individual bands were cut out and subjected to Edman amino acid sequence analysis using an Applied Biosystems Procise HT protein sequencer with on-line identification of phenylthiohydantoin derivatives.
Proteolytic Activity Assays
Cell supernatants were concentrated ∼60 times, diluted 30 times in reaction buffer (50 mm Hepes (pH 7.5), 100 mm NaCl, 1 mm CaCl2, 10 μm ZnCl2), and concentrated again to a final concentration ∼60 times that of the supernatant using Amicon Ultra 10,000 molecular weight cut-off centrifugal filters (Millipore). The relative concentrations of ADAMDEC1 variants were evaluated by densitometric analysis of Coomassie Brilliant Blue-stained SDS-PAGE using the ImageJ software (20). An added amount of cell supernatant was adjusted in an iterative fashion until sample normalization was obtained. Supernatants from cells with added transfection reagents without DNA vector (Mock) were used as background control.
Identical amounts of concentrated ADAMDEC1 variant proteins, corresponding to ∼400 μl of ADAMDEC1 WT in cell supernatant, and an excess amount of Mock cell supernatant were added to azocasein solubilized in reaction buffer to a final concentration of 2 mg/ml (∼94 μm). Reaction mixtures were incubated at 37 °C for 3 days. The proteolysis was terminated by the addition of TCA to a final concentration of 6% (w/v) followed by a 30-min incubation on ice and removal of undigested protein by centrifugation at 10,000 × g and 4 °C for 10 min. NaOH was added to the supernatants to a final concentration of 0.26 mm, and the absorbance was measured at 440 nm using a SpectraMax 190 (Molecular Devices). Absorption data were subtracted from buffer contribution and compared with the data of Mock using one-way analysis of variance analysis with Bonferroni's adjustment. Individual sample pairs were additionally compared using an unpaired, two-tailed t test.
Proteolysis of human plasma-derived α2M was carried out by incubating ADAMDEC1 variant proteins (corresponding to 150 μl of cell supernatant) with α2M (final concentration: 0.9 mg/ml (1.25 μm)) in reaction buffer for ∼65 h at 37 °C. The result was visualized by reducing SDS-PAGE, and cleavage of α2M was observed by appearance of an 85-kDa fragment (21, 22). Proteolysis of α2M in human plasma was followed by adding identical amounts of ADAMDEC1 variants (equal to 200 μl of cell supernatant) to human plasma stabilized with 200 nm tick anticoagulant peptide and 1 μm hirudin. After ∼40 h of incubation at 37 °C, α2M fragments were visualized by Western blot analysis using 1 μg/ml mouse anti-human α2M antibody (2D9, Abcam ab36995) and 1.3 μg/ml HRP-labeled rabbit anti-mouse IgG (Dako). Cross-linking of ADAMDEC1 to plasma-derived α2M was visualized by Western blot analysis using 1.31 μg/ml anti-HPC4 tag antibody and 1 μg/ml HRP-labeled goat anti-human IgG.
RESULTS
ADAMDEC1 Is Expressed and Secreted as a Mature and Glycosylated Protein
ADAMDEC1 protein was detected in the cell supernatant of HEK293 cells transfected with pCI-ADAMDEC1, encoding full-length human wild-type ADAMDEC1 (WT) (residues 1–470, Fig. 1A). SDS-PAGE and Western blot analysis both showed expression by transfected cells, and no endogenous ADAMDEC1 expression was detected in the supernatant of Mock-transfected cells (Fig. 1, B and C). The observed molecular mass of ADAMDEC1 is ∼33 kDa, corresponding to the proADAMDEC1 being processed and secreted as the mature protein. A decreased mobility of reduced ADAMDEC1 indicates the presence of disulfides in the secreted protein (Fig. 1C).
FIGURE 1.
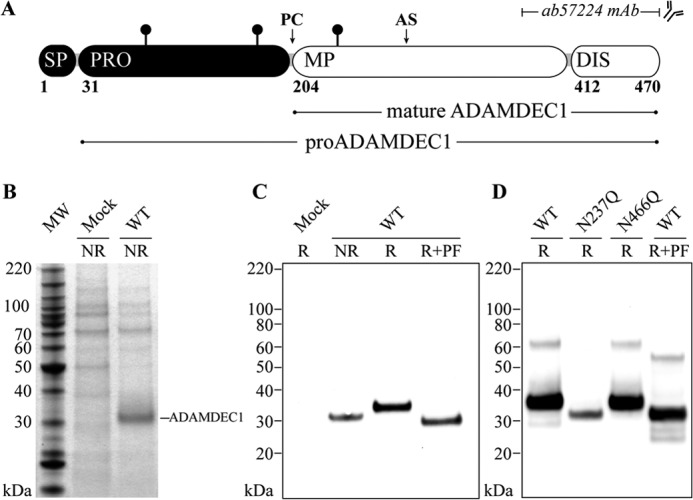
Expression and glycosylation status of mature ADAMDEC1. A, domain structure of full-length ADAMDEC1 consisting of a signal peptide (SP, res. 1–30), a prodomain (PRO, res. 31–203) predicted to be processed at a proprotein convertase recognition site (PC), a metzincin metalloprotease domain (MP, res. 204–411) containing the putative zinc-binding active site (AS, res. 352–362), and a short disintegrin-like domain (DIS, res. 412–470) roughly two-thirds the length of the disintegrin-like domain in other ADAMs (1, 2). The mAb used for Western blot analysis is raised against a C-terminal fragment (res. 361–470, indicated on the figure). Confirmed N-linked glycosylation sites (Asn-61, Asn-184, and Asn-237) are indicated by black lollipops. B, secretion of recombinant human ADAMDEC1 WT to mammalian cell supernatant shown in nonreducing (NR) SDS-PAGE loaded with ∼19 μl of cell supernatant. MW, molecular weight markers. C, anti-ADAMDEC1 Western blot analysis of nonreduced (NR), reduced (R), and reduced + PNGase F-treated (R+PF) ADAMDEC1 WT. D, electrophoretic mobility of ADAMDEC1 variants containing substitutions in the two predicted sequons in the mature protein (Asn-237 and Asn-466) analyzed in anti-ADAMDEC1 Western blot and compared with reduced (R) and reduced + PNGase F-treated (R+PF) ADAMDEC1 WT.
