Background: The histone demethylase KDM4B is overexpressed in several tumor types and is oncogenic upon overexpression.
Results: Kdm4b-EGFP recruits to DNA damage induced by laser micro-irradiation. Kdm4b-EGFP overexpression enhanced double-strand break repair and increased survival following γ-irradiation.
Conclusion: Kdm4b enhances the DNA damage response.
Significance: Kdm4b overexpression may contribute to cytotoxic anti-cancer treatment resistance.
Keywords: Chromatin Regulation, Chromatin Remodeling, Chromatin Structure, DNA Damage Response, DNA Repair, Heterochromatin, Histone Methylation
Abstract
DNA damage evokes a complex and highly coordinated DNA damage response (DDR) that is integral to the suppression of genomic instability. Double-strand breaks (DSBs) are considered the most deleterious form damage. Evidence suggests that trimethylation of histone H3 lysine 9 (H3K9me3) presents a barrier to DSB repair. Also, global levels of histone methylation are clinically predictive for several tumor types. Therefore, demethylation of H3K9 may be an important step in the repair of DSBs. The KDM4 subfamily of demethylases removes H3K9 tri- and dimethylation and contributes to the regulation of cellular differentiation and proliferation; mutation or aberrant expression of KDM4 proteins has been identified in several human tumors. We hypothesize that members of the KDM4 subfamily may be components of the DDR. We found that Kdm4b-enhanced GFP (EGFP) and KDM4D-EGFP were recruited rapidly to DNA damage induced by laser micro-irradiation. Focusing on the clinically relevant Kdm4b, we found that recruitment was dependent on poly(ADP-ribose) polymerase 1 activity as well as Kdm4b demethylase activity. The Kdm4 proteins did not measurably accumulate at γ-irradiation-induced γH2AX foci. Nevertheless, increased levels of Kdm4b were associated with decreased numbers of γH2AX foci 6 h after irradiation as well as increased cell survival. Finally, we found that levels of H3K9me2 and H3K9me3 were decreased at early time points after 2 gray of γ-irradiation. Taken together, these data demonstrate that Kdm4b is a DDR protein and that overexpression of Kdm4b may contribute to the failure of anti-cancer therapy that relies on the induction of DNA damage.
Introduction
One of the hallmarks of cancer is the acquisition of genomic instability (1). Genomic instability includes not only discrete mutations and gross chromosomal alterations, but also instability at the level of chromatin or epigenomic instability. Global histone modifications have demonstrated predictive value in esophageal cancer (2, 3), non-small cell lung cancer (4, 5), pancreatic cancer (6, 7), liver cancer (8), prostate cancer (9), breast cancer (7, 10), and ovarian cancer (7). For example, low levels of dimethylated lysine 9 on histone 3 (H3K9me2)2 are predictive of earlier relapse and decreased survival in both prostate and kidney cancer (11). This reduction in H3K9me2 was associated with the global genome and repetitive DNA elements, but not specific gene promoters. Generally, low levels of repressive histone marks (i.e., H3K9me2/3, H3K4me3, and H3K27me3) are associated with poor prognostic indicators (5–10, 12), although the reverse relationship also has been reported (2, 13, 14). Histone methyltransferase levels also are predictive of patient outcomes. For example, levels of the H3K9 dimethyltransferase G9a are increased in lung adenocarcinoma and are predictive of poor prognosis (15). It is notable that overexpression of G9a in adenocarcinoma cell lines resulted in increased invasive capacity due to the gene silencing of Ep-CAM, but that global levels of H3K9me2 were not increased (15). These observations underline the complex balance of histone methyltransferases and demethylases as well as the importance of their regulation to epigenomic stability.
Evidence suggests that control of histone methylation plays an important role in the maintenance of genomic stability. For example, Suv39h1/Suv39h2 double knock-out (Suv39h1/h2-dn) mice display a tumor-prone phenotype, and cells from these mice display chromosomal instability (16–18). Similarly, reduction of the histone methyltransferase G9a results in chromosomal instability (19). Suv39h1/h2 and G9a exert their function through H3K9 methylation. Suv39h1/h2 generates H3K9me3, and G9a generates H3K9me1 and H3K9me2 (20–22). The association of disrupted H3K9 methylation with both chromosomal instability and poor patient prognosis points to a role for the regulation of H3K9 methylation in the suppression of tumorigenesis via the maintenance of genomic stability.
The repair of DNA damage is an important component of the maintenance of genomic stability. Upon the detection of DNA damage, a complex and highly regulated DNA damage response is evoked (23). The DDR includes the induction of cell cycle arrest, DNA repair, and/or apoptosis. In addition, the chromatin structure must be relaxed and histones evicted at the site of DNA damage before DNA repair proceeds. Once repair is complete, the histones and histone post-translational modifications must be faithfully reinstated (23, 24). The chromatin of Suv39h1/h2 double-null mouse embryonic fibroblasts (MEFs) exhibit a significantly less dense chromatin structure than their isogenic wild-type counterparts (25). This observation is not surprising as H3K9me3 is a heterochromatin-associated mark and provides binding sites for proteins responsible for the maintenance of heterochromatin structure. Reducing the degree of chromatin compaction before the induction of DNA damage has been associated with an amplified DDR (26). Therefore, H3K9 demethylases may be an important component of the DDR.
Double-strand breaks (DSBs) are considered the most deleterious form of DNA damage. If left unrepaired they can give rise to the gross chromosomal instability that typifies malignant cells (e.g. translocations). ATM (ataxia telangiectasia mutated) is the main signaling kinase after the induction of DSBs. Ataxia telangiectasia is characterized by cerebellar ataxia, immune defects, small dilated blood vessels, as well as predisposition to malignancy (27). Cells from ataxia telangiectasia patients are abnormally sensitive to ionizing radiation, and tumors from ataxia telangiectasia patients display chromosomal breakage (27). The phenotype of ataxia telangiectasia illustrates the importance of ATM function in the maintenance of genomic stability and the suppression of tumorigenesis.
Goodarzi et al. (28) demonstrated that inhibition of ATM results in the failure to resolve heterochromatin-associated DSBs induced by γ-irradiation. However, it was observed that this ATM dependence could be circumvented through the depletion of H3K9me3 or the constitutive unmasking of H3K9me3 through the depletion of HP1 isoforms (28). These data suggest that the requirement for ATM activation is reduced in cells with reduced levels of H3K9me3 and further support the concept that H3K9 demethylation is key to the maintenance of genomic stability in the wake of DNA damage.
Until the identification of the LSD1 demethylase in 2004 (29), H3K9 methylation was thought to be a permanent post-translational modification. More recently the Jumonji D2 (JMJD2/KDM4) subfamily of proteins has been characterized as histone demethylases. These proteins belong to a larger superfamily of demethylases defined by the presence of the JmjC domain, which is responsible for the conferring of demethylase activity (30). A major branch of this superfamily also contains a JmjN domain. When present, the JmjN domain interacts with the JmjC domain, forming the catalytic component (31). All KDM4 subfamily members contain both JmjC and JmjN domains, and KDM4A-C subfamily members contain two plant homeodomain-type zinc fingers (PHD domains) and two Tudor domains in their carboxyl termini. KDM4A-D demethylate H3K9me3, H3K9me2, and to a lesser extent, H3K36me3 (32).
KDM4 proteins have been shown to exert oncogenic pressure upon overexpression (for review, see Ref. 32). KDM4A-C are overexpressed in several human tumor types (32). In prostate and breast cancer, KDM4 proteins bind androgen and estrogen receptors. For breast cancer, data indicate that KDM4/ERα binding expands the number of ERα transcriptional targets and drives cellular proliferation (33, 34). Additionally, Kdm4 proteins have been shown to negatively regulate p53 (32, 35). Finally, KDM4C overexpression confers pleuripotency and is sufficient to induce transformation (36). The importance of H3K9 demethylation during DSB repair, together with the KDM4 demethylase profiles, point the KDM4 proteins as strong candidates for participants in the DDR. Moreover, the oncogenic capacity of KDM4 proteins and the association of decreased H3K9 methylation with poor patient outcomes suggests that KDM4 overexpression is clinically relevant to cancer progression.
Based on these observations, we investigated the potential role of KDM4A-D as DDR proteins. We found that both Kdm4b and KDM4D were recruited rapidly to DNA damage induced by laser micro-irradiation. Of these two KDM4 proteins, only KDM4B is strongly supported in the literature as having a role in cancer progression. Focusing on Kdm4b, we found that moderate overexpression of Kdm4b-EGFP resulted in enhanced early double-strand break repair and conferred a survival and growth advantage after γ-irradiation.
EXPERIMENTAL PROCEDURES
Cell Culture, Vectors, and Transfection
Cell lines were maintained in DMEM supplemented with 10% FBS at 37 °C and 5% CO2. The Suv39h1/h2-double null (dn) MEFs and isogenic control MEFs were provided by Dr. Thomas Jenuwein (Vienna, Austria) (16). Kdm4b-EGFP, Kdm4bΔJmjC-EGFP, and Kdm4bΔJmjN-EGFP expression vectors were provided by Dr. Jenuwein (Vienna, Austria) (37). Kdm4bH189A-EGFP was generated using the QuikChange site-directed mutagenesis kit (Stratagene) as per the manufacturer's instructions. Kdm4a, Kdm4c, and KDM4D Gateway donor vectors were constructed using clones from OriGene (Rockville, MD) as template cDNA. The Gateway modified monomeric EGFP expression vector was provided by Dr. Gordon Chan (Edmonton, AB, Canada). Transfections were performed using Effectene Transfection Reagent as per the manufacturer's protocol (Qiagen, Toronto ON). The Kdm4b-EGFP stable cell line was generated by transfecting the U2OS human osteosarcoma cell line with a non-linearized Kdm4-EGFP expression vector. Transfected cells were selected on the basis of the neomycin resistance gene (neo) contained in the expression vector. The resulting non-clonal cell line was then sorted to obtain a low expression stable cell line on the basis of EGFP signal.
Inhibitors
ATM was inhibited using 10 μm concentrations of the specific inhibitor KU0055933 (KuDos Pharmaceuticals, Cambridge UK). DNA-PK was inhibited with 50 nm of NU7441/KU 57788 (Tocris Bioscience, Bristol UK). PARP-1 was inhibited with 2.5 μm AG14361 (Selleckchem, Houston, TX). To reduce H3K9me2 levels, cells were treated with 800 nm concentrations of the G9a inhibitor UNC0638 (Sigma) for 3 days before experimentation. Control cells were grown under the same conditions in DMSO.
