Background: Hsp110s are considered as mere nucleotide exchange factors of the Hsp70s.
Results: Human cytosolic Hsp110s can use ATP to unfold misfolded polypeptides and act as equal partner with Hsp70 to solubilize stable protein aggregates.
Conclusion: Hsp110s are Hsp70-like polypeptide unfolding chaperones.
Significance: Hsp110s are powerful disaggregating chaperones that can collaborate with Hsp70s to detoxify misfolding proteins in degenerative diseases.
Keywords: ATPases, Heat Shock Protein, Molecular Chaperone, Protein Aggregation, Protein Misfolding
Abstract
Structurally and sequence-wise, the Hsp110s belong to a subfamily of the Hsp70 chaperones. Like the classical Hsp70s, members of the Hsp110 subfamily can bind misfolding polypeptides and hydrolyze ATP. However, they apparently act as a mere subordinate nucleotide exchange factors, regulating the ability of Hsp70 to hydrolyze ATP and convert stable protein aggregates into native proteins. Using stably misfolded and aggregated polypeptides as substrates in optimized in vitro chaperone assays, we show that the human cytosolic Hsp110s (HSPH1 and HSPH2) are bona fide chaperones on their own that collaborate with Hsp40 (DNAJA1 and DNAJB1) to hydrolyze ATP and unfold and thus convert stable misfolded polypeptides into natively refolded proteins. Moreover, equimolar Hsp70 (HSPA1A) and Hsp110 (HSPH1) formed a powerful molecular machinery that optimally reactivated stable luciferase aggregates in an ATP- and DNAJA1-dependent manner, in a disaggregation mechanism whereby the two paralogous chaperones alternatively activate the release of bound unfolded polypeptide substrates from one another, leading to native protein refolding.
Introduction
Under optimal in vitro conditions, such as low protein concentrations and low temperature, the primary amino acid sequences may contain all the necessary information for small unfolded polypeptide chains to spontaneously reach their native three-dimensional structure (1). However, the cytoplasm of human cells may contain as much as 200 mg/ml proteins (2, 3), a highly crowded environment that may interfere with the productive native folding pathway of de novo synthesized or translocated multidomain polypeptides and with the proper refolding of stress-unfolded polypeptides (4–6). When a new polypeptide chain exits the ribosome or an organellar import pore or when a labile native protein becomes transiently heat- or cold-denatured, it may transiently unfold and expose hydrophobic segments to the aqueous environment (7, 8). It may then seek higher stability either by refolding to the native state or by generating non-native intramolecular misfolded structures, predominantly short β-strands with exposed hydrophobic residues (8). Depending on the intensity and duration of a stress and the degree of hydrophobic exposure, the misfolded monomers may then further proceed into the misfolding pathway by seeking intermolecular hydrophobic associations, leading to the formation of various insoluble disordered complexes, generally termed aggregates, that may further condense in vitro into compact protofilaments and insoluble fibrils, or in vivo, into tangles, protofibrils, and extracellular amyloids (7, 9–11). Stable misfolded monomers, such as freeze-thaw-inactivated luciferase (FTluc)2 (12) or freeze-thaw-inactivated rhodanese (8, 13), may thus serve as the start points of an alternative protein misfolding and aggregation pathway, which, at variance with the native folding pathway, may be toxic to animal cells, neurons in particular (10, 14, 15). Because of their high surface-to-volume ratio, intermediate misfolded proteins, such as stable soluble aggregates (16) and protofibrils, may be among the most toxic species (17), causing mammalian cell leakage, inflammation-induced oxidative stress, apoptosis, and tissue loss in neurodegenerative diseases such as Alzheimer, Huntington, and Parkinson diseases, amyotrophic lateral sclerosis, and aging in general (10, 17, 18).
Opportunely, both prokaryotes and eukaryotes have evolved a complex network of chemical and protein chaperones (19, 20), which can prevent the conversion of stress-labile native proteins into toxic aggregates. Moreover, proteotoxic species that already formed can be actively converted by disaggregating and unfolding chaperones into harmless natively refoldable proteins or be degraded by chaperone-gated proteases into reusable amino acids (14). Therefore, stress-induced or crowding-induced protein misfolding and aggregation can be efficiently counteracted by a network of so-called “holding” chaperones that upon binding to exposed hydrophobic residues on the surface of the misfolded polypeptides can prevent the formation of large aggregates (21–24). However, with the exception of the small Hsps, the other canonical families of molecular chaperones are ATPases with demonstrated abilities to act as polypeptide unfolding enzymes. Thus, even without ATP, apoGroEL and apoCCT chaperonins can act as effective enzymes, unfolding metastable misfolded monomeric species and thus averting further proceeding into the misfolding pathway and instead promoting native refolding (10, 12, 13, 24). Other molecular chaperones, such as Hsp104 and Hsp70, are strict ATP-dependent polypeptide unfoldases that can disentangle stable protein aggregates and unfold stable misfolded monomeric polypeptides species. Thus, Hsp70 (DnaK; the names of the Escherichia coli proteins are shown in parentheses), co-chaperoned by Hsp40 (DnaJ), regulated by nucleotide exchange factors (GrpE), and the disaggregating co-chaperone Hsp104 (ClpB) can use the energy of ATP hydrolysis to scavenge stable insoluble aggregates and unfold stably misfolded polypeptides by binding, clamping, and pulling apart entangled misfolded structures and whole polypeptides (12, 25–27).
Whereas in bacteria, chloroplasts, mitochondria, and the cytosol of plants and yeast, Hsp104/ClpB type chaperones can use their hexameric ring structure to pull apart stably entangled polypeptides in aggregates and collaborate with Hsp70 to recover native proteins, animal cells lack obvious Hsp104 orthologs. However, animal cells are not devoid of disaggregating chaperones, because recent in vivo and in vitro data showed that the mammalian Hsp110 (HSPH2, Apg-2) can act in synergy with Hsp70 (HSPA8, Hsc70) and Hsp40 (Ydj1, DNAJB1) to catalyze the ATP-dependent disentanglement, solubilization, and native refolding of various stable protein aggregates (28, 29).
The human cytosol contains three distinct Hsp110s: HSPH2 (Apg-2), HSPH1 (Hsp105), and HSPH3, present in 800, 510, and 130 copies per cubic micron of HeLa cell, respectively (28). Amounts and stoichiometries indicate that ∼15% of the cytosolic Hsp70 pool may be involved in the formation of Hsp70-Hsp110-dependent disaggregating machineries (3). Here, we tested with stringent in vitro refolding assays the chaperone activity of purified recombinant human HSPH1 in various combinations with HSPA1A and DNAJA, which are abundant cytosolic members of the Hsp70/110/40 chaperone family in human cells (3). This allowed us to assess the precise mechanistic contribution of Hsp110, individually as an independent molecular chaperone and as a possible chaperone that reciprocally collaborate with Hsp70 in the in vitro unfolding and reactivation of stable misfolded species and the disaggregation of extensively damaged protein substrates preformed in the absence of chaperones (12, 28, 29). We found that Hsp110 is by itself a bona fide ATP-dependent unfolding chaperone that can catalyze the unfolding of stable misfolded polypeptides and thus favor the conversion of stable, high affinity misfolded substrates in to stable low affinity native products of the chaperone reaction. Interestingly, even without ATP, Hsp110 could activate the release of a prebound luciferase substrate from Hsp70, and reciprocally Hsp70 could activate the release of a prebound luciferase substrate from Hsp110. Titration of the ATP- and HSP40-dependent luciferase refolding activity in the presence of various relative amounts of Hsp110 and Hsp70 showed optimal disaggregation activity at a 1:1 ratio. Confirming earlier in vivo and in vitro evidence with human HSPH2, yeast YDJ1, or the other major cytosolic human DNAJB1 (28, 29), our in vitro results mainly with HSPH1 (Hsp105) and DNAJA1 show that the cytosol and likewise the endoplasmic reticulum of mammalian cells contain powerful Hsp110/Hsp70 bichaperone machineries that can unfold and solubilize stably misfolded and aggregated protein species to combat the onset of protein conformational diseases.
EXPERIMENTAL PROCEDURES
Cloning
HSPA1A (HSP70, HsCD00002890) and DNAJA1 (HSP40, HsCD00002610) were amplified from cDNA, using following set of primers: HSPA1A forward, 5′-ATGATGGGCTCTTCTCATCATCATCATCATCATTCTTCTGGCCTGGTTCCGCGTGGCTCTCATATGGCCAAAGCCGCGGCGATCGGCATCG-3′; HSPA1A reverse, 5′-CGTGGCCACTAGTCTAATCCACCTCCTCAATGG-3′; DNAJA1 forward, 5′-ATGATGGGCTCTTCTCATCATCATCATCATCATTCTTCTGGCCTGGTTCCGCGTGGCTCTCATATGGTGAAAGAAACAACTTACTACG-3′; and DNAJA1 reverse, 5′-CGTGGCCACTAGTTTAAGAGGTCTGACACTGAACACCACCTC-3′. The PCR product with the N-terminal His6 tag and the thrombin cleavage site was cloned in between restriction sites NcoI and SpeI of the pSE420 expression vector (Invitrogen) using BspHI and SpeI at the 5′ and 3′ ends of the amplified PCR product.
