Background: The chitobiose region of nematode N-glycans can be modified with three fucose residues.
Results: Glycan arrays and other analytical techniques facilitated the definition of the biologically relevant activity of Caenorhabditis FUT-6.
Conclusion: The concerted action of Caenorhabditis FUT-1, FUT-6, and FUT-8 is required for trifucosylation of worm N-glycan cores.
Significance: New approaches for studying glycans from parasitic nematodes are now possible.
Keywords: Glycosyltransferases, Invertebrates, Mass Spectrometry (MS), Microarray, Oligosaccharide, Glycan
Abstract
Fucose is a common monosaccharide component of cell surfaces and is involved in many biological recognition events. Therefore, definition and exploitation of the specificity of the enzymes (fucosyltransferases) involved in fucosylation is a recurrent theme in modern glycosciences. Despite various studies, the specificities of many fucosyltransferases are still unknown, so new approaches are required to study these. The model nematode Caenorhabditis elegans expresses a wide range of fucosylated glycans, including N-linked oligosaccharides with unusual complex core modifications. Up to three fucose residues can be present on the standard N,N′-diacetylchitobiose unit of these N-glycans, but only the fucosyltransferases responsible for transfer of two of these (the core α1,3-fucosyltransferase FUT-1 and the core α1,6-fucosyltransferase FUT-8) were previously characterized. By use of a glycan library in both array and solution formats, we were able to reveal that FUT-6, another C. elegans α1,3-fucosyltransferase, modifies nematode glycan cores, specifically the distal N-acetylglucosamine residue; this result is in accordance with glycomic analysis of fut-6 mutant worms. This core-modifying activity of FUT-6 in vitro and in vivo is in addition to its previously determined ability to synthesize Lewis X epitopes in vitro. A larger scale synthesis of a nematode N-glycan core in vitro using all three fucosyltransferases was performed, and the nature of the glycosidic linkages was determined by NMR. FUT-6 is probably the first eukaryotic glycosyltransferase whose specificity has been redefined with the aid of glycan microarrays and so is a paradigm for the study of other unusual glycosidic linkages in model and parasitic organisms.
Introduction
Glycan arrays have begun to revolutionize the way in which we study carbohydrate-protein interactions (1) and, in combination with modern glycoanalytical and chemical glycobiological approaches (2), have transformed the experimental tools available for modern structural and functional glycobiology. However, in comparison with the examination of the binding of antibodies or lectins with glycan arrays, the determination of enzyme activities, especially of glycosyltransferases, with these platforms is not so well established, and generally only previously studied enzymes have been assessed (3–7). One exception is a recent study on the specificity of “new” glycosyltransferases from bacteria toward simple saccharide structures (8); nevertheless, the actual in vivo substrates for these enzymes remain unknown.
To date, because the focus of glycan arrays has been on mammalian glycans (9), a huge number of possible glycan structures in nature, especially non-mammalian glycans (e.g. those of model organisms or parasites) are underrepresented on existing platforms. These organisms also have or are predicted to have a variety of glycosyltransferases that have previously been unstudied or only incompletely studied; thereby, non-mammalian glycomes and enzymes represent an untapped resource as well as an underestimated challenge.
One of these model organisms with a particularly rich glycomic potential is the nematode Caenorhabditis elegans (10); this species has not only become well established as a model for studies of developmental biology and innate immunity but is also related to parasitic nematodes that represent a biological burden for millions of human beings and livestock worldwide as well as for plant crops. Because nematode glycoconjugates have immunomodulative properties (11) or are relevant in attempts to produce vaccines (12), there is a need for new approaches to study biosynthesis, binding partners, and functions of these molecules in order to identify new therapeutic targets.
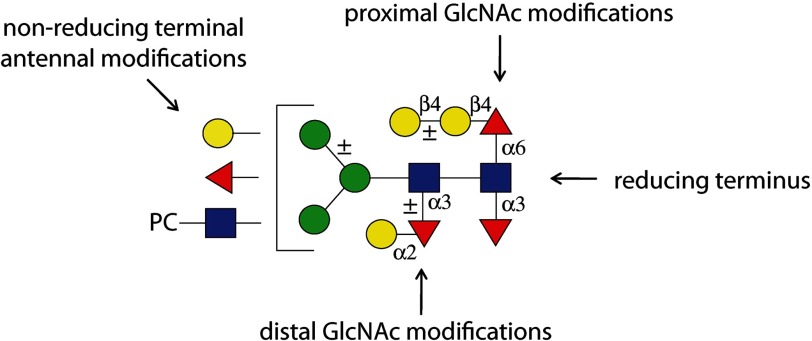
Indeed, the core region of nematode N-glycans is particularly unique due to the range of so-called complex core modifications (13, 14), which represent a set of targets for lectins and potential therapeutics; not just α1,6-fucosylation of the reducing terminal (proximal) asparagine-bound N-acetylglucosamine occurs as in mammals, but also α1,3-fucosylation of both GlcNAc residues (proximal and distal) of the chitobiose core (Fig. 1). Thereby, nematodes express both the core α1,3-fucose on the proximal residue as also present in plants, slime molds, and other invertebrates (15) as well as a novel form of α1,3-fucosylation of the distal N-acetylglucosamine. Furthermore, core fucose residues can be capped with hexose; for instance, substituted and unsubstituted galactose is found linked β1,4 to the core α1,6-fucose (GalFuc)2 and is also present in planaria and cephalopods (16), whereas α1,2-galactosylation of the distal fucose has been recently discovered in nematodes (17). These unusual modifications occur not just in C. elegans but also in a number of parasitic worms, such as Ascaris suum, Hemonochus contortus, and Oesophagostomum dentatum (17, 18), and represent epitopes particular to a subset of nematodes.
FIGURE 1.
Modifications of nematode N-glycans. Various modifications of C. elegans N-glycans are depicted, including those of the core chitobiose and the antennae, according to the nomenclature of the Consortium for Functional Glycomics (red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose). PC, phosphorylcholine.
