Abstract
Retinoid X receptor α–null (RXRα-null) mutants exhibit hypoplasia of their ventricular myocardium and die at the fetal stage. In the present study, we wished to determine whether transgenic re-expression of RXRα in mutant cardiac myocytes could rescue these defects. Two transgenic mouse lines specifically overexpressing an RXRα protein in cardiomyocytes were generated, using the cardiac α-myosin heavy chain (α-MHC) promoter. Breeding the high copy number transgenic line onto an RXRα-null genetic background did not prevent the myocardial hypoplasia and fetal lethality associated with the RXRα–/– genotype, even though the transgene was expressed in the ventricles as early as 10.5 days post-coitum. These data suggest that the RXRα function involved in myocardial growth may correspond to a non–cell-autonomous requirement forsignal orchestrating the growth and differentiation of myocytes. Interestingly, the adult transgenic mice developed a dilated cardiomyopathy, associated with myofibrillar abnormalities and specific deficiencies in respiratory chain complexes I and II, thus providing an additional model for this genetically complex disease.
Introduction
Retinoic acids are the active metabolites that exert most of the physiological functions of vitamin A. The cellular effectors that transduce the effects of these ligands belong to 2 families of nuclear receptors, the retinoic acid receptors (RARα, β, and γ) and the retinoid X receptors (RXRα, β, and γ) (1, 2). These receptors are retinoid-dependent transcriptional regulators that bind to their target sequence as RAR/RXR heterodimers. Extensive mutagenesis studies of RAR and RXR genes in the mouse have confirmed their role as transducers of vitamin A actions in vivo, in particular during embryonic and fetal development, and have assigned a large number of developmental functions to specific RXR/RAR heterodimers (3–5).
Mice bearing null mutations in the RXRα gene die during fetal development (6, 7). This death is at least in part due to myocardial defects: as the ventricular wall remains abnormally thin, the heart cannot accommodate the increased workload demanded by a growing fetus (8). In wild-type (WT) embryonic myocardia, the cardiac myocytes located beneath the epicardium (subepicardial myocytes) are cuboidal, tightly associated, do not show morphological features of differentiation (i.e., organized sarcomeres and sarcoplasmic reticulum), and proliferate at a higher rate than the cells located at a more internal location within the myocardium (6, 9, 10). All of these features are essentially lacking in RXRα mutant subepicardial myocytes, which exhibit an elongated morphology, are loosely associated, contain well-organized myofilaments, and divide at a lower frequency than WT cells (6, 9).
In the present study, we addressed the question of whether cardiomyocytes are primarily affected by the RXRα mutation. We generated transgenic mice specifically expressing an RXRα transgene in cardiomyocytes, at least from 10.5 days post-coitum (dpc) onward. When introduced by breeding into an RXRα-null background, this transgene failed to rescue the hypoplasia of the mutant ventricular myocardium. Interestingly, the cardiomyocytes of the transgenic mice were abnormal postnatally, as a significant proportion of these mice developed a dilated cardiomyopathy (DCM) and died from heart failure.
Methods
Generation of transgenic mice.
To direct heart-specific expression, a plasmid containing 5.5 kb from the myosin heavy chain α (MyHCα) gene (including the promoter and the first 3 untranslated exons) was used (11). A SalI fragment (2.2-kb mRXRα cDNA fragment linked to a 1.3-kb fragment from the human progesterone receptor 3′ untranslated region [incorporated in the construct to be used as a tag for in situ hybridization studies] and the SV40 polyadenylation sequence) was subcloned into the SalI site of the MyHCα promoter plasmid. The resulting construct was purified as a 9-kb NotI-XhoI fragment and used for pronuclear microinjection.
Mice were routinely genotyped by Southern blotting of EcoRI-restricted tail DNA with probe A (Figure 1b). Copy number was estimated by comparing the intensities (determined by PhosphorImager; Fuji) of the transgene fragment (1.5 kb) with that of the 2.8-kb fragment corresponding to the endogenous MyHCα locus.
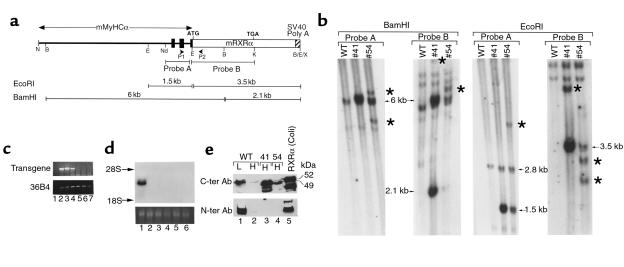
Figure 1.