Purification of His-tagged HSPA1A, DNAJA1, Luciferase, and HSPH1
E. coli DH5α cells transformed with pSE420 vector carrying HSPA1A and DNAJA1 were grown in LB medium with 100 μg/ml ampicillin at 37 °C. 0.6 mm isopropyl β-d-thiogalactopyranoside (Sigma-Aldrich) was added at OD 0.6, and 12 h later, the cells were harvested by centrifugation at 6000 rpm. Cell pellet was resuspended and sonicated in Buffer A (20 mm sodium phosphate buffer, pH 7.5, and 500 mm NaCl, supplemented with phenylmethanesulfonyl fluoride (Sigma-Aldrich). The lysate was centrifuged at 16,000 rpm (JA 25.50 rotor, Beckman J2-21 centrifuge) for 30 min, and supernatant was filtered though a 0.2-μm membrane and loaded onto a nickel affinity column (HisPrep FF 16/10; GE Healthcare) with buffer A. After loading, column was extensively washed (30–40 column volumes). The bound protein was eluted with linear gradient of 10–500 mm imidazol in buffer A. The purity of the fractions was confirmed on SDS-PAGE. Proteins were concentrated and stored in 20 mm Tris-HCl, pH 7.5, 10% glycerol, and 200 mm NaCl at −80 °C.
Cells harboring pET28a-HSP110 (Hsp105) plasmid (a gift from Dr. E. Lafer Department of Biochemistry, The University of Texas Health Science Center) were preincubated overnight at 37 °C on LB kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml) plates. Cells from the plate were transferred to autoinduction media (for 1 liter: 10 g of tryptone, 5 g of yeast extract, 5 g of glycerol, 0.5 g of glucose, 2 g of lactose, 26.8 ml of 0.1 g/ml NH4Cl, 2 ml of 1 m MgSO4), then 100 ml of sterilized 10× salt solution (stock: 35.49 g of Na2HPO4, 34.02 g of KH2PO4, 7.1 g of Na2SO4) and kanamycin (50 μg/ml) were added, and cells were grown at 30 °C for 24 h. Afterward, the cells were harvested and sonicated in 20 mm sodium phosphate buffer, pH 8, and 300 mm NaCl, 0.2% Triton X-100, PMSF, and centrifuged at 16,000 rpm; supernatant was filtered and loaded onto previously equilibrated nickel affinity column. After loading, the column was extensively washed (30–40 column volumes) with wash buffer (20 mm sodium phosphate buffer at pH 8.0, 20 mm imidazole, and 500 mm NaCl), and the pure protein was eluted using elution buffer (20 mm sodium phosphate buffer at pH 8, 100 mm imidazole, and 500 mm NaCl). After confirming on SDS-PAGE, pure fractions were pooled, buffer was exchanged into storage buffer (20 mm Tris-HCl at pH 8, 100 mm NaCl, 5% glycerol, and 3 mm DTT), concentrated, and stored at −80 °C. DnaK and Luciferase was purified as in Ref. 12. DnaJ and GrpE of E. coli were a gift from Dr. H. J. Schönfeld (Pharmaceutical Research, F. Hoffman-La Roche Ltd., Basel, Switzerland).
Luciferase Assays
Luciferase activity was measured as described previously (12) using a Victor Light 1420 Luminescence Counter from PerkinElmer Life Sciences.
Immunoblotting
FTluc samples were incubated with chaperones in the presence of ATP; aliquots were taken at different time intervals and treated with trypsin for 2 min and immediately mixed with loading dye; and samples were incubated at 95 °C for 2 min, run on SDS-PAGE, and electrotransferred onto a 0.2-μm nitrocellulose membrane (Bio-Rad). Following seven 5-min rinses in TTBS, the membrane was incubated in primary goat luciferase antibody in TTBS (1:20,000; Sigma-Aldrich) for overnight, followed by secondary donkey HRP-goat antibody. The membrane was washed and then developed using the chemiluminescent ImmunstarTM kit (Bio-Rad) according to the manufacturer's instructions.
Measurement of ATP Consumption
The ATP content ([2,5′,8-3H]ATP from Amersham Biosciences) of the refolding solution supplemented with 0.5 μm Hsp70, 0.5 μm Hsp110, 0.25 μm Hsp40, 100 μm ATP, and 0.5 μm luciferase (FTLuc) was determined as a function of time at 25 °C. At 50–60-s intervals over a time period of 7 min, 5-μl aliquots were taken and processed as explained elsewhere (12).
Thioflavin T Binding
Thioflavin T (ThT) binding of luciferase was measured in the presence of 60 μm ThT as in Ref. 12 (excitation, 435 nm; and emission, 465 nm) using a PerkinElmer Life Sciences LS55 fluorescence spectrometer.
Protein Structural Analysis
Protein structures of Hsc70-Sse1 (Protein Data Bank (PDB) code 3C7N) complex and DnaK/Hsp70 (PDB code 4B9Q, 2KHO) were taken from PDB and by using PyMOL molecular graphics system, Hsc70 NBD was structurally aligned with the structure of ADP-bound DnaK (PDB code 2KHO) or ATP-bound DnaK/Hsp70 (PDB code 4B9Q). Based on the analogy of DnaK structure, the substrate-binding domain (SBD) of Sse1 was superimposed to match the SBD structure of ADP-bound DnaK/Hsp70.
RESULTS
Human Hsp110 Cooperates with Hsp40 to Prevent Protein Aggregation in an ATP-dependent Manner
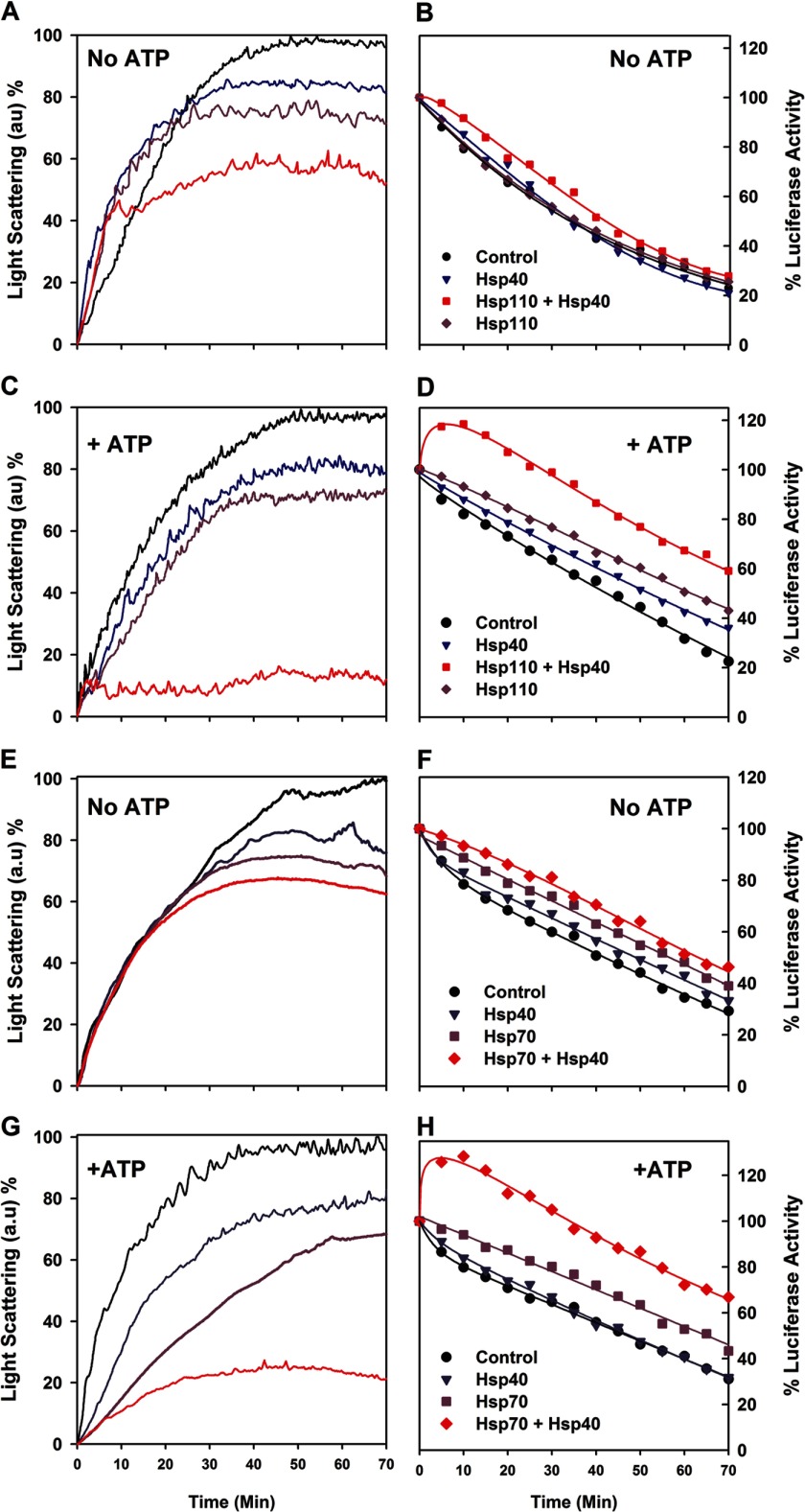
We tested the ability of purified recombinant Hsp110 (HSPH1) to prevent the aggregation of native firefly luciferase, which is mildly thermolabile. When native luciferase was incubated at 34 °C without chaperones and ATP or with equimolar Hsp40 or Hsp110, it lost activity at similar rates of 2.4 ± 0.2% per minute. The presence of both Hsp40 and Hsp110 slightly slowed down luciferase denaturation, as expected from a mild nonspecific protective effect caused by the presence of native proteins (Fig. 1, A and B). Whereas ATP alone did not stabilize the heat-denaturing native luciferase, in the presence of ATP and Hsp40 (DNAJA1), Hsp110 drove the transient net reactivation of up to 120% of the initial luciferase activity during the first 10 min, indicating that the original native luciferase stock contained at least 20% of misfolded inactive chaperone-amenable luciferase species (13). Thereafter, the luciferase activity flattened and steadily decreased at the same rate, yet at a constant higher level by 35%, compared with without chaperones and ATP (Fig. 1D). Online light scattering measurements showed that without ATP, the presence of Hsp110, or of Hsp40 was mildly yet significantly effective at passively preventing the aggregation of the denaturing luciferase species. Together, their effect was additive (Fig. 1A), confirming that like Hsp70, the protein-binding domain of Hsp110 can also bind aggregating polypeptides. In the presence of ATP, Hsp110 or Hsp40 individually remained mildly effective at preventing the luciferase aggregation as without ATP. Remarkably, in the presence of ATP, Hsp110 together with Hsp40 became strongly synergic at preventing luciferase aggregation (Fig. 1C). A similar synergic ATP-dependent prevention of aggregation has been previously shown in the case of bacterial DnaK and DnaJ (24) and was also confirmed here with human Hsp70 (HSPA1A) and Hsp40 (Fig. 1G). Thus, Hsp110 is functionally indistinguishable from its structurally related Hsp70 and DnaK paralogs. This is also strong evidence that the Hsp40 co-chaperone undergoes an ATP-driven specific interaction with Hsp110, as it does with Hsp70.