Two-photon Laser Micro-irradiation
Cells were plated on 35-mm glass bottom MatTec plates (Ashland, MA) and transfected as required. Twenty-four hours after plating and/or transfection, cells were incubated with 0.5 μg/ml Hoechst 33258 (Molecular Probes, Invitrogen) for 15 min. After the Hoechst-containing media was replaced, the dish was placed on the heated stage of a Zeiss LSM510 NLO laser-scanning confocal microscope equipped with a Plan-Neofluar 40×/1.3 N.A. oil immersion objective. DNA damage was generated by exciting the Hoechst 33258 using a near-infrared 750-nm titanium-sapphire laser line in a defined 1-μm band across the nucleus. The laser output was set to 10% power and 7 iterations. This combination of Hoechst sensitization and micro-irradiation generates a strong γH2AX signal that remains within the irradiated area. Time-lapse imaging was used to record the recruitment of EGFP-tagged protein to the DNA damage track using a 488-nm argon laser excitation and a 515–540-nm band-pass filter. For transfected cells, only low to moderate EGFP signals were selected for micro-irradiation. For immunofluorescence after micro-irradiation, cells were permitted to recover in a 37 °C humidified incubator for the indicated times and then were fixed as per indirect immunofluorescence staining, detailed below.
Fluorescence Recovery after Photo-bleaching
Fluorescence recovery after photo-bleaching (FRAP) was used to compare the relative chromatin binding of Kdm4b-EGFP both on and off the DNA damage track induced by two-photon laser micro-irradiation. Live U2OS cells transiently transfected with Kdm4b-EGFP were subjected to laser micro-irradiation as described above. FRAP was performed 120 s after micro-irradiation, a time by which Kdm4b-EGFP recruitment to the DNA damage tracks was shown to plateau (Fig. 1). A 1-μm band across the nucleus was photo-bleached using a 25-milliwatt argon laser (488 nm) with a band-pass filter of 505–530; this photo-bleached band was perpendicular to the laser micro-irradiation track. The high energy pulse resulted in permanent photo-beaching of EGFP in the 1-μm band. For proteins that are not tightly associated with the chromatin and diffuse freely throughout the nucleus, the EGFP signal in the photo-bleached region is recovered rapidly. Conversely, for a protein that is tightly chromatin-bound, the recovery of EGFP-tagged protein to the photo-bleached area takes substantially longer. For the resulting images, the EGFP intensity was measured in two 1-μm2 areas. The placement of the first square corresponded to the area at the intersection of the 1 μm FRAP and micro-irradiation tracks, and the second square was located on the FRAP track. Normalization was performed to account for global and/or cumulative photo-bleaching that may occur during the initial photo-bleaching and during imaging. EGFP intensity was expressed relative to pre-bleach intensity. Non-linear regression and curve comparison was performed using GraphPad Prism.
FIGURE 1.
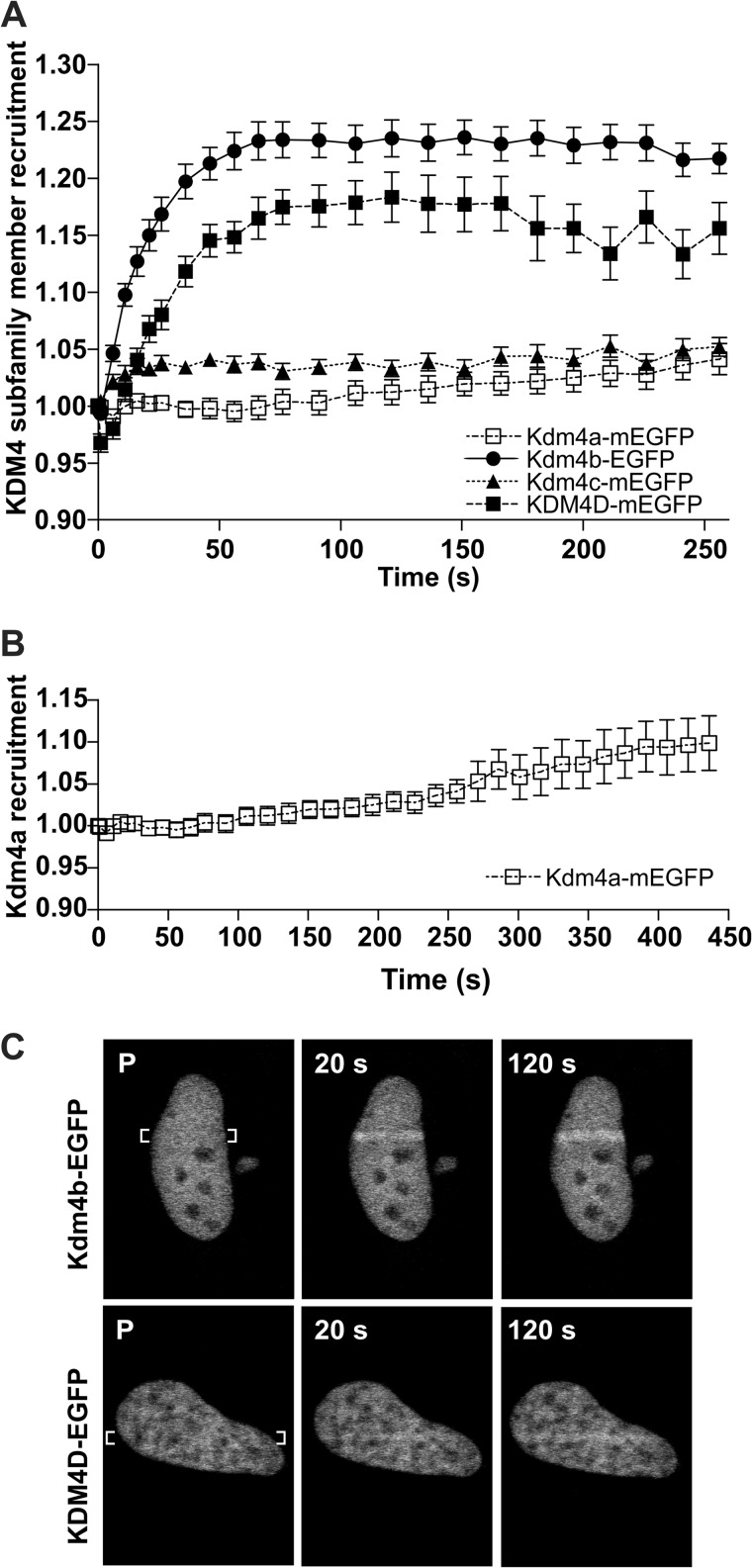
Kdm4b- and KDM4D-EGFP recruit to DNA damage. U2OS osteosarcoma cells were transfected with Kdm4b-EGFP. 24 h after transfection 2-photon laser micro-irradiation was used to measure the recruitment of EGFP-tagged Kdm4 demethylases to DNA damage. A, both Kdm4b-EGFP and KDM4D-EGFP recruited readily to the DNA damage tracks. Graphs represent the mean values for 18, 30, 25, and 16 nuclei ± S.E. (Kdm4a–c, respectively). B, Kdm4a-EGFP also recruited to the DNA damage tracks but at later time points. C, representative images show the accumulation of Kdm4b-EGFP (C, top) and KDM4D-EGFP (C, bottom) on the DNA damage tracks.
Indirect Immunofluorescence Microscopy and Quantification
Cells were plated on either glass coverslips or glass-bottom 35-mm tissue culture plates. After the desired treatment, cells were fixed with 4% paraformaldehyde and then permeabilized for 5 min in 0.5% Triton X-100, PBS, incubated with primary antibody for 1 h, washed with 0.1% Triton X-100, PBS, incubated with fluorescence-labeled secondary antibodies for 1 h, washed in 0.1% Triton X-100, PBS, and mounted in media containing 200 ng/ml DAPI in PBS, 90% glycerol and 1 mg/ml paraphenylenediamine. Antibodies used were as follows: γH2AX (Millipore 05-636 and Bethyl A300-081A), phospho-ATM Ser-1981 (Abcam ab13767), H3K9me3 (Upstate 07-442 and Active Motif 39162 (with H3K9me2 peptide added), H3K9me2 (Abcam ab1220), and H3K36me3 (Abcam ab9050). In experiments utilizing the Bethyl γH2AX primary antibody for γH2AX foci counting, cells were treated with CSK buffer (10 mm Pipes (pH 6.8), 300 mm sucrose, 50 mm NaCl, 3 mm MgCl2, 1 mm EGTA, and 0.5% Triton X-100) for 2 min before fixation to reduce background staining. Foci counting and staining quantification were performed on the resulting images using the CellProfiler open-source cell image analysis software (38–40). The ATM phosphorylation (pATM) foci were quantified using a high content analysis MetaXpress Micro XL system (Molecular Devices, Sunnyvale CA) using a 20 × 0.75 Plan Apo lens. Data were analyzed using MetaXpress software Version 5.0 (Molecular Devices) running on a Powercore system with 48 processors using a spot counting algorithm (Transfluor module) to quantify foci/nuclei and a cell masking algorithm (Cell Scoring module) to quantify KDM4-GFP expression.
Colony Formation Assay
Cells were plated at a density of 300 cells/60-mm tissue culture plate, allowed to settle for 24 h, and then irradiated at the stated conditions. Plates were incubated until the colony size in the untreated control plates was >50 cells, and then the plates were stained with crystal violet, and the colonies were counted. Each experiment was performed in triplicate. The MEFs were plated at half the density of the U2OS cells; the individual MEF colonies were less dense and physically larger than those of the U2OS cells.
RESULTS
Kdm4b and KDM4D Recruit to DNA Damage Induced by Laser Micro-irradiation
Human osteosarcoma U2OS cells were transfected with EGFP-tagged members of the KDM4 subfamily. Utilizing the EGFP tag to monitor localization, individual nuclei were subjected to laser micro-irradiation, and recruitment of the tagged proteins to the resulting DNA damage track was recorded using time-lapse microscopy. We found that Kdm4b-EGFP and KDM4D-mEGFP both recruited to laser micro-irradiation tracks, with maximum recruitment achieved by 90 s (Fig. 1, A and C). Kdm4b-EGFP recruited more readily than KDM4D-EGFP. Kdm4a also recruited to the laser micro-irradiation tracks, but at a later time point and at a lower relative abundance (Fig. 1B). There was no detectable recruitment of Kdm4c-mEGFP. KDM4B is overexpressed in several human cancers, displays oncogenic properties upon overexpression (32), and was recruited at higher levels to the DNA damage tracks (Fig. 1). Therefore, subsequent experiments focused on the impact for Kdm4b overexpression on the DNA damage response.
Kdm4b Recruitment Is Dependent on Demethylase Activity
To determine the functional requirement of Kdm4b for recruitment to DNA damage, laser micro-irradiation was performed using variants of Kdm4b-EGFP lacking either the JmjC or JmjN domains as well as a catalytically dead variant (Kdm4bH189A) (37). Both the ΔJmjN and ΔJmjC variants failed to accumulate on the micro-irradiation tracks, and Kdm4bH189A demonstrated at least an 80% decrease in recruitment (Fig. 2A). These data indicate that the demethylase activity of Kdm4b is required for retention at DNA damage sites induced by laser micro-irradiation.