ADAMDEC1 has two predicted N-linked glycosylation sites in the prodomain and two in the mature protein. Deglycosylation using PNGase F resulted in an increased electrophoretic mobility and an apparent molecular mass of 30 kDa, which correlates with the predicted molecular mass of 29.4 kDa for the mature protein without glycans and shows that mature ADAMDEC1 is modified with at least one N-linked glycan. We individually removed the glycosylation sites in the metalloprotease domain (Asn-237) and in the truncated disintegrin-like domain (Asn-466) by site-directed mutagenesis to determine which sites are utilized. Because the N237Q variant exhibits increased mobility when compared with WT, identical to that of PNGase F-treated WT, and the N466Q variant displays no change in electrophoretic mobility when compared with WT, we conclude that the Asn-237 glycosylation site is occupied, whereas the Asn-466 site is unmodified (Fig. 1D).
Recombinant ADAMDEC1 Is Proteolytically Active
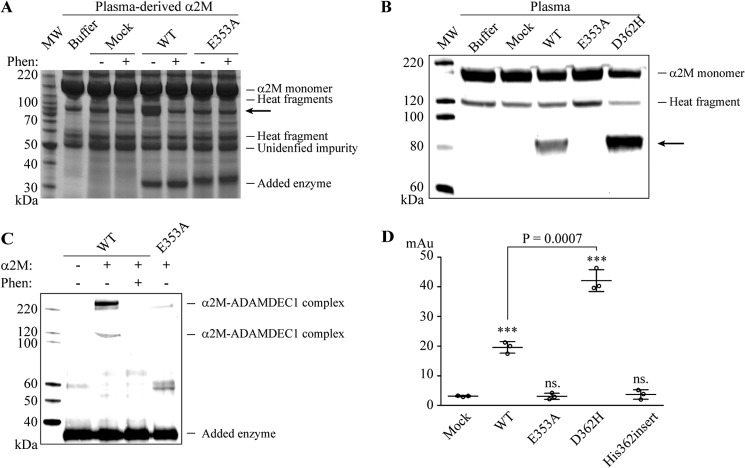
To determine whether ADAMDEC1 has proteolytic activity, despite a noncanonical ADAM zinc-binding motif, we tested whether ADAMDEC1 was able to cleave α2M, an important regulator of a broad spectrum of proteases including several ADAMs (23–25). We incubated ADAMDEC1 with plasma-derived α2M (Fig. 2A) and stabilized human plasma (Fig. 2B). α2M proteolysis was analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining and anti-α2M Western blot analysis, respectively. We found that ADAMDEC1 cleaves α2M, demonstrated by generation of an immunoreactive 85-kDa fragment (marked by arrow in Fig. 2, A and B). In addition, SDS-stable ADAMDEC1-α2M complexes were observed by Western blot analysis specific for the recombinant ADAMDEC1, demonstrating thioester-mediated cross-linking (Fig. 2C). Further, cleavage of α2M by ADAMDEC1 was inhibited by 5 mm (1,10)-phenanthroline, confirming metal ion dependence (Fig. 2, A and C). Substitution of the predicted catalytic glutamic acid residue (E353A, Table 2) completely abrogated the proteolytic activity, confirming the identity of the ADAMDEC1 active site (Fig. 2, A–C). Incubation of α2M with supernatant from Mock-transfected cells did not induce α2M proteolysis.
FIGURE 2.
Proteolytic activity of ADAMDEC1. A, incubation of plasma-derived α2M with supernatants from Mock-, WT-, and HPC4-tagged E353A-transfected cells, respectively, in the absence or presence of 5 mm (1,10)-phenanthroline (Phen). Proteolysis of α2M is demonstrated by generation of an 85-kDa α2M fragment indicated by the arrow in reducing SDS-PAGE. Intact 180-kDa α2M monomeric subunit, 120- and 60-kDa α2M heat fragments (47), and the added ADAMDEC1 variants (∼33 kDa) are indicated, as well as a 50-kDa unidentified impurity. MW, molecular weight markers. B, anti-α2M Western blot analysis of stabilized human plasma incubated with supernatants from Mock-, WT-, E353A-, and D362H-transfected cells, respectively. The arrow marks the immunoreactive 85-kDa α2M proteolytic fragment. C, covalent cross-linking between HPC4-tagged ADAMDEC1 WT and plasma-derived α2M, in the absence or presence of 5 mm (1,10)-phenanthroline, visualized by anti-HPC4 Western blot analysis. D, proteolytic activity assay of ADAMDEC1 WT and variants using azocasein as a substrate. The sample means are shown by a horizontal line. ***, p < 0.0001; ns., not significant relative to Mock.
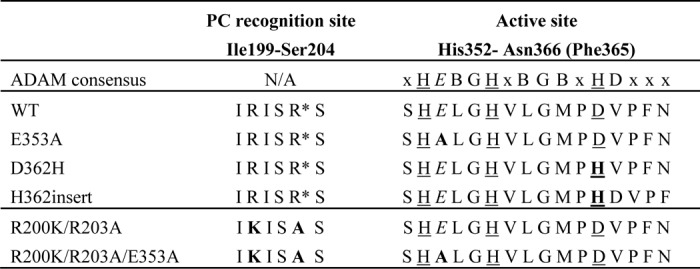
TABLE 2.
List of selected ADAMDEC1 variants
Sequences surrounding the furin-like proprotein convertase recognition site (Ile-99–Ser-204, predicted processing site marked by asterisk) and the predicted active site (Ser-351–Phe-365 (Asn-366)) for a subset of the produced ADAMDECI variants. The predicted catalytic residue (Glu-353) is in italic font, predicted zinc-coordinating ligands are underlined, and mutations are in bold. The predicted active site sequence is compared with the ADAM family consensus sequence (gray box) where X is a less conserved residues and B is a bulky hydrophobic residues. Prodomain processing among the ADAMs is subject to great variation and is therefore not included in this comparison.