Molecular characterization of MyHCα-RXRα transgenic mice. (a) Schematic representation of the transgene construct (not at scale). Black boxes indicate exons 1–3 of the MyHCα gene, which correspond to 5′-untranslated sequences. P1 and P2 are the RT-PCR primer used in the experiment shown in Figure 2a. Probe A corresponds to a EcoRI-NdeI fragment from the MyHCα gene, and probe B corresponds to a EcoRI-KpnI fragment containing all RXRα coding sequences. Fragments detected by these probes after EcoRI or BamHI digestion are shown. B, BamHI; E, EcoRI; K, KpnI; N, NotI; Nd, NdeI; X, XhoI. (b) Southern blot analysis. A total of 10 μg of BamHI- or EcoRI-restricted genomic DNA from WT mice or mice from lines 41 or 54 were analyzed by Southern blotting with probes A and probe B, as indicated. Arrows point to the bands of the expected size, whereas stars point to additional bands having undergone some deletions or rearrangements. (c) RT-PCR analysis of tissue samples from line 41. Lanes 1–7 correspond to: (1) adult ventricle with the reverse transcriptase omitted in the RT step, (2) adult ventricle, (3) adult atrium, (4) lungs, (5) skeletal muscle, (6) liver, and (7) brain. The top panel corresponds to transgene transcript amplification with the transgene-specific primers depicted in a; the bottom panel corresponds to the control amplification of 36B4 gene transcripts (39). (d) Northern blot analysis of transgene expression in line 41. A total of 20 μg of total RNA from adult heart (lane 1), lungs (lane 2), spleen (lane 3), skeletal muscle (lane 4), kidney (lane 5), and liver (lane 6) were analyzed by Northern blotting with a cDNA probe corresponding to a 2-kb fragment containing the RXRα coding sequences. The bottom panel corresponds to the ethidium bromide staining of ribosomal 28S RNA of the same RNA sample separated on an identical gel. (e) Western blot analysis. A total of 50 μg of nuclear extracts prepared from WT liver (L) or heart (H) and from heart from transgenic mice from lines 41 or 54, as well as a control extract of bacterially produced RXRα (lane 5), was separated on a 8% SDS-polyacrylamide gel and revealed with polyclonal antibodies directed against either COOH-terminal RXRα epitopes (top panel) or an NH2-terminal RXRα epitope (bottom panel). Note that transgenic hearts express an RXRα polypeptide that is smaller than WT RXRα and not detected by the NH2-terminal antibody. Smaller fragments seen in lanes 3 and 5 likely correspond to proteolytic degradation products.
RT–PCR, in situ hybridization, and Western blot analysis.
RNA was prepared from adult tissues by the single-step guanidinium-isothiocyanate-phenol technique (12). For RT-PCR, RT was performed using avian myeloblastosis virus reverse transcriptase at 50°C for 15 minutes, followed by a PCR reaction (35 cycles each for 1 minute at 94°C, 30 seconds at 55°C, and 1 minute at 72°C; PCR primers: P1 5′-CTGCGCCACTGTGGTGCCTC, P2 5′-AAGCCCAATGTGGGTGTGG; Figure 1a). For Northern blot analysis, 20 μg of total RNA was used per lane. In situ hybridization analysis of transgene expression was performed with an RXRα antisense riboprobe (13).
For detection of the RXRα protein, nuclear extracts were prepared from tissues according to Andrews and Faller (14). A total of 50 μg of extracts was separated by SDS-PAGE. After electrotransfer to a nitrocellulose membrane, RXRα polypeptides were detected with either the polyclonal antibody RPRXα(A), detecting an NH2-terminal epitope, or with the mAb 4RX-3A2, detecting an epitope in the RXRα COOH terminal D-E region (15).
Electron microscopy and histology.
Histological and electron microscopic analyses of embryos and tissues were performed according to standard protocols (9, 16).
Analysis of mitochondrial enzymatic activities.
Determination of the activities of the respiratory chain complexes I–IV and of citrate synthase were performed as described (17).
Results
Transgenic mice overexpressing RXRα in the heart.
With the aim of re-expressing RXRα in cardiac myocytes of RXRα-null mutants, we generated transgenic mice harboring a construct containing the RXRα cDNA placed under the control of regulatory sequences from the MyHCα that were previously shown to specifically direct gene expression to cardiomyocytes (11; Figure 1a). Two independent lines of mice expressing this transgene were obtained (MyHCα-RXRα 41 and 54). Southern analysis (Figure 1b) with MyHCα- and RXRα-derived probes revealed the presence of DNA fragments of expected sizes (arrows). Several additional fragments of unexpected sizes were also detected (stars in Figure 1b), indicating that a subset of the transgene copies had undergone rearrangements. Copy number was estimated to be 4 and 16 in lines 54 and 41, respectively. Most of the experiments described here were performed with mice from line 41. All transgenic mice used in this study were heterozygous for the transgene.
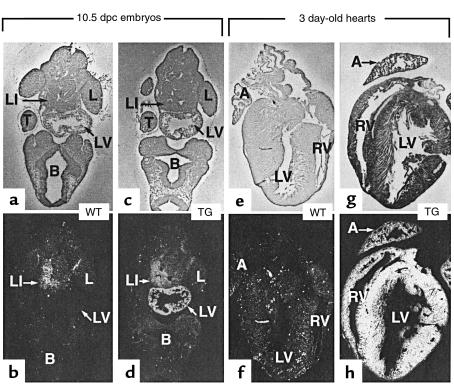
Northern blot analysis of total RNA of several adult tissues from line 41 revealed a high level of transgene expression in the heart, but not in several other adult tissues (Figure 1d; under these experimental conditions, the endogenous 5-kb RXRα transcripts were not detectable). A more sensitive RT-PCR assay also failed to detect transgene expression in noncardiac tissues, with the exception of lungs (Figure 1c). This pulmonary expression most likely occurs in “pulmonary myocardium,” a discrete population of myocytes lining pulmonary veins in which the MyHCα promoter used here was shown to be active (11). In situ hybridization analysis of histological sections from 3-day-old hearts showed transgene expression throughout the ventricular and atrial myocardium (Figure 2h; compare with Figure 2f). Transgene expression was initiated early during embryogenesis, as it was detected by RT-PCR on RNA extracted from 9.5 dpc transgenic embryos (not shown). In situ hybridization analysis of histological sections from 10.5 dpc embryos showed that this expression was confined to the heart and was strong in both ventricles and atria (Figure 2d, and data not shown). Thus, the onset of the transgene expression occurs early in the ventricular myocardium, the tissue that is affected in RXRα-null mutants.
