Background: Allophanate hydrolase (AH) is essential for urea utilization in many organisms.
Results: We determined the crystal structure of the Kluyveromyces lactis AH and performed mechanistic studies.
Conclusion: Our work revealed that the AH N and C domains catalyze sequential reactions and provided insights into their catalysis.
Significance: The catalytic mechanism of the C domain might expand the knowledge of decarboxylation reactions.
Keywords: Decarboxylase, Enzyme Catalysis, Enzyme Structure, Nitrogen Metabolism, X-ray Crystallography, Allophanate Hydrolase, Amidase Signature Family, Urea Utilization
Abstract
Allophanate hydrolase converts allophanate to ammonium and carbon dioxide. It is conserved in many organisms and is essential for their utilization of urea as a nitrogen source. It also has important functions in a newly discovered eukaryotic pyrimidine nucleic acid precursor degradation pathway, the yeast-hypha transition that several pathogens utilize to escape the host defense, and an s-triazine herbicide degradation pathway recently emerged in many soil bacteria. We have determined the crystal structure of the Kluyveromyces lactis allophanate hydrolase. Together with structure-directed functional studies, we demonstrate that its N and C domains catalyze a two-step reaction and contribute to maintaining a dimeric form of the enzyme required for their optimal activities. Our studies also provide molecular insights into their catalytic mechanism. Interestingly, we found that the C domain probably catalyzes a novel form of decarboxylation reaction that might expand the knowledge of this common reaction in biological systems.
Introduction
Nitrogen is an essential element in all life forms (1). Most organisms cannot directly utilize the relatively inert nitrogen gas abundant in the atmosphere as a nitrogen source; biologically active forms of nitrogen are constantly being recycled (2). Urea is the degradation product of a wide range of nitrogen-containing biomolecules. In mammals, urea is excreted. Many plants, bacteria, algae, and fungi can use urea for their anabolism, reintegrating its nitrogen into the biosphere. The first step of their urea utilization is converting it to ammonium and carbon dioxide (3–5), and two enzymes catalyzing such a reaction, urease and urea amidolyase (UA),2 have been reported (6).
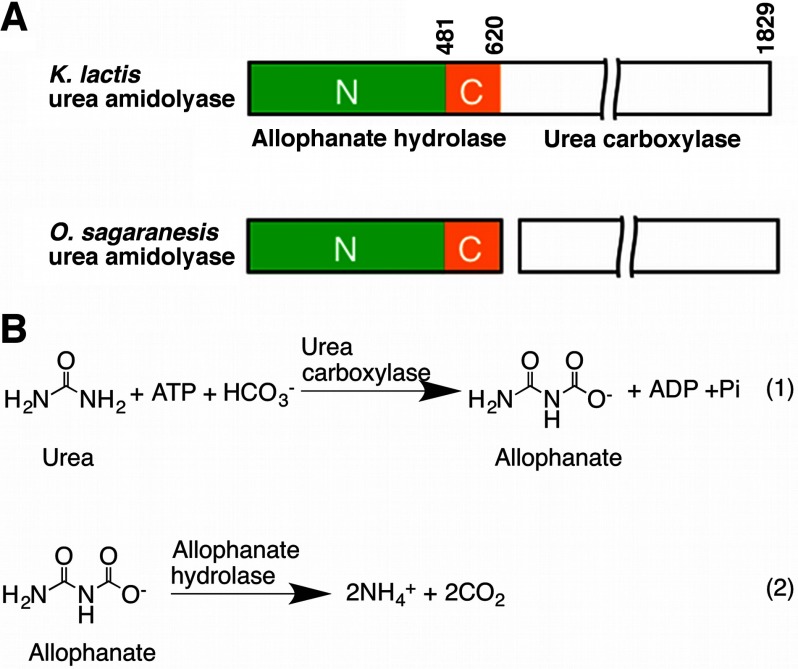
UA has two activities: the urea carboxylase activity carboxylates urea to allophanate, and the allophanate hydrolase (AH) activity converts it to ammonium and carbon dioxide (7). In some organisms, the urea carboxylase and AH activities are carried out by separate polypeptides (Fig. 1, A and B) (8, 9). UA is widely distributed in bacteria and fungi (10, 11) and is found in algae (12). However, the Ni2+-dependent urease and nickel/cobalt transporter are not found in many of the UA-containing fungal species. They might have dropped all Ni2+-dependent metabolisms and as a result might have a selective advantage because the cellular concentration of Ni2+ has to be tightly regulated (10). Besides its function in urea utilization, UA is an essential component of a recently identified eukaryotic pyrimidine nucleic acid precursor degradation pathway (13). In the human pathogen Candida albicans, UA plays an important role in its yeast-hypha transition, a mechanism to escape the host defense (14, 15).
FIGURE 1.
Domain architecture and reactions catalyzed by UA. A, domain organization of UA. Domain boundaries are indicated for the K. lactis UA. The N and C domains of the AH component are colored in green and orange, respectively. This color scheme is used throughout the figures unless otherwise indicated. B, reactions catalyzed by the urea carboxylase and AH components of UA. O., Oleomonas.
Besides working with urea carboxylase in converting urea to ammonium and carbon dioxide, AH also has an important function in a newly evolved s-triazine herbicide degradation pathway. Since their introduction more than half a century ago, the environmental half-life of the commercial s-triazine herbicides have decreased dramatically because of the emergence of such a pathway in many soil bacteria (16). In this pathway, the s-triazine compounds are ultimately degraded to allophanate, which is converted to ammonium and carbon dioxide by AH (16–19).
A group of proteins and protein complexes, including the Mycobacterium smegmatis Msmeg0435-Msmeg0436 complex and the Thermus thermophilus TTHA0988 protein, were annotated as allophanate hydrolase. Structures of the Msmeg0435-Msmeg0436 complex and TTHA0988 (20, 21) have been determined. These proteins and complexes show no sequence homology to the allophanate hydrolases discussed here, and attempts to demonstrate the AH activity of TTHA0988 were not successful (21). They are instead homologous to the KipI-KipA complex involved in the Bacillus subtilis sporulation regulation (21) and the urea carboxylase carboxyltransferase domain (22) and are probably not true allophanate hydrolases.
We have previously reported the crystal structure of the urea carboxylase component of UA (22). However, the structure and catalytic mechanism of its AH component are not clear. We present here the crystal structure of the Kluyveromyces lactis AH (KlAH). The structure revealed that AH is composed of N and C domains. Structure-directed functional studies indicate that these domains catalyze sequential reactions: the N domain converts allophanate to N-carboxycarbamate, and the C domain converts it to carbon dioxide and ammonium. They also contribute to maintaining a dimeric form of the enzyme that is essential for their optimal activities. Whereas the N domain catalyzes an amide hydrolysis reaction typical for the amidase signature (AS) family members, our data indicate that the reaction catalyzed by the C domain probably represents a novel kind of decarboxylation reaction.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The DNA segment corresponding to the AH domain of the K. lactis UA (residues 1–621) was amplified from the K. lactis genome and inserted into vector pET28a (Novagen). The Escherichia coli BL21 Star (DE3) cells transformed with this plasmid were cultured in LB medium supplemented with 50 mg/liter kanamycin and induced with 0.3 mm of isopropyl β-d-thiogalactopyranoside (Bio Basic Inc.) at 16 °C for 12 h. Cells were collected by centrifugation and lysed with a high pressure homogenizer (ATS Engineering Inc.). KlAH was purified with nickel-nitrilotriacetic acid (Qiagen) and size exclusion (Sephacryl S300 HR, GE Healthcare) columns; concentrated to 10 mg/ml in a buffer containing 20 mm Tris/HCl, pH 7.5, 200 mm NaCl, 2 mm DTT, and 5% (v/v) glycerol; flash cooled in liquid nitrogen; and stored at −80 °C.
The K. lactis UA (KlUA; residues 1–1829) cDNA was cloned into vector pET28a. The resulting plasmid was co-transformed with a pTYB12 (New England Biolabs)-based plasmid containing the E. coli biotin ligase gene birA into the E. coli strain BL21 Star (DE3). Cells were cultured in LB medium supplemented with 50 mg/liter kanamycin and 100 mg/liter ampicillin and induced with 0.3 mm isopropyl β-d-thiogalactopyranoside at 16 °C for 12 h. 30 min before the induction, 15 mg/liter d-biotin was added to the medium. KlUA was purified following the same protocol used for KlAH.