When applied to the broadly used substrate azo-labeled casein (azocasein), HEK293 cell supernatant containing recombinant ADAMDEC1 WT exhibited caseinolytic activity, as observed by a significant increase in release of azo-labeled peptides, whereas supernatant from Mock-transfected cells showed no activity (Fig. 2D). Again, the E353A substitution completely abrogated the proteolytic activity.
Immediately downstream of the long zinc-binding consensus sequence, metzincins have a so-called family-specific residue (5), which for adamalysins/ADAMs in general comprises an aspartic acid. Interestingly, it is a valine in ADAMDEC1 (Table 2). In addition, the third zinc-binding residue of the consensus sequence is an aspartic acid, whereas it is generally a histidine in other adamalysin/ADAMs and other metzincins with the exception of snapalysins and, possibly, thuringilysins (5). This feature could have resulted from an evolutionary substitution event or alternatively from a deletion event. To reconstitute an intact zinc binding motif, two new variants were constructed: D362H, substituting the predicted zinc-coordinating aspartic acid with histidine, and His-362insert, inserting a histidine residue at position 362, thereby creating the complete ADAM zinc-coordinating sphere (HEXXHXXGXXHD) followed by the ADAM family-specific aspartic acid residue (Table 2). When tested as described above, the D362H variant demonstrated significantly enhanced proteolytic activity toward both α2M and azocasein, whereas no proteolytic activity could be detected for the His-362insert variant (Fig. 2, B and D). Combination of the D362H substitution and the inactivating E353A mutation completely abrogated the proteolytic activity (data not shown), thus confirming the reconstitution of the zinc-binding site in ADAMDEC1.
Processing of proADAMDEC1
N-terminal sequencing of the recombinant ADAMDEC1WT protein by Edman degradation confirmed processing at the furin-like proprotein convertase recognition site: RISR203-SLKS (Fig. 3A). To enable studying of the ADAMDEC1 pro-form in regard to post-translational modification, we generated a variant with two mutations substituting residues Arg-200 and Arg-203 with lysine and alanine, respectively (R200K/R203A, Table 2), rendering the protein impervious to proprotein convertase processing. This variant was expected to be secreted as the full-length proADAMDEC1 form after signal peptide cleavage, starting with residue Ile-31 (Fig. 1A). Surprisingly, R200K/R203A was found present in the medium as three distinct species with apparent molecular masses of 55, 50, and 42 kDa, respectively, rather than as a single unprocessed pro-form (Fig. 3A, lane RR). Using Edman degradation, the N-terminal sequence of the 42-kDa species was identified as LKSTDE, corresponding to cleavage in the ADAMDEC1 prodomain after residue Pro-161 (IKP161-LKS, Fig. 3A). The N-terminal sequence of the 50-kDa fragment could not be determined, but the 55-kDa species had the N-terminal sequence EIKNNQTE, corresponding to cleavage after Arg-56 (HKR56-EIK, Fig. 3A). Next, we investigated whether the observed prodomain processing was due to an auto-catalytic event by introducing the inactivating E353A substitution (R200K/R203A/E353A, Table 2). This variant was exclusively found in the medium as a secreted 55-kDa species, with an N-terminal sequence demonstrating proteolytic processing after Arg-56. These results demonstrate that the ADAMDEC1 prodomain is cleaved by an unknown protease at Arg-56 and partially processed by two ADAMDEC1-dependent proteolytic events, giving rise to the 50-kDa fragments and the predominant 42-kDa proADAMDEC1 fragments, the latter resulting from cleavage after Pro-161.
FIGURE 3.
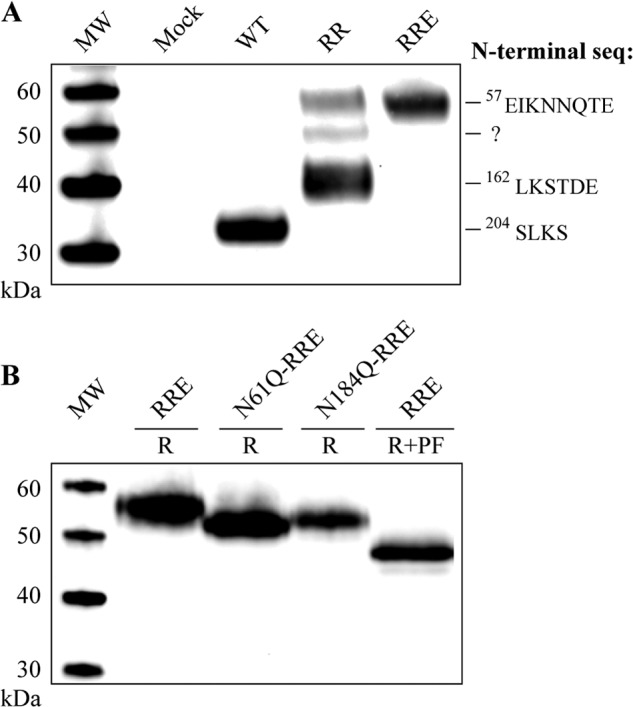
Prodomain modification and processing visualized by reducing anti-ADAMDEC1 Western blot analysis. A, analysis of cell supernatants from expression of ADAMDEC1 WT, R200K/R203A (RR), and R200K/R203A/E353A (RRE). The N-terminal sequences were determined by Edman degradation. MW, molecular weight markers. B, electrophoretic mobility of R200K/R203A/E353A (RRE)-based variants containing substitutions in the two predicted N-linked glycosylation sites of the prodomain (Asn-61 and Asn-184) when compared with reduced (R) and reduced + PNGase F-treated (R+PF) R200K/R203A/E353A.