These N-glycan core motifs have been found to be targets of one endogenous C. elegans galectin (LEC-6) and two nematoxic lectins (CCL2 and CGL2) derived from fungi (19–21). However, because nematode glycans are poorly represented by current glycan arrays, misleading results can be obtained regarding which glycans are the true binding partners. For instance, CCL2 was observed to bind mammalian glycans of types (fucosylated on the antennae) absent from most nematodes (20), whereas CGL2 bound mammalian glycans with (sub)terminal galactose (22); in contrast, the biological data indicated that the respective in vivo targets are core α1,3-fucosylated glycans and GalFuc epitopes actually found in nematodes (20, 21).
The enzymology of the nematode core modifications is only partly understood. Indeed, the genome of C. elegans encodes nearly 30 potential fucosyltransferases, of which the activity of only two α1,2-, one α1,6-, and four α1,3-fucosyltransferases has been demonstrated. Initially the FUT-1 enzyme and then later three other α1,3-fucosyltransferase homologues (including FUT-6) were suggested to synthesize Lewis-type epitopes, including Lex and fucosylated LacdiNAc (LDNF) (i.e. Gal(NAc)β1,4(Fucα1,3)GlcNAc) (23, 24); however, such epitopes have not, to date, been detected in C. elegans and only occur in few nematode species (25–27). Later work showed that FUT-1 is actually a core α1,3-fucosyltransferase with an unusual substrate requirement (28), whereas FUT-8, a homologue of the mammalian core α1,6-fucosyltransferase, displays a substrate specificity typical for such enzymes (29). The identity of the β1,4-galactosyltransferase encoded by the galt-1 gene, which modifies the core α1,6-fucose residue, thus forming the GalFuc epitope, was first revealed by a screen for mutants resistant to the fungal CGL2 lectin (21, 30); this indicates that non-standard techniques are essential for examining glycosylation-relevant enzymes in these organisms.
The definition of these three enzymes (FUT-1, FUT-8, and GALT-1) still left the molecular basis for the other core modifications unsolved. Neither the fucosyltransferase modifying the distal N-acetylglucosamine nor other glycosyltransferases modifying α1,3-linked fucose or the galactosylated core α1,6-fucose have, to date, been studied. There were clues from glycomic studies of mutant worms, because not only fut-1 and fut-8 mutants are deficient in certain fucosylated N-glycans (21, 28), but also fut-6 mutant worms have an altered glycomic profile (28). Therefore, we suspected that FUT-6 may have a role in N-glycan biosynthesis independent of its ability to generate Lewis-type epitopes in vitro. Using a mixture of array- and glycomic-based approaches, we now show that FUT-6 is the enzyme that generates the distal Fucα1,3GlcNAc unit in C. elegans, a result then exploited in the chemoenzymatic synthesis of a complete trifucosylated N-glycan core. In the process, we were able to recreate the biosynthetic pathway leading in nematodes to a multiply fucosylated core recently shown to have relevance to nematoxic lectin binding.
EXPERIMENTAL PROCEDURES
Enzyme and Lectin Preparation
HisFLAG-tagged soluble forms of C. elegans FUT-1, FUT-6, and FUT-8 were expressed in Pichia pastoris. The constructs were prepared directly from RT-PCR fragments in the case of FUT-1 and FUT-6, also known as CEFT1 and CEFT3 (24), or by reamplification from a previously described expression vector in the case of FUT-8 (28, 31) into a reconstructed form of pPICZα vectors named pPICZαHisFLAG. First, the modified expression vector was obtained after two rounds of inverse PCR using KOD polymerase (Takara) to incorporate a region encoding a His tag and a FLAG tag between the region encoding the α-factor signal sequence and the ClaI, PstI, and EcoRI restriction sites. Truncated open reading frames for the three fucosyltransferases (excluding the cytoplasmic and transmembrane domains) were then isolated after PCR using the following forward and reverse primers for FUT-1 (AACTGCAGAAATCTGAACAAAAGGATTGG with GCTCTAGACTAATCTAACGGAATAGAATC), FUT-6 (AACTGCAGAGGAGTAAACATAAAGATTCC with GCTCTAGACAACTACAAATATTTCGAAGC) and FUT-8 (TCTGGAAAAAGAAAGACAAGAAC with CGGGTACCTAATCTAAAAGAGCTTCG). The PCR products were cut with the relevant restriction enzymes and then ligated into the pPICZαHisFLAG vector. Linearized constructs were integrated into the Pichia genome by electroporation. All recombinant FUTs were expressed at 18 °C for 96 h prior to His tag purification using nickel affinity chromatography; purified FUTs were rebuffered and stored in 25 mm Tris-HCl, 150 mm NaCl, pH 7.0, at 4 °C. Expression of the enzymes was verified by SDS-PAGE as well as Western blotting with an anti-FLAG antibody (for FUT-6, a single band of ∼45 kDa was observed upon Coomassie Blue staining; data not shown). C. elegans GALT-1 (30) and Coprinopsis cinerea lectin CCL2 (20) were the kind gifts of Dr. Markus Künzler.
Glycan Microarray
Microarrays were printed on Nexterion® H N-hydroxysuccinimide-activated glass slides (Schott AG, Mainz, Germany) employing a robotic non-contact spotter, sciFLEXARRAYER S11, from Scienion AG (Berlin, Germany). Droplets (2.5 nl) of each glycan solution (50 μm; glycans 1–17, Fig. 3) in sodium phosphate buffer (300 mm, pH 8.5, 0.005% Tween 20) were spatially arrayed with a spot pitch of 550 μm. Each glycan was spotted in six replicates (2 different glycans/row), producing a 12 × 11 spot array, which was printed in 14 copies onto each slide. After printing, the slides were placed in a 75% humidity chamber (saturated NaCl solution) at 25 °C for 18 h. Unreacted N-hydroxysuccinimide groups were quenched with 50 mm ethanolamine in 50 mm sodium borate buffer, pH 9.0, for 1 h. The slides were washed with PBST (PBS solution containing 0.5% Tween 20), PBS, and water and dried in a slide spinner.