FIGURE 1.
Hsp110 (HSPH1) and Hsp40 (DNAJA1) are synergic at preventing aggregation and transiently reactivating thermo-labile luciferase. A–D, native luciferase (1 μm) was incubated at 34 °C without or with Hsp110 (1 μm) and/or Hsp40 (0. 5 μm), without (A and B) or with (C and D) 5 mm ATP. Light scattering at 550 nm (A and C) and luciferase activity (B and D) were measured online during exposure to 34 °C. E–H, similarly, Hsp70 and Hsp40 are synergic at preventing aggregation and transiently reactivating thermolabile luciferase at 34 °C.
The transient luciferase reactivation that we observed during the first 10 min of the reaction with the particular Hsp110 + Hsp40 + ATP combination (Fig. 1D) suggested that Hsp110 is a bona fide Hsp40- and ATP-dependent chaperone that can drive the transient renaturation of a misfolding protein against the general tendency of the substrate at 34 °C to remain inactive and become increasingly aggregated. Hsp70, Hsp40, and ATP presented the same ability to transiently protect and reactivate the heat-denaturing luciferase (Fig. 1F), implying that under the conditions of the in vitro assay, the Hsp110 was an Hsp40-dependent, ATP-fueled antiaggregation reactivating chaperone, functionally indistinguishable from its structurally related Hsp70 paralog.
Human Hsp110 Is a Bona Fide Polypeptide Unfolding/Refolding Chaperone
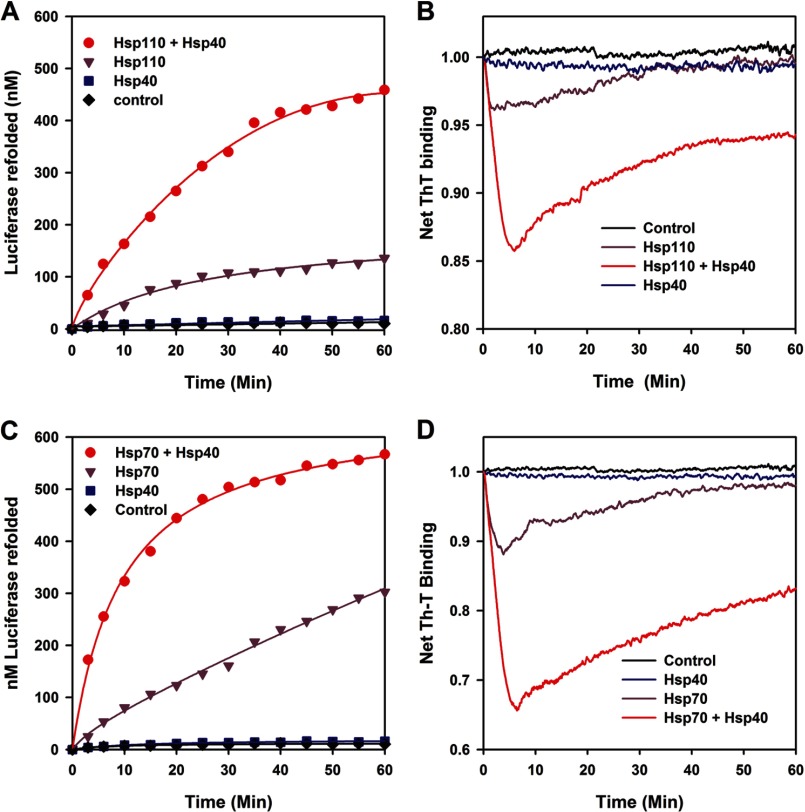
We next addressed the ability of Hsp110 (HSPH1) to act upon stable inactive misfolded luciferase monomers (FTluc), which were prepared beforehand without chaperones by iterative cycles of freeze-thawing (12) and convert them at 22 °C into native luciferase. Whereas ATP and/or Hsp40 (DNAJA1) alone did not cause a significant net native refolding of FTluc (Fig. 2A), the presence of equimolar Hsp110 with ATP regenerated a net 12% (120 nm) of native luciferase in 60 min. Remarkably, in the presence of Hsp40 (0.5 μm) and ATP, Hsp110 (1 μm) produced a net 44% (440 nm) of native luciferase, demonstrating that alone but preferably when assisted by Hsp40, Hsp110 can use the energy of ATP hydrolysis to convert stable preformed misfolded polypeptide species into their stable native state. Similar specific assisted refolding results were obtained when we used urea predenatured malate dehydrogenase or stable pre-heat-denatured G6PDH aggregates (Fig. 3, A and B). Thus, the chaperone activity of Hsp110 (Hsp105, HSPH1) is not limited to the special case of stable inactive FTluc monomers. Noticeably, using FTluc as substrate, Apg-2 (HSPH2) and DNAJB1, which are respectively the other most abundant human cytosolic Hsp110 and Hsp40, were found to be as effective refolding chaperones as HSPH1 and DNAJA1 (Fig. 3C). This was not unexpected, given that the human Apg-2 (HSPH2), together with DNAJB1 or yeast YDJ1 have recently been shown in vivo and in vitro to act in synergy with Hsc70 (HSPA8) at disaggregating preformed protein aggregates (28, 29). Thus, we show here that Apg-2 is as a bona fide ATP-fueled chaperone on its own, as HSPH1. Without Hsc70, Apg-2 may also use ATP to drive the conversion of stable misfolded polypeptide substrates into natively refolded products.
FIGURE 2.
Hsp110 (HSPH1) is an Hsp40- and ATP-dependent polypeptide unfolding/refolding chaperone similar to Hsp70. A and B, the activity (A) and the net ThT fluorescence (B) of stable misfolded FT luciferase (1 μm) were measured online at 22 °C in the presence of ThT (60 μm) at various time points as indicated, in the absence (black) or presence of ATP (5 mm) with Hsp110 alone (1 μm, purple), Hsp40 (DNAJA1) alone (0.5 μm, blue), or both Hsp110 + Hsp40 (red). C and D, the activity (C) and the net ThT fluorescence (D) of stable misfolded FT luciferase (1 μm) measured online without (black) or with ATP as in A and B, in the presence of Hsp70 alone (1 μm, purple), Hsp40 alone (0.5 μm, blue), or both Hsp70 and Hsp40 (red).
FIGURE 3.
Various Hsp110s and Hsp40s can collaborate to reactivate various misfolded protein substrates. A, time-dependent refolding of thermally predenatured G6PDH following 30 min of incubation at 30 °C without or with ATP, 6 μm Hsp70 or Hsp110 (HSPH1), without or with 3 μm DNAJA1, as indicated. B, refolding yields of urea-preunfolded MDH (600 nm final concentration) at 34 °C. 60 μm MDH was incubated 30 min at 34 °C in 8 m urea and diluted 100-fold in refolding buffer also containing 5% glycerol, without or with 4 μm Hsp70, or Hsp110 (Hsp105) and 2 μm DNAJA1, as indicated. C, time-dependent refolding of 1 μm FTluc without or with ATP, 1 μm Apg-2 or Hsp105, and 0.5 μm DNAJA1 or DNAJB1. The protein combinations are indicated.
ThT is a fluorescent dye that specifically binds cross-β structures, both in native proteins, but more strongly and specifically in misfolded polypeptides entangled within dense protein aggregates, fibrils, and amyloids (30, 31). Because ThT does not interfere with chaperone activity (12, 24), this allows online measurements of the net time-dependent changes in the misfolded β-structures of the substrates during the chaperone reaction. When the Hsp110-mediated Hsp40-dependent refolding reaction as in Fig. 2A was carried in the presence of excess ThT, online fluorescence showed that addition of ATP at T = 0 min caused a sharp decrease in the net amount of ThT-binding sites in the misfolded FTluc, indicating that the chaperone caused a massive ATP-dependent loss of misfolded structures and the unfolding, at least in part of the FTluc (Fig. 2B). After 5 min, during which 130 nm luciferase became natively refolded, the decrease of ThT fluorescence leveled, and a net regain of ThT-binding structures was observed, alongside a parallel regain of up to 440 nm of native luciferase. The reactivation profiles confirmed that Hsp110 alone with ATP (but not Hsp40 alone with ATP) had a significant unfolding activity, whose maximal yield at 5 min was ∼4.5 times lower than when Hsp40 was also present. When in the same chaperone assay Hsp110 was replaced by Hsp70, slightly faster reactivation rates but overall similar refolding yields (Fig. 2C) and time-dependent ThT changes in the luciferase substrate were observed (Fig. 2D). Thus, as already shown with bacterial DnaK (12), both human cytoplasmic Hsp70 and Hsp110 can independently act as efficient polypeptide unfolding enzymes that use ATP hydrolysis to convert stable misfolded polypeptide substrates into stable natively refolded protein products.