FIGURE 2.
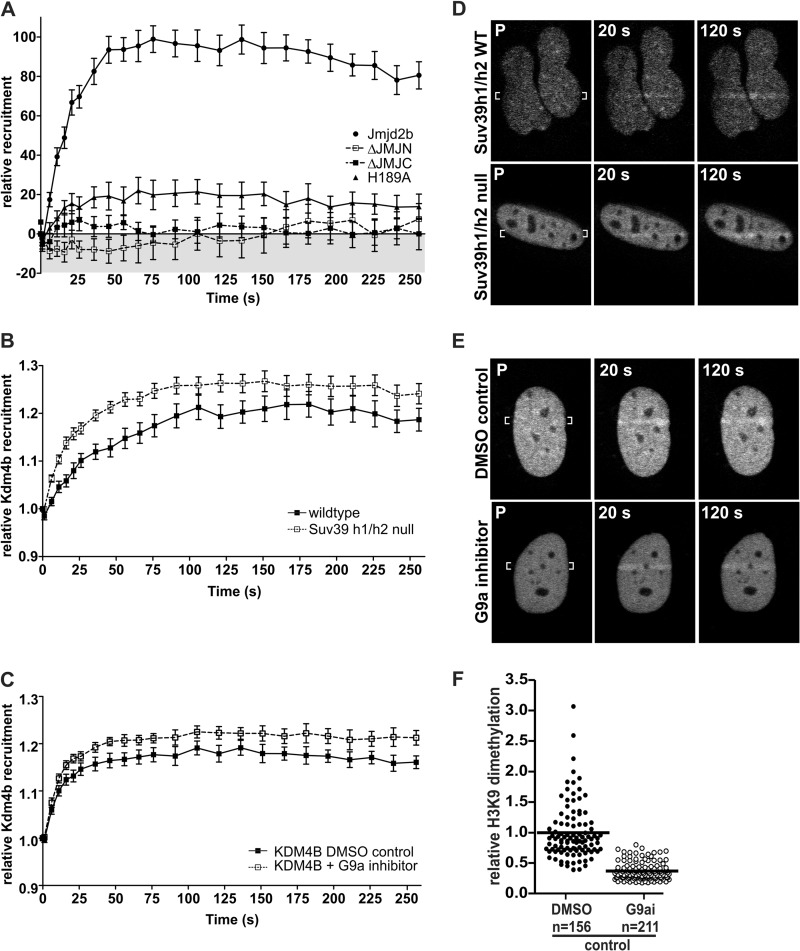
The influence of demethylase activity on the recruitment of Kdm4b-EGFP to DNA damage. A, deletion of either the JmjN domain or the JmjC domain abolished Kdm4b-EGFP recruitment to the DNA damage tracks. Mutational inactivation of Kdm4b-EGFP (H189A mutation in the JmjC domain) resulted in at least an 80% reduction in recruitment. Graphs represent the mean values for at least 12 nuclei ± S.E. B and D, Kdm4b-EGFP recruited more readily to the DNA damage tracks in the Suv39H1/H3-double null MEFS (n = 25) than in the isogenic wild type control MEFs (n = 15). P, pre-irradiation. C and E, similarly, reduction of H3K9me2 levels through the inhibition of the G9a methyltransferase (n = 21) resulted in enhanced Kdm4b-EGFP recruitment to the DNA damage tracks, as compared with DMSO control cells (n = 18). F, shown is an example of the decreased H3K9me2 methylation observed after 3 days of treatment with 800 nm G9a inhibitor UNC0638. Mean decreases in H3K9me2 between experiments ranged from 60 to 70%.
Kdm4b Recruitment Is Not Dependent on H3K9 Methylation
We next assessed the requirement of H3K9me3 and H3K9me2 for the recruitment of Kdm4b-EGFP to DNA damage induced by laser micro-irradiation. The Suv39h1/h2-dn cells are deficient in both the H1 and H2 homologues of the Su(var)3-9 histone methyltransferase, which methylates H3K9me1 to H3K9me3 (21). Using Suv39h1/h2-dnMEFs, we found that recruitment of Kdm4b-EGFP to laser micro-irradiation tracks was not impaired compared with isogenic wild type control MEFs. Indeed, the recruitment of Kdm4b-EGFP was both accelerated and increased in the Suv39h1/h2-dn cells (Fig. 2, B and D).
H3K9me2 levels were reduced using the G9a methyltransferase inhibitor UNC0638. Pretreatment of U2OS cells for 3 days resulted in a 60–70% reduction in H3K9me2 (Fig. 2F) but did not reduce the recruitment of Kdm4b-EGFP to the DNA damage tracks (Figs. 2, C and E). As in the Suv39h1/h2-dn cells, reduction of H3K9me2 enhanced the recruitment of Kdm4b-EGFP to the laser micro-irradiation tracks.
Kdm4b-EGFP Recruitment to Laser Micro-irradiation Tracks Is Dependent on PARP-1 but Not ATM, ATR, DNA-PK, or γH2AX
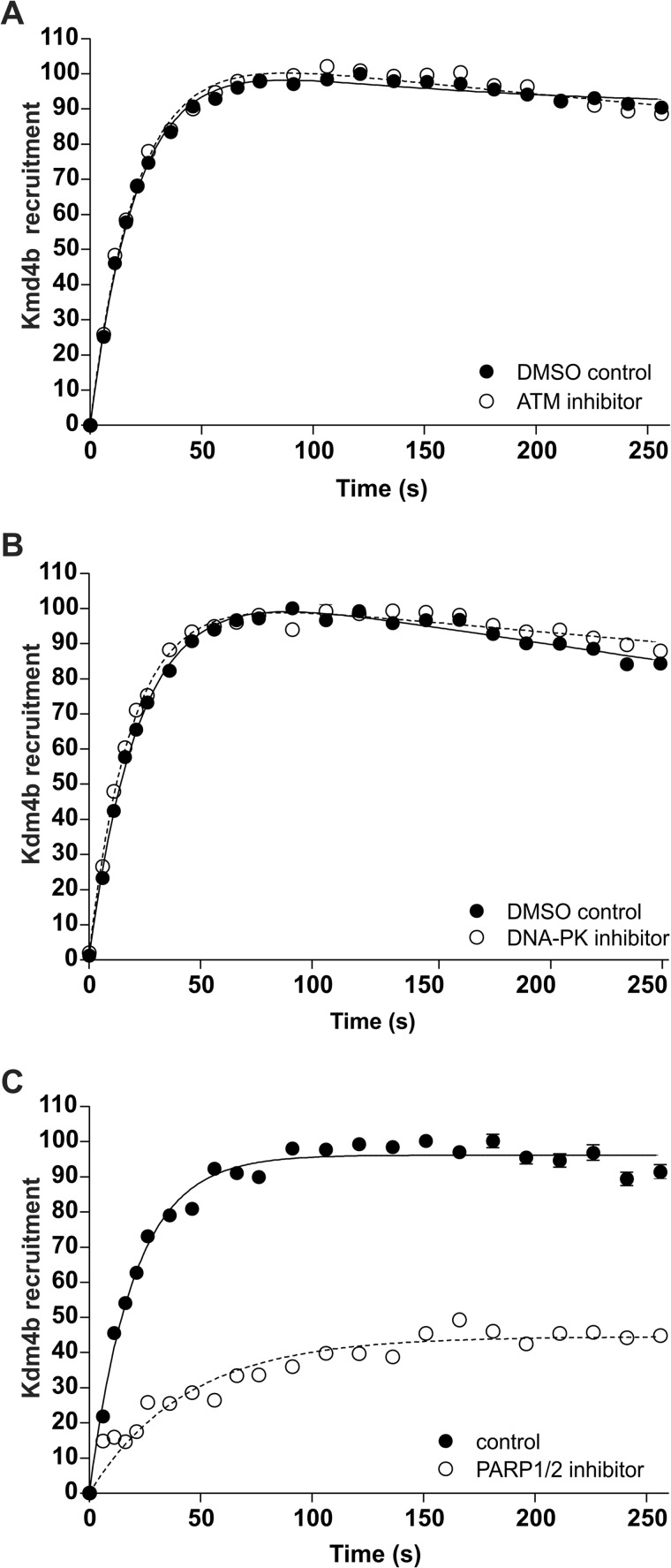
Laser micro-irradiation was performed for Kdm4b-EGFP in the presence of inhibitors specific for ATM, DNA-PK, and PARP-1. Inhibitor effects on recruitment were determined relative to appropriate solvent controls. We found that neither ATM inhibitor (10 μm) nor DNA-PK inhibitor (50 nm) had any measureable effect on the recruitment of Kdm4b-EGFP to the laser micro-irradiation tracks (Fig. 3, A and B). Control experiments verified that the ATM inhibitor abolished the phosphorylation of ATM on the DNA damage track (data not shown). In contrast, 2.5 μm PARP-1 inhibitor AG14361 resulted in a 60% decrease in Kdm4b-EGFP recruitment to laser micro-irradiation tracks (Fig. 3C).
FIGURE 3.
Kdm4b-EGFP is dependent on PARP-1 for recruitment to DNA damage. U2OS cells were transiently transfected with Kdm4b-EGFP. 24 h after transfection two-photon micro-irradiation was used to determine the requirement of Kdm4b-EGFP on key DDR proteins for the recruitment to DNA damage. The recruitment of Kdm4b-EGFP to the DNA damage tracks was not affected by ATM inhibition (A) or DNA-PK inhibition (B). C, inhibition of PARP-1 resulted in a 30% decrease in Kdm4b-EGFP recruitment. Cells were treated for 1 h before micro-irradiation. Graphs represent the mean values for at least 18 nuclei ± S.E.