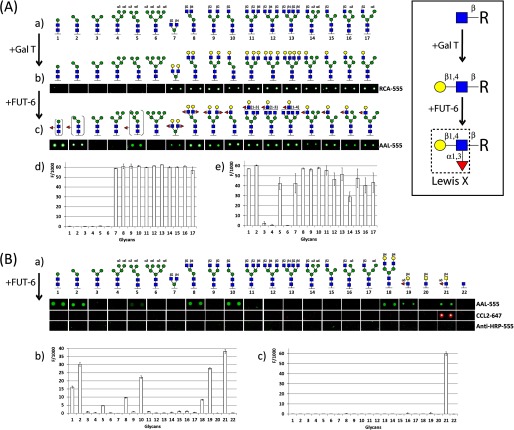
FIGURE 3.
Glycan array screening for FUT-6 substrates. A, N-glycan microarray after enzymatic reaction with bovine milk β1,4-galactosyltransferase followed by C. elegans α1,3-fucosyltransferase (FUT-6) to generate Lewis x type epitopes (boxed inset) on the antennae of complex and hybrid N-glycans. a, non-modified N-glycan structures originally printed on the microarray. b, N-glycan microarray images after galactosylation incubated with the galactose-recognizing lectin from R. communis (RCA-555). c, images of the galactosylated N-glycan microarray after FUT-6 reaction and incubation with the fucose-recognizing lectin from A. aurantia (AAL-555). d, fluorescence intensities of the galactosylated array after incubation with RCA-555. e, fluorescence intensities with AAL-555 after GalT and FUT-6 incubation. B, N-glycan microarray after enzymatic reaction with CeFUT-6 without preincubation with galactosyltransferase. a, N-glycan microarray images after incubation with fucose-recognizing lectins (AAL and CCL2) and anti-HRP-555 antibody. b, fluorescence intensities after incubation with AAL-555. c, fluorescence intensities after incubation with CCL2–647. Compounds 19 and 21, already containing fucose, were included as controls; however, only the latter (LDNF) reacted with CCL2. Each histogram represents the average relative fluorescent unit values for six replicates with the S.D. Red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose.
Printed synthetic glycans 1–17 were further derivatized by on-chip enzymatic modifications with recombinant glycosyltransferases. One subarray was galactosylated with a mixture containing bovine milk β1,4-galactosyltransferase (10 milliunits), alkaline phosphatase (25 milliunits), MnCl2 (5 mm), Hepes buffer (50 mm, pH 7.4), and UDP-Gal (2 mm) at 37 °C for 48 h. The introduced galactose was detected with fluorescently labeled Ricinus communis agglutinin RCA-555 (50 μg/ml), a lectin that recognizes β-linked galactose. Afterward, the galactosylated subarrays were incubated with a reaction mixture containing purified recombinant C. elegans FUT-6 (8.5 μg), MnCl2 (20 mm), MES buffer (80 mm, pH 6.5; compatible with data on the pH optimum), and GDP-Fuc (1 mm). The subarrays were then probed with fluorescently labeled Aleuria aurantia lectin AAL-555 (50 μg/ml), which has a broad affinity against fucose. In addition, a non-galactosylated glycan subarray was incubated with FUT-6 as above, and the introduced fucose was probed with fluorescently labeled A. aurantia lectin AAL-555 (50 μg/ml) and with fluorescently labeled forms of CCL2-647 (50 μg/ml) and anti-HRP-555 (50 μg/ml), which recognize core α1,3-fucose.
Fluorescence was measured using an Agilent G265BA microarray scanner system (Agilent Technologies, Santa Clara, CA) and quantified with ProScanArray® Express software (PerkinElmer Life Sciences), employing an adaptive circle quantitation method from 50 μm (minimum spot diameter) to 300 μm (maximum spot diameter). Average relative fluorescent unit values with local background subtraction of six spots and S.D. were reported as histograms.
Glycome Preparation
The N-glycomes of selected double mutant worm strains were prepared as described (17); two-dimensional HPLC of pyridylaminated C. elegans N-glycans was performed on a normal phase column (TSKgel Amide-80, Tosoh Bioscience) in the first dimension and reversed phase in the second (Hypersil ODS, Agilent). Selected purified and characterized glycans were among the substrates tested with FUT-6.
MALDI-TOF MS
Mass spectra were recorded using either UltrafleXtreme III or Autoflex Speed MALDI-TOF mass spectrometers, respectively, equipped with a pulsed N2 laser (337 nm) or a SmartBEAM solid state laser and controlled by FlexControl version 3.3 software (Bruker Daltonics). MS/MS was performed by laser-induced dissociation of selected parent ions.
In-solution Assays
For natural pyridylaminated glycans or chemically synthesized compounds with the alkylamine linker, the reaction was carried on in a minimal mixture (2.5 μl) of glycan candidate, MnCl2 (20 mm), MES buffer (80 mm, pH 6.5), GDP-Fuc (2 mm), and recombinant C. elegans FUT-6, at room temperature or 37 °C overnight. Aliquots of the reaction mixture were examined by MALDI-TOF MS using 6-aza-2-thiothymine as matrix (3 mg/ml), and MS/MS (laser-induced dissociation) was performed on the products to assign the structure. As required these compounds were preincubated with FUT-1, FUT-8, and/or GALT-1 at room temperature with the requisite nucleotide sugar and Mn(II) and/or with jack bean β-hexosaminidase at 37 °C.