Prodomain Glycosylation
The prodomain of ADAMDEC1 contains two predicted N-linked glycosylation sites (Asn-61 and Asn-184, Fig. 1A). Availability of a single species pro-form containing all predicted glycosylation sites (R200K/R203A/E353A) allowed us to evaluate these by site-directed mutagenesis (Fig. 3B). Substitution of either Asn-61 or Asn-184 with glutamine resulted in increased electrophoretic mobility when compared with R200K/R203A/E353A, demonstrating that both sites are modified by N-linked glycosylation. The apparent molecular masses of R200K/R203A/E353A, N61Q/R200K/R203A/E353A, and N184A/R200K/R203A/E353A were determined to be 55.0, 52.0, and 52.4 kDa, respectively, showing that the effect of one N-linked glycan is a decrease in mobility corresponding to a difference in molecular mass of 2.6–3 kDa. Enzymatically deglycosylated R200K/R203A/E353A migrated with an apparent molecular mass of 47.5 kDa, ∼7.5 kDa less than the untreated protein, consistent with removal of the three N-linked glycans at Asn-61, Asn-184, and Asn-237, respectively (Fig. 3B).
Sequential Proteolytic Events Remove N-terminal Parts of the ADAMDEC1 Prodomain
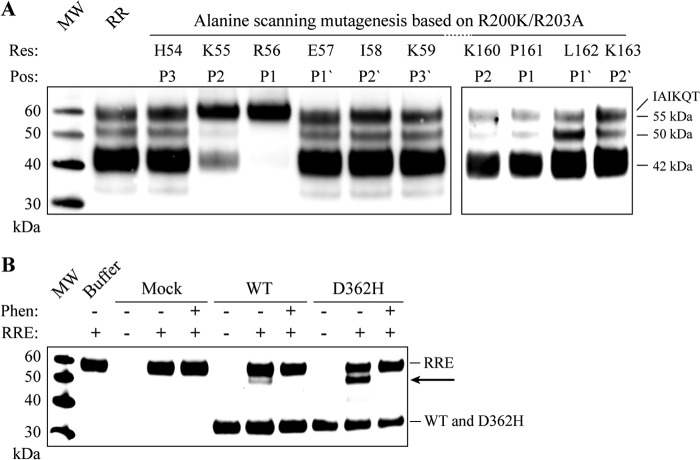
An alanine-scanning mutagenesis of the P3-P3′ residues constituting the Arg-56 cleavage site (HKR56-EIK), using the R200K/R203A variant as template, demonstrated that the P1 Arg-56 to alanine substitution (R56A) inhibited all observed proteolytic processing of the prodomain, resulting in the secretion of a single species pro-form. Reduced proteolysis at all cleavage sites was observed with the K55A P2 variant (Fig. 4A, left). This surprisingly demonstrates that the two cleavage events resulting in the 50- and 42-kDa species depend on initial cleavage after Arg-56. None of the other substitutions had any visible effect on the distribution between proADAMDEC1 species. N-terminal sequencing of the R56A/R200K/R203A secreted protein (Fig. 4A, lane R56) resulted in the sequence IAIKQT, demonstrating that the cleavage after residue 56 was inhibited and confirming the predicted signal peptidase cleavage site in proADAMDEC1 at Ala-30 (TQA30-IAI) (Fig. 1A).
FIGURE 4.
ADAMDEC1 prodomain auto-catalysis and mutational analysis. A, secretion pattern of cells transfected with ADAMDEC1 R200K/R203A-based variants subjected to alanine-scanning mutagenesis around the identified prodomain cleavage sites at position Arg-56 (left) and Pro-161 (right). The alanine-scanned residues (res) and their individual positions (pos) relative to the scissile bond are indicated above the lanes. R56A/R200K/R203A is identified as full-length proADAMDEC1 by Edman sequencing. MW, molecular weight markers. B, co-incubation of R200K/R203A/E353A (RRE) with Mock, ADAMDEC1 WT, or D362H in the absence or presence of 5 mm (1,10)-phenanthroline (Phen). Proteolytic cleavage resulting in an ∼50-kDa species is indicated with an arrow.
Alanine-scanning mutagenesis of P2-P2′ residues around the Pro-161 cleavage site did not inhibit cleavage at Pro-161. However, the distribution of the individual processing fragments seems to vary. All four variants give rise to the ∼42-kDa species, produced from cleavage at Pro-161, but mutagenesis of the P1′ Leu residue appears to result in a slight accumulation of the 50-kDa species (Fig. 4A, right).
Processing of the prodomain at an unidentified position between Arg-56 and Pro-161 and at Pro-161, giving rise to the 50- and 42-kDa species, respectively, is dependent on the cleavage at residue Arg-56 and ADAMDEC1 proteolytic activity (Figs. 3A and 4A). To investigate whether the ADAMDEC1-dependent processing can take place in trans, we co-incubated proteolytically inactive R200K/R203A/E353A with supernatants from Mock-, ADAMDEC WT-, and D362H-transfected cells, respectively, in the absence or presence of (1,10)-phenanthroline (Fig. 4B). ADAMDEC1 WT and, more efficiently, D362H were able to generate an ∼50-kDa proADAMDEC1 fragment (Fig. 3A). This proteolysis was completely inhibited by the addition of (1,10)-phenanthroline, and no cleavage was observed after incubation with Mock cell supernatant. These observations indicate that ADAMDEC1 can perform auto-cleavage of its own prodomain in trans, giving rise to an ∼50-kDa species.