Kdm4b-EGFP Displays Increased Turnover on the Laser Micro-irradiation Tracks
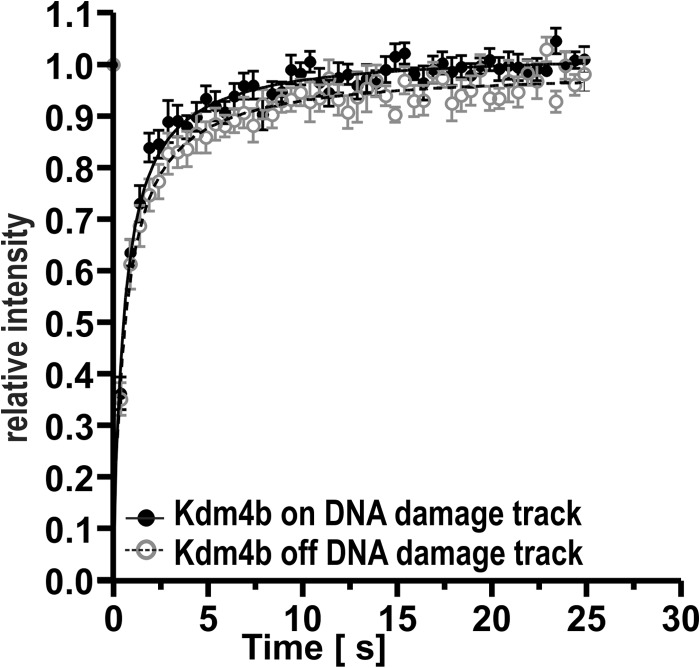
FRAP was performed on individual cells expressing Kdm4b-EGFP 2 min after laser micro-irradiation. The recovery of the EGFP-tagged Kdm4b was quantified both on and off the DNA damage tracks. We found that Kdm4b-EGFP recovered more rapidly on the DNA damage tracks, indicating that Kdm4b-EGFP was less tightly bound in the context of DNA damage induced by two-photon laser micro-irradiation (Fig. 4). This difference was small but significant (p < 0.0001, curve-fitting analysis GraphPad Prism). The more rapid recovery of Kdm4b-EGFP is indicative of a more rapid turnover on the DNA damage track or increased Kdm4b-EGFP re-recruitment. This suggests that the binding site in laser micro-irradiated chromatin is distinct from that in undamaged chromatin.
FIGURE 4.
FRAP. FRAP was used to compare the chromatin binding of Kdm4b-EGFP both on and off the DNA damage tracks induced by 2-photon micro-irradiation (n = 10). Kdm4b-EGFP recovered significantly more rapidly on the DNA damage track (p < 0.001, curve-fitting analysis GraphPad Prism).
KDM4 Proteins Do Not Form γ-Irradiation-induced Foci at DSBs
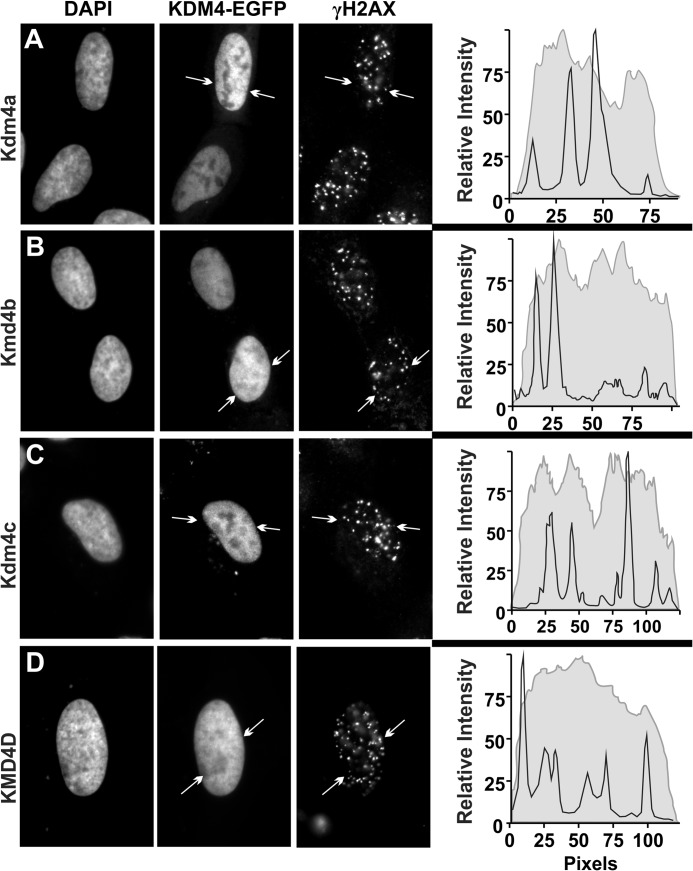
Twenty-four hours after transfection with one of the four Kdm4 EGFP-tagged proteins, U2OS cells were exposed to 2Gy of γ-irradiation and were fixed immediately or allowed to respond for up to 6 h (a 1-h recovery is shown in Fig. 5). Using immunofluorescence for the DSB biomarker γH2AX, relative abundance of EGFP-tagged KDM4 demethylases at γ-irradiation-induced foci (IRIF) was examined. We found no evidence of increased abundance of the KDM4 subfamily members at IRIF at any time point examined. This observation is not without precedent. The DSB repair proteins Ku70/80 also do not form IRIF (41).
FIGURE 5.
Kdm4-EGFP does not accumulate at IRIF. U2OS cells were plated on glass coverslips and transfected with EGFP-tagged Kdm4a (A), Kdm4b (B), Kdm4c (C), or KDM4D (D). 24 h after transfection, cells were treated with 2 Gy of γ-irradiation and fixed at 60 min post-irradiation. Cells were stained for γH2AX, and the nuclei were counterstained with DAPI. Line scans show the fluorescence intensity for the EGFP-tagged Kdm4 protein (gray line, curve area filled) and the Cy3-stained γH2AX (black line). The line scans shown were performed on the lines indicated in the Kdm4-EGFP panels (delineated by arrows).
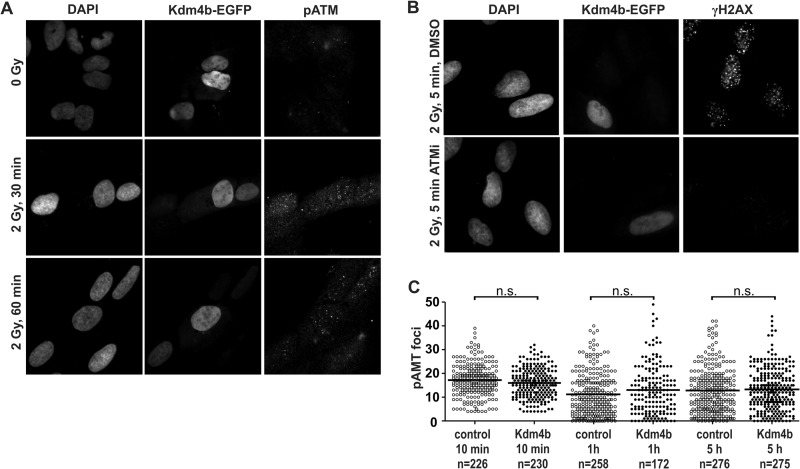
ATM Activation Is Not Abrogated by Reduced H3K9 Methylation
Previous studies have linked H3K9 methylation, ATM activation (pATM) and DSB repair. It has been reported previously that ATM is not activated in cells overexpressing KDM4A or KDM4D after 5 Gy irradiation (42). In contrast, we found that ATM activation and pATM foci formation was proficient in Kdm4b-EGFP-overexpressing cells after 2 Gy of γ-irradiation (Fig. 6, A and C). This observation was confirmed using γH2AX foci formation as a readout for ATM activation (43) in the presence or absence of ATM inhibitor. At 5 min post-irradiation, the overexpression of Kdm4b-EGFP efficiently generated γH2AX foci in an ATM-dependent manner (Fig. 6B), supporting our observation that Kdm4b did not abrogate ATM activation (Fig. 6, A and C).
FIGURE 6.
ATM activation is proficient in Kdm4b-overexpressing cells. U2OS cells were plated on glass coverslips and transfected with Kdm4b-EGFP. 24 h after transfection the plated cells were exposed to 2 Gy γ-irradiation and then fixed at 30 and 60 min recovery. Indirect immunofluorescence was used to stain phosphorylated ATM. Nuclei were counterstained with DAPI. Images were obtained and processed to ensure quantitative micrographs. Exposure times were held constant, and the resulting images were scaled identically. B, γH2AX foci formation 5 min after 2 Gy γ-irradiation as a readout for ATM activation. Kdm4b-EGFP cells were proficient at the formation of ATM-dependent γH2AX foci. C, shown is quantification of γ-irradiation-induced pATM foci in Kdm4b-EGFP-positive and -negative cells. Overexpression of Kdm4b-EGFP resulted in no significant change in pATM foci numbers. n.s., not significant.
Kdm4b Overexpression Enhances the Repair of γ-Irradiation-induced DSBs
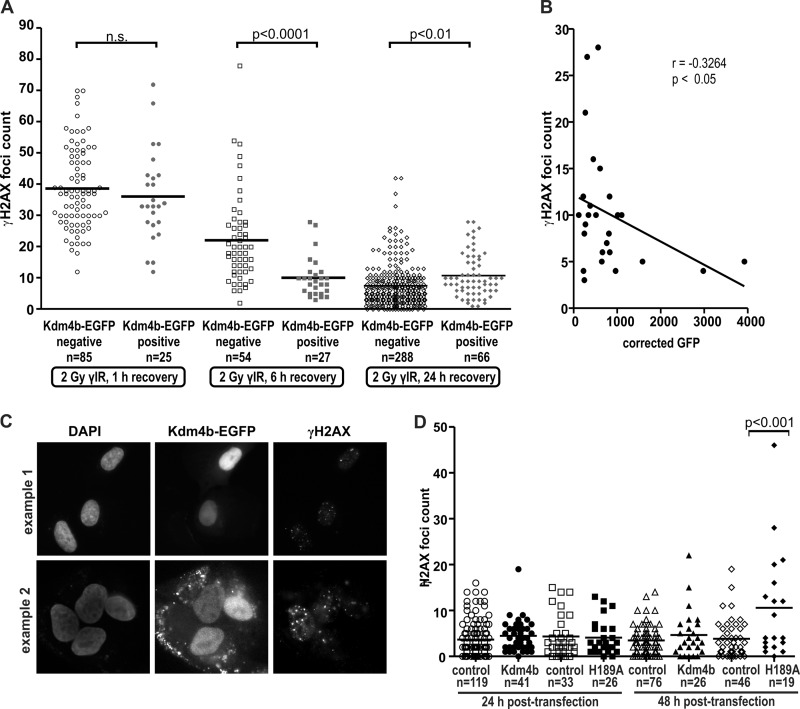
Twenty-four hours after transfection of Kdm4b-EGFP, U2OS cells were exposed to 2 Gy of γ-irradiation. Cells were fixed at 1, 6, and 24 h post-irradiation and then analyzed for numbers of γH2AX foci compared with non-irradiated controls. At 1 h after irradiation, we found that the number of γH2AX foci in the EGFP-positive cells was comparable to the foci numbers in EGFP-negative cells (Fig. 7A). However, 6 h after irradiation the Kdm4b-EGFP-positive cells displayed a significantly lower number of γH2AX foci than the non-overexpressing cells (Fig. 7A). After 24 h of recovery the Kdm4b-overexpressing cells displayed no significant change in the number of γH2AX foci compared with the 6-h time point, whereas the cells not overexpressing Kdm4b (EGFP-negative) continued to resolve the γH2AX foci. There was a small but significant difference between the EGFP-positive and -negative cells at 24 h post-irradiation. The EGFP-positive cells displayed an increased mean number of γH2AX foci. Correlation analyses at 6 h post-irradiation confirmed that there was a negative correlation between the levels of EGFP and γH2AX foci (Fig. 7, B and C). Therefore, increased Kdm4b levels enhanced early repair of DSBs, as assayed by the biomarker γH2AX.