Pyridylaminated lacto-N-neotetraose (6 pmol) was incubated with FUT-6 under similar reaction conditions prior to isocratic reversed phase HPLC (32). For Lewis-type fucosylation, dabsylated Gly-Glu-Asn-Arg-glycopeptides derived from bovine fibrin (0.25 nmol), either the asialo GalGal glycopeptide or the asialoagalacto glycopeptide incubated with bovine galactosyltransferase in the presence of UDP-GalNAc (to form dabsyl-βGNβGN with terminal GalNAc residues), were also incubated under the same conditions prior to product analysis by MALDI-TOF MS using α-cyanohydroxycinamic acid as matrix. For assessment of core fucosylation activity, 5 nmol of the dabsyl asialoagalacto glycopeptides were remodeled with FUT-8 and then hexosaminidase to yield dabsyl-MMF6 (Man3GlcNAc2Fuc1), prior to α-mannosidase digestion to yield dabsyl-00F6 (Man1GlcNAc2Fuc1) and incubation with FUT-6 (see also Fig. 5).
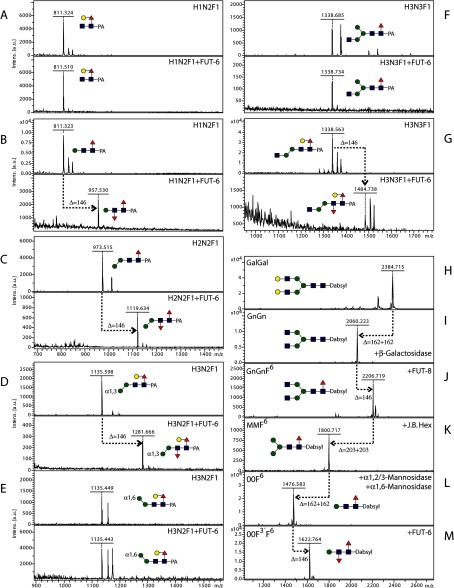
FIGURE 5.
In-solution modification of pyridylaminated natural glycans and a remodeled dabsylated glycopeptide with C. elegans FUT-6. A–G, selected two-dimensional HPLC fractions of N-glycans from double mutant strains of C. elegans were analyzed by MALDI-TOF MS before and after incubation with recombinant FUT-6; the analyzed glycans were detected as [M + H]+, and transfer of fucose to four of the glycans (B, C, D, and G) is indicated by a Δm/z of 146. H–M, MALDI-TOF MS analysis of a dabsylated asialoglycopeptide derived from bovine fibrin (GalGal) after treatment with glycosidases and with recombinant C. elegans core α1,6-fucosyltransferase FUT-8 to yield 00F6 (H–L) prior to incubation with recombinant C. elegans FUT-6 (M); whereas GalGal itself is a substrate for the Lewis-type fucosylation by FUT-6 (Fig. 2), the remodeling to 00F6 is necessary to detect the core fucosylation activity because GnGn and MM glycopeptides are not substrates for this enzyme (data not shown). Red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose.
For fucosylation of trisaccharide 1 (see Scheme 1) on a large scale for NMR, 500 μl of a solution of trisaccharide 1 (1.0 mg, 1.5 μmol), GDP-Fuc (1.2 mg, 1.9 μmol), C. elegans FUT-6 (50 μg), and MnCl2 (20 mm) in MES buffer (80 mm, pH 6.5) were incubated at room temperature for 72 h. The resulting mixture was heated at 95 °C for 5 min and centrifuged, and the solution was purified on graphitized carbon (SupelCleanTM ENVITMCarb cartridges, Sigma-Aldrich). The fucosylated tetrasaccharide 23 was freeze-dried to obtain the title compound as a white powder (1.1 mg, 1.34 μmol, 89%).
SCHEME 1.
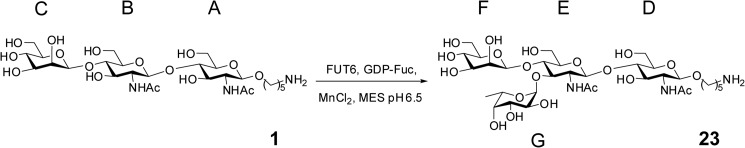
Structure of compound 1 and its fucosylation to form compound 23. The individual sugar residues are annotated with the letters A–G.
HPLC Analysis of Pyridylaminated Products
The examination of FUT-6 modified pyridylaminated lacto-N-neotetraose was carried out on the reversed phase HPLC. A Hypersil ODS column (250 × 4.0 mm; Agilent) was used with 0.1 m ammonium acetate, pH 4.0 (buffer A) and 30% methanol (buffer B). The gradient of buffer B was applied as follows: 0–11 min, 5% B; 11–12, 5–50% B; 12–15 min, 50% B; 15–16 min, 50–0% B; 16–21 min, 0% B.
NMR
Compounds were freeze-dried and dissolved in deuterium oxide (D2O) for recording 1H NMR spectra. Nuclear magnetic resonance experiments were acquired on a Bruker 500-MHz spectrometer, and chemical shifts (δ) are given in ppm relative to the residual signal of the solvent used (D2O 4.79 ppm).
RESULTS
Array-based Screening of Fucosyltransferase Specificities
As initial glycomic evidence from single mutants (28) suggested that the C. elegans α1,3-fucosyltransferase FUT-6 might have an activity other than its known ability to synthesize Lex as well as fucosylated LacdiNAc (LDNF) in vitro (24), a finding that was reproduced by our own assays (Fig. 2), we considered new approaches to reveal the in vivo specificity of this enzyme. We have previously tested a new N-glycan array with two fucosyltransferases with known specificity, the core α1,3-fucosyltransferase FucTA from Arabidopsis thaliana, and the core α1,6-fucosyltransferase FUT-8 from C. elegans (7). On-array fucosylation by both these enzymes was easily assessed using the fucose-specific lectin from A. aurantia (AAL). Furthermore, human asialotransferrin fucosylated on the LacNAc antennae in vitro by FUT-6 was previously shown to bind AAL (33). Therefore, we investigated the impact of His tag-purified recombinant FUT-6 on AAL binding to the array with and without prior incubation with β1,4-galactosyltransferase (Fig. 3) because pregalactosylation is a requirement for synthesis of Lex; thereby, FUT-6 was incubated with galactosylated and non-galactosylated forms of the N-glycan array, which contained paucimannosidic glycan cores lacking non-reducing GlcNAc as well as mono-, bi-, tri-, and tetra-antennary N-glycans, the latter representing also galactosyltransferase substrates. The efficiency of pregalactosylation was assessed using the galactose-specific lectin from R. communis (RCA).