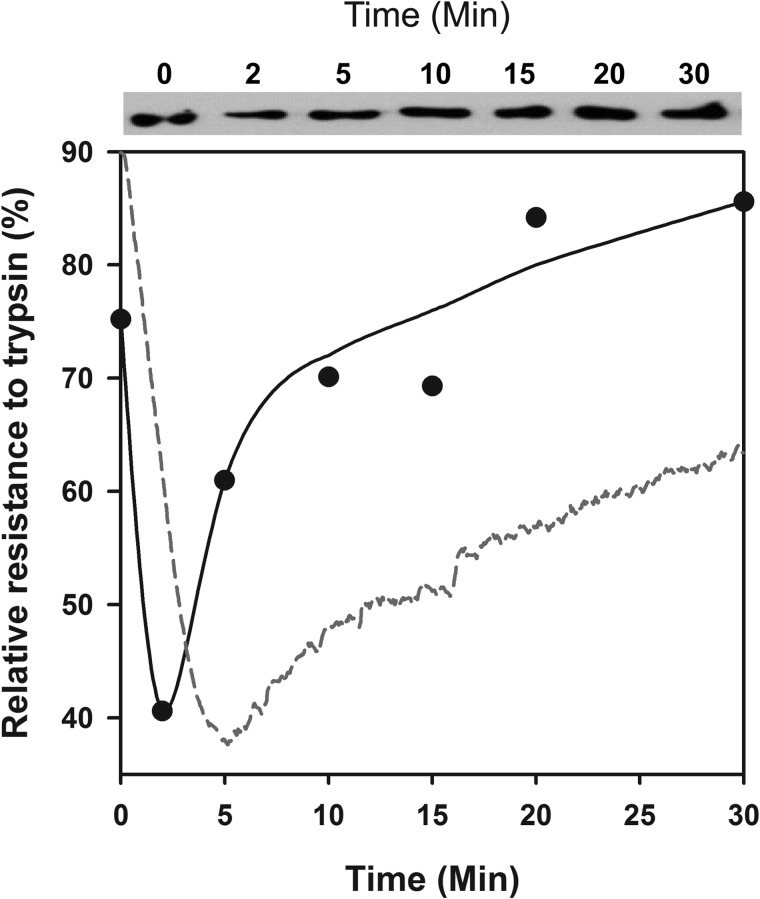
Partial trypsin digestions for 2 min at various time points of the reaction and Western blots of SDS gels (Fig. 4) confirmed that Hsp110 is an ATP-dependent polypeptide-unfolding chaperone. Whereas at T = 0 min, FTluc was 75.2% trypsin-resistant, following addition of Hsp110 + Hsp40 + ATP, trypsin resistance dropped at T = 5 min to 40.6% and thereafter recovered to 85.6% within 30 min. Thus, confirming the chaperone-mediated transient loss of ThT binding misfolded β-sheets in the FTLuc substrate, partial trypsin digestions showed a transient loss of the FTLuc compactness under the combined ATP-fueled action of Hsp110, Hsp40, despite the additional chaperones that were also competing for the limiting amounts of trypsin. The subsequent regain of luciferase trypsin resistance, which paralleled that of ThT rebinding and regained luciferase activity (Fig. 2B), confirmed that the natively refolded chaperone product was, as expected, more compact than the chaperone-unfolded intermediate.
FIGURE 4.
Trypsin treatment of misfolded monomeric luciferase revealed unfolding by Hsp110/Hsp40. Upper panel, Western blots of trypsin-treated luciferase. At the indicated time points of the Hsp110 + Hsp40 + ATP-dependent refolding reaction as in Fig. 2A, the samples were treated for 3 min with 0.04 mg/ml trypsin, separated on SDS gel, and detected on Western blots with luciferase antibodies. Lower panel, the relative semiquantitative luciferase signal at 60 kDa from the Western blot (plain circles). The relative net ThT fluorescence signal of FTluc under the same conditions from Fig. 2B is shown for comparison (red dotted line).
Equimolar Hsp110 and Hsp70 Form an Optimal Bi-chaperone Disaggregating Machine
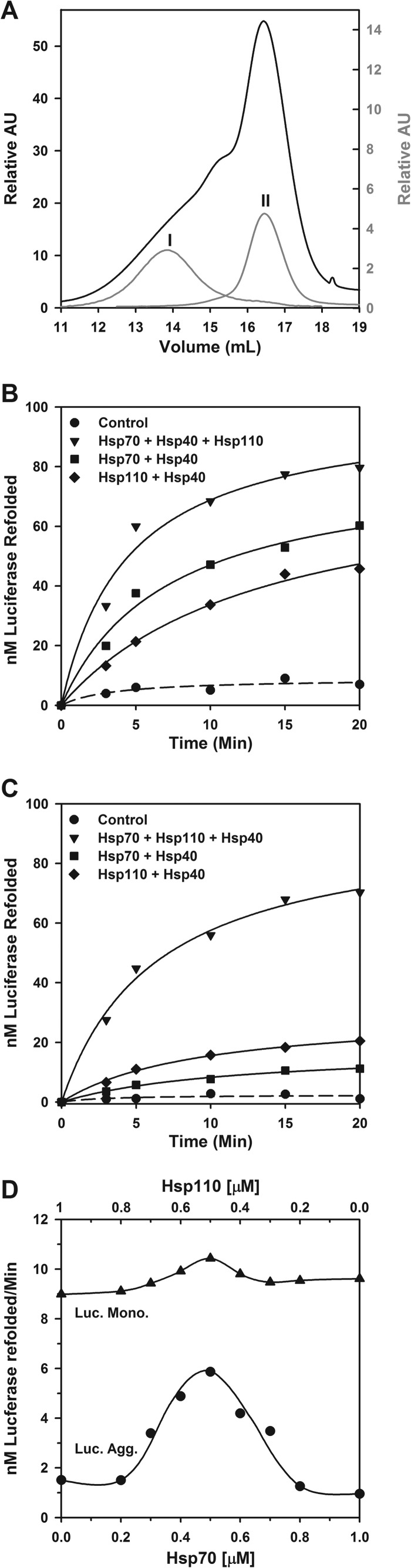
A slow significant disaggregating activity has been demonstrated for mammalian Hsp110 (Apg-2, HSPH2) and Hsc70 (HSPA8), which dissolved large disordered aggregates but failed to act on α-synuclein fibrils (29). We thus next addressed the ATP-dependent chaperone activity of both human cytoplasmic Hsp110 (HSPH1) and Hsp70 (HSPA1A) in the presence of Hsp40 (DNAJA1), using two different substrates: stable oligomeric FTluc aggregates, which eluted on gel filtration with an apparent molecular weight of soluble 4–12-mers (fraction I; Fig. 5A), or misfolded FTluc monomers that eluted last (fraction II; Fig. 5A). At equal protomer concentrations, light scattering and ThT fluorescence of the various gel filtration fractions (data not shown) showed that, as initially demonstrated by analytical ultracentrifugation, the monomeric FTluc fraction was composed mostly of stable misfolded luciferase monomers that bound 1.8 times more ThT than native luciferase (12) but, similar to the native monomeric luciferase, did not scatter light. In contrast, the oligomeric fraction, although containing as many ThT binding misfolded structures, scattered light five times more than the monomeric species (data not shown). When 2 h after their first elution the 10-fold diluted oligomers were reinjected on the same column, at least 95% eluted at the same positions as the original oligomers, and the remaining 5% eluted at the same position as monomer (Fig. 5A, peak I), confirming that although soluble, the luciferase aggregates were stable and mostly dilution-resistant.
FIGURE 5.
Hsp110 (HSPH1) collaborates with Hsp70 (HSPA1A) to convert stable luciferase aggregates into native species. A, purification and confirmation of freeze-thawed luciferase aggregates (fraction I) and monomers (fraction II). B and C, soluble monomeric misfolded FTluc (0.5 μm) from fraction II (B) or soluble oligomeric aggregates of FTluc monomers from fraction II (C) was incubated at 22 °C as in the presence of 5 mm ATP, 125 nm Hsp40, and either 250 nm Hsp70 alone (■) or 250 nm Hsp110 alone (♦) or 125 nm Hsp70 together with 125 nm Hsp110 (▾), as indicated. D, the rates of reactivation of monomeric FTluc (▴) or oligomeric FTluc (●) in the presence of 5 mm ATP, 0.5 μm Hsp40, and increasing concentrations of Hsp70 (0–1 μm) supplemented by corresponding decreasing concentrations of Hsp110 maintaining the sum of Hsp70 and Hsp110 molecules at 1 μm. Luc., luciferase; Mono., monomer; Agg., aggregate.
When the monomeric misfolded luciferase species (fraction II) were incubated with Hsp70 (HSPA1A), Hsp40 (DNAJA1), and ATP, a net minor amount of 50 nm native Luc was produced within 20 min, and similarly, Hsp110 (HSPH1) with ATP and Hsp40 produced 40 nm native Luc (Fig. 5B). In contrast, an equimolar mixture of half as concentrated Hsp70 and Hsp110 with Hsp40 and ATP produced 75 nm luciferase, i.e., nearly twice more than the average yield of 45 nm expected if the two chaperones were acting independently from one another. Hsp70, Hsp40, and ATP produced only 5 nm of net native luciferase from stable aggregated species, as expected from less amenable aggregated substrates. Hsp110, Hsp40, and ATP produced twice more native protein than Hsp70, suggesting that Hsp110, by itself, may be slightly more competent a chaperone to process large oligomeric misfolded polypeptides than Hsp70 (Fig. 5C). Remarkably, an equimolar mixture of half as concentrated Hsp70 and Hsp110 molecules produced 70 nm native luciferase, which was nine times higher than the expected average yield of 7.5 nm if the two chaperones were acting independently from one another. This strongly suggests that Hsp110 forms a functional complex with Hsp70, combining their individual chaperone unfolding abilities into a unique synergic polypeptide disentangling and unfolding mechanism to convert stable protein aggregates in native proteins.