Mutagenesis was carried out with a QuikChange kit (Agilent Technologies) and verified by DNA sequencing. The mutants were expressed and purified following the same protocol for the wild type proteins.
The selenomethionine (SeMet)-substituted KlAH was expressed by growing cells in M9 medium supplemented with specific amino acids to block endogenous methionine synthesis and supplementing with SeMet (23). Purification of the SeMet-substituted KlAH was the same as for the native protein except that the DTT concentration was increased to 10 mm.
Protein Crystallization
Rod-shaped crystals of the SeMet-substituted KlAH and the KlAH S177A mutant were obtained with the sitting drop method at 20 °C. The reservoir liquid contained 16% PEG 8000, 20% glycerol, and 0.04 m potassium phosphate monobasic. Prior to crystallization, sodium/potassium tartrate (Hampton Research) was added to the protein solution to a final concentration of 100 mm. Crystals were flash cooled and stored in liquid nitrogen before data collection.
Data Collection, Structure Determination, and Refinement
Data collection was performed at 100 K. Diffraction data were collected on an ADSC Q315 charge-coupled device detector at the Shanghai Synchrotron Radiation Facility beamline BL17U. A single wavelength anomalous diffraction data set was collected at the selenium K-edge on a SeMet-substituted crystal, and a native data set was collected at the same energy on a crystal of the S177A mutant. Diffraction data were scaled with mosflm and integrated with scala, and the intensities were converted to structure factors with ctruncate.
The KlAH structure was determined with a combination of molecular replacement and single wavelength anomalous diffraction methods with phaser using coordinates of the Aquifex aeolicus glutamyl-tRNAGln amidotransferase (Protein Data Bank code 3H0L) α subunit as a search model. The electron density map was improved by solvent flattening, histogram averaging, and noncrystallographic symmetry averaging with dm. Manual building and inspection of the structure were carried out with Coot (24) and O (25). Refinement was carried out with refmac.
The structure of the S177A mutant was determined by molecular replacement with molrep using the structure of the wild type protein as a search model. The structure was refined with refmac.
Data processing and refinement statistics are summarized in Table 1. mosflm, scala, ctruncate, phaser, refmac, and molrep are programs in the CCP4 suite (26).
TABLE 1.
Data collection and refinement statistics
| SeMet-substituted wild type | S177A | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 93.1, 107.7, 150.8 | 89.9, 107.4, 152.3 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 0.979 | 0.979 |
| Resolution (Å) | 46.5-2.5 (2.64-2.50)a | 40.0-2.6 (2.64-2.60) |
| Rsym (%) | 9.3 (43.2) | 9.4 (47.0) |
| I/σI | 6.1 (1.6) | 30.0 (5.1) |
| Completeness (%) | 99.9 (100.0) | 93.2 (89.0) |
| Redundancy | 6.0 (6.0) | 5.7 (5.9) |
| Wilson B factor (Å2) | 45.8 | 52.6 |
| Refinement | ||
| Resolution (Å) | 46.5-2.5 (2.57-2.50) | 40.0-2.6 (2.66-2.60) |
| No. reflections | 50,331 (3,387) | 40,953 (2,600) |
| Rwork/Rfree (%) | 17.6 (26.2)/24.7 (36.3) | 18.4 (26.2)/25.6 (34.9) |
| No. atoms | ||
| Protein | 9,479 | 9,494 |
| Ligand/ion | 16 | 19 |
| Water | 273 | 139 |
| B factors (Å2) | ||
| Protein | 37.8 | 48.2 |
| Ligand/ion | 58.5 | 59.7 |
| Water | 33.4 | 37.1 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.015 | 0.013 |
| Bond angles (°) | 1.995 | 1.827 |
| ESUb (Å) | 0.197 | 0.247 |
| Protein Data Bank code | 4ISS | 4IST |
a Numbers enclosed in parentheses are for the highest resolution shells.
b Estimated overall coordinate error based on maximum likelihood.
Kinetic Assays
Potassium allophanate was prepared and quantified as described (9). The AH activity was measured by coupling ammonium release to NADH to NAD conversion through glutamate dehydrogenase (9). The resulting absorption change at 340 nm was monitored on an Ultrospec 2100 pro spectrophotometer (GE Healthcare). Kinetic assays were carried out at room temperature. The reaction mixture contained 100 mm Tris/HCl, pH 8.0, 1 μm EGTA, 3 mm magnesium chloride, 19.5 mm potassium chloride, 800 units/ml glutamate dehydrogenase (Roche Applied Science), 50 mm oxoglutaric acid, 0.3 mm NADH, 0.5 μm KlAH, and variable concentrations of potassium allophanate.
The UA activity was measured similarly. The reaction mixture was the same as for KlAH except that KlAH was replaced by 0.75 μm KlUA, potassium allophanate was replaced by urea, and 20 mm ATP and 200 mm sodium bicarbonate were added to the reaction.
Mass Spectrometry
Mass spectrometric analysis of the AH and UA reaction mixtures was performed on an MDS SCIEX 4000 Q-TRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems). Both reaction mixtures contained 100 mm Tris/HCl, pH 8.0, 1 μm EGTA, 3 mm magnesium chloride, and 19.5 mm potassium chloride. For the AH reaction, 0.15 m potassium allophanate and 2.5 μm KlAH were added. For the UA reaction, 100 mm urea, 200 mm sodium bicarbonate, 20 mm ATP, and 2 μm KlUA were added. Samples of the reaction mixture were mixed 1:100 with 80% acetonitrile and analyzed by direct liquid infusion in the negative ion mode of electrospray ionization mass spectrometry. For reactions performed in the presence of 18O-labeled water, 50% 18O-labeled water was supplemented to the reaction.
Dynamic Light Scattering Experiments
Dynamic light scattering experiments were performed at 20 °C on a DynaPro Titan dynamic light scattering instrument (Wyatt Technologies). KlAH was measured at a concentration of 5 mg/ml in a buffer containing 20 mm Tris/HCl, pH 7.5, 200 mm NaCl, and 1 mm DTT. Data were collected and analyzed with DYNAMICS V6 software (Wyatt Technologies).
Data Deposition
Crystal structures of the wild type KlAH and the S177A mutant have been deposited into the Protein Data Bank with accession codes 4ISS and 4IST, respectively.
RESULTS AND DISCUSSION
Structure Determination
KlAH was expressed in E. coli, purified, and crystallized with the sitting drop vapor diffusion method. The crystals belong to space group P212121 with two monomers in the asymmetric unit. The structure was determined with a combination of molecular replacement and single wavelength anomalous diffraction methods and refined to a resolution of 2.5 Å. The refined structure agrees well with the diffraction data and expected geometric values (Table 1) with 87.3% of the residues in the most favored region of the Ramachandran plot and 11.9% in the additionally allowed region.
Overall Structure
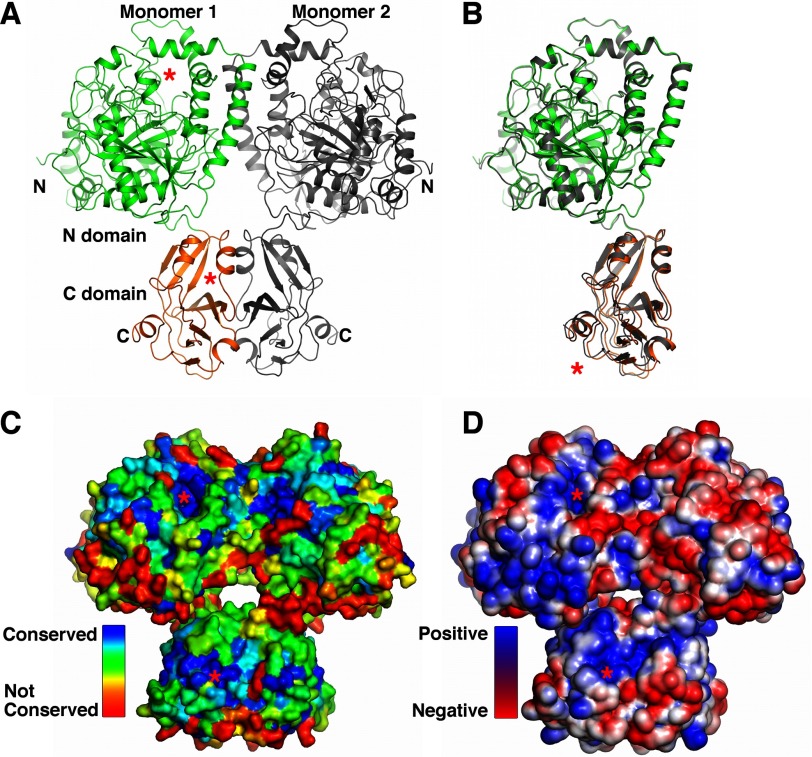
KlAH residues 1–614 are clearly defined by electron density maps for both monomers in the crystal, and the few residues at the C terminus of the expression construct are probably disordered. The KlAH monomer adopts an elongated structure and consists of N (residues 1–481) and C (residues 482–614) domains (Fig. 2A and supplemental Fig. S1). The structures of the two KlAH monomers in the crystal are highly similar. After alignment, the root mean square (r.m.s.) deviations for Cα atoms are 0.35 Å for the N domain and 0.44 Å for the C domain. With the N domains aligned, the C domains are related by a small rotation of 3.8° (Fig. 2B) that is probably caused by crystal packing interactions. A surface pocket is found on each of the domains. They face the same direction and are separated by a distance of 50 Å (Fig. 2A). Residues in these pockets are highly conserved (Fig. 2C), and both regions are significantly positively charged (Fig. 2D).