Figure 2.
Detection of transgene expression in heterozygote transgenic mice (TG) from line 41 by in situ hybridization with an RXRα antisense riboprobe. The top panels correspond to bright-field micrographs, and the lower panels are the corresponding dark-field micrographs. (a–d) Frontal sections of 10.5 dpc WT (a and b) and transgenic (c and d) embryos. (e–h) Sections of hearts from 3-day-old WT (e and f) and transgenic (g and h) mice. A, atrium; B, brain; L, limb bud; LI, liver; LV, left ventricle; RV, right ventricle; T, tail. ×20 (a–h).
Western blot analysis with an antibody recognizing an epitope in the RXRα COOH-terminal region revealed an abundant RXRα polypeptide in adult heart extracts from transgenic animals from both lines (Figure 1e, upper panel, lanes 3 and 4). The expression level was higher in line 41 than in line 54. The level of the transgenic protein greatly exceeded that of endogenous heart RXRα (lane 2) but was comparable to that of endogenous RXRα in the liver (the adult tissue expressing the highest level of RXRα transcript; compare lanes 3 and 4 to lane 1). However, the transgenic polypeptide had a slightly increased electrophoretic mobility when compared with WT RXRα (apparent molecular weight 49 kDa instead of 52 kDa for the latter, irrespective of whether it was endogenously expressed in liver or heart, or produced in Escherichia coli [lane 5]). Furthermore, an antibody directed against the RXRα 27 NH2-terminal amino acids failed to detect the transgenic protein (Figure 1e, lower panel), whereas it revealed endogenous liver and heart RXRα. Therefore, the transgenically expressed RXRα protein appears to be NH2-terminally truncated, most probably because of abnormal initiation of translation from an internal AUG codon. As no mutation was detected in the DNA of the construct used to produce the transgenic mice, the reason for this failure to correctly initiate translation remains unclear but could reflect complex interactions between 5′-untranslated MyHCα gene sequences and 5′ RXRα sequences.
Despite the production of an NH2-terminally truncated protein, the results described here are nevertheless most probably pertinent for the role of RXRα in heart development for 2 reasons: (i) an NH2-terminally truncated RXRα retains most of its functional properties (DNA binding, ligand binding, and dimerization) and appears to be only minimally affected in its transactivation properties, as judged from transient transfection assays (18); and, importantly, (ii) deletion, in the mouse, of the sequences encoding the entire RXRα NH2-terminal region preceding the DNA binding domain does not lead to abnormal cardiac development (B. Mascrez, M. Mark, P. Kastner, and P. Chambon, unpublished study), indicating that this region is dispensable for the function of RXRα that specifies normal heart development.
The transgenic RXRα does not prevent the ventricular hypoplasia of RXRα-null mutants.
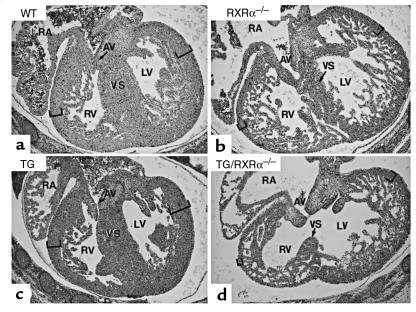
To determine whether the expression of the RXRα transgene could rescue the RXRα-null mutant myocardial defects, the transgene from the high copy number mouse line 41 was bred into the RXRα-null background, and the resulting fetuses were analyzed by histology at 14.5 dpc. A similar ventricular myocardial hypoplasia was observed in RXRα–/– and TG/RXRα–/– fetuses (Figure 3; see also ref. 6). The presence of the transgene did also not extend the life span of RXRα–/– mutants, as no viable TG/RXRα–/– fetus could be recovered after 16.5 dpc (not shown). Thus, efficient expression of RXRα in ventricular myocytes as early as 10.5 dpc cannot prevent the cardiac defects and fetal lethality of RXRα–/– mutants.
Figure 3.
Failure of the RXRα-expressing transgene to rescue the ventricular myocardial hypoplasia phenotype of RXRα–/– mutants. Frontal sections of 14.5 dpc fetuses at the level of the right atrioventricular canal (AV). Genotypes are as indicated; TG indicates heterozygosity for the MyHCα-RXRα transgene (line 41). The thickness of the compact layer in the left and the right ventricles is indicated by brackets. AV, atrio-ventricular canal; RA, right atrium; VS, ventricular septum; LV, left ventricle; RV, right ventricle. ×50 (a–d).
DCM in adult transgenic mice expressing RXRα in the heart.