ADAMDEC1 Prodomain Confers Partial Catalytic Latency
Several ADAM proteases have been shown to be synthesized as zymogens, and removal of the prodomain is required for proteolytic activity (26–33). To determine whether a similar regulatory mechanism applies to ADAMDEC1, we investigated the proteolytic activity of three ADAMDEC1 variants with different prodomain lengths, but with intact active sites: mature ADAMDEC1 WT (33 kDa, residues 204–470), the R200K/R203A variant predominately found as the 42-kDa species (residues 162–470) cleaved after Pro-161, and the R56A/R200K/R203A variant, exclusively found in the full-length pro-form (57 kDa, residues 31–470) (Fig. 5A). We used azocasein as substrate and included the inactive R200K/R203A/E353A variant as a control (Fig. 5B). Interestingly, the full-length R56A/R200K/R203A pro-form shows significantly reduced activity when compared with mature ADAMDEC1 WT, indicating substantial inhibition of ADAMDEC1 by the intact prodomain (Fig. 5B). Even the 42-kDa R200K/R203A variant exhibited significantly reduced proteolytic activity when compared with the mature WT enzyme, although to a much lower degree than the full-length proenzyme, demonstrating that the 42 remaining residues of the prodomain (res. 162–203) are partially inhibitory to ADAMDEC1. As expected, the R200K/R203A/E353A variant did not cleave azocasein (Fig. 5B).
FIGURE 5.
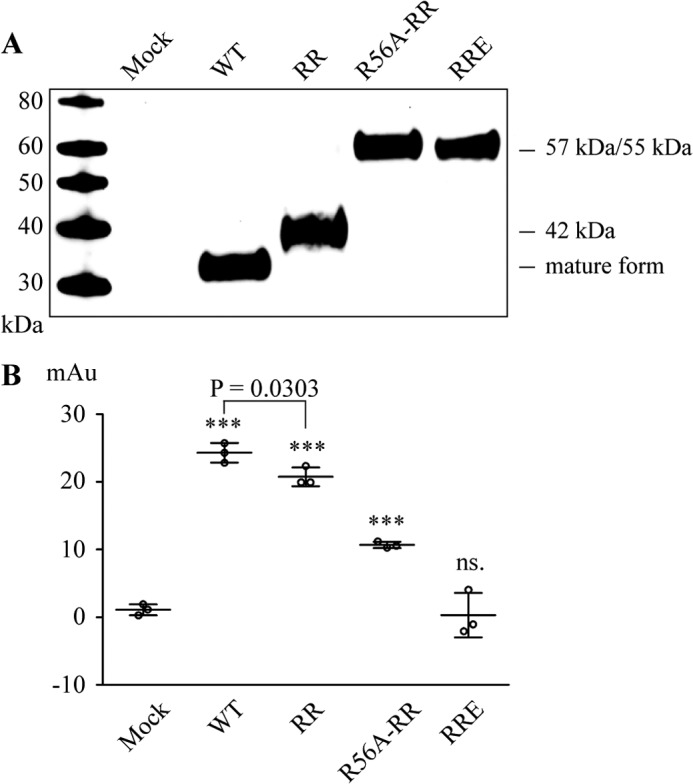
Proteolytic activity of proADAMDEC1. A, reducing anti-ADAMDEC1 Western blot of ADAMDEC1 variants including mature wild-type (WT, 33 kDa, res. 204–470), R200K/R203A (RR, predominately 42-kDa species, res. 162–470), R56A/R200K/R203A (R56A-RR, 57 kDa, res. 31–470), and the inactivated R200K/R203A/E353A (RRE, 55 kDa, res. 57–470). MW, molecular weight markers. B, activity assay of ADAMDEC1 variants with different prodomain lengths using azocasein as a substrate. Sample means are shown by horizontal lines. ***, p < 0.0001; ns., not significant relative to Mock.
DISCUSSION
ADAMDEC1 is a putative ADAM-like metzincin metalloprotease with a rare zinc-binding consensus sequence and an unusually short domain structure. It is expressed during differentiation of dendritic cells and is constitutively expressed in macrophages (1, 12). Until now, ADAMDEC1 has primarily been studied on a transcriptional level, where it has been found differently regulated in a number of pathologies (13–18).
We expressed recombinant, human ADAMDEC1 in HEK293 cells and detected the mature and glycosylated protein secreted into the cell supernatant. Other ADAM family members are membrane proteins due to their type-1 transmembrane domain, but secretion of ADAMDEC1 into the cell medium was expected because the short domain structure does not contain a transmembrane domain (1).
Mature ADAMDEC1 produced in HEK293 cells cleaves α2M, resulting in the appearance of a characteristic 85-kDa protein fragment consistent with bait region cleavage (21). We were able to demonstrate covalent trapping of ADAMDEC1 by α2M, as is seen for e.g. ADAM-12 (28), resulting from activation of the α2M cross-linking thioester. Both covalent cross-linking of ADAMDEC1 to α2M and the observed molecular mass of the α2M proteolytic fragment are supportive of cleavage of α2M inside the bait region (34). This also indicates that α2M could be a physiological inhibitor of ADAMDEC1. ADAMDEC1 also cleaves azocasein, resulting in release of azo-labeled peptides.
The amino acid sequence of human ADAMDEC1 contains four predicted N-linked glycosylation sites, which are conserved for higher primates; however, more distant orthologs lack one or more modification site(s). Mutational analysis of the four predicted sites demonstrated modification by N-linked glycosylation at Asn-61, Asn-184, and Asn-237. N184Q and N237Q both exhibited reduced expression levels when compared with mature and pro-form ADAMDEC1, respectively, whereas the expression level of N61Q was comparable with full-length ADAMDEC1. This is similar to ADAMTS-13, where substitutions of some N-linked glycosylation sites leads to decreased expression levels, whereas others do not (35). The varying expression levels may reflect decreased stability upon removal of certain N-linked glycosylations. In contrast, we demonstrated that the fourth predicted N-glycosylation site, Asn-466 in the disintegrin-like domain, is not modified in the present expression system. The ADAMDEC1 protein terminates four residues downstream to Asn-466, and the proximity to the C-terminal tail likely explains the lack of glycan modification (36, 37).