FIGURE 2.
Overall structure of KlAH. A, structure of the KlAH dimer. The KlAH dimer observed in the crystal is colored in green and orange for one monomer and gray for the other. Structures in A, C, and D are shown in the same orientation. The red stars indicate surface pockets on the N and C domains. B, superimposition of the KlAH monomers. One monomer in the KlAH dimer is colored in green and orange, and the other is in gray. Their N domains are aligned, and the red star indicates differences in their C domain position. C, conservation of residues in the KlAH dimer. The KlAH dimer is shown in surface representation and colored according to the conservation of individual residues, which was calculated with the ConSurf server (46). D, surface charge distribution of the KlAH dimer. The KlAH dimer is shown in surface representation and is colored in blue and red for positively and negatively charged regions, respectively. Structure figures were prepared with PyMOL.
In the KlAH dimer observed in the crystal, interactions are found between the N domains and the C domains, and N and C domains from different monomers do not interact. The dimer adopts the shape of a butterfly with the N domain pair located on top of the C domain pair. The surface pockets on the N and C domains of different monomers face opposite directions (Fig. 2A).
AH Is Dimeric in Solution
The KlAH dimer interface observed in the crystal buries 5000 Å2 of surface area with 2600 Å2 contributed by the N domains and 2400 Å2 contributed by the C domains. Such an extensive interface suggests that KlAH is dimeric in solution. Consistently, dynamic light scattering measurements indicated a molecular mass of 172 kDa for KlAH in solution, and in gel filtration experiments, the elution volume of KlAH corresponds to a molecular mass of 125 kDa (Fig. 3C; the molecular mass of the KlAH monomer is 70 kDa).
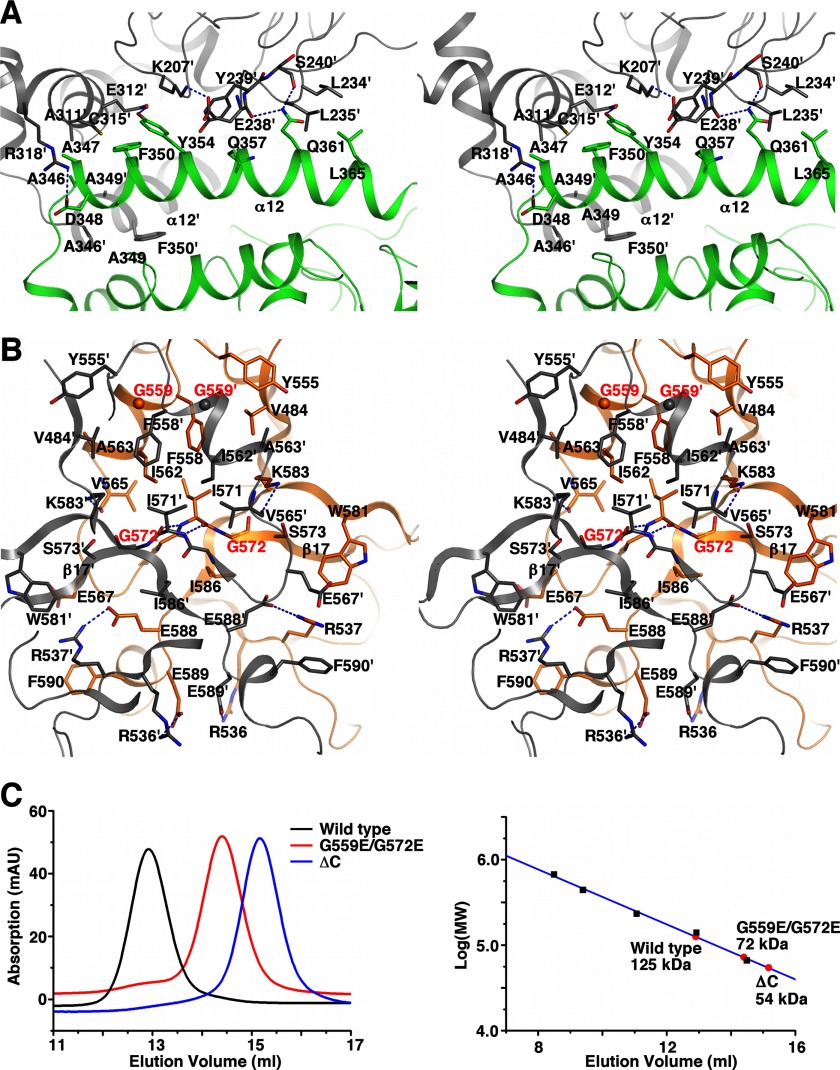
FIGURE 3.
The KlAH dimer interface. A, stereoview of the N domain dimer interface. The two N domains are colored in green and gray, respectively. For clarity, only interface residues on helix α12 of monomer 1 and those they interact with on monomer 2 are shown. B, stereoview of the C domain dimer interface. The two C domains are colored in orange and gray, respectively. Red labels highlight residues Gly559 and Gly572 at the interface. C, gel filtration analysis of the wild type KlAH and the G559E/G572E and ΔC mutants. Experiments were performed on a Superdex 200 10/30 column (GE Healthcare) with a buffer containing 20 mm Tris/HCl, pH 7.5 and 200 mm NaCl (left panel). The column was calibrated with albumin (66 kDa), lactate dehydrogenase (140 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa; black squares in the right panel). Apparent molecular weights of the wild type and mutant forms of KlAH are indicated (red circles in the right panel). mAU, milliabsorbance units.
At the dimer interface observed in the crystal, helix α12 plays an important role in mediating interactions between the N domains. The first turn of α12 is surrounded on three sides by residues from the same region and α10 of the other monomer. Ala346, Ala347, and Ala349 in this region form hydrophobic interactions with Ala311′ (the ′ signs indicate the other monomer), Cys315′, Ala346′, Ala349′, and Phe350′; and the Asp348 side chain in this region ion pairs with the Arg318′ side chain. Residues Phe350, Tyr354, Gln357, Gln361, and Leu365 along α12 as well as residues Lys207′, Leu234′, Leu235′, Glu238′, Tyr239′, and Glu312′ contribute to additional van der Waals interactions. Hydrogen bonds are formed between the side chains of Tyr354 and Glu312′ and between the Gln361 side chain and the main chain carbonyl groups of Glu238′ and Ser240′ (Fig. 3A). Interactions mediated by the first turn of α12 and the Tyr354-Glu312′ hydrogen bond are generally conserved among AHs from different species (supplemental Fig. S1).
At the center of the interface between the C domains, the Gly570-Ile571 peptide on β17 forms antiparallel β-sheet interactions with the same region in the other monomer. Gly570, Ile571, and Gly572 in this region together with Val484, Tyr555, Phe558, Gly559, Ile562, Ala563, Val565, and Ile586 form a hydrophobic surface patch that interacts with the same region on the other monomer. Additional buried surface area is contributed by Arg537, Glu567, Ser573, Trp581, Lys583, Glu588, Glu589, and Phe590. The C domain dimer interface contains two pairs of salt bridges between side chains of Arg536 and Glu589 and side chains of Arg537 and Glu588 from different monomers. Hydrogen bonds are formed between the Lys583 side chain amine and the Val565′ main chain carbonyl (Fig. 3B). Many of the residues at the C domain dimer interface are conserved among AHs from different species (supplemental Fig. S1).