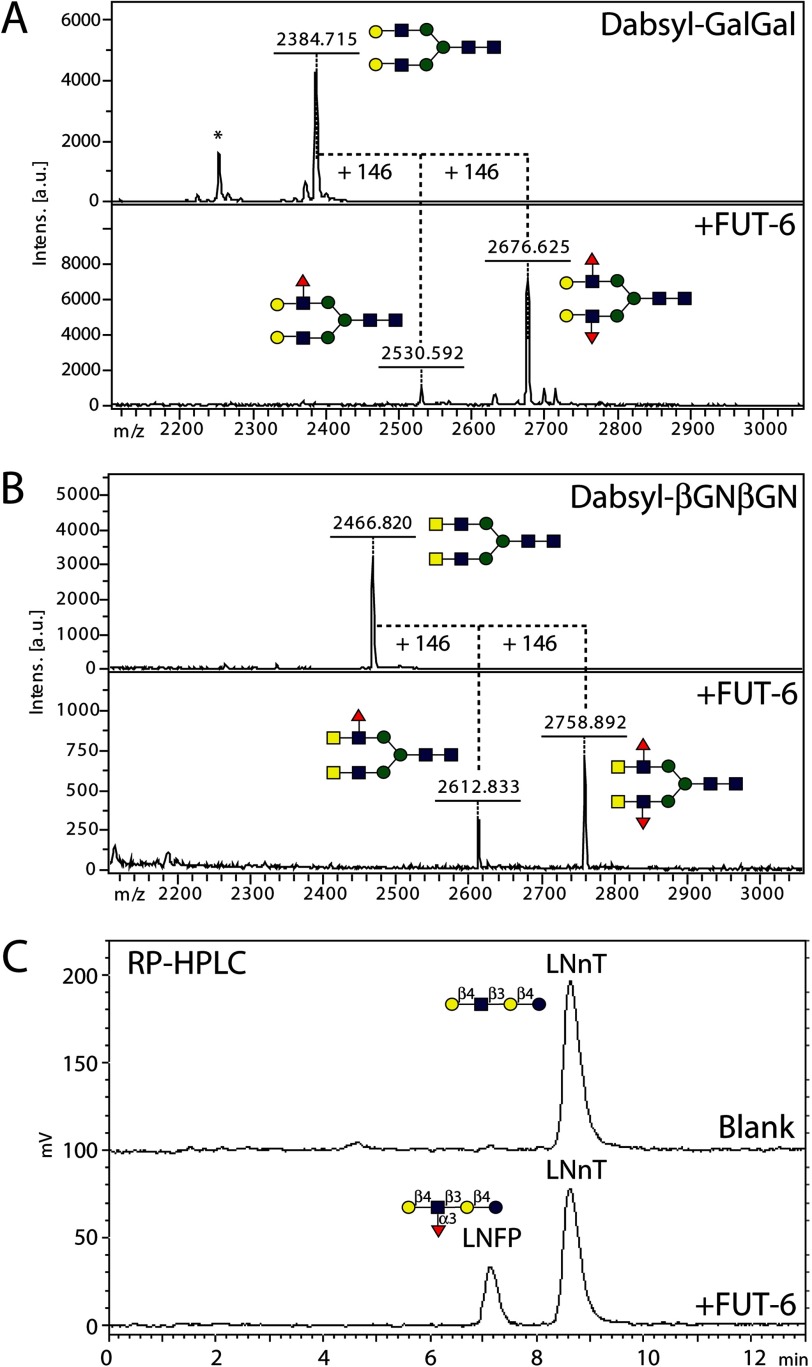
FIGURE 2.
Reappraisal of the Lewis-type activity of C. elegans FUT-6. Previous data indicated that FUT-6 can generate Galβ1,4(Fucα1,3)GlcNAc (LeX) and GalNAcβ1,4(Fucα1,3)GlcNAc (LeX-like; LDNF) epitopes. As shown here, FUT-6 is capable of introducing up to two fucoses on both antennae of the dabsylated GalGal (A) and βGNβGN (B) glycopeptides as judged by the increased molecular weight of products on MALDI-TOF MS spectra, whereas fucosylation (C) of pyridylaminated Galβ1,4GlcNAcβ1,3Galβ1,4Glc (lacto-N-neotetraose; LNnT) resulted in an earlier eluting product (lacto-N-fucopentaose III; LNFP) on isocratic reversed phase HPLC. Red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose.
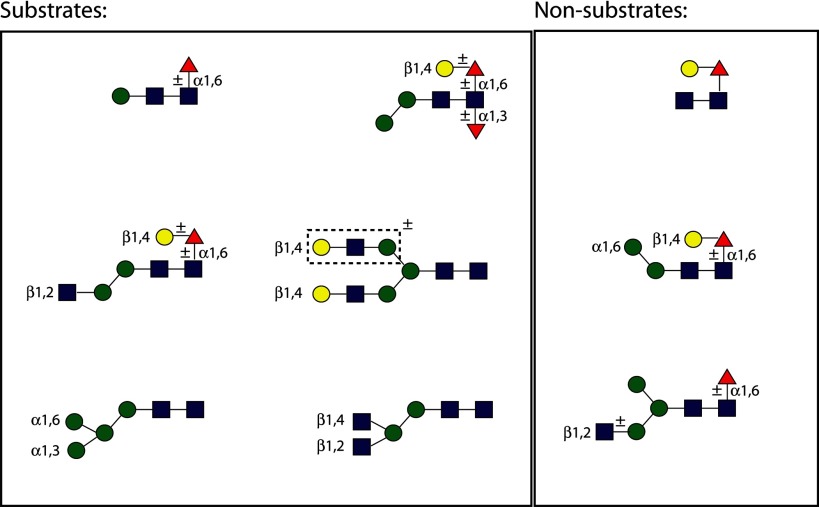
As expected from the previous in vitro data on FUT-6, galactosylated structures 7–17 (whose galactosylation status was proven by RCA binding) gained AAL reactivity upon incubation with FUT-6 (Fig. 3A). Furthermore, non-galactosylated paucimannosidic structures (compounds 1, 2, and 5 on the arrays either with or without preincubation with galactosyltransferase; Fig. 3, A and B) lacking the α1,6-arm were surprisingly also AAL-positive. On the non-galactosylated array, additional FUT-6 substrates were those with non-reducing GlcNAc on the α1,3-arm but lacking the α1,6-mannose linked to the core β1,4-mannose (8 and 10; Fig. 3B). The spot corresponding to galactosylated N-glycan 18, included as a positive substrate control on an otherwise non-galactosylated array, was recognized by AAL after incubation with FUT-6 due to the expected formation of antennal Lex. The multiantennary non-galactosylated glycans and the hybrid-like structures with an α1,6-mannose were not modified by FUT-6. None of the products of FUT-6 were bound by anti-HRP or by the fungal CCL2 lectin, both of which can recognize core α1,3-fucose; only the preformed LDNF trisaccharide 21 was recognized by CCL2. Therefore, we concluded that FUT-6 not only generates Lex epitopes on the antennae of glycan substrates in vitro but transfers fucose to another position on selected N-glycans. However, it cannot form the anti-HRP epitope, which is dependent on α1,3-fucosylation of the proximal core GlcNAc.