FIGURE 7.
Kdm4b overexpression promotes DSB repair. A, U2OS cells were plated on glass coverslips and transfected with Kdm4b-EGFP. 24 h after transfection, the plated cells were treated with 2 Gy of γ-irradiation (γIR) and fixed at 1, 6, and 24 h post-irradiation. Cells were stained by immunofluorescence for γH2AX, and the nuclei were counterstained with DAPI. The numbers of γH2AX foci were counted after 1, 6, and 24 h recovery in both Kdm4b-EGFP-positive cells and neighboring untransfected (control) cells. The mean number of foci is indicated by the horizontal bar. Statistical differences were determined by using a two-tailed t test. n.s., not significant. B, levels of Kdm4b-EGFP were negatively correlated with the number of remaining γH2AX foci after 6 h recovery. Correlation was measured using one-tailed Pearson correlation calculation. The mean EGFP intensity was measured for individual masked nuclei using CellProfiler. C, two examples show the inverse correlation between Kdm4b-EGFP levels and γH2AX foci at 6 h post-irradiation. D, U2OS cells were plated onto glass coverslips and transfected with either wild type Kdm4b-EGFP or inactive Kdm4bH189A-EGFP. At 24 and 48 h post-transfection, coverslips were fixed and stained for γH2AX foci, and the nuclei were counterstained with DAPI. Foci numbers were counted for the transfected cells as well as the EGFP-negative control cells. Kdm4H189A-EGFP/48h had a significantly (p < 0.001) higher number of γH2AX foci than any other experimental condition (one way analysis of variance); all other comparisons did not show significant differences.
To verify that the overexpression of Kdm4b-EGFP did not induce an increase in spontaneous damage that would confound data interpretation, γH2AX foci numbers were analyzed 24 and 48 h post-transfection in non-irradiated cells. We found that overexpression of Kdm4b-EGFP did not result in increased γH2AX foci numbers, whereas overexpression of the demethylase dead mutant (H189A) resulted in the induction of spontaneous γH2AX foci by 48 h (Fig. 7D). This suggests that the catalytic dead mutant has a dominant-negative effect on DNA repair.
DNA Damage-induced H3K9 Demethylation; Laser Micro-irradiation
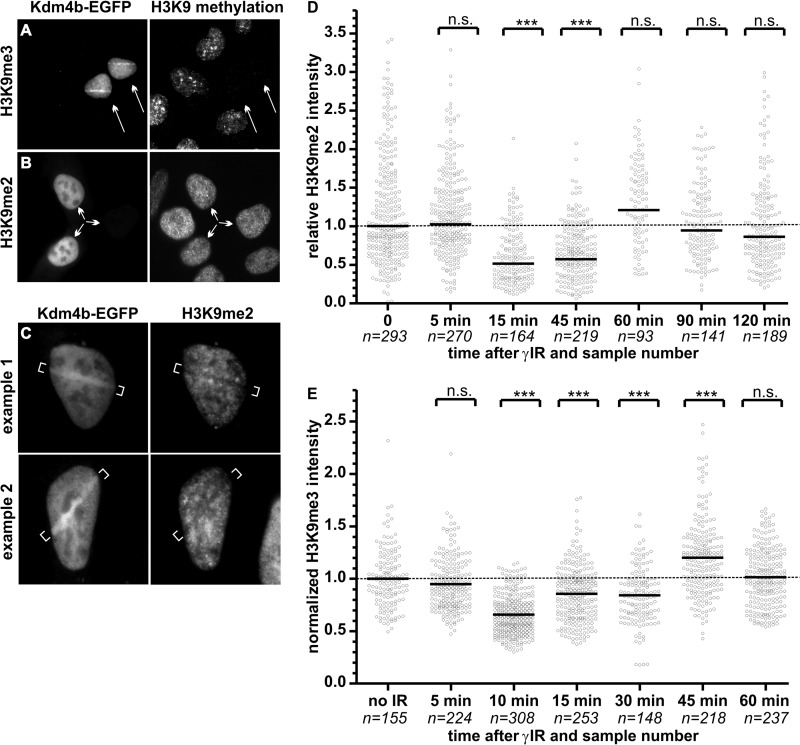
Using U2OS cells transiently transfected with Kdm4b-EGFP, we performed laser micro-irradiation followed by 10 min of recovery, timed from irradiation of the first cell. The actual recovery time ranged from 6 to 10 min, and Kdm4b was still present on the DNA damage track (Fig. 8, A and C). Residual Kdm4b remained on the DNA damage tracks for as long as 20 min (data not shown). Fixed cells were then stained for H3K9me3 or H3K9me2 in order to investigate the degree of demethylation on the DNA damage tracks. For the experiments reported in this manuscript, only cells displaying low to moderate Kdm4b-EGFP levels were used. Using this level of transient transfection in U2OS cells, we observed that cells overexpressing Kdm4b-EGFP displayed a complete loss of H3K9me3 by immunofluorescence (Fig. 8A). In contrast, the H3K9me2 signal was retained for the levels of Kdm4b-EGFP used in these experiments (Fig. 8B). Cells with high levels of Kdm4b-EGFP overexpression did display a reduction in H3K9me2 (data not shown), which is consistent with the established Kdm4b demethylase profile (44).
FIGURE 8.
DNA damage-induced changes in H3K9 methylation. 24 h after transfection, U2OS cells expressing low level Kdm4b-EGFP demonstrated a complete loss of H3K9me3 (A) but not H3K9me2 (B). C, U2OS cells were plated on glass-bottom dishes and transfected with Kdm4b-EGFP. 24 h after transfection, DNA damage was induced in EGFP-positive cells using two-photon laser micro-irradiation. Ten minutes after micro-irradiation of the first cell, the plates were fixed and stained for H3K9me2 by immunofluorescence. Two examples are shown. D and E, U2OS plated on glass coverslips were treated with two Gy of γ-irradiation (γIR). Plated cells were fixed at denoted time-points after irradiation. Cells were stained by immunofluorescence for H3K9me3 (D) or H3K9me2 (E), and the nuclei were counterstained with DAPI. H3K9me2 levels were quantified for each nuclei (sample numbers are shown on the graph) using CellProfiler and expressed relative to the mean H3K9me3 levels of the non-irradiated control cells. The mean methylation level was compared with the non-irradiated control using an unpaired t test with Welch's correction for unequal variance. n.s., not significant. p > 0.05. ***, p < 0.0001.
We observed evidence of reduced H3K9me2 on the DNA damage track induced by laser micro-irradiation in Kdm4b-overexpressing cells (Fig. 8C). In non-transfected cells (i.e. no Kdm4b-EGFP expression), we did not find evidence of decreased H3K9me2 or H3K9me3 on the DNA damage tracks (data not shown). Therefore, overexpression of Kdm4-EGFP resulted in a global loss of H3K9me3 and a DNA damage-induced decrease in H3K9me2 after micro-irradiation.
DNA damage-induced H3K9 Demethylation: γ-Irradiation
As outlined in the Introduction, evidence suggests that demethylation of H3K9 is important to the DDR. Based on our observations of decreased H3K9me2 on the micro-irradiation-induced DNA damage tracks (Fig. 8C), we measured the global alterations in H3K9me2 and H3K9me3 levels in non-transfected cells at early time points after 2 Gy irradiation. In the absence of Kdm4b overexpression, we found that global levels of H3K9me2 were rapidly reduced by 50% and remained at this level for approximately 1 h after γ-irradiation (Fig. 8D). This 50% decrease in dimethylation was reproducible between four independent experiments. Inter-experimental variability affected the time at which H3K9me2 levels began to return to base-line levels. Similarly, global levels of H3K9me3 rapidly decreased after 2Gy γ-irradiation (Fig. 8E). However, unlike the decrease in H3K9me2, the observed decrease in H3K9me3 was transient. The irradiation-induced decreases in H3K9 di- and trimethylation were validated by Western blot analysis with decreased methylation levels of 45 and 58% observed, respectively.
The Effect of H3K9 Demethylation before Irradiation
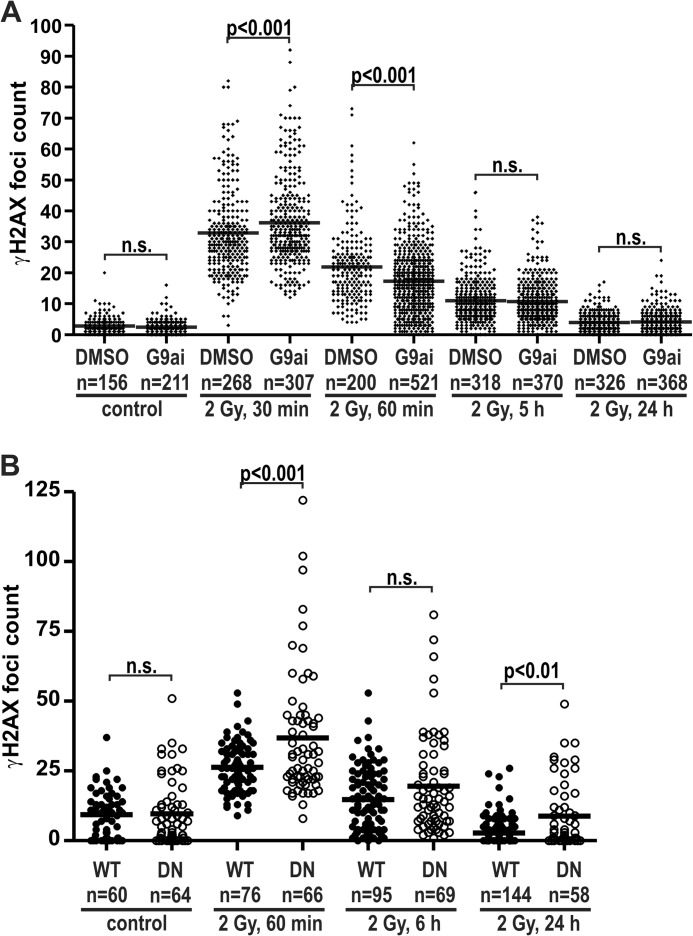
U2OS cells were cultured in 800 nm G9a inhibitor for 3 days before irradiation, resulting in at least a 60% decrease in H3K9me2 (Fig. 2F). On day 3 of G9a inhibition, cells plated on glass coverslips were exposed to 2 Gy γ-irradiation and allowed to recover for the specified time. Immunofluorescence was performed for γH2AX, and the IRIF foci were counted. The reduction of H3K9me2 resulted in significantly higher numbers of γH2AX foci at 30 min post-irradiation. However, 30 min later the H3K9me2-defficient cells had significantly fewer γH2AX foci, thus demonstrating accelerated repair. At later time points no repair advantage was conferred by the constitutive reduction of H3K9me2 (Fig. 9A).