To test the physiological relevance of the observed interface, we introduced a G559E/G572E double mutation into the interface and analyzed the oligomeric state of the mutant by gel filtration experiments. As expected, this mutant is monomeric in solution (Fig. 3C). The ΔC mutant (containing residues 1–482) lacking the entire C domain, which contributes almost half of the observed buried surface area, is also monomeric in solution (Fig. 3C).
The N Domain Catalyzes an Amide Hydrolysis Reaction
The N domain contains a twisted, mixed 11-strand β sheet surrounded by 16 α helices (Fig. 2A and supplemental Fig. S1). Sequence analysis suggested that AH is an AS family member (27). Consistently, a search with the Dali server (28) indicated that the N domain structure is homologous to the structures of AS family members, including fatty acid amide hydrolase (29), malonamidase E2 (30), glutamyl-tRNAGln amidotransferase α subunit (GatA) (31–34), peptide amidase (35), 6-aminohexanoate-cyclic-dimer hydrolase (36), and an amidase from Rhodococcus sp. N771 (37). The structures of the N domain and these enzymes can be superimposed with r.m.s. deviations for Cα atoms between 2.4 and 3.1 Å. Their sequence identities are between 19 and 29%.
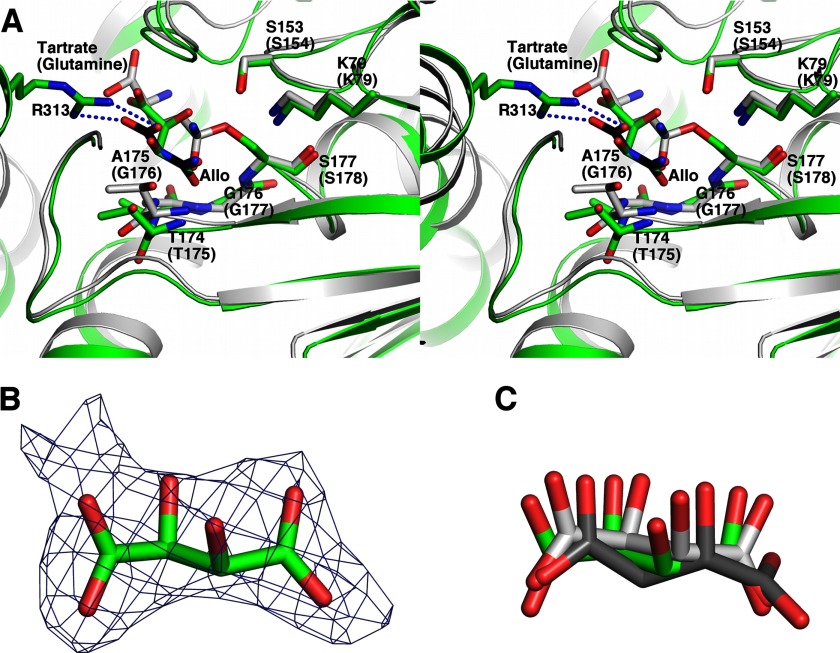
Malonamidase E2 (30, 38), GatA (31, 32), and other well studied AS family members hydrolyze the substrate amide. A Ser-cis-Ser-Lys triad in these enzymes catalyzes the reaction. The serine side chain hydroxyl performs the initial nucleophilic attack on the substrate carbonyl carbon, generating a tetrahedral intermediate covalently linked to the serine side chain hydroxyl and stabilized by an oxyanion hole in the enzyme. The cis-serine subsequently protonates the leaving ammonium group on the intermediate, prompting the C–N bond breakage, and the lysine facilitates this process. Finally, a water molecule attacks the carbonyl carbon of the acyl-enzyme intermediate, regenerating the catalytic serine (30, 38). Their active sites correspond to the conserved surface pocket on the N domain. In this region of KlAH, the Ser177-cis-Ser153-Lys79 triad and the main chain amides of Thr174, Ala175, Gly176, and Ser177 occupy locations identical to their catalytic triad and the oxyanion hole (Fig. 4A). In addition, a tartrate molecule probably introduced through the crystallization process is found in this pocket (Fig. 4, B and C). One of its carboxyl groups occupies a location similar to that of the substrate (glutamine) amide in the Staphylococcus aureus GatA active site (Fig. 4A) (31) and probably mimics the amide group of the N domain substrate. This structural evidence indicates that the surface pocket is the active site of the N domain, and it catalyzes an amide hydrolysis reaction similar to other AS family amidases.
FIGURE 4.
Structure of the N domain active site. A, stereoview of the N domain active site. Structures of the KlAH N domain (green for the carbon atoms) and the S. aureus GatA (Protein Data Bank code 2F2A; gray for the carbon atoms) are superimposed and shown in stereo. Important catalytic residues conserved between them are highlighted. Labels enclosed in parentheses are for GatA. A model of allophanate (Allo) at the N domain active site is shown in black for its carbon atoms. Arg313, which is potentially important for the AH substrate specificity, is highlighted. The dashed lines indicate potential ion pair interactions between the allophanate molecule and the Arg313 side chain. B, difference density map for the tartrate molecule found at the wild type KlAH N domain active site. The map was calculated before tartrate and solvent molecules were included and contoured at 2 σ. C, superimposition of the tartrate molecules found at the N domain active site. Tartrate molecules were found at the active sites of one of the N domains in the wild type KlAH structure and both N domains in the S177A mutant structure. These N domains are aligned, and the tartrate molecules are shown. Their carbon atoms are colored in green for the wild type KlAH structure and different shades of gray for the S177A mutant structure.
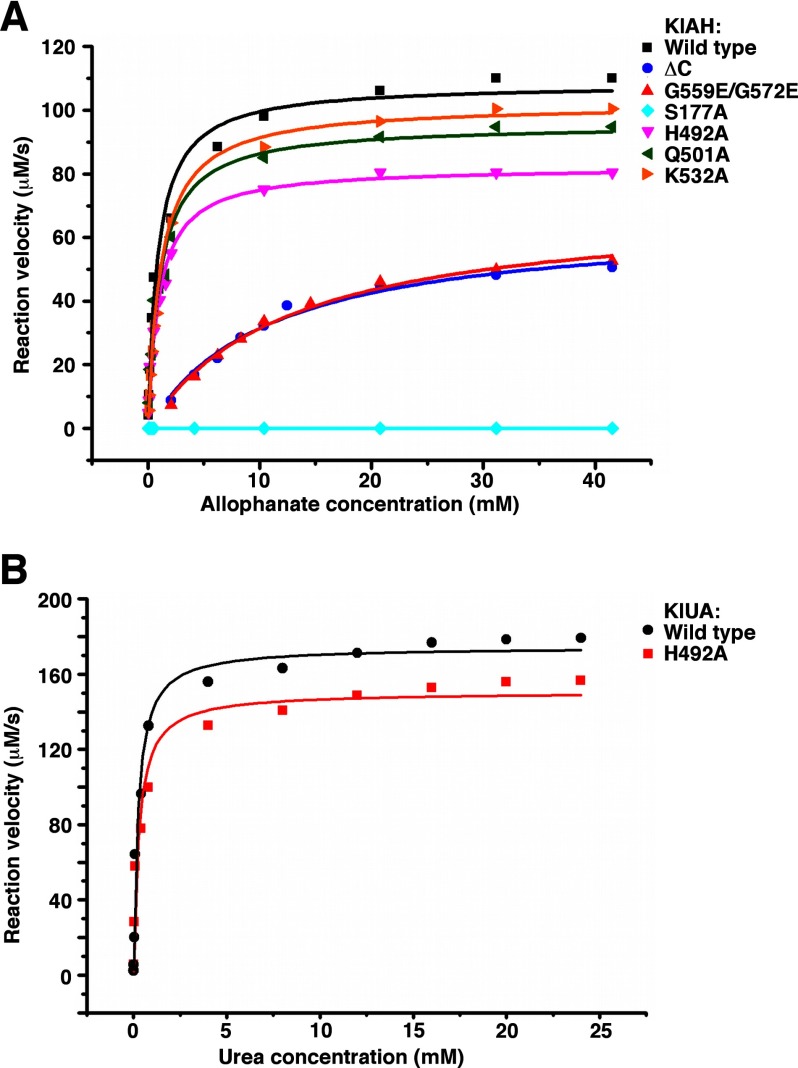
To verify the functional importance of the N domain active site, we substituted Ser177 with alanine and measured the activity of the mutant. The S177A mutant can no longer convert allophanate to ammonium at a detectable rate (Table 2 and Fig. 5A). The structure of the S177A mutant was determined with molecular replacement and refined to a resolution of 2.6 Å (Table 1). Except for the point mutation, it is almost identical to the wild type structure: after alignment, the r.m.s. deviation for Cα atoms between them is 0.42 Å. The loss of activity is therefore due to the loss of the Ser177 hydroxyl, which performs a nucleophilic attack on the substrate carbonyl carbon (30, 38).