Glycomic Analysis of Fucosyltransferase Mutants
Additional clues regarding the specificity of FUT-6 were expected by analyzing C. elegans mutants lacking this enzyme. The fut-6 single mutant was previously shown to lack tetrafucosylated glycans in the peptide:N-glycanase A-released pool but still possessed trifucosylated glycans (28). Therefore, various double mutants lacking two fucosyltransferases each were also prepared: fut-1;fut-6 (previously shown to be completely resistant to CCL2 (20)), fut-6;fut-8, and fut-1;fut-8. Glycomic analyses indicated that the latter two mutants had maximally two fucose residues on their N-glycans (Fig. 4), whereas the fut-1;fut-6 still had traces of trifucosylated N-glycans. This is a further suggestion of an N-glycan-modifying activity of FUT-6.
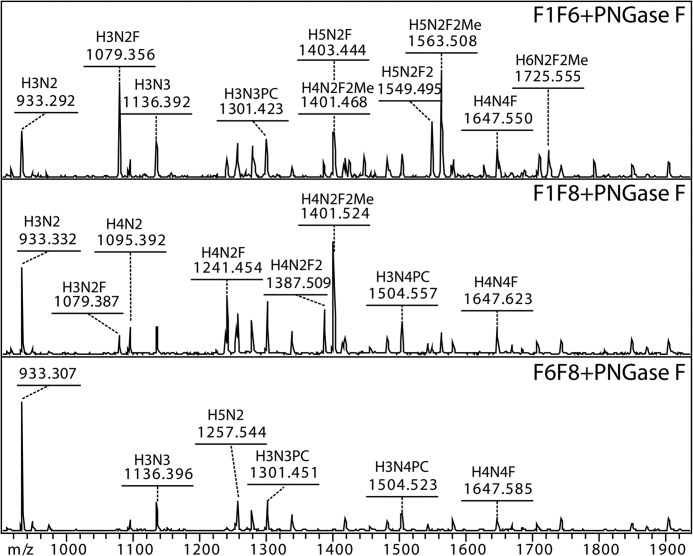
FIGURE 4.
Glycomic data of double fucosyltransferase mutants. Peptide:N-glycanase F (PNGase F)-released glycan pools of fut-1;fut-6 (F1F6), fut-1;fut-8 (F1F8), and fut-6;fut-8 (F6F8) mutants were examined by MALDI-TOF MS. Peptide:N-glycanase A digestion, after peptide:N-glycanase F treatment, of F1F6 and F1F8 resulted in only trace amounts of the same structures. Glycan compositions are abbreviated in the form HxNyFz (i.e. HexxHexNAcyFucz).
In-solution Assays with Various Substrates
Although the in vitro lectin-based assay was a first indication of the unique specificity of FUT-6 and the glycomic analyses were in accordance with a role of this enzyme in the modification of N-glycans in vivo, further verification was required in order to localize the new glycosidic linkage formed by this enzyme. Therefore, a range of substrates suitable for “in-solution” studies were examined: specifically, pyridylaminated forms of glycans isolated from C. elegans, remodeled dabsylated glycopeptides, and chemically synthesized glycans of the form (i.e. functionalized with an alkylamine spacer) also used for printing the N-glycan array.
A working hypothesis was that FUT-6 transfers fucose to the distal GlcNAc of N-glycans; this type of fucose modification was presumed missing from fut-1;fut-6 mutants but was previously shown to be overrepresented in a hex-2;hex-3 strain (17). Therefore, N-glycans from these mutants presumed to be biosynthetic intermediates were incubated with purified FUT-6. In particular, two isomers of core α1,6-fucosylated glycans with the compositions Hex1HexNAc2Fuc1 and Hex3HexNAc3Fuc1 as well as two structures of the form Hex2–3HexNAc2Fuc1 (one of which is an α1,3-mannosidase digestion product of a natural glycan) were tested (Fig. 5, A–G). Only four of these seven glycans were actual acceptor substrates for the in vitro enzymatic reaction; the common element in these substrates was an absence of an α1,6-mannose, but the presence of the β1,4-mannose, on the core region. Unfortunately, MS/MS of these difucosylated products did not result in the appearance of a key fragment for the transferred fucose (data not shown). Notably, a glycan treated with α1,3-mannosidase but retaining the α1,6-mannose is not accepted by FUT-6 (Fig. 5E).