Further addressing the optimal stoichiometry in the 1:1 Hsp110:Hsp70 disaggregating machinery, we measured the initial rates of luciferase reactivation, either with stable misfolded FTluc monomers as substrates or with stable misfolded FTluc oligomers, in the presence of increasing concentrations of Hsp70 (0–1 μm), supplemented with the corresponding decreasing concentrations of Hsp110 (1–0 μm) to keep a constant chaperone concentration of 1 μm. As initially observed in Fig. 5A, the reactivation rates of misfolded FTluc monomers by individual Hsp70, or individual Hsp110 were nearly equally high: ∼9 nm min−1 with a minor optimal rate of 10.3 nm min−1 observed at equimolar Hsp70:Hsp110 (Fig. 5D, upper line). Whereas the reactivation rates of misfolded Luc oligomers by individual Hsp70 or Hsp110 was expectedly low in both cases (∼1–1.5 nm min−1), as the stoichiometry between Hsp70 and Hsp110 approached 1:1, the reactivation rates increased 5-fold, to reach up to 55% of the optimal rates obtained with misfolded FTluc monomers (Fig. 5D, lower line). Thus, corroborating the crystal 1:1 heterocomplexes between bovine Hsc70 and yeast Hsp110 (SSE, 32), the human Hsp110 (Hsp105) can form in solution with Hsp70 a functional 1:1 bichaperone complex to disaggregate proteins.
Hsp70 and Hsp110 Mutually Catalyze Substrate Release from One Another
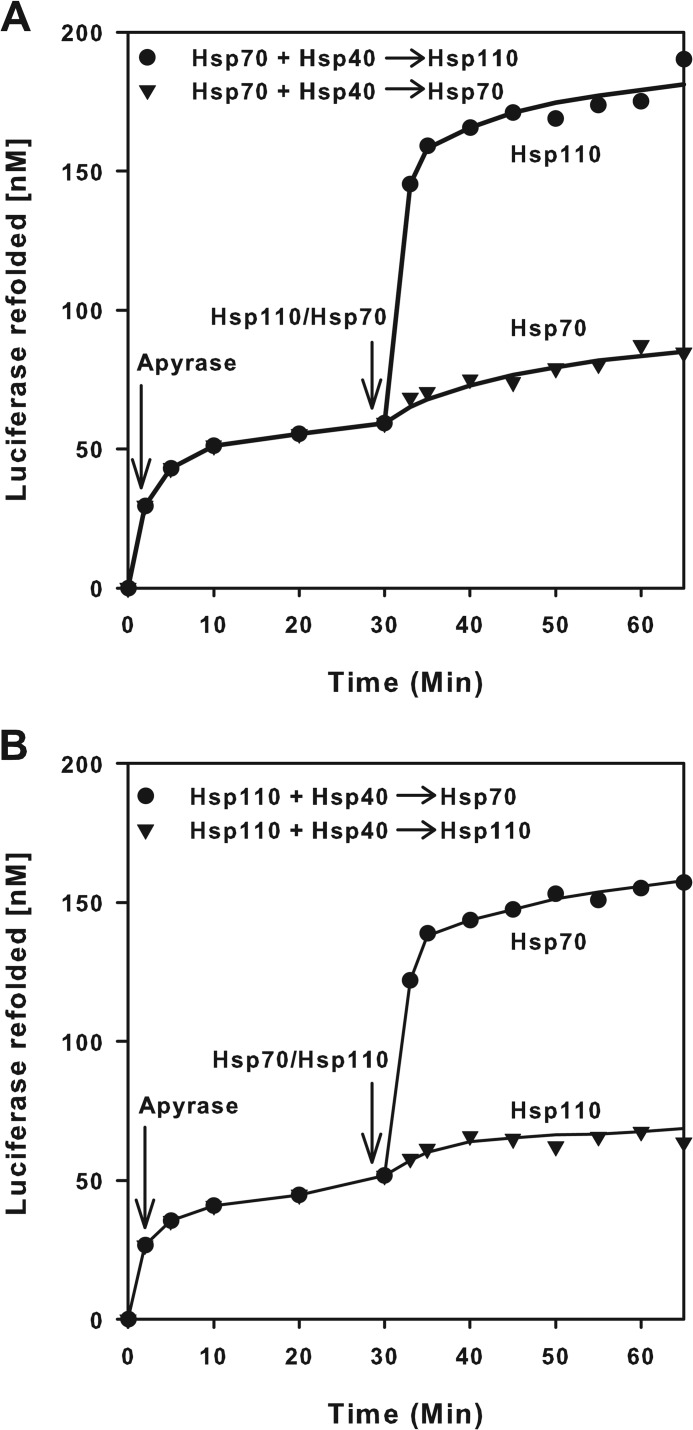
We next assessed the ability of Hsp110 (HSPH1) to act as a presumed nucleotide exchange factor (NEF) of Hsp70 (33, 34). Because a NEF, by definition, should catalyze the ATP-dependent release of ADP and of polypeptide substrates from stable preformed (ADP-Hsp70-substrate) complexes, we use a simplified protocol from Ref. 12 in which FTluc was first incubated for 2 min with Hsp40, ATP, and Hsp70 (Fig. 6A). Because of the high ATPase activity of Hsp70 with Hsp40 even in the absence of a NEF, Luc expectedly started to refold at ∼15 nm min−1. Addition at T = 2 min of apyrase sharply reduced by 10-fold the rate of luciferase refolding to 0.2 nm min−1. Thus, the instantaneous destruction of the ATP molecules reduced as expected the release and refolding of the unfolded luciferase substrate from the so-called “locking” state of the ADP-bound Hsp70 chaperone (12). Whereas the subsequent addition at T = 30 min of an equimolar amount of free Hsp70 did not change the slow ATP-independent rate of luciferase release and refolding from the preformed ADP-Hsp70-Luc complex, addition of equimolar Hsp110 caused a dramatic release and spontaneous refolding of the Hsp70-bound FTluc, at a rate that was three times faster than the initial rate with ATP, although release and refolding at T = 30 min took place in the total absence of ATP (Fig. 6A). Thus, justifying in part why it was initially described as a NEF of Hsp70, we found that Hsp110 is not a true NEF, because ATP binding was apparently not necessary for its action upon Hsp70 as a substrate release factor. Surprisingly, swapping the roles between Hsp70 and Hsp110 in this experiment produced the same results. The addition at T = 30 min of equimolar amounts of free Hsp70 to apyrase-treated preformed Hsp110-Luc complexes caused a strong release and native refolding of the unfolded luciferase, whereas addition of substrate-free Hsp110 did not significantly affect the slow rate of luciferase release from Hsp110 (Fig. 6B). Thus, Hsp110 is symmetrically as much a substrate release factor for the Hsp70 unfolding chaperone as Hsp70 is a substrate release factor for the Hsp110 unfolding chaperone.
FIGURE 6.
Hsp70 (HSPA1A) and Hsp110 (HSPH1) mutually activate substrate release. Misfolded luciferase monomers (1 μm) were supplemented with 0.1 mm ATP and Hsp70 (1 μm), Hsp40 (0.5 μm) (A), or Hsp110 (1 μm) and Hsp40 (0.5 μm) (B), and luciferase activity was followed in time. At T = 2 min, apyrase (2 μm) was added, which readily destroyed the ATP. At T = 30 min, 1 μm of Hsp110 in A and of Hsp70 in B was added, and luciferase activity was further measured up to T = 65 min.
ATP Hydrolysis Assays Show Functional Interactions between Hsp110, Hsp70, Hsp40, and the Substrate
In the absence of substrate polypeptides, the highly conserved J-domain of Hsp40 spontaneously associates with the nucleotide-binding domain (NBD) of nucleotide-less or ATP-bound Hsp70 molecules, near the hinge region that connects the NBD to the SBD (35). Thus, even without a substrate polypeptide, DnaJ can activate futile ATP hydrolysis by bacterial Hsp70 (12, 36). Measures of steady state rates of ATP hydrolysis (Table 1) showed that the presence of Hsp40 expectedly nearly doubled the rate of ATP hydrolysis, both by Hsp70 alone and by Hsp110 alone. Thus, as previously shown in the case of SSE1 and Ydj1 (37), with regard to J-domain-induced ATPase activity, human Hsp110 is functionally indistinguishable from other bona fide Hsp70 chaperones.
TABLE 1.