TABLE 2.
Summary of kinetic parameters
| kcat | Kma | kcat/Km | |
|---|---|---|---|
| s−1 | mm | s−1 mm−1 | |
| KlAH | |||
| Wild type | 216.8 ± 5.8 (1.0)b,c | 0.88 ± 0.12 (1.0) | 246 ± 40 (1.0) |
| ΔC | 131.6 ± 5.8 (0.61) | 10.9 ± 1.2 (12) | 12.1 ± 1.9 (0.049) |
| G559E/G572E | 141.0 ± 6.0 (0.65) | 12.4 ± 1.3 (14) | 11.4 ± 1.7 (0.046) |
| S177A | NDd | ND | |
| H492A | 164.6 ± 3.4 (0.76) | 0.98 ± 0.08 (1.1) | 168 ± 17 (0.68) |
| Q501A | 191.2 ± 5.2 (0.88) | 1.07 ± 0.11 (1.2) | 179 ± 23 (0.73) |
| K532A | 204.0 ± 3.2 (0.94) | 1.16 ± 0.07 (1.3) | 176 ± 13 (0.72) |
| KlUA | |||
| Wild type | 232 ± 5.3 (1.0) | 0.24 ± 0.04 (1.0) | 967 ± 183 (1.0) |
| H492A | 200 ± 6.4 (0.86) | 0.27 ± 0.06 (1.1) | 740 ± 188 (0.77) |
a Km values are for allophanate (KlAH) and urea (KlUA).
b Numbers in parentheses are ratios to the wild-type values.
c Standard deviations were obtained from fitting the experimental data to the Michaelis-Menten equation.
d ND, not detectable.
FIGURE 5.
Kinetic measurements. A, kinetic measurements of the wild type and mutant forms of KlAH. B, kinetic measurements of the wild type KlUA and its H492A mutant.
The N Domain Catalyzes the First Step of the AH Reaction
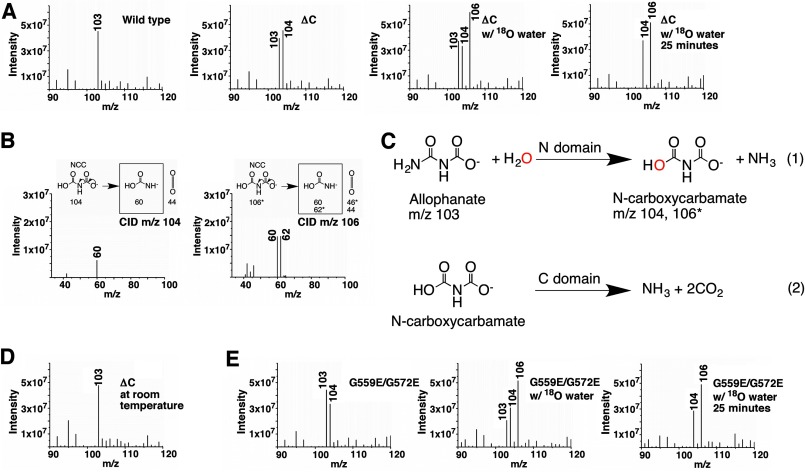
Simply hydrolyzing an amide bond will not accomplish the overall AH reaction (Fig. 1B). It is likely that the C domain also possesses an enzymatic activity, and the N and C domains catalyze different steps of the AH reaction. To understand the order of these reactions and what their substrates and products are, we used mass spectrometry to identify intermediates produced between them. A molecule with m/z 104 was detected in the reaction catalyzed by the KlAH ΔC mutant on ice but not in the reaction catalyzed by the wild type KlAH at the same temperature (Fig. 6A, left two panels). It is therefore most likely the product of the N domain reaction that is degraded by the C domain in the wild type KlAH. If this is the case, then the N domain catalyzes the first reaction, amide hydrolysis of allophanate. N-Carboxycarbamate is produced by such reaction, and its molecular mass is 104 Da, which is consistent with our observations. We further tested this hypothesis by performing the reaction in the presence of 18O-labeled water. Hydrolyzing the allophanate amide with 18O-labeled water will produce N-carboxycarbamate molecules with an oxygen atom substituted by 18O with a molecular mass of 106 Da. As expected, in the reaction catalyzed by the ΔC mutant in the presence of 18O-labeled water on ice, we observed a predominant peak at m/z 106 in the mass spectrum (Fig. 6A, right two panels). Furthermore, breaking the m/z 104 and 106 molecules by collision-induced dissociation (CID) produced fragments consistent with the molecular structure of N-carboxycarbamate (Fig. 6B). Collectively, this evidence indicates that the N domain catalyzes the first step of the AH reaction, hydrolysis of the allophanate amide, and the C domain is essential in converting the product N-carboxycarbamate to ammonium and carbon dioxide (Fig. 6C).
FIGURE 6.
The N domain catalyzes the first step of the AH reaction. A, mass spectrometric analysis of reactions catalyzed by the wild type KlAH and its ΔC mutant. In the presence of 18O-labeled water, the reaction catalyzed by the ΔC mutant was sampled at 10 and 25 min. Reactions analyzed by mass spectrometry presented throughout the text were performed on ice and allowed to proceed for 10 min before being analyzed unless otherwise indicated. The m/z 103 peak corresponds to allophanate. B, CID analysis of the m/z 104 and 106 molecules. In the CID experiments described here and in Fig. 8D, the fragmentation of N-carboxycarbamate (NCC) and the m/z values (or molecular weights for the uncharged molecules) are shown in the upper panels. The number of stars indicates the number of oxygen atoms substituted by 18O. Fragments observed by mass spectrometry are enclosed in black boxes. C, reactions catalyzed by the N and C domains. The oxygen atom colored in red in N-carboxycarbamate comes from water. m/z values for allophanate and N-carboxycarbamate are indicated. The number of stars indicates the number of oxygen atoms substituted by 18O. D, mass spectrometric analysis of the reaction catalyzed by the ΔC mutant at room temperature. E, mass spectrometric analysis of reactions catalyzed by the G559E/G572E mutant. In the presence of 18O-labeled water, the reaction was sampled at 10 and 25 min.
Additional evidence comes from a study of the N-carboxycarbamate stability. Abe et al. (39) found that sodium N-carboxycarbamate is unstable at room temperature. Upon dissolving in water, it is instantaneously hydrolyzed to ammonium and probably carbon dioxide but is stable at 0 °C in water. Consistently, we were only able to detect the m/z 104 molecules in reactions performed on ice but not in those performed at room temperature (Fig. 6D).
Identifying allophanate as the N domain substrate provided insights into substrate recognition at its active site. At physiological pH, the N domain active site and allophanate are positively and negatively charged, respectively, so electrostatic interactions might play a role in facilitating substrate binding. When we modeled the allophanate molecule into the N domain active site using the structure of the tartrate molecule found at the active site as a guide, we found that the allophanate carboxyl is positioned to form ion pair interactions with the Arg313 side chain guanidinium (Fig. 4A). Among the AS family members, Arg313 is conserved in AHs from different species, so it is probably responsible for the substrate specificity for their N domains.
Understanding what reactions the N and C domains catalyze enabled us to analyze them individually. The Km for allophanate indicates substrate affinity of the N domain, and the amount of N-carboxycarbamate accumulated in the reaction reflects the C domain activity. Such analysis revealed that both the N and the C domains require dimerization for their optimal activities. The Km values for allophanate for the monomeric mutants ΔC and G559E/G572E are 12- and 14-fold higher than that of the wild type KlAH, respectively (Table 2 and Fig. 5A). Despite reduced substrate binding at the N domain active site, in the reaction catalyzed by the G559E/G572E mutant, significant amounts of N-carboxycarbamate were detected (Fig. 6E). Active sites of both domains are located close to the dimer interface (Fig. 2A). The loss of the dimer interactions might distort their structure, resulting in reductions in their activities observed for the monomeric mutants.