FIGURE 9.
Constitutively reduced H3K9 methylation reduced DSB repair. A, U2OS cells were pretreated with 800 nm concentrations of the G9a inhibitor (G9ai in DMSO) UNC0638 or DMSO alone (control) and plated onto glass coverslips. Three days after treatment initiation, the cells were exposed to 2 Gy of γ-irradiation. The cells were fixed at the time points indicated and stained for γH2AX and H3K9me2 by immunofluorescence. γH2AX foci and H3K9me2 levels were quantified by CellProfiler software. The mean foci number is denoted by the horizontal bar. Total numbers of nuclei assays are indicated on the graph. The number γH2AX foci were compared for each time point using analysis of variance with Bonferroni's correction for multiple comparisons. G9a inhibition resulted in a 63% reduction in H3K9me3 levels at the time of γ-irradiation (Fig. 2F). B, Suv39h1/h2 double null MEFs (DN) and isogenic wild type control MEFs (WT) were assayed for γH2AX foci numbers, as above. n.s., not significant.
Similarly, Suv39h1/h2-dn and wild type cells were used to compare the repair of IRIF in the context of constitutive H3K9me3 reduction. The Suv39h1/h2-dn cells harbored significantly higher numbers of γH2AX foci 1 h after irradiation (Fig. 9B). Normalization of these data to the 1-h time point confirmed that there was no significant difference in the degree of repair 6 h after irradiation. However, 24 h post-irradiation the H3K9me3-deficient cells (DN) demonstrated a mean number of γH2AX foci that was ∼2-fold higher than that of the isogenic wild type MEFs (WT, Fig. 9B). This could reflect increased formation of spontaneous foci that have been found in heterochromatin in the absence of this lysine 9 methylation (45) and is also observed in untreated Suv39h1/h2-dn MEFs (data not shown).
Kdm4b-EGFP Overexpression Confers Increased γ-Irradiation Survival
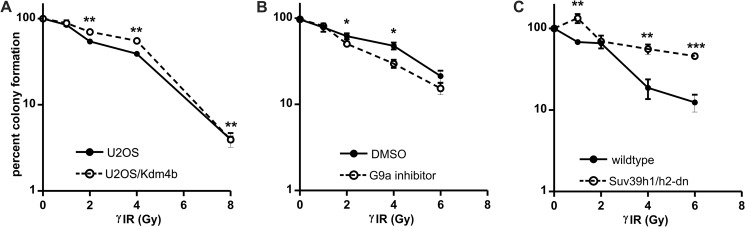
The survival of Kdm4-EGFP stable cells after γ-irradiation was compared with the survival of the parental U2OS cell line. Using the colony formation assay, we found that low level overexpression of Kdm4b-EGFP conferred a moderate, but significant, survival advantage (Fig. 10A). Moreover, the colonies for the Kdm4b-EGFP-overexpressing cells were larger than for the control U2OS cells. Next, we performed the colony formation assay with U2OS cells that had been pretreated with the G9a inhibitor for 3 days, reducing global H3K9me2 levels by 60–70% (see Fig. 2F). Reduction of H3K9me2 levels before γ-irradiation resulted in decreased cell survival (Fig. 10B). Colony formation assays for the Suv39h1/h2-dn and isogenic wild type MEFs were difficult to analyze, as these cells do not form dense colonies. However, data collected indicate that the Suv39h1/h2-dn cells may be less sensitive to γ-irradiation than their wild type counterparts (Fig. 10C). Therefore, the low level of Kdm4b overexpression conferred both a survival and proliferative advantage after the induction of DNA damage by γ-irradiation and may be related to the constitutive loss of H3K9me3. It should be noted that in the cases of G9a inhibition, reduced H3K9me2 levels were induced before irradiation, whereas H3K9me2 levels are not markedly altered by Kdm4b overexpression.
FIGURE 10.
Low level Kdm4b overexpression confers decreased sensitivity to γ-irradiation. A, stable U2OS/Kdm4b-EGFP cells were sorted for low levels of Kdm4b-EGFP expression. The resulting U2OS/Kdm4b-EGFPlow and parental U2OS cells were plated at low levels for the colony formation assay. 24 h after plating, the cells were irradiated and then were allowed to recover until the resulting colonies reached at least 50 cells. The plates were then stained with crystal violet to allow visualization of the colonies; both cell lines were plated and stained on the same day. The experiment was performed in triplicate. Colonies were counted and expressed relative to untreated controls. B, U2OS cells were treated with G9a inhibitor or DMSO (control). On day 2 of treatment the pretreated cells were plated for the colony formation assay, as above. C, the sensitivity of the Suv39h1/h2-dn and isogenic wild type MEFs was compared using the colony formation assay, as above γIR, γ-irradiation.
DISCUSSION
The induction of DNA damage elicits a complex cellular response termed the DDR. Evidence supports the concept that reduction of H3K9 methylation may be a key component of the DDR. The association of constitutively reduced H3K9 methylation with genomic instability, tumorigenesis, and poor patient prognoses illustrates the requirement for tight control over H3K9 methylation. KDM4A-C demethylate H3K9me3, H3K9me2, and H3K36me3 (44) and are overexpressed in several tumor types (32). Based on these observations, we investigated the potential contribution of KDM4 subfamily members to the DDR. We show here that the clinically relevant Kdm4b demethylase is a component of the DDR and that overexpression of Kdm4b confers a survival and growth advantage after γ-irradiation-induced DNA damage.
Using two-photon laser micro-irradiation, we found that both Kdm4b- and KDM4D-EGFP were rapidly recruited to the DNA damage tracks. Kdm4a-EGFP also recruited but at much later time points. KDM4A and KDM4B are overexpressed in several human tumors, and their overexpression is associated with increased proliferation, invasive capacity, and anchorage-independent cell growth (for review, see Ref. 32). KDM4C also is overexpressed in human tumors, but its overexpression is associated with the promotion of pluripotency (46, 47). In keeping with this different phenotype, Kdm4c showed no recruitment to DNA damage tracks. KDM4D has not been reported as overexpressed in clinical samples. Therefore, we focused on the role of KDM4B in the DDR, as it both recruits to DNA damage induced by laser micro-irradiation and is clinically relevant.
We next investigated the biochemical requirements for Kdm4b-EGFP recruitment to DNA damage tracks. Inhibition of the key DDR signaling kinases (ATM, ATR, and DNA-PK) did not diminish Kdm4b-EGFP recruitment. In contrast, inhibition of PARP-1 resulted in a 60% decrease in Kdm4b-EGFP recruitment to the DNA damage tracks. One of the earliest proteins recruited to DNA adducts, the catalytic activity of PARP-1 increases 10–500-fold upon DNA damage detection, and poly-ADP-ribose chains can be detected 15–30 s after the induction of DNA damage (48). These poly-ADP-ribose chains in turn recruit hundreds of additional DDR proteins. In addition to an established role in response to single-strand breaks, PARP-1 has been identified as one of the first proteins recruited to DSBs (49). PARP-1 is in a complex with MRE11 and is required for the rapid recruitment of the MRE11/RAD50/NBS1 (MRN) complex to DSBs (49), underlining the importance of PARP-1 in DSB repair. The remaining recruitment of Kdm4b-EGFP in the presence of PARP-1 inhibitor may be attributable to PARP-3. Although PARP-2 is not a main contributor to the DDR provoked by DSBs (for review, see Ref. 50), PARP-3 has been reported to function in synergy with PARP-1 in response to DSBs (51). The requirement of Kdm4b-EGFP on PARP-1, but not ATM, ATR, or DNA-PK, places Kdm4b as an early responder in the DDR.
We found that the demethylase function of Kdm4b was required for accumulation on the micro-irradiation-induced DNA damage tracks, but that neither H3K9me3 nor H3K9me2 was obligatory (Fig. 2). Fodor et al. (37) reported that Kdm4b-EGFP was not localized to the H3K9me3-rich chromocenters in mouse cells. However, removal of the PHD and Tudor targeting domains resulted in relocalization of the truncated Kdm4b1–424 to the chromocenters (37). This aberrant localization was relieved upon addition of the H189A inactivating mutation in the JmjC domain. These data suggest that the catalytic domain participates in the recruitment of Kdm4b to heterochromatin, but that this binding is normally masked by higher affinity binding outside of heterochromatin. Thus, our data showing a requirement for the catalytic domain in retention at DSBs is consistent with this domain being involved in targeting to some sites in chromatin.
Having demonstrated by laser micro-irradiation that Kdm4b is a DDR protein, we next sought to determine the impact of Kdm4b overexpression on the repair of γ-irradiation induced DSBs. We found that the Kdm4 proteins did not accumulate at sufficient levels to form IRIF (Fig. 5 and Ref. 52). These observations parallel the characteristics of DSB repair proteins Ku70 and Ku80. Ku70 and Ku80 recruit to micro-irradiation tracks (41, 53) but do not accumulate at sufficient levels to form IRIF (54). Given the more rapid turnover of Kdm4b-EGFP on the micro-irradiation DNA damage track and the localization patterns observed by Fodor et al. (Ref. 37 and summarized above), the failure of Kdm4 to accumulate at IRIF was not unexpected.
In keeping with our identification of Kdm4b as a DDR protein, overexpression of Kdm4b-EGFP enhanced the repair of DSBs, as monitored by γH2AX foci. Moreover, the overexpression of inactive Kdm4b resulted in a significant increase in the number of spontaneous DSBs, supporting a role for Kdm4b in facilitating the repair of DSBs. Although the numbers of the remaining γ-irradiation-induced γH2AX foci were equivalent by 24 h (Fig. 7), low level overexpression of Kdm4b-EGFP conferred both a survival and proliferative advantage after 2Gy of γ-irradiation (Fig. 10). Therefore, H3K9 demethylases may be an important component of the DDR. This is consistent with observations that reducing the degree of chromatin compaction before the induction of DNA damage is associated with an amplified DDR (26). Sun et al. (42) reported that loss of the H3K9me3 mark resulted in a failure to activate ATM. Loss of H3K9me3 was modeled through the overexpression of Kdm4a or Kdm4d as well as in Suv39h1/h2-dn MEFs. ATM-deficient cells demonstrate DSB repair defects and radio-sensitivity (55). Our observations of Kdm4b-associated increased survival and enhanced DSB repair kinetics are inconsistent with the concept of a Kdm4b-induced defect in ATM activation. Goodarzi et al. (28) observed that loss of H3K9me3 circumvented the requirement for ATM in the repair of DSB, suggesting that the requirement for ATM in DSB repair is reduced in the context of reduced H3K9me3 levels. In testing ATM activation, we found that pATM foci formation was intact in Kdm4b-overexpressing cells (Fig. 6) despite the loss of H3K9me3. Moreover, ATM inhibition did not impact the recruitment of Kdm4b-EGFP to micro-irradiation-induced DNA damage, and ATM inhibition did not alter the Kdm4b-EGFP binding characteristics on the DNA damage track (Fig. 3 and data not shown). However, ATM inhibition did reduce the chromatin binding characteristics of Kdm4b-EGFP in the absence of exogenous DNA damage (data not shown). Also, we observed an increase in spontaneous DSBs with the overexpression of demethylase-dead Kdm4bH189A-EGFP (Fig. 7D). These data support a role for Kdm4b in the suppression of genomic instability in the absence of exogenous DNA damage as well as a role in the DSB-induced DDR.