The sequence of human ADAMDEC1 contains a proprotein convertase site (RISR203) (1). Our findings confirm processing of human ADAMDEC1 at RISR203-SLKS consistent with furin-like proprotein convertase activity. In comparison, murine ADAMDEC1 contains two proprotein convertase-like recognition sites, RDQR156 and RTSR203, where in vitro furin processing at the latter site was demonstrated to generate the mature protein (11). Moreover, we demonstrate that full-length proADAMDEC1 has reduced proteolytic activity, and thus the prodomain maintains partial catalytic latency of the protease. A number of ADAMs have been shown to be synthesized as zymogens; however, prodomain processing varies among the members. ADAM-9 (26), -10 (27), -12 (28), -15 (29), -17 (30), and -19 (31) are all activated by a subtilisin/Kex2p-like proprotein convertase (e.g. furin) at a paired basic recognition site (e.g. RXXR), whereas ADAM-8 (32) and -28 (33) are activated by auto-catalytic events, inhibited by substitution of the catalytic glutamic acid residue.
Interestingly, removal of the proprotein convertase recognition site in ADAMDEC1 revealed extensive catalytic processing of the prodomain at three positions. First, proADAMDEC1 is cleaved after Arg-56 by an unidentified protease. This cleavage site has positively charged residues at the P1, P2, and P6 positions relative to the scissile bond and could therefore likely be a substrate for a proprotein convertase family member (38). Prevention of this cleavage, by introducing the P1 variant R56A, inhibited catalytic processing of the prodomain at the two additional sites and resulted in secretion of full-length proADAMDEC1, with the predicted N-terminal sequence after signal peptidase cleavage at Ala-30. However, if the Arg-56 cleavage event is allowed to occur, proADAMDEC1 is further cleaved at two positions. One cleavage occurs after Pro-161, resulting in a 42-kDa species, and the other occurs at an unidentified site N-terminal to Pro-161, resulting in an ∼50-kDa ADAMDEC1 species. These events both depend on ADAMDEC1 proteolytic activity because the inactivated pro-form R200K/R203A/E353A does not undergo proteolysis at these sites. Further, this ADAMDEC1-dependent catalysis can occur in trans, demonstrated by co-incubating the inactivated R200K/R203A/E353A with mature ADAMDEC1 WT. This in trans cleavage may be a partial activating event for ADAMDEC1, as is the case for ADAM-8 (32) and ADAM-28 (33). However, in trans cleavage is only observed at the unknown site, giving rise to the 50-kDa species, potentially reflecting differences in the kinetics between cis and trans cleavage of the two ADAMDEC1-dependent proteolysis events.
ADAMDEC1 displays an unusual active site with a non-histidine zinc coordination ligand, which is conserved within all sequenced ADAMDEC1 orthologs available in the Merops database (39). Despite this rare feature, we were able to demonstrate proteolytic activity of the wild-type protein. Interestingly, substituting the aspartic acid constituting the third zinc-binding ligand with histidine (D362H), thereby reconstructing the ADAM active site consensus sequence (HEXXHXXGXXH), increases the proteolytic activity significantly. These data indicate that the change of the zinc ligand chemical properties, from a “medium/soft,” polarizable imidazole nitrogen to a “hard” negatively charged carboxyl oxygen, significantly influences the effectiveness of the zinc ion as a catalyst of hydrolysis. We speculate that this could be due to a reduced potency to activate the coordinated water molecule for nucleophilic attack or a lowered ability to polarize the carbonyl of the scissile bond. Also, the presence of a negatively charged ligand would be expected to reduce the stabilization of the negatively charged transition state (40). Introduction of an aspartic acid residue as the third zinc-coordinating residue also potentially changes the zinc binding geometry of ADAMDEC1, leading to a reduced zinc binding affinity (41, 42). The significant attenuation of the proteolytic activity may have evolved to compensate for the short disintegrin domain and loss of C-terminal auxiliary domains, which confers substrate specificity through exo-site interactions in other ADAMS, e.g. ADAM-17 (43) and ADAM-10 (7). Interestingly, inserting a histidine at position 362 (His-362insert), thereby creating an ADAM-like elongated zinc-binding motif followed by an ADAM family-specific aspartic acid residue (HEXXHXXGXXHD), completely abrogates the proteolytic activity. We hypothesize that the His-362 insertion may perturb the structure immediately C-terminal to the active site, thereby inhibiting catalytic activity, and suggest that the appearance of the aspartic acid is most likely a result of an evolutionary substitution event. Two distantly related bacterial metzincin families, the snapalysins (ScNP, Merops M7 family) and the thuringilysins (Merops M6 family) (1, 2, 8, 39), share the HEXXHXXGXXD active site sequence with ADAMDEC1. The low overall sequence identity between ADAMDEC1 and members of the M6 and M7 families, however, suggests convergent evolution of this particular zinc-binding sequence (2). Snapalysin from Streptomyces has been shown to be an active protease (44, 45), and a high resolution crystal structure of snapalysin (Protein Data Bank (PDB) 1C7K) shows that the aspartic acid residue functions as the third zinc-coordinating ligand, in an identical manner to what we expect in ADAMDEC1 (46).
In conclusion, ADAMDEC1 is a proteolytic active metzincin metalloprotease despite the rare zinc-coordinating site and C-terminal truncated domain structure. It appears to have evolved with a dampened proteolytic activity because substituting the zinc-ligating aspartic acid with a histidine increases the activity significantly. Decreasing ADAMDEC1 proteolytic activity may have been evolutionarily important to reduce unspecific proteolysis, which we suggest would be the consequence of lacking the C-terminal auxiliary domains.
Acknowledgments
We thank Professor Lars Sottrup-Jensen and Henrik Østergaard for providing plasma-derived α2M.
Footnotes
- ADAM
- a disintegrin and metalloprotease
- α2M
- α2-macroglobulin
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- PNGase F
- N-glycosidase F
- res.
- residues.