Based on these data, a dabsylated glycopeptide derived from bovine fibrin was remodeled by degalactosylation and core α1,6-fucosylation followed by removal of the antennal GlcNAc residues and of the α1,3/6-mannose residues (Fig. 5, H–L). The resultant glycopeptide carrying a Man1GlcNAc2Fuc1 glycan was a substrate for recombinant FUT-6 (Fig. 5M); MS/MS showed a low intensity fragment of m/z 512, indicative that the transferred fucose is associated with the Manβ1,4GlcNAc region but not with the reducing terminal GlcNAc (data not shown).
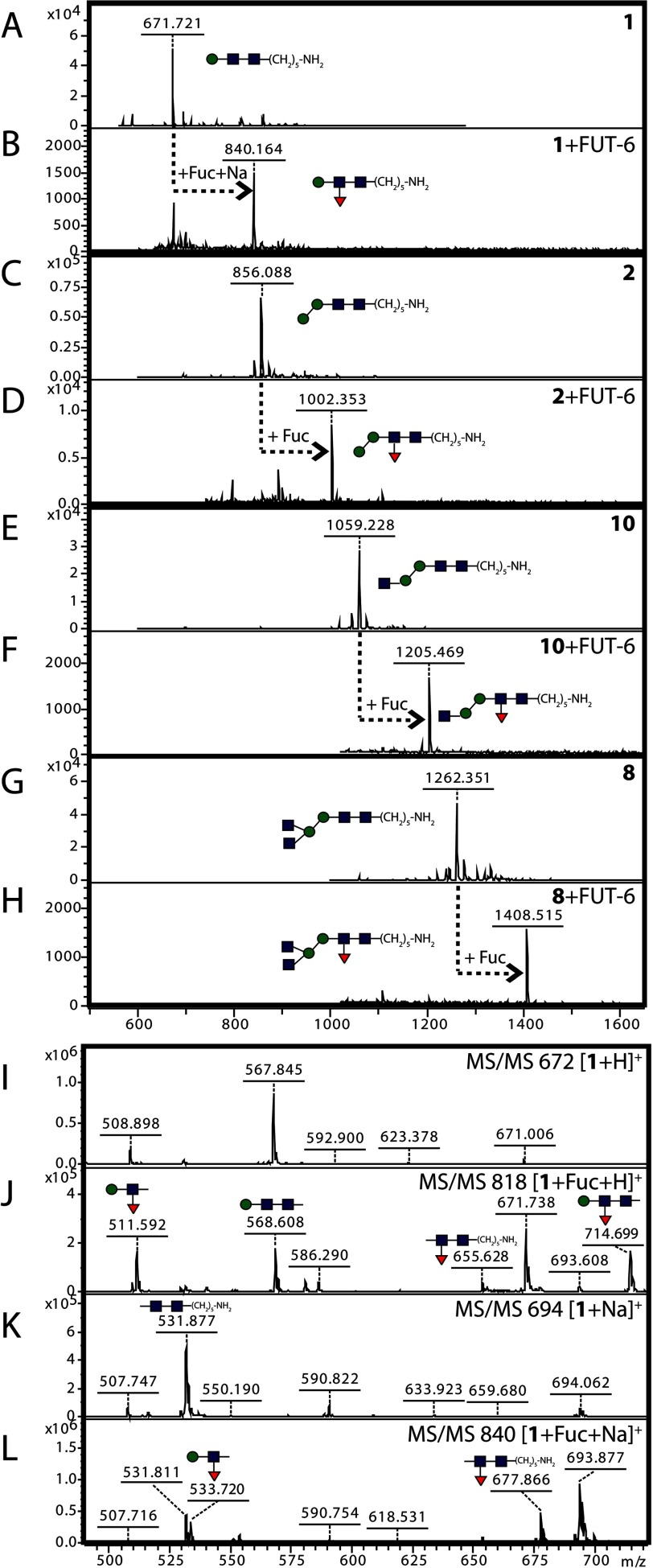
For exact analysis of the position of the transferred fucose, a selection of glycans with an alkylamine linker on the reducing end (used also for printing the glycan arrays) was employed for reactions in solution. In total, four substrates (compounds 1, 2, 8, and 10), chosen on the basis of a positive result with the glycan array (Fig. 3), were tested, and all were found to be fucosylated by FUT-6 in vitro as shown by mass spectral data (Fig. 6). MS/MS of the simplest structure resulted in a set of fragments that were more informative than those of the pyridylaminated and dabsylated products; thereby, the localization of the transferred fucose to the distal GlcNAc rather than to the β-linked mannose was more certain. In particular, the simultaneous appearance of Hex1HexNAc1Fuc1 (m/z 511 as [M + H]+ and m/z 533 as [M + Na]+) and alkylaminated HexNAc2Fuc1 (m/z 655 as [M + H]+ and m/z 677 as [M + Na]+) ions as fragments of the fucosylated “monoantennary” compound 1 was the most promising piece of evidence that the fucosylation by FUT-6 took place on the distal GlcNAc (Fig. 6, J and L).
FIGURE 6.
In-solution modification by FUT-6 of chemically synthesized glycans. Alkylamine-modified glycans 1, 2, 8, and 10 (Hex1–2HexNAc2–4) were incubated with recombinant C. elegans FUT-6 and GDP-Fuc; the substrates and products were analyzed by MALDI-TOF MS (A–H) and MS/MS (I–L). The analyzed glycans were generally observed as [M + Na]+, except for unmodified compound 1, which was detected as [M + H]+; the transfer of fucose by FUT-6 is indicated by +Fuc, as shown by an increase in m/z of 146. Red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose.