Rate of ATP hydrolysis and ATP cost by various combinations of Hsp110, Hsp70, and Hsp40, without or with equimolar aggregated (Agg.) or misfolded monomeric (Mono.) luciferase (Luc.) substrates
| Sample | ATP consumed by 500 nm chaperones only | ATP consumed by chaperones in the presence of substrates | ATP | Net ATP cost to refold a luciferase |
|---|---|---|---|---|
| min−1 | min−1 | chaperone−1 min−1 | ||
| Hsp70 | 312 | 0.62 | ||
| Hsp110 | 252 | 0.50 | ||
| Hsp70 + Hsp40 | 624 | 1.25 | ||
| Hsp110 + Hsp40 | 516 | 1.03 | ||
| Hsp70 + Hsp110 + Hsp40 | 924 | 1.85 | ||
| Hsp70 + Hsp40 + Luc. Mono. | 1392 | 2.78 | 77.45 | |
| Hsp110 + Hsp40 + Luc. Mono. | 1644 | 3.29 | 168.6 | |
| Hsp70 + Hsp110 + Hsp40 + Luc. Mono | 2772 | 5.54 | 148.9 | |
| Hsp70 + Hsp40 + Luc. Agg. | 1536 | 3.07 | 650.7 | |
| Hsp110 + Hsp40 + Luc. Agg. | 1890 | 3.8 | 485.4 | |
| Hsp70 + Hsp110 + Hsp40 + Luc. Agg. | 2388 | 4.8 | 228.9 |
Whereas, theoretically, half amounts of Hsp70 and Hsp110 (in the presence of constant Hsp40) should hydrolyze 1.1 ATP min−1, they effectively hydrolyzed 1.8 ATP min−1 (Table 1). This 1.6-fold activation was in agreement with the refolding rates, suggesting that Hsp110 and Hsp70 form a functional 1:1 complex, and with the crystal structure of SSE1 with Hsc70 that showed that the two chaperones interact though their distal lobes in their respective NBDs, far away from where the J-domain of Hsp40 can bind Hsp110 or Hsp70.
Hsp110 ATPase Is Activated by FT Misfolded and Aggregated Luciferase Substrates
The ATPase activity of DnaK, with and without DnaJ and GrpE, has been shown to become activated severalfold upon binding of various short, mostly hydrophobic peptides (22, 38) and of artificially unfolded polypeptide substrates (39, 40), as well as of native polypeptide substrates, such as the σ32 (41). Here, we found that the ATPase rate of Hsp70 in the presence of Hsp40 was activated 2.2- or 2.5-fold by stable misfolded luciferase monomers or oligomers, respectively (Table 1). Similarly, the ATPase rate of Hsp110 (HSPH1) in the presence of Hsp40 (DNAJA1) was activated 3.2- and 3.7-fold by stable misfolded luciferase monomers or oligomers, respectively. Thus, with regard to substrate-induced ATPase activation, Hsp110 was functionally indistinguishable from a bona fide Hsp70 chaperone.
Although the luciferase aggregates were less efficiently refolded, they triggered the ATPase activity to a higher extent than the misfolded luciferase monomers, suggesting that there is more futile hydrolysis and that despite strong interactions with the chaperone, the misfolded oligomers are more resistant to ATP-fueled Hsp70- or Hsp110-mediated unfolding than the misfolded monomers. Theoretically, if equimolar Hsp70 and Hsp110 molecules were not collaborating with one another, 568 ATPs should be hydrolyzed while reactivating a single luciferase polypeptide entangled within a stable luciferase aggregate. We found that only 229 ATPs were hydrolyzed, indicating that the two chaperones collaborate in a way that increases the efficiency of ATP hydrolysis to unfold/refold stable protein aggregates (Table 1). In contrast, compared with the theoretical average of 123 ATPs, the equimolar mixture of Hsp110 and Hsp70 consumed 21% more ATP (149 ATPs) while reactivating a misfolded monomer, suggesting that the ATP-fueled unfolding/refolding of misfolded monomers does not require that the two chaperones collaborate.
DISCUSSION
The chaperone mechanism of Hsp70 has been extensively studied (42). Bacterial DnaK was shown to use ATP to apply an unfolding force on stable misfolded polypeptides and small aggregates by two complementary mechanisms likely involving both direct “clamping” by individual DnaK molecules that may unfold non-native β-structures in stable misfolded polypeptides (12, 43) and indirect “entropic pulling” by several concomitantly clamped DnaK molecules on the same misfolded polypeptide, cooperating to unfold by thermal movements local misfolded regions in between chaperone-bound segments (44). NEFs can accelerate both effects by promoting the dissociation of ADP and the consequent release of the unfolded polypeptides from the stable ADP-Hsp70-substrate clamped complexes, allowing the unfolded products to spontaneously refold to their native state (12). In eukaryotes, at least four classes of Hsp70 NEFs have been identified, all acting on the same distal NBD lobes (IB and IIB) of Hsp70s. Three conserved classes of the GrpE-like, HspBP1-like, and BAG1–5-like NEFs cannot bind misfolded polypeptides. Although they are also unable to bind ATP, they are considered to be ATP-dependent ADP release catalysts (20, 45–48). The Hsp110s have been categorized as a fourth class of NEFs (32, 48–51), despite the fact that sequence-wise and structure-wise, they are close orthologs of their acknowledged Hsp70 co-chaperone partners in disaggregation. The Hsp110s have a very similar NBD and C-terminal SBD, in which the peptide binding β-sandwich subdomain (SBDβ) and the α-helical lid subdomain (SBDα) can be clearly recognized (52). In vitro, purified human Hsp110 (HSPH1) can drive the ATP-, Hsc70-, and DNAJC6-dependent disaggregation of clathrin baskets (32). Like Hsp70s, Hsp110 can hydrolyze ATP (33) and is activated by J-domain co-chaperones (34). Hsp110 can also bind nascent polypeptide chains (33, 34, 53) and small peptides with different specificities (54). By doing so, Hsp110 may act as a “holding” chaperone that passively prevents the aggregation of stress- or mutation-induced misfolding and aggregating proteins (55). Moreover, yeast Hsp110 (Sse1) has been co-crystallized in 1:1 complex with the NBD of bovine Hsp70 (Hsc70), showing a pseudosymmetrical dimer with many tight interactions between the two NBDs, mostly in the distal lobes (IB and IIB), precisely at the same locations where the other NEFs also interact with Hsp70 and DnaK (32). However, at variance with the DnaK structure, the crystal structure of Sse1 showed the SBDα lid in an extensively wide open conformation. Although this initially questioned the ability of Hsp110 to close on its base and consequently to act as unfolding chaperones on its own (28), the same extensively wide open conformation was observed with functional DnaK mutants that crystallized as dimers associated through their NBDs as in the case of the SSE1 and Hsp70 crystals (56).
Hsp70s are polypeptide-unfolding enzymes on their own with a significant disaggregase and reactivation activities, which need to be in a large stoichiometric excess to cooperatively solubilize misfolded polypeptides from small soluble aggregates (12, 16). Generalizing the initial finding that Hsp105, together with Hsc70, can efficiently disaggregate purified clathrin cages (32), less specific in vitro disaggregating assays recently showed that mammalian cytosol prepared from different sources possess a potent, ATP-dependent activity requiring the apparent NEF activity of mammalian Hsp110 (Apg-2), the chaperone activity of Hsc70, and the co-chaperoning of Hdj1, to slowly dissolve large disordered aggregates and recover natively refolded proteins. A similar significant disaggregase activity was also observed in the case of yeast Sse1 acting upon Ssa1 together with the Sis1 or Ydj1 J-domain proteins (29). Likewise, Hsp110 (Apg-2) promotes the ATP- and Hdj1-dependent disaggregation activity of Hsc70 on stable heat-denatured aggregates in vitro and in vivo (28). Thus, contrary to other co-chaperones, Hsp110 is a unique NEF by being able to turn Hsc70 into an effective disaggregating machinery that can solubilize and reactivate large stable proteins aggregates (33), thereby possibly reducing the need for a large molar excess of Hsp70 to obtain some disaggregation by cooperative entropic pulling (16, 25, 44).
Noticeably, prior in vitro chaperone assays have reported that Hsp110 (Apg-2) lacks a chaperone unfolding/refolding activity of its own (28). In contrast, using various sensitive misfolded substrates, we found here in vitro that HSPH1 (Hsp105) or HSPH2 (Apg-2), assisted by DNAJA1 or DNAJB1, can act as bona fide ATP-dependent unfolding/refolding chaperones, even when Hsp70 is not present. This difference may be attributed to the use of slightly different substrates: predominantly stable freeze-thawed misfolded luciferase monomers rather than urea preaggregated luciferase.
Our finding that human Hsp110 alone can act as a chaperone using ATP hydrolysis to drive the catalytic unfolding of stable misfolded polypeptide substrates and to efficiently convert them into natively refolded products suggests that in solution, the substrate-binding domain of Hsp110 can effectively “clamp” upon its misfolded substrate and apply a pressure leading to unfolding, as in the case of DnaK (43). We therefore suggest a model accounting for the polypeptide unfoldase activity of Hsp110, in which the α-helical lid may depart from its stable position alongside the nucleotide-binding domain observed in the crystal structures and adopt during ATP hydrolysis an extensively opened position capable of binding to large bulky hydrophobic misfolded domains of a substrate polypeptide and, upon ATP hydrolysis, operate a large clamping movement toward the β-sheet base subdomain, as in the case of DnaK (57), that may in turn cause the unfolding of the clamped misfolded polypeptide segment (43).
An allosteric communication between the NBD and the substrate-binding domain of Hsp110 has been recently reported (54). Our dose responses further showed that, as suggested from the crystal structure of yeast Hsp110 with bovine Hcp70 NBD, equimolar Hsp70 and Hsp110 can optimally refold stable aggregated luciferase, likely by binding to orthologous topological sites in their respective NBDs and by effective reciprocal allosteric signals of similar quality and intensities between the two orthologous SBDs (42). The presence of such symmetrical allosteric signals was confirmed by the reciprocal release of the unfolded luciferase that we observed in the presence of apyrase, upon addition of Hsp110 to stable preformed ADP-Hsp70-Luc complexes or symmetrically of Hsp70 to stable preformed ADP-Hsp110-Luc complexes.