Global changes in histone methylation in response to DNA damage are not well documented, and studies are difficult to compare. Because our data indicate that the demethylase activity of Kdm4b contributes to the DDR and the repair of DSBs, we chose to investigate the levels of H3K9me2 and H3K9me3 immediately after γ-irradiation in cells that do not overexpress Kdm4b. Fnu et al. (56) observed an increase in H3K9me2 after DSB-induction, whereas others have reported increased H3 acetylation (57) or no change in H3K9me3 (42, 58). However, these studies did not survey time points immediately after the induction of DNA. Changes in H3K9me2 in response to DNA damage have not been reported previously. In human U2OS cells (no Kdm4b overexpression), we observed a rapid decrease in both H3K9me2 and H3K9me3 after DNA damage induced by 2Gyγ-irradiation (Fig. 8, D and E). The decrease in H3K9me3 was transient, whereas H3K9me2 levels were suppressed for 1 h after irradiation. This DNA-damage-induced reduction in H3K9 methylation is supported by reports of DNA damage-induced chromatin expansion in Drosophila that persisted for up to 1 h and was partially ATM-dependent (59). The authors concluded that the chromatin expansion enabled extraction of DSBs from within the heterochromatin, thus facilitating repair. It is notable that DNA damage-induced reductions in H3K9 methylation are consistent with the global chromatin relaxation that occurs after DNA damage (24).
It has been shown previously that the H3K9me3-deficient Suv39h1/h2-dn cells repair DSBs more slowly than their wild type counterparts (28), and our data are consistent with this observation. Moreover, we demonstrated that despite reduced DBS repair, as assayed by γH2AX foci, the Suv39h1/h2-dn cells display an increased tolerance to γ-irradiation-induced DNA damage. This is consistent with a recent report by Zheng et al. (60) demonstrating that overexpression of Suv39h1 increased sensitivity to γ-irradiation. Taking into account the increased genomic instability and tumor-prone phenotype associated with Suv39h1/h2 deficiency (16–18), it is possible that reductions in H3K9me3 confer an increased tolerance to DNA damage. Comparatively little is known about the impact of H3K9me2 deficiency on the DDR. Using U2OS cells pretreated with a specific G9a inhibitor, we found that reduced H3K9me2 levels were associated with enhanced repair DBS at 1 h after irradiation (Fig. 9). However, inhibition of G9a conferred increased sensitivity to γ-irradiation (Fig. 10B), suggesting that the DNA damage-induced reduction in H3K9me2 may not be sufficient to account for the resistance associated with Kdm4b overexpression (Fig. 10A). It is important to note that in the case of G9a inhibition, reduced H3K9me2 levels were present before irradiation, whereas overexpression of Kdm4b did not result in a constitutive loss of H3K9me2.
It has previously been shown that Kdm4a regulates DNA repair through competition with 53BP1 for H4K20 (52). We and Mallette et al. (52) demonstrated that overexpression of either Kmd4a or Kdm4b reduced the numbers of DNA damage induced-53BP1 foci, although the reduction was greater for Kdm4a. Of interest is their observation that blocking 53BP1 foci formation was not dependent on the demethylase activity of Kdm4a, whereas we show in this manuscript that the recruitment of Kdm4b-EGFP to DNA damage was dependent on the catalytic capacity of Kdm4b. Furthermore, the overexpression of Kdm4a (wild type or demethylase dead) or Kdm4b sensitized U2OS cells to the cytotoxic effects of etoposide (52). One key difference between these two manuscripts is that this manuscript focuses exclusively on cells with low levels of Kdm4b-EGFP overexpression, which would not be expected to abrogate the formation of 53BP1 foci3. Examination of the Oncomine database reveals KDM4B overexpression in cancer to be in the range 1.5-fold. Also, we found that cells expressing high levels of Kdm4b-EGFP were rapidly lost during cell culture and were associated with cells undergoing cell death. Indeed, a second stable cell line with moderate-to-high levels of Kdm4b-EGFP was more sensitive to the cytotoxic effects of γ-irradiation (data not shown). However, this stable cell line lost Kdm4-EGFP signal over time, whereas the cell line selected for low levels of Kdm4-EGFP (Fig. 10) remained stable. Therefore, these data suggest that Kdm4b has two opposing effects on the DDR, depending on the degree of overexpression.
Finally, we addressed whether the changes induced by moderate Kdm4b overexpression were biologically relevant. We found that moderate overexpression of Kdm4b-EGFP induced a 1.3–1.4-fold increase in survival after 2–4 Gy of γ-irradiation. During the revision of this manuscript, Zheng et al. (60) reported that p53 was required for H3K9me3 down-regulation after 5 Gy γ-irradiation and that p53 was required for the regulation of Kdm4b. This association between p53 and Kdm4 family members has been reported previously (61–63). Zheng et al. (60) also reported that Kdm4b knockdown increased sensitivity to DSBs. Our data support and extend these observations by demonstrating that reductions in either H3K9me3 or H3K9me2 accelerated DSB repair and increased survival. In addition, our data place the H3K9 histone demethylase Kdm4b as an early component of the DDR. Moreover, we examined the impact of Kdm4b overexpression at clinically relevant levels, which would be expected to result in a complete removal of the H3K9me3 mark. Kdm4b has not been reported as having decreased expression in clinical tumor samples. Therefore, the data reported here and by Zheng et al. (60) address two different clinical scenarios, KDM4B overexpression and p53 deficiency, respectively. It might be expected that combined p53 deficiency and KDM4B overexpression would have an even greater impact on the response to DNA damage-based anticancer therapies.
In summary, we identified Kdm4b as a DDR protein that is dependent on its inherent catalytic capacity and PARP-1 function but not key DDR kinases. Overexpression of Kdm4b has demonstrated oncogenicity (32), and we show here that Kdm4b also confers resistance to γ-irradiation and enhances the repair of DSBs. This increased tolerance to exogenous DNA damage may be related to the constitutive reduction in H3K9me3 and possibly the DNA damage-induced reduction in H3K9me2. These data suggest that Kdm4b overexpression may contribute to the failure of cytotoxic anti-cancer treatment.
Acknowledgments
We thank Dr. X. Sun and Geraldine Barron of the Cross Cancer Institute Imaging Facility.
This work was supported by the Alberta Cancer Foundation and the Canadian Institutes of Health Research.
F. Mallette, personal communication.
- H3K9
- lysine 9 on histone 3
- me2 and me3
- di- and trimethyl
- MEF
- mouse embryonic fibroblast
- DDR
- DNA damage response
- DSB
- double-strand breaks
- FRAP
- fluorescence recovery after photo-bleaching
- ATM
- ataxia telangiectasia mutated
- IRIF
- γ-irradiation-induced foci
- pATM
- ATM phosphorylation
- Gy
- gray
- EGFP
- enhanced GFP
- PARP-1
- poly(ADP-ribose) polymerase 1
- PK
- protein kinase
- dn
- double null.
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer. The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 2. Tzao C., Tung H. J., Jin J. S., Sun G. H., Hsu H. S., Chen B. H., Yu C. P., Lee S. C. (2009) Prognostic significance of global histone modifications in resected squamous cell carcinoma of the esophagus. Mod. Pathol. 22, 252–260 [DOI] [PubMed] [Google Scholar]
- 3. I H., Ko E., Kim Y., Cho E. Y., Han J., Park J., Kim K., Kim D. H., Shim Y. M. (2010) Association of global levels of histone modifications with recurrence-free survival in stage IIB and III esophageal squamous cell carcinomas. Cancer Epidemiol. Biomarkers Prev. 19, 566–573 [DOI] [PubMed] [Google Scholar]
- 4. Barlési F., Giaccone G., Gallegos-Ruiz M. I., Loundou A., Span S. W., Lefesvre P., Kruyt F. A., Rodriguez J. A. (2007) Global histone modifications predict prognosis of resected non small-cell lung cancer. J. Clin. Oncol. 25, 4358–4364 [DOI] [PubMed] [Google Scholar]
- 5. Song J. S., Kim Y. S., Kim D. K., Park S. I., Jang S. J. (2012) Global histone modification pattern associated with recurrence and disease-free survival in non-small cell lung cancer patients. Pathol. Int. 62, 182–190 [DOI] [PubMed] [Google Scholar]
- 6. Manuyakorn A., Paulus R., Farrell J., Dawson N. A., Tze S., Cheung-Lau G., Hines O. J., Reber H., Seligson D. B., Horvath S., Kurdistani S. K., Guha C., Dawson D. W. (2010) Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma. results from RTOG 9704. J. Clin. Oncol. 28, 1358–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei Y., Xia W., Zhang Z., Liu J., Wang H., Adsay N. V., Albarracin C., Yu D., Abbruzzese J. L., Mills G. B., Bast R. C., Jr., Hortobagyi G. N., Hung M. C. (2008) Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol. Carcinog. 47, 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tamagawa H., Oshima T., Shiozawa M., Morinaga S., Nakamura Y., Yoshihara M., Sakuma Y., Kameda Y., Akaike M., Masuda M., Imada T., Miyagi Y. (2012) The global histone modification pattern correlates with overall survival in metachronous liver metastasis of colorectal cancer. Oncol. Rep. 27, 637–642 [DOI] [PubMed] [Google Scholar]
- 9. Ellinger J., Kahl P., von der Gathen J., Rogenhofer S., Heukamp L. C., Gütgemann I., Walter B., Hofstädter F., Büttner R., Müller S. C., Bastian P. J., von Ruecker A. (2010) Global levels of histone modifications predict prostate cancer recurrence. Prostate. 70, 61–69 [DOI] [PubMed] [Google Scholar]
- 10. Elsheikh S. E., Green A. R., Rakha E. A., Powe D. G., Ahmed R. A., Collins H. M., Soria D., Garibaldi J. M., Paish C. E., Ammar A. A., Grainge M. J., Ball G. R., Abdelghany M. K., Martinez-Pomares L., Heery D. M., Ellis I. O. (2009) Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 69, 3802–3809 [DOI] [PubMed] [Google Scholar]
- 11. Seligson D. B., Horvath S., McBrian M. A., Mah V., Yu H., Tze S., Wang Q., Chia D., Goodglick L., Kurdistani S. K. (2009) Global levels of histone modifications predict prognosis in different cancers. Am. J. Pathol. 174, 1619–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ellinger J., Kahl P., Mertens C., Rogenhofer S., Hauser S., Hartmann W., Bastian P. J., Büttner R., Müller S. C., von Ruecker A. (2010) Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. Int. J. Cancer 127, 2360–2366 [DOI] [PubMed] [Google Scholar]
- 13. He C., Xu J., Zhang J., Xie D., Ye H., Xiao Z., Cai M., Xu K., Zeng Y., Li H., Wang J. (2012) High expression of trimethylated histone H3 lysine 4 is associated with poor prognosis in hepatocellular carcinoma. Hum. Pathol. 43, 1425–1435 [DOI] [PubMed] [Google Scholar]