REFERENCES
- 1. Mueller C. G. F., Rissoan M. C., Salinas B., Ait-Yahia S., Ravel O., Bridon J. M., Briere F., Lebecque S., Liu Y. J. (1997) Polymerase chain reaction selects a novel disintegrin proteinase from CD40-activated germinal center dendritic cells. J. Exp. Med. 186, 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bates E. E., Fridman W. H., Mueller C. G. (2002) The ADAMDEC1 (decysin) gene structure: evolution by duplication in a metalloprotease gene cluster on chromosome 8p12. Immunogenetics 54, 96–105 [DOI] [PubMed] [Google Scholar]
- 3. Long J., Li M., Ren Q., Zhang C., Fan J., Duan Y., Chen J., Li B., Deng L. (2012) Phylogenetic and molecular evolution of the ADAM (A Disintegrin And Metalloprotease) gene family from Xenopus tropicalis, to Mus musculus, Rattus norvegicus, and Homo sapiens. Gene 507, 36–43 [DOI] [PubMed] [Google Scholar]
- 4. Bode W., Gomis-Rüth F. X., Stöckler W. (1993) Astacins, serralysins, snake venom, and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins’. FEBS Lett. 331, 134–140 [DOI] [PubMed] [Google Scholar]
- 5. Gomis-Rüth F. X. (2003) Structural aspects of the metzincin clan of metalloendopeptidases. Mol. Biotechnol. 24, 157–202 [DOI] [PubMed] [Google Scholar]
- 6. Primakoff P., Myles D. G. (2000) The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 16, 83–87 [DOI] [PubMed] [Google Scholar]
- 7. Janes P. W., Saha N., Barton W. A., Kolev M. V., Wimmer-Kleikamp S. H., Nievergall E., Blobel C. P., Himanen J. P., Lackmann M., Nikolov D. B. (2005) Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 123, 291–304 [DOI] [PubMed] [Google Scholar]
- 8. Gomis-Rüth F. X. (2009) Catalytic domain architecture of metzincin metalloproteases. J. Biol. Chem. 284, 15353–15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kester W. R., Matthews B. W. (1977) Crystallographic study of the binding of dipeptide inhibitors to thermolysin: implications for the mechanism of catalysis. Biochemistry 16, 2506–2516 [DOI] [PubMed] [Google Scholar]
- 10. Klein T., Bischoff R. (2011) Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. J. Proteome Res. 10, 17–33 [DOI] [PubMed] [Google Scholar]
- 11. Mueller C. G. F., Cremer I., Paulet P. E., Niida S., Maeda N., Lebeque S., Fridman W. H., Sautès-Fridman C. (2001) Mannose receptor ligand-positive cells express the metalloprotease decysin in the B cell follicle. J. Immunol. 167, 5052–5060 [DOI] [PubMed] [Google Scholar]
- 12. Fritsche J., Müller A., Hausmann M., Rogler G., Andreesen R., Kreutz M. (2003) Inverse regulation of the ADAM-family members, decysin and MADDAM/ADAM19 during monocyte differentiation. Immunology 110, 450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Papaspyridonos M., Smith A., Burnand K. G., Taylor P., Padayachee S., Suckling K. E., James C. H., Greaves D. R., Patel L. (2006) Novel candidate genes in unstable areas of human atherosclerotic plaques. Arterioscler. Thromb. Vasc. Biol. 26, 1837–1844 [DOI] [PubMed] [Google Scholar]
- 14. Crouser E. D., Culver D. A., Knox K. S., Julian M. W., Shao G., Abraham S., Liyanarachchi S., Macre J. E., Wewers M. D., Gavrilin M. A., Ross P., Abbas A., Eng C. (2009) Gene expression profiling identifies MMP-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. Am. J. Respir. Crit. Care Med. 179, 929–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dommels Y. E., Butts C. A., Zhu S., Davy M., Martell S., Hedderley D., Barnett M. P., McNabb W. C., Roy N. C. (2007) Characterization of intestinal inflammation and identification of related gene expression changes in mdr1a−/− mice. Genes Nutr. 2, 209–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu J., Liu L., Zheng X., You C., Li Q. (2012) Expression and inhibition of ADAMDEC1 in craniopharyngioma cells. Neurol. Res. 34, 701–706 [DOI] [PubMed] [Google Scholar]
- 17. Lucchesi W., Brady G., Dittrich-Breiholz O., Kracht M., Russ R., Farrell P. J. (2008) Differential gene regulation by Epstein-Barr virus type 1 and type 2 EBNA2. J. Virol. 82, 7456–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Macartney-Coxson D. P., Hood K. A., Shi H. J., Ward T., Wiles A., O'Connor R., Hall D. A., Lea R. A., Royds J. A., Stubbs R. S., Rooker S. (2008) Metastatic susceptibility locus, an 8p hot-spot for tumour progression disrupted in colorectal liver metastases: 13 candidate genes examined at the DNA, mRNA, and protein level. BMC Cancer 8, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baran N., Kelly P. A., Binart N. (2003) Decysin, a new member of the metalloproteinase family, is regulated by prolactin and steroids during mouse pregnancy. Biol. Reprod. 68, 1787–1792 [DOI] [PubMed] [Google Scholar]
- 20. Abramoff M. D., Magalhães P. J., Ram S. J. (2004) Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 21. Harpel P. C. (1973) Studies on human plasma α2-macroglobulin-enzyme interactions. Evidence for proteolytic modification of the subunit chain structure. J. Exp. Med. 138, 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sottrup-Jensen L., Hansen H. F., Pedersen H. S., Kristensen L. (1990) Localization of ϵ-lysyl-γ-glutamyl cross-links in five human α2-macroglobulin-proteinase complexes: nature of the high molecular weight cross-linked products. J. Biol. Chem. 265, 17727–17737 [PubMed] [Google Scholar]
- 23. Loechel F., Gilpin B. J., Engvall E., Albrechtsen R., Wewer U. M. (1998) Human ADAM 12 (meltrin α) is an active metalloprotease. J. Biol. Chem. 273, 16993–16997 [DOI] [PubMed] [Google Scholar]