Despite their optimal 1:1 functional association, Hsp110 and Hsp70 apparently form loose heterodimers, because gel filtration, dynamic light scattering, chemical cross-linking, and electrophoresis on native gels thus far failed to reveal the unambiguous presence of stable Hsp110-Hsp70 heterodimers. Noticeably, proteomic analysis showed that in the cytoplasm and the endoplasmic reticulum of human cells, the Hsp70s are respectively 7- and 14-fold more abundant than the Hsp110s (3). Thus, despite their low affinity, most of the Hsp110 molecules are expected to form loose but potent disaggregating heterocomplexes with Hsp70, leaving the remaining excess Hsp70 molecules free to carry Hsp70-specific functions, such as the catalytic unfolding of amenable misfolded monomers (12) and the pulling apart of native clathrin cages (32) or of active IκB complexes (58), the activation of steroid hormone receptors, the inhibition of the heat shock transcription factor (59), or mediating proteasomal and lysosomal degradation (60).
Analysis of the x-ray structures from various Hsp70s associated with NEFs (DnaK-GrpE, Hsp70-Bag1, and Hsp70-Bag2) shows a common region of interaction (data not shown) in the IB lobe of the Hsp70 NBD, which is structurally similar as in the reciprocal binding of the SSE1 to bovine Hsc70 NBD. This may justify initial description of Hsp110 acting as an apparent NEF of Hsp70 (33, 61). However, our results with apyrase suggested that Hsp110 does not need ATP to catalyze the release of a locked polypeptide clamped into an ADP-bound Hsp70. Moreover, Hsp110 was found to be as much an ATP-independent substrate release factor for Hsp70, as Hsp70 was a substrate release factor for Hsp110.
A Possible Disaggregation Mechanism
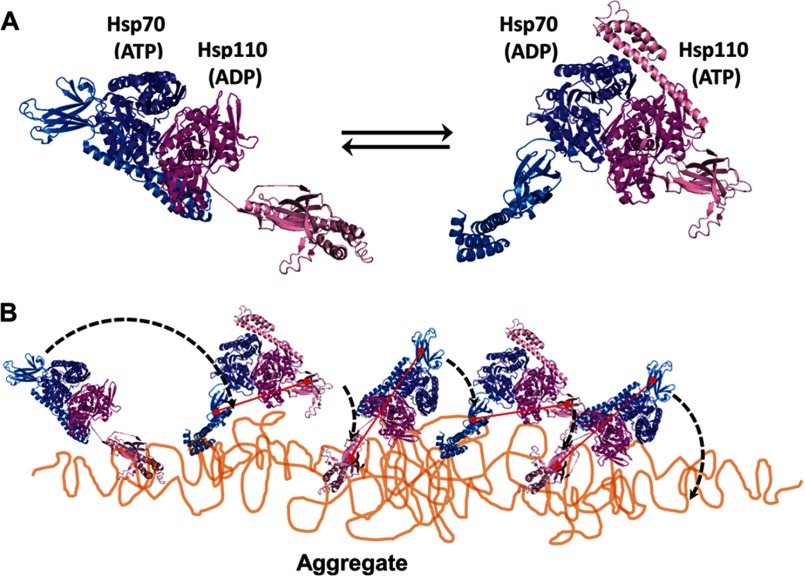
The molecular mechanism by which Hsp110-Hsp70 heterodimers may utilize ATP to disentangle stable protein aggregates and fibrils is unclear. The core mechanism should include basic unfolding by individual Hsp70 and, as we found, also of Hsp110 molecules, amplified by the heterodimeric complex into a cooperative disaggregating machine. Inspired from the partial crystal structure of the heterodimer showing SSE1 and bovine Hsc70 in different nucleotide binding states (32), one could propose a “clamping and walking on the aggregate” mechanism, in which both SBDs of the Hsp110-Hsp70 heterodimer would side by side face the aggregate. The cycles of ATP hydrolysis would alternate between the ADP-Hsp110 (clamped):ATP-Hsp70 (open) state and the ATP-Hsp110 (open):ADP-Hsp70 (clamped) state (Fig. 7). Although one SBD would be firmly anchored onto the aggregate surface by way of an ADP-bound subunit clamping and unfolding a misfolded polypeptide segment, the flexible hinge between the SBD and the NBD would allow for the other ATP-binding chaperone molecule with its wide open SBD to scan the surrounding surfaces for new exposed hydrophobic surfaces to clamp upon (Fig. 7). Triggered by this “substrate-probing” chaperone subunit, ADP release from the NBD of the first subunit would “unclamp” its SBD and thus allow the heterodimer to “walk” on the aggregate surface, thus leaving behind a trail of unfolded polypeptide loops that would thereafter spontaneously seek more native-like conformations and thus gradually dissociate and refold into soluble native polypeptides.
FIGURE 7.
Model for Hsp110-Hsp70-mediated disaggregation. A, model of the two alternating functional HSPA8-SSE1 heterodimers, respectively, in the ATP-ADP state (left panel) and the ADP-ATP state (right panel), reconstructed by structural alignments of the SBDs from HSPA8 (blue tones) and SSE1 (magenta tones), either with the closed ADP-bound form of DnaK (PDB code 2KHO) or with the opened ATP-bound form of DnaK (PDB code 4B9Q). The association of the HSPA8 NBD with the SSE1 NBD was from PDB code 3C7N. The substrate binding sites of HSPA8 and SSE1 SBDs are shown as red circles. The connecting red line shows the distances between binding sites in the heterodimer. B, model for the disaggregation mechanism. The crystal structure of the HSPA8-SSE1 heterodimer (32) suggests that when one chaperone is ADP-bound, its SBD can be locked on an unfolded substrate, whereas the other chaperone is in the ATP-bound state with a widely opened SBD. During disaggregation, SSE1 and Hsp70 may thus alternate between a closed locked state, anchoring the whole chaperone heterodimer to the aggregate surface at a single misfolded loop (left panel), becoming unfolded by entropic pulling (44), and an opened state that can scan far from the anchor for new misfolded structures to bind to. Then, upon ATP hydrolysis, the second chaperone may bind and apply an unfolding pressure on the newly targeted misfolded loops (right panel). When successful, locking may unfold the misfolded structure, while concomitantly, an allosteric signal is sent from the NBD of the second locking chaperone to the NBD of the first chaperone to release ADP and thus unlock the SBD from a now newly enlarged unfolded anchoring loop, which may then refold to the native state.
In Vivo Relevance of Hsp110
The cytoplasm and endoplasmic reticulum of animal cells lack ClpB/Hsp104-like co-chaperones and may use only Hsp110-Hsp70 to disaggregate potential proteotoxic conformers. Thus, Hsp110 was co-localized to GFP-luciferase aggregates in human U2OS cells, and Hsp110 RNAi dramatically increased the number of heat-induced aggregates of Luciferase-YFP in whole Caenorhabditis elegans animals. Moreover, knockdown of Hsp110 decreased the nematode lifespan, particularly upon heat shock (28). Thus, functional Hsp70-Hsp110 complexes in the cytoplasm and the endoplasmic reticulum of human cells likely form a powerful unfolding/disaggregating machinery to counteract age- and mutation-induced toxic protein misfolding and aggregations, causing the onset of degenerative protein conformational diseases.
Acknowledgments
We thank Bernd Bukau for a gift of Apg-2 and DNAJB1, Rui Sousa and Eileen M. Lafer for expression plasmids encoding for human HSPH1, and Anika Braune and América Farina Henriquez Cuendet for technical assistance.
This work was supported in part by the Faculty of Biology and Medicine of Lausanne University of Lausanne and by Grant 31003A-140512/1 from the Swiss National Fund.
- FTluc
- freeze-thaw inactivated luciferase
- PDB
- Protein Data Bank
- SBD
- substrate-binding domain
- ThT
- thioflavin T
- Luc
- luciferase
- NEF
- nucleotide exchange factor
- NBD
- nucleotide-binding domain.