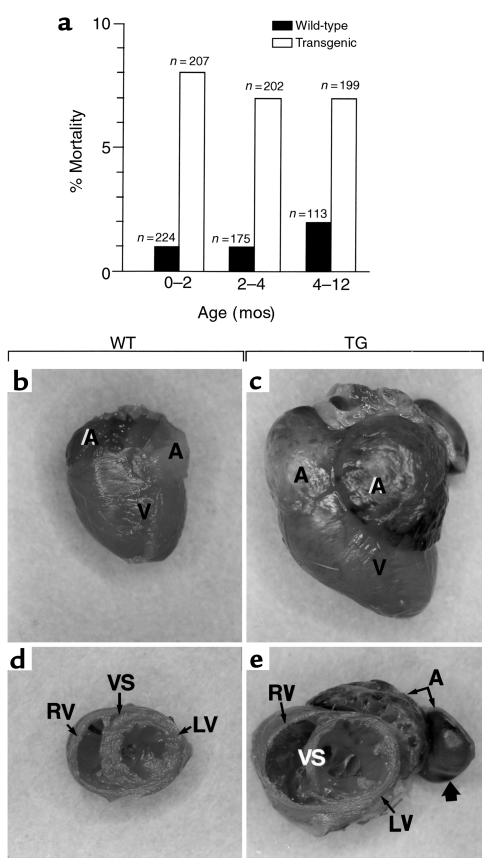
Adult transgenic mice from line 41 had a mortality rate that was significantly higher than that of their nontransgenic littermates (Figure 4a). Over a period of 1 year, about 20% of the transgenic animals, but only 3% of their WT littermates, died. In many cases, the death occurred suddenly, without prior symptoms of morbidity, whereas in other cases, the death was preceded for several days by general weakness, dyspnea, and lack of food and water intake. No increased mortality was observed with the lower-expressing transgenic line 54 (data not shown).
Figure 4.
DCM in mice from line 41. (a) Increased lethality of heterozygous mice from line 41. Mice were subdivided within 3 age groups, and the percentages of deaths occurring within the corresponding life periods are indicated. WT mice were the littermates of the transgenic ones. (b–e) Cardiac dilation in heterozygous transgenic mice from line 41. External views of the hearts from a WT mouse and of a dilated heart from a transgenic littermate (TG) (2 months old) are shown in b and c, and transverse sections through the same hearts, in d and e. Note that the transgenic mouse from which the heart was taken displayed obvious signs of weakness. All the chambers of the transgenic heart contained organized thrombi that were removed during dissection, except one in the atrium (large arrow). A, atria; V, ventricles; VS, ventricular septum; LV, left ventricle; RV, right ventricle.
At autopsy, the hearts of the sick animals were markedly enlarged, and all 4 heart chambers were dilated (Figure 4, b and c). The thickness of the ventricular wall and ventricular septum appeared either normal or decreased (compare LV and VS in Figure 4, d and e). The cardiac valves were normal, and intracavitary thrombi were always present in the atria and/or ventricles (large arrow in Figure 4e). These sick animals also showed increases in heart and lung/body weight ratios accompanied by a pleural effusion (data not shown). Such a cardiac dilation was observed in all sick transgenic animals, as well as in about 10% of healthy-looking transgenic littermates, but not in any of the WT littermates sacrificed in the course of this study (n = 35). The association of a dilation of the heart with lung weight increase and pleural effusion suggests that the death of the transgenic animals was secondary to heart failure. No gross heart abnormalities were observed in mice from the lower-expressing line 54.
Analysis of the myocardium from sick transgenic mice revealed the presence of many necrotic cardiomyocytes showing a loss of myofibrils (Figure 5, c, e, and f, asterisks) and large, pale, nuclear profiles indicative of nuclear destruction (N in Figure 5c, asterisks). Hypertrophy of cardiac myocytes was not observed. These observations, together with the presence of specific cellular defects in cardiomyocytes (see later here), indicates that mice from line 41 develop a form of DCM.
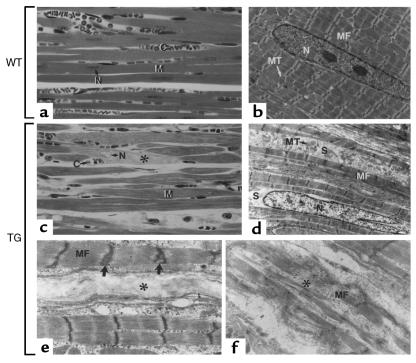
Figure 5.
Abnormal myocytes in dilated hearts. (a and c) Semithin sections through the ventricles of a WT and a dilated transgenic heart (2-month-old mice). Note the degenerating cardiomyocytes (asterisks) in the transgenic heart. (b, d–f) Electron microscopy of longitudinally sectioned cardiac muscle cells from ventricles of a WT (b) and dilated (d–f) transgenic heart. In d, note the swelling of the transgenic cardiomyocytes, manifested by the absence of compaction of the myofibrils (MF) and the loss of myofibrils resulting in a relative increase of intervening sarcoplasm (S) and a more dispersed arrangement of mitochondria (MT). e and f illustrate 2 different aspects of cardiomyocytes undergoing necrolysis (asterisks), as well as the abnormal, duplicated aspect of some Z lines (large arrows). C, capillaries; M, myofibers; N, nuclei; MT, mitochondria; MF, myofibrils; S, sarcoplasm. ×340 (a and c); ×4,200 (b and d); ×10,000 (e and f).