The C Domain Probably Catalyzes a Novel Form of Decarboxylation Reaction
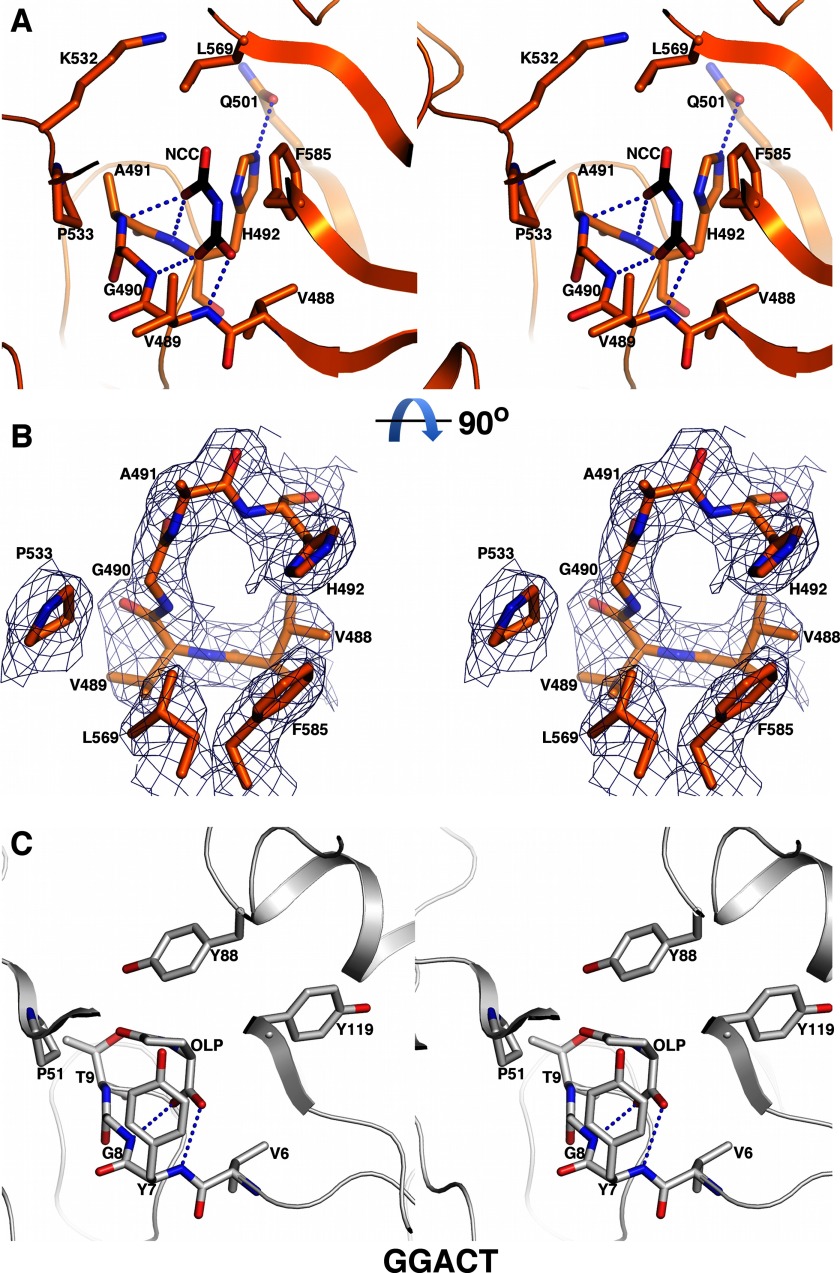
The C domain adopts a mixed α/β structure. It is composed of four α helices and eight β strands, and strands β12–β18 form a barrel (Fig. 2A and supplemental Fig. S1). A search with the Dali server indicated that this structure is homologous to the structures of γ-glutamyl cyclotransferase (40) and γ-glutamylamine cyclotransferase (GGACT) (41), and a number of proteins without known functions (Protein Data Bank codes 2QIK, 1V30, 2KL2, 2G0Q, 2I5T, 2Q53, 1XHS, and 2JQV). After alignment, the r.m.s. deviations between Cα atoms in KlAH and in γ-glutamyl cyclotransferase and GGACT are 2.8 and 2.3 Å, respectively. The sequence identities shared by KlAH and these enzymes are 10% for γ-glutamyl cyclotransferase and 15% for GGACT.
The active sites of γ-glutamyl cyclotransferase and GGACT correspond to the surface pocket on the C domain formed by the β12-α16 peptide, Val488-Val489-Gly490-Ala491, and the side chains of Pro533, Leu569, and Phe585 (Fig. 7A). It is well resolved in the electron density map (Fig. 7B). Despite a low overall sequence homology between KlAH and GGACT, these residues are quite conserved among them (Fig. 7, A and C). Side chains of His492 and Lys532 are located at the opening of the pocket, and a hydrogen bond is formed between the side chains of His492 and Gln501 (Fig. 7A). This pocket is probably the C domain active site as residues in this pocket are conserved among AHs from different species (Fig. 2C). The positive electrostatic potential around this pocket (Fig. 2D) could facilitate binding of N-carboxycarbamate, which is negatively charged at physiological pH. The catalytic glutamate in the active sites of γ-glutamyl cyclotransferase (Glu98) and GGACT (Glu82) is not conserved in the C domain (Val565), indicating that it catalyzes a very different reaction.
FIGURE 7.
Structure of the C domain active site. A, stereoview of the C domain active site. A model of N-carboxycarbamate (NCC) at the C domain active site is shown in black for its carbon atoms. The dashed lines indicate potential hydrogen bonding interactions. B, electron density map for the C domain active site. The map was contoured at 1 σ. Important active site residues are highlighted. A and B are roughly related by a 90° rotation along the horizontal axis. C, stereoview of the GGACT active site. The reaction product 5-oxo-l-proline (OLP) and hydrogen bonds between its carboxyl group and main chain amides are shown.
A complex structure of GGACT with its reaction product, 5-oxo-l-proline, has been reported. At the GGACT active site, hydrogen bonds are formed between the 5-oxo-l-proline carboxyl and the main chain amides of Tyr7 and Gly8 (Fig. 7C) (41). Similarities between the active sites of GGACT and the C domain prompted us to model N-carboxycarbamate into the C domain active site with one of its carboxyl groups forming similar hydrogen bonds with main chain amides of Val489 and Gly490. In such a model, the other carboxyl group of N-carboxycarbamate is located close to the opening of the pocket with one of its oxygen atoms forming hydrogen bonds with the main chain amides of Ala491 and His492, and the amine group is located close to the His492 side chain (Fig. 7A).
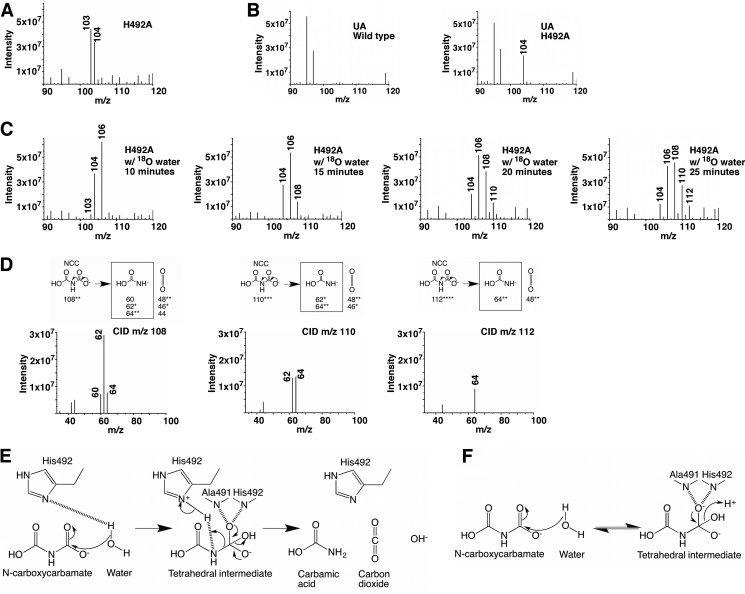
To address the physiological relevance of this putative C domain active site, we introduced several mutations into this region and analyzed whether they impair the C domain activity. Significant amounts of N-carboxycarbamate were detected in the reaction catalyzed by the H492A mutant on ice (Fig. 8A) but not in reactions catalyzed by the Q501A and K532A mutants, indicating that His492 plays an important role in the catalysis. In our kinetic experiments performed at room temperature, the H492A mutation also caused an appreciable reduction in the overall AH reaction rate (Table 2 and Fig. 5A). Similar phenomena were observed when we introduced the H492A mutation into the full-length KlUA (Figs. 5B and 8B and Table 2).