- 14. Park Y. S., Jin M. Y., Kim Y. J., Yook J. H., Kim B. S., Jang S. J. (2008) The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann. Surg. Oncol. 15, 1968–1976 [DOI] [PubMed] [Google Scholar]
- 15. Chen M. W., Hua K. T., Kao H. J., Chi C. C., Wei L. H., Johansson G., Shiah S. G., Chen P. S., Jeng Y. M., Cheng T. Y., Lai T. C., Chang J. S., Jan Y. H., Chien M. H., Yang C. J., Huang M. S., Hsiao M., Kuo M. L. (2010) H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 70, 7830–7840 [DOI] [PubMed] [Google Scholar]
- 16. Peters A. H., O'Carroll D., Scherthan H., Mechtler K., Sauer S., Schöfer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., Opravil S., Doyle M., Sibilia M., Jenuwein T. (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 [DOI] [PubMed] [Google Scholar]
- 17. McManus K. J., Biron V. L., Heit R., Underhill D. A., Hendzel M. J. (2006) Dynamic changes in histone H3 lysine 9 methylations. Identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J. Biol. Chem. 281, 8888–8897 [DOI] [PubMed] [Google Scholar]
- 18. Heit R., Rattner J. B., Chan G. K., Hendzel M. J. (2009) G2 histone methylation is required for the proper segregation of chromosomes. J. Cell Sci. 122, 2957–2968 [DOI] [PubMed] [Google Scholar]
- 19. Kondo Y., Shen L., Ahmed S., Boumber Y., Sekido Y., Haddad B. R., Issa J. P. (2008) Downregulation of histone H3 lysine 9 methyltransferase G9a induces centrosome disruption and chromosome instability in cancer cells. PLoS ONE 3, e2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tachibana M., Sugimoto K., Fukushima T., Shinkai Y. (2001) Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276, 25309–25317 [DOI] [PubMed] [Google Scholar]
- 21. Peters A. H., Kubicek S., Mechtler K., O'Sullivan R. J., Derijck A. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Martens J. H., Jenuwein T. (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 22. Robin P., Fritsch L., Philipot O., Svinarchuk F., Ait-Si-Ali S. (2007) Post-translational modifications of histones H3 and H4 associated with the histone methyltransferases Suv39h1 and G9a. Genome Biol. 8, R270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rossetto D., Truman A. W., Kron S. J., Côté J. (2010) Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin. Cancer Res. 16, 4543–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cann K. L., Dellaire G. (2011) Heterochromatin and the DNA damage response. The need to relax. Biochem. Cell Biol. 89, 45–60 [DOI] [PubMed] [Google Scholar]
- 25. Casas-Delucchi C. S., van Bemmel J. G., Haase S., Herce H. D., Nowak D., Meilinger D., Stear J. H., Leonhardt H., Cardoso M. C. (2012) Histone hypoacetylation is required to maintain late replication timing of constitutive heterochromatin. Nucleic Acids Res. 40, 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murga M., Jaco I., Fan Y., Soria R., Martinez-Pastor B., Cuadrado M., Yang S. M., Blasco M. A., Skoultchi A. I., Fernandez-Capetillo O. (2007) Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 178, 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lavin M. F. (2008) Ataxia-telangiectasia. From a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 9, 759–769 [DOI] [PubMed] [Google Scholar]
- 28. Goodarzi A. A., Noon A. T., Deckbar D., Ziv Y., Shiloh Y., Löbrich M., Jeggo P. A. (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 31, 167–177 [DOI] [PubMed] [Google Scholar]
- 29. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 30. Whetstine J. R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Shi Y. (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 [DOI] [PubMed] [Google Scholar]
- 31. Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T. G., Simpson M., Mao Q., Pan C. H., Dai S., Hagman J., Hansen K., Shi Y., Zhang G. (2006) Structural insights into histone demethylation by JMJD2 family members. Cell 125, 691–702 [DOI] [PubMed] [Google Scholar]
- 32. Young L. C., Hendzel M. J. (2012) The oncogenic potential of Jumonji D2 (JMJD2/KDM4) histone demethylase overexpression. Biochem. Cell Biol. 10.1139/bcb-2012-0054 [DOI] [PubMed] [Google Scholar]
- 33. Kawazu M., Saso K., Tong K. I., McQuire T., Goto K., Son D. O., Wakeham A., Miyagishi M., Mak T. W., Okada H. (2011) Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS ONE 6, e17830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang J., Jubb A. M., Pike L., Buffa F. M., Turley H., Baban D., Leek R., Gatter K. C., Ragoussis J., Harris A. L. (2010) The histone demethylase JMJD2B is regulated by estrogen receptor α and hypoxia and is a key mediator of estrogen-induced growth. Cancer Res. 70, 6456–6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mallette F. A., Richard S. (2012) JMJD2A promotes cellular transformation by blocking cellular senescence through transcriptional repression of the tumor suppressor CHD5. Cell Rep. 2, 1233–1243 [DOI] [PubMed] [Google Scholar]
- 36. Liu G., Bollig-Fischer A., Kreike B., van de Vijver M. J., Abrams J., Ethier S. P., Yang Z. Q. (2009) Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 28, 4491–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fodor B. D., Kubicek S., Yonezawa M., O'Sullivan R. J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T. (2006) Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20, 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., Golland P., Sabatini D. M. (2006) CellProfiler. image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jones T. R., Kang I. H., Wheeler D. B., Lindquist R. A., Papallo A., Sabatini D. M., Golland P., Carpenter A. E. (2008) CellProfiler Analyst. Data exploration and analysis software for complex image-based screens. BMC Bioinformatics 9, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kamentsky L., Jones T. R., Fraser A., Bray M. A., Logan D. J., Madden K. L., Ljosa V., Rueden C., Eliceiri K. W., Carpenter A. E. (2011) Improved structure, function, and compatibility for CellProfiler. Modular high-throughput image analysis software. Bioinformatics 27, 1179–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koike M., Koike A. (2008) Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp. Cell Res. 314, 1061–1070 [DOI] [PubMed] [Google Scholar]
- 42. Sun Y., Jiang X., Xu Y., Ayrapetov M. K., Moreau L. A., Whetstine J. R., Price B. D. (2009) Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 11, 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. (2001) ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276, 42462–42467 [DOI] [PubMed] [Google Scholar]
- 44. Hillringhaus L., Yue W. W., Rose N. R., Ng S. S., Gileadi C., Loenarz C., Bello S. H., Bray J. E., Schofield C. J., Oppermann U. (2011) Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J. Biol. Chem. 286, 41616–41625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peng J. C., Karpen G. H. (2009) Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet 5, e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Loh Y. H., Zhang W., Chen X., George J., Ng H. H. (2007) Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang J., Zhang M., Zhang Y., Kou Z., Han Z., Chen D. Y., Sun Q. Y., Gao S. (2010) The histone demethylase JMJD2C is stage-specifically expressed in preimplantation mouse embryos and is required for embryonic development. Biol Reprod. 82, 105–111 [DOI] [PubMed] [Google Scholar]
- 48. Rouleau M., Patel A., Hendzel M. J., Kaufmann S. H., Poirier G. G. (2010) PARP inhibition. PARP1 and beyond. Nat. Rev. Cancer 10, 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haince J. F., McDonald D., Rodrigue A., Déry U., Masson J. Y., Hendzel M. J., Poirier G. G. (2008) PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208 [DOI] [PubMed] [Google Scholar]
- 50. Sousa F. G., Matuo R., Soares D. G., Escargueil A. E., Henriques J. A., Larsen A. K., Saffi J. (2012) PARPs and the DNA damage response. Carcinogenesis 33, 1433–1440 [DOI] [PubMed] [Google Scholar]
- 51. Boehler C., Gauthier L. R., Mortusewicz O., Biard D. S., Saliou J. M., Bresson A., Sanglier-Cianferani S., Smith S., Schreiber V., Boussin F., Dantzer F. (2011) Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. U.S.A. 108, 2783–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mallette F. A., Mattiroli F., Cui G., Young L. C., Hendzel M. J., Mer G., Sixma T. K., Richard S. (2012) RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31, 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hong Z., Jiang J., Lan L., Nakajima S., Kanno S., Koseki H., Yasui A. (2008) A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 36, 2939–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mirzoeva O. K., Petrini J. H. (2001) DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kühne M., Riballo E., Rief N., Rothkamm K., Jeggo P. A., Löbrich M. (2004) A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 64, 500–508 [DOI] [PubMed] [Google Scholar]
- 56. Fnu S., Williamson E. A., De Haro L. P., Brenneman M., Wray J., Shaheen M., Radhakrishnan K., Lee S. H., Nickoloff J. A., Hromas R. (2011) Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. U.S.A. 108, 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yin D. T., Wang Q., Chen L., Liu M. Y., Han C., Yan Q., Shen R., He G., Duan W., Li J. J., Wani A., Gao J. X. (2011) Germline stem cell gene PIWIL2 mediates DNA repair through relaxation of chromatin. PLoS ONE 6, e27154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tjeertes J. V., Miller K. M., Jackson S. P. (2009) Screen for DNA damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 28, 1878–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chiolo I., Minoda A., Colmenares S. U., Polyzos A., Costes S. V., Karpen G. H. (2011) Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zheng H., Chen L., Pledger W. J., Fang J., Chen J. (2013) p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression. Oncogene 10.1038/onc.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li W., Zhao L., Zang W., Liu Z., Chen L., Liu T., Xu D., Jia J. (2011) Histone demethylase JMJD2B is required for tumor cell proliferation and survival and is overexpressed in gastric cancer. Biochem. Biophys. Res. Commun. 416, 372–378 [DOI] [PubMed] [Google Scholar]
- 62. Kim T. D., Shin S., Berry W. L., Oh S., Janknecht R. (2012) The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J. Cell. Biochem. 113, 1368–1376 [DOI] [PubMed] [Google Scholar]
- 63. Kim T. D., Oh S., Shin S., Janknecht R. (2012) Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS ONE 7, e34618. [DOI] [PMC free article] [PubMed] [Google Scholar]