Abnormal myofibril organization in transgenic cardiomyocytes.
Electron microscopic analysis was also carried out on cardiac muscle of apparently healthy mice from both lines (Figure 6). WT myofibrils exhibited a striated arrangement with clearly distinguishable Z lines and M lines, as well as I bands and A bands (Figure 6, a, c and e). In contrast, the myofibrils from transgenic cardiomyocytes were thinner than their WT counterparts; the sarcomeric length was often reduced, I bands were undistinguishable (Figure 6, b, d and f), and M lines were poorly defined. In many instances, Z lines were duplicated (Figure 6d, arrows; see also Figure 5e). No cardiomyocyte hypertrophy was observed. Together, these observations show that the structure of the myofibrils is abnormal in transgenic cardiomyocytes. Interestingly, this phenotype was completely penetrant in both transgenic lines and was not correlated with either symptoms of cardiac failure, cardiac dilation, or age (note that the abnormal myofibrils illustrated in Figure 6f are from the heart of a healthy-looking 15-day-old mouse from the lower-expressing line 54). In addition to these sarcomeric abnormalities, the shape of the nuclei in transgenic myocytes of both lines was often altered, as it showed numerous deep indentations (Figure 6f; see also Figure 5d). Together, these observations show that, even though it does not systematically lead to DCM, the transgenic RXRα expression consistently perturbs the structure of cardiomyocytes.
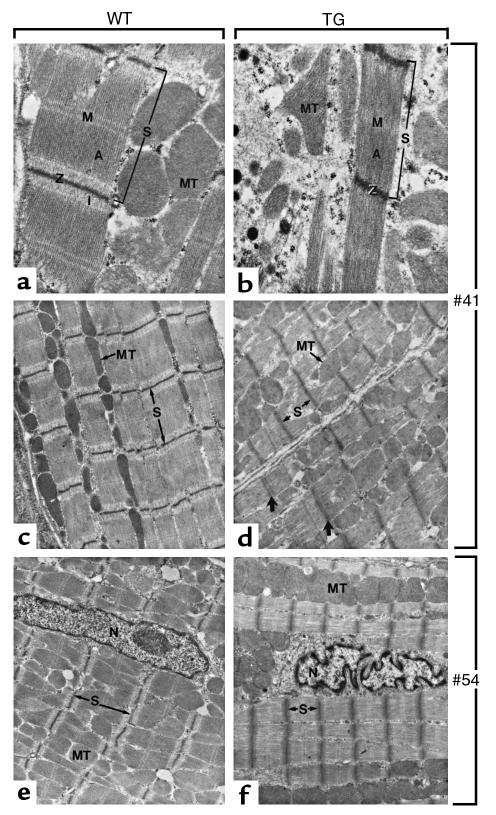
Figure 6.
Early cytological abnormalities in the myocardium of transgenic mice. Electron microscopic analysis of the myocardium of wild-type (WT) and transgenic mice (TG) at 15 days postpartum. (a, c, and e) In WT cardiac muscle cells, the sarcomeres (S) show clearly defined A and I bands and Z and M lines (A, I, Z, and M, respectively); mitochondria (MT) are disposed in rows along myofibrils. (b) and (d) correspond to transgenic line 41, whereas (f) corresponds to line 54. In transgenic samples, the length of the sarcomeres (S) is in general reduced, the M lines and I bands are almost indistinguishable, and the Z lines are occasionally duplicated (large arrows). The shape of nuclei (N) in transgenic myocytes is often irregular (f). Large areas of the myocardium (d) display a complete disorganization of myofibril and mitochondrial arrangements. ×30,000 (a and b) and ×12,000 (c–f).
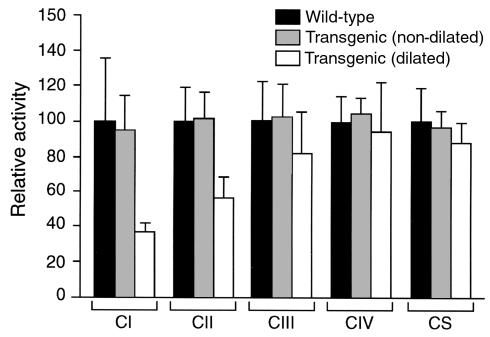
Decreased activity of respiratory chain complexes I and II in diseased hearts.
Deficits in mitochondrial function are often associated with cardiomyopathies (see ref. 19 for review). Mitochondria of all transgenic animals appeared ultrastructurally normal. We analyzed the activities of several mitochondrial enzymatic complexes in dilated transgenic hearts (obtained from healthy-looking mice), nondilated transgenic hearts, and WT hearts. Interestingly, the activity of the respiratory chain complex I (NADH-ubiquinone dehydrogenase) was specifically reduced in dilated transgenic hearts (Figure 7). Similarly, the activity of complex II (succinate dehydrogenase) was significantly reduced in these hearts (Figure 7). In contrast, the activities of complexes III (cytochrome bc1) and IV (cytochrome c oxidase), as well as that of citrate synthase, were normal. The DCM syndrome of the RXRα-overexpressing transgenic mice is therefore associated with specific mitochondrial dysfunctions.
Figure 7.