FIGURE 8.
Molecular insights into the C domain catalyzed reaction. A, mass spectrometric analysis of the reaction catalyzed by the KlAH H492A mutant. B, mass spectrometric analysis of reactions catalyzed by the wild type KlUA and its H492A mutant. In both KlAH and KlUA, the H492A mutation caused a significant accumulation of N-carboxycarbamate (m/z 104). C, mass spectrometric analysis of the reaction catalyzed by the KlAH H492A mutant in the presence of 18O-labeled water. The reaction was sampled at 10, 15, 20, and 25 min. D, CID analysis of the m/z 108, 110, and 112 molecules. Refer to the legend of Fig. 6B for a detailed description. E, proposed reaction scheme for the C domain. F, possible reaction catalyzed by the C domain of the KlAH H492A mutant.
Interestingly, in the presence of 18O-labeled water, reactions catalyzed by the H492A mutant produced additional molecules with m/z values of 108, 110, and 112. These molecules appeared sequentially: those with low m/z values appeared earlier followed by those with high m/z values. Over time, the amounts of the molecules with high m/z values increased, and the amounts of those with low m/z values decreased (Fig. 8C). The m/z values of these molecules and their fragments generated by CID (Fig. 8D) indicate that they are N-carboxycarbamate molecules with two, three, and four oxygen atoms substituted by 18O. The change in their amounts over time suggests that those containing more 18O are converted from those containing less. This is probably due to an oxygen exchange process between N-carboxycarbamate and environmental 18O-labeled water molecules. The appearance of these molecules was only observed in reactions catalyzed by the H492A mutant. In reactions catalyzed by the ΔC (Fig. 6A) and G559E/G572E (Fig. 6E) mutants (both produced significant amounts of N-carboxycarbamate), these molecules were not detected despite prolonged incubation with 18O-labeled water.
Based on our model of N-carboxycarbamate binding at the C domain active site and the results of the mutagenesis studies, we propose that the C domain catalyzes a decarboxylation reaction. A water molecule performs an initial nucleophilic attack on the N-carboxycarbamate carboxyl carbon, generating a tetrahedral intermediate stabilized by the oxyanion hole formed by main chain amides of Ala491 and His492. The His492 side chain subsequently protonates the amine group of the intermediate, prompting breakage of its C–N bond, producing carbon dioxide and carbamic acid (Fig. 8E). Carbamic acid is very unstable and spontaneously decomposes to ammonium and carbon dioxide. Without the His492 side chain, the C–N bond breakage cannot take place, and in reactions catalyzed by the H492A mutant, the conversion of N-carboxycarbamate to the tetrahedral intermediate becomes reversible, and in the process oxygen atoms between N-carboxycarbamate and the attacking water molecule are exchanged (Fig. 8F). This proposed C domain reaction is unlike any biological decarboxylation reactions reported to date (42). It bears similarities to reactions catalyzed by serine proteases. Serine proteases use a Ser-His-Asp triad to cleave peptide bonds. The serine residue performs the initial nucleophilic attack, generating a tetrahedral intermediate, and the histidine residue protonates the amine group of the intermediate, prompting its C–N bond breakage (43). The proposed functions of the catalytic water and His492 in KlAH are similar to the functions of the catalytic serine and histidine in serine proteases. The aspartate residue in the Ser-His-Asp triad in serine proteases forms a hydrogen bond with the catalytic histidine, which stabilizes its conformation and facilitates the proton transfer (43). In the C domain, a similar hydrogen bond is formed between the side chains of Gln501 and His492 (Fig. 7A). However, in reactions performed on ice, substituting Gln501 with alanine did not cause a detectable accumulation of N-carboxycarbamate; likewise at room temperature, the Q501A mutation caused only a small decrease in the AH reaction rate (Table 2 and Fig. 5A). Further studies are necessary to address the function of the Gln501-His492 hydrogen bond.
At room temperature, the spontaneous decomposition of N-carboxycarbamate releases ammonium and carbon dioxide, same as the C domain catalysis, making the latter somewhat redundant. Nevertheless, our data indicated that at room temperature the C domain enzymatic activity makes the AH and UA catalysis faster, indicating that it contributes to the net conversion rate of N-carboxycarbamate to ammonium and carbon dioxide. At low temperature, the rate of N-carboxycarbamate spontaneous decomposition drops toward zero, and the C domain activity becomes important in converting it to ammonium and carbon dioxide. The C domain activity therefore is essential for the AH and UA catalysis at low temperature as indicated by our mass spectrometric studies of these reactions. UA-containing organisms are able to survive or even grow at low temperatures. Yarrowia lipolytica (Candida lipolytica), for instance, contains two copies of UA (11) and is able to grow at a temperature as low as 5 °C (44). It remains to be seen whether in those organisms UA is expressed at low temperature and still plays roles in urea utilization, pyrimidine nucleic acid precursor degradation, etc. If so, the enzymatic activity of the AH C domain is essential for their fitness at low temperature.
As our manuscript was being prepared, the crystal structure of the Granulibacter bethesdensis AH was reported (45). This work provided molecular insights into the G. bethesdensis AH N domain catalysis that are similar to what we have reported here for the KlAH N domain. However, although G. bethesdensis AH appeared to be intact in the crystal, in the structure, its entire C domain is missing. An enzymatic activity for the G. bethesdensis AH C domain was not discovered by the authors. The C domains of AHs from different species, especially their active sites, are quite conserved. It is very likely that the AH C domains from other species possess the same enzymatic activity as the KlAH C domain. However, as this activity is apparent at low temperature, it might have escaped these researchers' attention.
In summary, our studies have provided molecular insights into AH catalysis and a framework to further understand important biological processes, including urea utilization, pyrimidine nucleic acid precursor degradation, and s-triazine herbicide degradation in many organisms, and to develop novel antifungal drugs. In addition, our data suggested that the C domain catalyzes a novel form of decarboxylation reaction. Decarboxylation reactions are among the most common reactions in biological systems (42). Further studies are required to verify our finding, which might expand the knowledge of this important biochemical process.
Acknowledgments
We thank Prof. Jianhua He and staff members of the Shanghai Synchrotron Radiation Facility beamline BL17U for setting up the beamline and assistance during diffraction data collection; Prof. Jianping Ding, Meilan Zhang, and Li Han at the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences for assistance with dynamic light scattering experiments; and Prof. Ying Yu at the Institute for Nutritional Sciences, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences for kindly sharing reagents.
This work was supported by grants from the Ministry of Science and Technology of China (973 Program Grants 2011CB910500 and 2010CB912502) and the One Hundred Talents Program of the Chinese Academy of Sciences.

This article contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 4ISS and 4IST) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- UA
- urea amidolyase
- AH
- allophanate hydrolase
- Kl
- K. lactis
- AS
- amidase signature
- r.m.s.
- root mean square
- CID
- collision-induced dissociation
- GGACT
- γ-glutamylamine cyclotransferase.