REFERENCES
- 1. Anfinsen C. B. (1973) Principles that govern the folding of protein chains. Science 181, 223–230 [DOI] [PubMed] [Google Scholar]
- 2. Geiger T., Wehner A., Schaab C., Cox J., Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11, M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Finka A., Goloubinoff P. (2013) Proteomic data from human cell cultures refine mechanisms of chaperone-mediated protein homeostasis. Cell Stress Chaperones DOI 10.1007/s12192-013-0413-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cabrita L. D., Dobson C. M., Christodoulou J. (2010) Protein folding on the ribosome. Curr. Opin. Struct. Biol. 20, 33–45 [DOI] [PubMed] [Google Scholar]
- 5. Ellis R. J. (2001) Macromolecular crowding. Obvious but underappreciated. Trends Biochem. Sci. 26, 597–604 [DOI] [PubMed] [Google Scholar]
- 6. Martin J., Hartl F. U. (1997) The effect of macromolecular crowding on chaperonin-mediated protein folding. Proc. Natl. Acad. Sci. U.S.A. 94, 1107–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dobson C. M. (2003) Protein folding and misfolding. Nature 426, 884–890 [DOI] [PubMed] [Google Scholar]
- 8. Natalello A., Mattoo R. U., Priya S., Sharma S. K., Goloubinoff P., Doglia S. M. (2013) Biophysical characterization of two different stable misfolded monomeric polypeptides that are chaperone-amenable substrates. J. Mol. Biol. 425, 1158–1171 [DOI] [PubMed] [Google Scholar]
- 9. Gidalevitz T., Ben-Zvi A., Ho K. H., Brignull H. R., Morimoto R. I. (2006) Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311, 1471–1474 [DOI] [PubMed] [Google Scholar]
- 10. Hinault M. P., Ben-Zvi A., Goloubinoff P. (2006) Chaperones and proteases. Cellular fold-controlling factors of proteins in neurodegenerative diseases and aging. J. Mol. Neurosci. 30, 249–265 [DOI] [PubMed] [Google Scholar]
- 11. Lashuel H. A., Hartley D., Petre B. M., Walz T., Lansbury P. T., Jr. (2002) Neurodegenerative disease. Amyloid pores from pathogenic mutations. Nature 418, 291. [DOI] [PubMed] [Google Scholar]
- 12. Sharma S. K., De los Rios P., Christen P., Lustig A., Goloubinoff P. (2010) The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat. Chem. Biol. 6, 914–920 [DOI] [PubMed] [Google Scholar]
- 13. Priya S., Sharma S. K., Sood V., Mattoo R. U., Finka A., Azem A., De Los Rios P., Goloubinoff P. (2013) GroEL and CCT are catalytic unfoldases mediating out-of-cage polypeptide refolding without ATP. Proc. Natl. Acad. Sci. U.S.A. 110, 7199–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hinault M. P., Farina-Henriquez-Cuendet A., Goloubinoff P. (2011) Molecular chaperones and associated cellular clearance mechanisms against toxic protein conformers in Parkinson's disease. Neurodegener. Dis. 8, 397–412 [DOI] [PubMed] [Google Scholar]
- 15. Priya S., Sharma S. K., Goloubinoff P. (2013) Molecular chaperones as enzymes that catalytically unfold misfolded polypeptides. FEBS Lett. 587, 1981–1987 [DOI] [PubMed] [Google Scholar]
- 16. Diamant S., Ben-Zvi A. P., Bukau B., Goloubinoff P. (2000) Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J. Biol. Chem. 275, 21107–21113 [DOI] [PubMed] [Google Scholar]
- 17. Bucciantini M., Giannoni E., Chiti F., Baroni F., Formigli L., Zurdo J., Taddei N., Ramponi G., Dobson C. M., Stefani M. (2002) Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 [DOI] [PubMed] [Google Scholar]
- 18. Selkoe D. J. (2004) Cell biology of protein misfolding. The examples of Alzheimer's and Parkinson's diseases. Nat. Cell Biol. 6, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 19. De Los Rios P., Goloubinoff P. (2012) Protein folding. Chaperoning protein evolution. Nat. Chem. Biol. 8, 226–228 [DOI] [PubMed] [Google Scholar]
- 20. Finka A., Mattoo R. U., Goloubinoff P. (2011) Meta-analysis of heat- and chemically upregulated chaperone genes in plant and human cells. Cell Stress Chaperones 16, 15–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fenton W. A., Kashi Y., Furtak K., Horwich A. L. (1994) Residues in chaperonin GroEL required for polypeptide binding and release. Nature 371, 614–619 [DOI] [PubMed] [Google Scholar]
- 22. Rüdiger S., Buchberger A., Bukau B. (1997) Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4, 342–349 [DOI] [PubMed] [Google Scholar]
- 23. Rüdiger S., Schneider-Mergener J., Bukau B. (2001) Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 20, 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sharma S. K., De Los Rios P., Goloubinoff P. (2011) Probing the different chaperone activities of the bacterial HSP70-HSP40 system using a thermolabile luciferase substrate. Proteins 79, 1991–1998 [DOI] [PubMed] [Google Scholar]
- 25. Ben-Zvi A., De Los Rios P., Dietler G., Goloubinoff P. (2004) Active solubilization and refolding of stable protein aggregates by cooperative unfolding action of individual hsp70 chaperones. J. Biol. Chem. 279, 37298–37303 [DOI] [PubMed] [Google Scholar]
- 26. Glover J. R., Lindquist S. (1998) Hsp104, Hsp70, and Hsp40. A novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 [DOI] [PubMed] [Google Scholar]
- 27. Goloubinoff P., Mogk A., Zvi A. P., Tomoyasu T., Bukau B. (1999) Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. U.S.A. 96, 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rampelt H., Kirstein-Miles J., Nillegoda N. B., Chi K., Scholz S. R., Morimoto R. I., Bukau B. (2012) Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 31, 4221–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shorter J. (2011) The mammalian disaggregase machinery. Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One 6, e26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hinault M. P., Cuendet A. F., Mattoo R. U., Mensi M., Dietler G., Lashuel H. A., Goloubinoff P. (2010) Stable α-synuclein oligomers strongly inhibit chaperone activity of the Hsp70 system by weak interactions with J-domain co-chaperones. J. Biol. Chem. 285, 38173–38182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Biancalana M., Makabe K., Koide A., Koide S. (2009) Molecular mechanism of thioflavin-T binding to the surface of β-rich peptide self-assemblies. J. Mol. Biol. 385, 1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., Sousa R. (2008) Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andréasson C., Fiaux J., Rampelt H., Mayer M. P., Bukau B. (2008) Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 283, 8877–8884 [DOI] [PubMed] [Google Scholar]
- 34. Raviol H., Sadlish H., Rodriguez F., Mayer M. P., Bukau B. (2006) Chaperone network in the yeast cytosol. Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25, 2510–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wisén S., Bertelsen E. B., Thompson A. D., Patury S., Ung P., Chang L., Evans C. G., Walter G. M., Wipf P., Carlson H. A., Brodsky J. L., Zuiderweg E. R., Gestwicki J. E. (2010) Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem. Biol. 5, 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liberek K., Marszalek J., Ang D., Georgopoulos C., Zylicz M. (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. U.S.A. 88, 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raviol H., Bukau B., Mayer M. P. (2006) Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett. 580, 168–174 [DOI] [PubMed] [Google Scholar]
- 38. Siegenthaler R. K., Christen P. (2006) Tuning of DnaK chaperone action by nonnative protein sensor DnaJ and thermosensor GrpE. J. Biol. Chem. 281, 34448–34456 [DOI] [PubMed] [Google Scholar]
- 39. Rüngeling E., Laufen T., Bahl H. (1999) Functional characterisation of the chaperones DnaK, DnaJ, and GrpE from Clostridium acetobutylicum. FEMS Microbiol. Lett. 170, 119–123 [DOI] [PubMed] [Google Scholar]
- 40. Zolkiewski M. (1999) ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem. 274, 28083–28086 [DOI] [PubMed] [Google Scholar]
- 41. Rodriguez F., Arsène-Ploetze F., Rist W., Rüdiger S., Schneider-Mergener J., Mayer M. P., Bukau B. (2008) Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol Cell 32, 347–358 [DOI] [PubMed] [Google Scholar]
- 42. Zhuravleva A., Gierasch L. M. (2011) Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc. Natl. Acad. Sci. U.S.A. 108, 6987–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baneyx F., Nannenga B. L. (2010) Chaperones. A story of thrift unfolds. Nat Chem Biol 6, 880–881 [DOI] [PubMed] [Google Scholar]
- 44. De Los Rios P., Ben-Zvi A., Slutsky O., Azem A., Goloubinoff P. (2006) Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc. Natl. Acad. Sci. U.S.A. 103, 6166–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brehmer D., Gässler C., Rist W., Mayer M. P., Bukau B. (2004) Influence of GrpE on DnaK-substrate interactions. J. Biol. Chem. 279, 27957–27964 [DOI] [PubMed] [Google Scholar]
- 46. Kabani M., Beckerich J. M., Brodsky J. L. (2002) Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22, 4677–4689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mayer M. P., Bukau B. (2005) Hsp70 chaperones. Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sondermann H., Scheufler C., Schneider C., Hohfeld J., Hartl F. U., Moarefi I. (2001) Structure of a Bag/Hsc70 complex. Convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291, 1553–1557 [DOI] [PubMed] [Google Scholar]
- 49. Packschies L., Theyssen H., Buchberger A., Bukau B., Goody R. S., Reinstein J. (1997) GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry 36, 3417–3422 [DOI] [PubMed] [Google Scholar]
- 50. Shomura Y., Dragovic Z., Chang H. C., Tzvetkov N., Young J. C., Brodsky J. L., Guerriero V., Hartl F. U., Bracher A. (2005) Regulation of Hsp70 function by HspBP1. Structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell 17, 367–379 [DOI] [PubMed] [Google Scholar]
- 51. Steel G. J., Fullerton D. M., Tyson J. R., Stirling C. J. (2004) Coordinated activation of Hsp70 chaperones. Science 303, 98–101 [DOI] [PubMed] [Google Scholar]
- 52. Knappe D., Zahn M., Sauer U., Schiffer G., Sträter N., Hoffmann R. (2011) Rational design of oncocin derivatives with superior protease stabilities and antibacterial activities based on the high-resolution structure of the oncocin-DnaK complex. Chembiochem 12, 874–876 [DOI] [PubMed] [Google Scholar]
- 53. Polier S., Hartl F. U., Bracher A. (2010) Interaction of the Hsp110 molecular chaperones from S. cerevisiae with substrate protein. J. Mol. Biol. 401, 696–707 [DOI] [PubMed] [Google Scholar]
- 54. Xu X., Sarbeng E. B., Vorvis C., Kumar D. P., Zhou L., Liu Q. (2012) Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J. Biol. Chem. 287, 5661–5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oh H. J., Chen X., Subjeck J. R. (1997) Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J. Biol. Chem. 272, 31636–31640 [DOI] [PubMed] [Google Scholar]
- 56. Kityk R., Kopp J., Sinning I., Mayer M. P. (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 48, 863–874 [DOI] [PubMed] [Google Scholar]
- 57. Bertelsen E. B., Chang L., Gestwicki J. E., Zuiderweg E. R. (2009) Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U.S.A. 106, 8471–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weiss Y. G., Bromberg Z., Raj N., Raphael J., Goloubinoff P., Ben-Neriah Y., Deutschman C. S. (2007) Enhanced heat shock protein 70 expression alters proteasomal degradation of IκB kinase in experimental acute respiratory distress syndrome. Crit. Care Med. 35, 2128–2138 [DOI] [PubMed] [Google Scholar]
- 59. Voellmy R. (2004) On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Muchowski P. J., Wacker J. L. (2005) Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 6, 11–22 [DOI] [PubMed] [Google Scholar]
- 61. Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. (2006) Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]