Mitochondrial enzymatic activities. The activities of respiratory chain complexes (CI–CIV) and of citrate synthase (CS) were determined in hearts from WT, nondilated transgenic hearts, or dilated transgenic hearts from 6-month-old mice, as indicated (3 mice in each group). Values are represented as arbitrary units, with 100 being the mean values measured for WT hearts.
Discussion
RXRα may not be required in cardiac myocytes during development.
We show here that overexpression of an RXRα protein in embryonic cardiomyocytes is not able to rescue, even partially, the hypoplasia of the ventricular myocardium of RXRα-null mutants. This observation suggests that the RXRα function required for normal ventricular muscle development is not exerted (or not exerted exclusively) in myocytes. It is unlikely that the failure to rescue the mutant cardiac phenotype is due to the absence of the NH2-terminal part of the overexpressed RXRα protein, as deletion of sequences encoding the RXRα NH2-terminal A/B region from the mouse genome does not lead to any obvious cardiac defects (B. Mascrez, M. Mark, P. Kastner, and P. Chambon, unpublished study). We cannot exclude that an RXRα function might be required in cardiomyocytes at very early stages. However, this possibility appears unlikely, as the transgene is already highly expressed in the ventricles at 10.5 dpc and probably earlier, transgene expression being readily detected by RT-PCR in 9.5 dpc embryos. The conclusion that RXRα expression is most likely not required in the myocytes themselves was independently reached in 2 other recent studies: Chen et al. (20) generated a ventricular myocyte-specific knockout of the RXRα gene, via specific deletion of a LoxP-flanked RXRα exon 4 (encoding most of the DNA binding domain) with the Cre recombinase expressed under the control of regulatory elements from the MLC2v gene. These authors documented myocyte-specific deletion of this exon as early as 8.5 dpc, which did not affect cardiac development and function. Tran and Sucov (21) generated chimeric fetuses between LacZ-marked RXRα–/– cells and WT cells and observed that RXRα mutant cardiomyocytes contribute normally to the ventricular wall in these chimeras. Thus, 3 different approaches (the present rescue attempt with a cardiomyocyte-specific RXRα transgene, ventricular myocyte specific knockout, and chimera analysis) all lead to the same conclusion: the primary defect in RXRα mutants does not lie within the myocyte lineage and is therefore a non–cell-autonomous event.
Which cells then require the RXRα function during cardiac development? The primary defect may occur in cells that control myocyte fate through paracrine signaling. The most likely candidates are the cardiac endothelial cells, as their signaling is known to be crucial for controlling the developmental behavior of the myocytes. For instance, endocardial cells secrete neuregulin, which is necessary for the formation of myocardial trabeculae (22). Epicardial cells are other possible candidate cells. However, a chief role for these latter cells in controlling myocyte development is unlikely, as (i) the absence of the epicardium, which results from an integrin α4 mutation, does not lead to ventricular hypoplasia (23); and (ii) defects in myocytes are already detected in RXRα mutants at 8.5 dpc, before the formation of the epicardium (9, 24). In fact, one cannot definitely rule out that RXRα exerts some function in myocytes, as the abnormal phenotype displayed by ventricular myocytes in RXRα mutants is complex and could possibly be the result of abnormalities affecting several cell types. In this respect, we note that the subepicardial myocytes from RARα–/– mutants display abnormal differentiation features similar to those of RXRα–/– mutants, while keeping a normal cellular shape (9). RXRα inactivation may therefore disrupt an RAR-dependent function in myocytes, as well as an RAR-independent function in another cell type.
Overexpression of RXRα results in cardiomyocyte abnormalities and DCM.
Unexpectedly, the present transgenic mice exhibit an abnormal organization of the contractile elements of their cardiomyocytes, and about 20% of the mice from the highest-expressing line succumb to heart failure. The molecular mechanisms underlying these effects are unclear. Elevated RXRα levels might result in the deregulation of genes that can normally be controlled by an RXR-dependent pathway (such as thyroid hormone receptor or RAR target genes). It is of interest in this respect that hypothyroidism has been linked to some cases of DCM (25). On the other hand, high RXR levels might squelch factors shared with other partners, thereby indirectly interfering with their function.
DCM represents the most common indication leading to cardiac transplantation worldwide (26, 27). Very few genes implicated in this condition have been identified so far, even though about 30% of the cases are estimated to have a clear autosomal dominant hereditary component (see ref. 28 for review). Characterization of affected genes by classic reverse genetic approaches proves difficult because of the large genetic heterogeneity of the disease and the small size of affected families. Thus, only a few chromosomal loci (1p1–q1, 1q32, 3p22–p25, 9q13–q22, and 10q21–q23) have been implicated in DCM, and the corresponding genes have not yet been identified (note that none of these loci corresponds to that of an RXR gene). The current genetic data, originating both from human and from several animal models of DCM, point to alterations of components of the contractile apparatus as an important causal group for DCM: mutations or decreased expression of dystrophin, cardiac actin, metavinculin, δ-sarcoglycan, titin, and the LIM sarcomere assembly protein have thus been implicated in DCM pathogenesis (see refs. 26, 29–31 for reviews). In transgenic mouse models, events mimicking overstimulation of the β-adrenergic receptor, such as overexpression of a dominant negative form of CREB (32) or of the cardiac stimulatory G-protein α subunit (33) have also been shown to produce DCM phenotypes. However, given the large genetic heterogeneity of DCM, it seems unlikely that abnormalities of the contractile apparatus or the adrenergic signaling pathway will fully explain DCM etiology. In this context, new genetically well-defined animal models of DCM, such as the one described in the present study, might be helpful to characterize other genes/factors contributing to human DCM. Most interestingly, expression of an RARα-βgalactosidase fusion protein under the control of the MyHCβ promoter was also shown to induce a severe DCM phenotype in transgenic mice (34). Although the molecular properties of the RARα-βgalactosidase fusion protein used in that study are not clear, the fact that its overexpression in cardiac myocytes leads to a disease similar to the one developed by the MyHCα-RXRα mice suggests that alterations of RXR/RAR signaling in the heart might be implicated in the etiology of DCM. It will therefore be interesting to investigate whether functional or genetic defects of the retinoid signaling pathway are associated with human cases of DCM.