- 24. Tortorella M. D., Arner E. C., Hills R., Easton A., Korte-Sarfaty J., Fok K., Wittwer A. J., Liu R. Q., Malfait A. M. (2004) α2-Macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. J. Biol. Chem. 279, 17554–17561 [DOI] [PubMed] [Google Scholar]
- 25. Kuno K., Terashima Y., Matsushima K. (1999) ADAMTS-1 is an active metalloproteinase associated with the extracellular matrix. J. Biol. Chem. 274, 18821–18826 [DOI] [PubMed] [Google Scholar]
- 26. Roghani M., Becherer J. D., Moss M. L., Atherton R. E., Erdjument-Bromage H., Arribas J., Blackburn R. K., Weskamp G., Tempst P., Blobel C. P. (1999) Metalloprotease-disintegrin MDC9: intracellular maturation and catalytic activity. J. Biol. Chem. 274, 3531–3540 [DOI] [PubMed] [Google Scholar]
- 27. Anders A., Gilbert S., Garten W., Postina R., Fahrenholz F. (2001) Regulation of the α-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 15, 1837–1839 [DOI] [PubMed] [Google Scholar]
- 28. Loechel F., Overgaard M. T., Oxvig C., Albrechtsen R., Wewer U. M. (1999) Regulation of human ADAM 12 protease by the prodomain: evidence for a functional cysteine switch. J. Biol. Chem. 274, 13427–13433 [DOI] [PubMed] [Google Scholar]
- 29. Lum L., Reid M. S., Blobel C. P. (1998) Intracellular maturation of the mouse metalloprotease disintegrin MDC15. J. Biol. Chem. 273, 26236–26247 [DOI] [PubMed] [Google Scholar]
- 30. Endres K., Anders A., Kojro E., Gilbert S., Fahrenholz F., Postina R. (2003) Tumor necrosis factor-α converting enzyme is processed by proprotein-convertases to its mature form which is degraded upon phorbol ester stimulation. Eur. J. Biochem. 270, 2386–2393 [DOI] [PubMed] [Google Scholar]
- 31. Kang T., Zhao Y. G., Pei D., Sucic J. F., Sang Q. X. (2002) Intracellular activation of human adamalysin 19/disintegrin and metalloproteinase 19 by furin occurs via one of the two consecutive recognition sites. J. Biol. Chem. 277, 25583–25591 [DOI] [PubMed] [Google Scholar]
- 32. Schlomann U., Wildeboer D., Webster A., Antropova O., Zeuschner D., Knight C. G., Docherty A. J. P., Lambert M., Skelton L., Jockusch H., Bartsch J. W. (2002) The metalloprotease disintegrin ADAM8: processing by autocatalysis is required for proteolytic activity and cell adhesion. J. Biol. Chem. 277, 48210–48219 [DOI] [PubMed] [Google Scholar]
- 33. Howard L., Maciewicz R. A., Blobel C. P. (2000) Cloning and characterization of ADAM28: evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem. J. 348, 21–27 [PMC free article] [PubMed] [Google Scholar]
- 34. Sottrup-Jensen L. (1989) α-Macroglobulins: structure, shape, and mechanism of proteinase complex formation. J. Biol. Chem. 264, 11539–11542 [PubMed] [Google Scholar]
- 35. Zhou W., Tsai H. (2009) N-Glycans of ADAMTS13 modulate its secretion and von Willebrand factor cleaving activity. Blood 113, 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shakin-Eshleman S. H., Wunner W. H., Spitalnik S. L. (1993) Efficiency of N-linked core glycosylation at asparagine-319 of rabies virus glycoprotein is altered by deletions C-terminal to the glycosylation sequon. Biochemistry 32, 9465–9472 [DOI] [PubMed] [Google Scholar]
- 37. Nilsson I., von Heijne G. (2000) Glycosylation efficiency of Asn-Xaa-Thr sequons depends both on the distance from the C terminus and on the presence of a downstream transmembrane segment. J. Biol. Chem. 275, 17338–17343 [DOI] [PubMed] [Google Scholar]
- 38. Seidah N. G., Chrétien M. (1999) Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 848, 45–62 [DOI] [PubMed] [Google Scholar]
- 39. Rawlings N. D., Barrett A. J., Bateman A. (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCall K. A., Huang C., Fierke C. A. (2000) Function and mechanism of zinc metalloenzymes. J. Nutr. 130, 1437S–1446S [DOI] [PubMed] [Google Scholar]
- 41. Ippolito J. A., Christianson D. W. (1994) Structural consequences of redesigning a protein-zinc binding site. Biochemistry 33, 15241–15249 [PubMed] [Google Scholar]
- 42. Kiefer L. L., Ippolito J. A., Fierke C. A., Christianson D. W. (1993) Redesigning the zinc binding site of human carbonic anhydrase II: structure of a His2Asp-Zn2+ metal coordination polyhedron. J. Am. Chem. Soc. 115, 12581–12582 [Google Scholar]
- 43. Reddy P., Slack J. L., Davis R., Cerretti D. P., Kozlosky C. J., Blanton R. A., Shows D., Peschon J. J., Black R. A. (2000) Functional analysis of the domain structure of tumor necrosis factor-α converting enzyme. J. Biol. Chem. 275, 14608–14614 [DOI] [PubMed] [Google Scholar]
- 44. Lampel J. S., Aphale J. S., Lampel K. A., Strohl W. R. (1992) Cloning and sequencing of a gene encoding a novel extracellular neutral proteinase from Streptomyces sp. strain C5 and expression of the gene in Streptomyces lividans 1326. J. Bacteriol. 174, 2797–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kurisu G., Sugimoto A., Harada S., Takagi M., Imanaka T., Kai Y. (1997) Characterization of a small metalloprotease from Streptomyces caespitosus with high specificity to aromatic residues. J. Ferment. Bioeng. 83, 590–592 [Google Scholar]
- 46. Kurisu G., Kai Y., Harada S. (2000) Structure of the zinc-binding site in the crystal structure of a zinc endoprotease from Streptomyces caespitosus at 1 Ä resolution. J Inorg Biochem. 82, 225–228 [DOI] [PubMed] [Google Scholar]
- 47. Harpel P. C., Hayes M. B., Hugli T. E. (1979) Heat-induced fragmentation of human α2-macroglobulin. J. Biol. Chem. 254, 8669–8678 [PubMed] [Google Scholar]