REFERENCES
- 1. Sterner R. W., Elser J. J. (2002) Ecological Stoichiometry: the Biology of Elements from Molecules to the Biosphere, pp. 44–79, Princeton University Press, Princeton, NJ [Google Scholar]
- 2. Canfield D. E., Glazer A. N., Falkowski P. G. (2010) The evolution and future of Earth's nitrogen cycle. Science 330, 192–196 [DOI] [PubMed] [Google Scholar]
- 3. Mobley H. L., Hausinger R. P. (1989) Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53, 85–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mobley H. L., Island M. D., Hausinger R. P. (1995) Molecular biology of microbial ureases. Microbiol. Rev. 59, 451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sirko A., Brodzik R. (2000) Plant ureases: roles and regulation. Acta Biochim. Pol. 47, 1189–1195 [PubMed] [Google Scholar]
- 6. Roon R. J., Levenberg B. (1968) An adenosine triphosphate-dependent, avidin-sensitive enzymatic cleavage of urea in yeast and green algae. J. Biol. Chem. 243, 5213–5215 [PubMed] [Google Scholar]
- 7. Whitney P. A., Cooper T. G. (1972) Urea carboxylase and allophanate hydrolase. Two components of adenosine triphosphate:urea amido-lyase in Saccharomyces cerevisiae. J. Biol. Chem. 247, 1349–1353 [PubMed] [Google Scholar]
- 8. Kanamori T., Kanou N., Atomi H., Imanaka T. (2004) Enzymatic characterization of a prokaryotic urea carboxylase. J. Bacteriol. 186, 2532–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanamori T., Kanou N., Kusakabe S., Atomi H., Imanaka T. (2005) Allophanate hydrolase of Oleomonas sagaranensis involved in an ATP-dependent degradation pathway specific to urea. FEMS Microbiol. Lett. 245, 61–65 [DOI] [PubMed] [Google Scholar]
- 10. Navarathna D. H., Harris S. D., Roberts D. D., Nickerson K. W. (2010) Evolutionary aspects of urea utilization by fungi. FEMS Yeast Res. 10, 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strope P. K., Nickerson K. W., Harris S. D., Moriyama E. N. (2011) Molecular evolution of urea amidolyase and urea carboxylase in fungi. BMC Evol. Biol. 11, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leftley J. W., Syrett P. J. (1973) Urease and ATP:urea amidolyase activity in unicellular algae. J. Gen. Microbiol. 77, 109–115 [Google Scholar]
- 13. Andersen G., Björnberg O., Polakova S., Pynyaha Y., Rasmussen A., Møller K., Hofer A., Moritz T., Sandrini M. P., Merico A. M., Compagno C., Akerlund H. E., Gojković Z., Piskur J. (2008) A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. J. Mol. Biol. 380, 656–666 [DOI] [PubMed] [Google Scholar]
- 14. Ghosh S., Navarathna D. H., Roberts D. D., Cooper J. T., Atkin A. L., Petro T. M., Nickerson K. W. (2009) Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect. Immun. 77, 1596–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vylkova S., Carman A. J., Danhof H. A., Collette J. R., Zhou H., Lorenz M. C. (2011) The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio 2, e00055-00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shapir N., Mongodin E. F., Sadowsky M. J., Daugherty S. C., Nelson K. E., Wackett L. P. (2007) Evolution of catabolic pathways: genomic insights into microbial s-triazine metabolism. J. Bacteriol. 189, 674–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng G., Shapir N., Sadowsky M. J., Wackett L. P. (2005) Allophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism. Appl. Environ. Microbiol. 71, 4437–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shapir N., Sadowsky M. J., Wackett L. P. (2005) Purification and characterization of allophanate hydrolase (AtzF) from Pseudomonas sp. strain ADP. J. Bacteriol. 187, 3731–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shapir N., Cheng G., Sadowsky M. J., Wackett L. P. (2006) Purification and characterization of TrzF: biuret hydrolysis by allophanate hydrolase supports growth. Appl. Environ. Microbiol. 72, 2491–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacques D. A., Langley D. B., Kuramitsu S., Yokoyama S., Trewhella J., Guss J. M. (2011) The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase. Acta Crystallogr. D Biol. Crystallogr. 67, 105–111 [DOI] [PubMed] [Google Scholar]
- 21. Jacques D. A., Langley D. B., Hynson R. M., Whitten A. E., Kwan A., Guss J. M., Trewhella J. (2011) A novel structure of an antikinase and its inhibitor. J. Mol. Biol. 405, 214–226 [DOI] [PubMed] [Google Scholar]
- 22. Fan C., Chou C. Y., Tong L., Xiang S. (2012) Crystal structure of urea carboxylase provides insights into the carboxyltransfer reaction. J. Biol. Chem. 287, 9389–9398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doublié S., Kapp U., Aberg A., Brown K., Strub K., Cusack S. (1996) Crystallization and preliminary x-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 384, 219–221 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 26. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chebrou H., Bigey F., Arnaud A., Galzy P. (1996) Study of the amidase signature group. Biochim. Biophys. Acta 1298, 285–293 [DOI] [PubMed] [Google Scholar]
- 28. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bracey M. H., Hanson M. A., Masuda K. R., Stevens R. C., Cravatt B. F. (2002) Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science 298, 1793–1796 [DOI] [PubMed] [Google Scholar]
- 30. Shin S., Lee T. H., Ha N. C., Koo H. M., Kim S. Y., Lee H. S., Kim Y. S., Oh B. H. (2002) Structure of malonamidase E2 reveals a novel Ser-cisSer-Lys catalytic triad in a new serine hydrolase fold that is prevalent in nature. EMBO J. 21, 2509–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakamura A., Yao M., Chimnaronk S., Sakai N., Tanaka I. (2006) Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science 312, 1954–1958 [DOI] [PubMed] [Google Scholar]
- 32. Wu J., Bu W., Sheppard K., Kitabatake M., Kwon S. T., Söll D., Smith J. L. (2009) Insights into tRNA-dependent amidotransferase evolution and catalysis from the structure of the Aquifex aeolicus enzyme. J. Mol. Biol. 391, 703–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ito T., Yokoyama S. (2010) Two enzymes bound to one transfer RNA assume alternative conformations for consecutive reactions. Nature 467, 612–616 [DOI] [PubMed] [Google Scholar]
- 34. Blaise M., Bailly M., Frechin M., Behrens M. A., Fischer F., Oliveira C. L., Becker H. D., Pedersen J. S., Thirup S., Kern D. (2010) Crystal structure of a transfer-ribonucleoprotein particle that promotes asparagine formation. EMBO J. 29, 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Labahn J., Neumann S., Büldt G., Kula M. R., Granzin J. (2002) An alternative mechanism for amidase signature enzymes. J. Mol. Biol. 322, 1053–1064 [DOI] [PubMed] [Google Scholar]
- 36. Yasuhira K., Shibata N., Mongami G., Uedo Y., Atsumi Y., Kawashima Y., Hibino A., Tanaka Y., Lee Y. H., Kato D., Takeo M., Higuchi Y., Negoro S. (2010) X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation. J. Biol. Chem. 285, 1239–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ohtaki A., Murata K., Sato Y., Noguchi K., Miyatake H., Dohmae N., Yamada K., Yohda M., Odaka M. (2010) Structure and characterization of amidase from Rhodococcus sp. N-771: Insight into the molecular mechanism of substrate recognition. Biochim. Biophys. Acta 1804, 184–192 [DOI] [PubMed] [Google Scholar]
- 38. Shin S., Yun Y. S., Koo H. M., Kim Y. S., Choi K. Y., Oh B. H. (2003) Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. J. Biol. Chem. 278, 24937–24943 [DOI] [PubMed] [Google Scholar]
- 39. Abe S., Takahashi S. (1959) Thermal decomposition of sodium carbamate. Nippon Kagaku Zasshi 80, 92–94 [Google Scholar]
- 40. Oakley A. J., Yamada T., Liu D., Coggan M., Clark A. G., Board P. G. (2008) The identification and structural characterization of C7orf24 as γ-glutamyl cyclotransferase. An essential enzyme in the γ-glutamyl cycle. J. Biol. Chem. 283, 22031–22042 [DOI] [PubMed] [Google Scholar]
- 41. Oakley A. J., Coggan M., Board P. G. (2010) Identification and characterization of γ-glutamylamine cyclotransferase, an enzyme responsible for γ-glutamyl-ϵ-lysine catabolism. J. Biol. Chem. 285, 9642–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li T., Huo L., Pulley C., Liu A. (2012) Decarboxylation mechanisms in biological system. Bioorg. Chem. 43, 2–14 [DOI] [PubMed] [Google Scholar]
- 43. Polgár L. (2005) The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 62, 2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arthur H., Watson K. (1976) Thermal adaptation in yeast: growth temperatures, membrane lipid, and cytochrome composition of psychrophilic, mesophilic, and thermophilic yeasts. J. Bacteriol. 128, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin Y., St Maurice M. (2013) The structure of allophanate hydrolase from Granulibacter bethesdensis provides insights into substrate specificity in the amidase signature family. Biochemistry 52, 690–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N. (2003) ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164 [DOI] [PubMed] [Google Scholar]