The present mice also illustrate the multistep nature of DCM. Although cardiomyocytes from all mice (from both transgenic lines) exhibit abnormal ultrastructural features (abnormal sarcomere organization and nuclear shapes), only some mice from the higher-expressing line reach the state of cardiac dilation. This observation is consistent with the proposal that DCM and heart failure are the final stage of a chronic condition characterized by an impaired mechanism of force transmission within the cardiac muscle (29). Events associated with the transition from this chronic weakness to the stage of cardiac dilation are only poorly understood. Interestingly, this disease stage in the present MyHCα-RXRα transgenic mice is characterized by specific mitochondrial abnormalities (significantly decreased activities of respiratory chain complexes I and II). These mitochondrial abnormalities are therefore not a primary consequence of an elevated RXRα level but are associated with the transition from a structurally abnormal heart muscle to a diseased muscle. Reduced activities of respiratory chain complexes has previously been associated with some cardiomyopathies, including cases with cardiac dilation (see refs. 35, 36 for reviews). Interestingly, decreased activity of respiratory chain complex I has been found in tissues from RXRα-null fetuses (37). However, it is not immediately clear whether this deficiency, which appears to be a direct consequence of the absence of RXRα, is mechanistically related to the complex I deficiency detected in the present study, which is a secondary consequence of elevated RXRα levels. Recently, Hirota et al. (38) have shown that DCM can result from a failure to activate a gp130-dependent cardiomyocyte survival program in response to biomechanical stress. Whether the high number of necrotic cardiomyocytes observed in the present transgenic dilated hearts corresponds to a similar failure to activate a survival pathway or is merely the consequence of biochemical deficiencies (such as dysfunctional respiratory chain enzymes) remains to be seen. In any event, the present mice may offer a valuable model for studying progression from a state of cellular abnormalities in myocytes to that of cardiac dilation and heart failure. In this respect, gene expression studies on a large scale (e.g., using DNA microarrays) might shed light on molecular events associated with these distinct steps of the disease.
Acknowledgments
We thank J. Robbins for the gift of the MyHCα promoter plasmid; C. Bronn, and S. Bronner for technical assistance; and the microinjection and animal staffs for help in the establishment and maintenance of the transgenic lines. This work was supported by the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the Collège de France, the Fondation pour la Recherche Médicale (FRM), and Bristol-Myers Squibb.
Footnotes
Vemparala Subbarayan’s present address is: M.D. Anderson Cancer Center, Department of Clinical Cancer Prevention, Box 236, 1515 Holcombe Boulevard, Houston, Texas 77030, USA.
References
- 1.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, Evans R. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 3.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 4.Kastner P, et al. Genetic evidence that the retinoid signal is transduced by heterodimeric RXR/RAR functional units during mouse development. Development. 1997;124:313–326. doi: 10.1242/dev.124.2.313. [DOI] [PubMed] [Google Scholar]
- 5.Mascrez B, et al. The RXRα ligand dependent activation function 2 (AF-2) is important for mouse development. Development. 1998;125:4691–4707. doi: 10.1242/dev.125.23.4691. [DOI] [PubMed] [Google Scholar]
- 6.Kastner P, et al. Genetic analysis of RXRα developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 7.Sucov HM, et al. RXRα mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 8.Dyson E, et al. Atrial-like phenotype is associated with embryonic ventricular failure in retinoid X receptor α –/– mice. Proc Natl Acad Sci USA. 1995;92:7386–7390. doi: 10.1073/pnas.92.16.7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kastner P, et al. Vitamin A deficiency and mutations of RXRα, RXRβ and RARα lead to early differentiation of embryonic ventricular cardiomyocytes. Development. 1997;124:4749–4758. doi: 10.1242/dev.124.23.4749. [DOI] [PubMed] [Google Scholar]
- 10.Rumyantsev PP. Interrelations of the proliferation and differentiation processes during cardiac myogenesis and regeneration. Int Rev Cytol. 1977;91:187–273. [PubMed] [Google Scholar]
- 11.Subramaniam A, et al. Tissue-specific regulation of the α-myosin heavy chain gene promoter in transgenic mice. J Biol Chem. 1991;266:24613–24620. [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. A single step method of RNA isolation by acid guanidinium thiocyanate–phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Dollé P, Fraulob V, Kastner P, Chambon P. Developmental expression of murine retinoid X receptor (RXR) genes. Mech Dev. 1994;45:91–104. doi: 10.1016/0925-4773(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 14.Andrews NC, Faller D. A rapid micropreparation technique for extraction of DNA binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochette-Egly C, et al. Detection of retinoid X receptors using specific monoclonal and polyclonal antibodies. Biochem Biophys Res Commun. 1994;204:525–536. doi: 10.1006/bbrc.1994.2491. [DOI] [PubMed] [Google Scholar]
- 16.Mark M, et al. Two rhombomeres are altered in Hoxa-1 null mutant mice. Development. 1993;119:319–338. doi: 10.1242/dev.119.2.319. [DOI] [PubMed] [Google Scholar]
- 17.Rustin P, et al. Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 18.Brocard J, Kastner P, Chambon P. Two novel RXRα isoforms from mouse testis. Biochem Biophys Res Commun. 1996;229:211–218. doi: 10.1006/bbrc.1996.1782. [DOI] [PubMed] [Google Scholar]
- 19.DiMauro S, Hirano M. Mitochondria and heart disease. Curr Opin Cardiol. 1998;13:190–197. [PubMed] [Google Scholar]
- 20.Chen J, Kubalak S, Chien K. Ventricular muscle-restricted targeting of the RXRα gene reveals a non-cell autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–1949. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- 21.Tran C, Sucov H. The RXRα gene functions in a non-cell-autonomous manner during mouse cardiac morphogenesis. Development. 1998;125:1951–1956. doi: 10.1242/dev.125.10.1951. [DOI] [PubMed] [Google Scholar]
- 22.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Rayburn H, Hynes R. Cell adhesion events mediated by α4 integrins are essential in placental and cardiac development. Development. 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- 24.Viragh S, Challice C. The origin of the epicardium and the embryonic myocardial circulation in the mouse. Anat Rec. 1981;201:157–168. doi: 10.1002/ar.1092010117. [DOI] [PubMed] [Google Scholar]
- 25.Ladenson PW, Sherman SI, Baughman KL, Ray PE, Feldman AM. Reversible alterations in myocardial gene expression in a young man with dilated cardiomyopathy and hypothyroidism. Proc Natl Acad Sci USA. 1992;89:5251–5255. doi: 10.1073/pnas.89.12.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schowengerdt G, Towbin J. Genetic basis of inherited cardiomyopathies. Curr Opin Cardiol. 1995;10:312–321. doi: 10.1097/00001573-199505000-00013. [DOI] [PubMed] [Google Scholar]
- 27.Keating MT, Sanguinetti MC. Molecular genetic insights into cardiovascular disease. Science. 1996;272:681–685. doi: 10.1126/science.272.5262.681. [DOI] [PubMed] [Google Scholar]
- 28.Mestroni L, Giacca M. Molecular genetics of dilated cardiomyopathy. Curr Opin Cardiol. 1997;12:303–309. doi: 10.1097/00001573-199705000-00012. [DOI] [PubMed] [Google Scholar]
- 29.Olson T, Michels V, Thibodeau S, Tai Y, Keating M. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 30.Hein S, Schaper J. Pathogenesis of dilated cardiomyopathy and heart failure: insights from cell morphology and biology. Curr Opin Cardiol. 1996;11:293–301. doi: 10.1097/00001573-199605000-00010. [DOI] [PubMed] [Google Scholar]
- 31.Towbin J. The role of cytoskeletal proteins in cardiomyopathies. Curr Opin Cell Biol. 1998;10:131–139. doi: 10.1016/s0955-0674(98)80096-3. [DOI] [PubMed] [Google Scholar]
- 32.Frenzke R, Korcarz C, Lang R, Leiden J. Dilated cardiomyopathy in transgenic mice expressing a dominant-negative CREB transcription factor in the heart. J Clin Invest. 1998;101:2415–2426. doi: 10.1172/JCI2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwase M, et al. Cardiomyopathy induced by cardiac Gs alpha overexpression. Am J Physiol. 1997;272:585–589. doi: 10.1152/ajpheart.1997.272.1.H585. [DOI] [PubMed] [Google Scholar]
- 34.Colbert MC, et al. Cardiac compartment-specific overexpression of a modified retinoic acid receptor produces dilated cardiomyopathy and congestive heart failure in transgenic mice. J Clin Invest. 1997;100:1958–1968. doi: 10.1172/JCI119727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marin-Garcia J, Goldenthal MJ. Mitochondrial cardiomyopathy: molecular and biochemical analysis. Pediatr Cardiol. 1997;18:251–260. doi: 10.1007/s002469900169. [DOI] [PubMed] [Google Scholar]
- 36.Antozzi C, Zeviani M. Cardiomyopathies in disorders of oxidative metabolism. Cardiovasc Res. 1997;35:184–199. doi: 10.1016/s0008-6363(97)00141-7. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Lozano P, et al. Energy deprivation and a deficiency in downstream metabolic target genes during the onset of embryonic heart failure in RXRα –/– embryos. Development. 1998;125:533–544. doi: 10.1242/dev.125.3.533. [DOI] [PubMed] [Google Scholar]
- 38.Hirota H, et al. Loss of a gp130 cardiac muscle cell survival pathway is a critical event in the onset of heart failure during biomechanical stress. Cell. 1999;97:189–198. doi: 10.1016/s0092-8674(00)80729-1. [DOI] [PubMed] [Google Scholar]
- 39.Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res. 1991;19:3998. doi: 10.1093/nar/19.14.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]