Characterization of an Enzymatic Product by NMR
In order to more definitively verify the linkage of the transferred fucose, the chemically synthesized trisaccharide 1 (Man1GlcNAc2) was fully fucosylated by FUT-6 (Scheme 1) and the fucosylated product 23 analyzed by 1H NMR; the chemical shifts for each proton of both compounds are listed in Table 1. The introduction of one fucose moiety was confirmed by the appearance of a new anomeric proton at δ 5.16 ppm with a coupling constant of JH-1,H-2 = 3.9 Hz, characteristic of an α-glycosidic bond. The characteristic signal of H-6 protons from the newly introduced fucose appeared as a doublet at δ 1.08 ppm with a coupling constant of 6.6 Hz and an integration for three protons. The complete assignment of the 1H NMR spectra of the fucosylated product and the trisaccharide 1 was achieved by one- and two-dimensional NMR experiments, using 1H–1H COSY, 1H–13C heteronuclear single quantum correlation spectroscopy, total correlation spectroscopy, and NOESY. A closer look at the protons of each sugar (Scheme 1, A–G) showed that the H-3 signal of the distal GlcNAc was shifted from δ 3.69 ppm (GlcNAc B) in trisaccharide 1 to δ 3.96 ppm (GlcNAc E) in the fucosylated product 23, indicating fucosylation at this position. Furthermore, an NOE effect was observed between the H-1 of the fucose at 5.16 ppm and the H-3 of the GlcNAc E at 3.96 ppm, confirming the α1,3 linkage.
TABLE 1.
1H NMR data for fucosylated tetrasaccharide 23
Data were collected after incubation of trisaccharide 1 with FUT-6; the letters A–G refer to the residues as annotated in Scheme 1. Highlighted in boldface type are the alterations in the chemical shift value for H3 of the distal GlcNAc, indicating the introduction of the fucose at this position.
| δ |
|||||||
|---|---|---|---|---|---|---|---|
| GlcNAc (A) | GlcNAc (B) | Man (C) | GlcNAc (D) | GlcNAc (E) | Man (F) | Fucose (G) | |
| ppm | |||||||
| H-1 | 4.42 | 4.53 | 4.69 | 4.42 | 4.54 | 4.68 | 5.16 |
| H-2 | 3.63 | 3.71 | 3.99 | 3.64 | 3.91 | 3.98 | 3.67 |
| H-3 | 3.62 | 3.69 | 3.58 | 3.62 | 3.96 | 3.58 | 3.94 |
| H-4 | 3.54 | 3.69 | 3.5 | 3.53 | 3.88 | 3.45 | 3.72 |
| H-5 | 3.43 | 3.53 | 3.35 | 3.43 | 3.59 | 3.29 | 4.49 |
| H-6a,b | 3.59, 3.77 | 3.70, 3.82 | 3.66, 3.86 | 3.58, 3.78 | 3.70, 3.85 | 3.59, 3.87 | |
| CH3 | 1.08 | ||||||
Biosynthetic Pathways for Complex Core Modifications
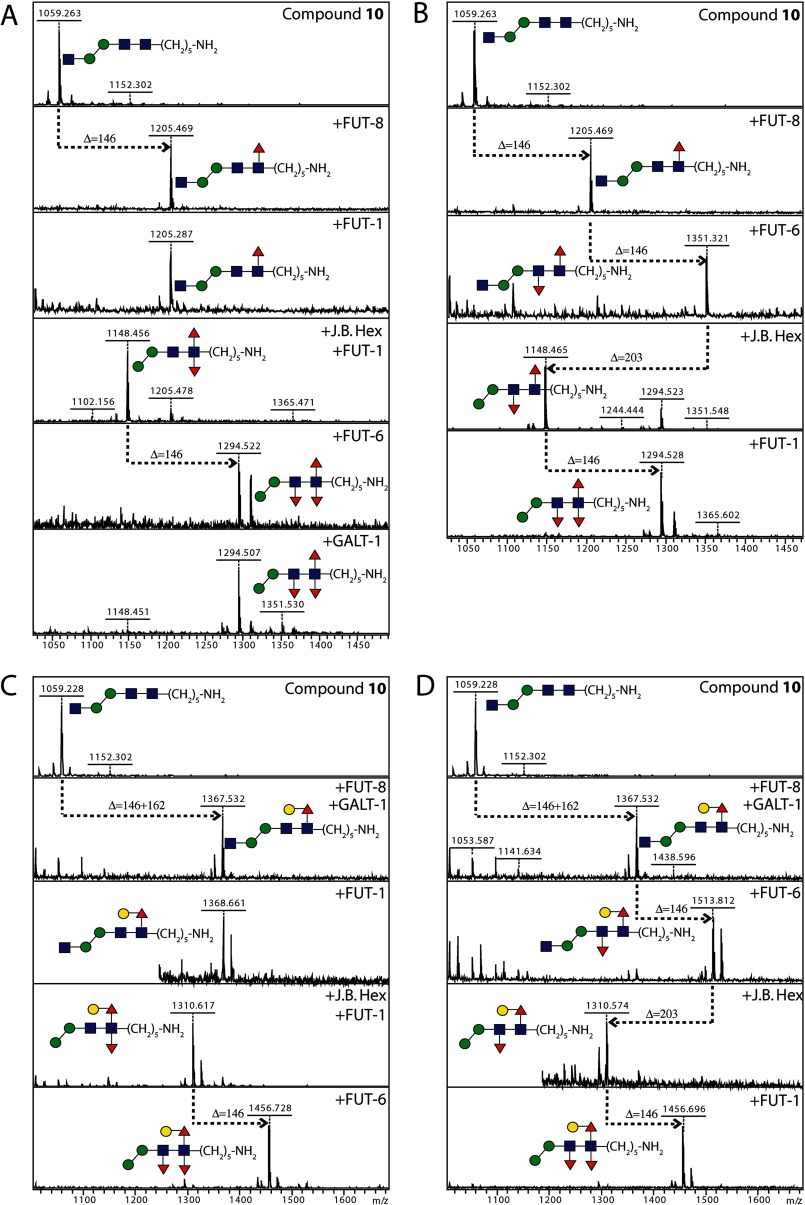
To investigate the basis for the multifucosylated core, the chemically synthesized compound 10 was first α1,6-fucosylated by C. elegans FUT-8 and sequentially remodeled using various recombinant glycosyltransferases from C. elegans as well as jack bean hexosaminidase. In conclusion, two biosynthesis pathways were revealed, leading to the final formation of a trifucosylated N-glycan core: (a) hexosaminidase → FUT-1 → FUT-6 and (b) FUT-6 → hexosaminidase → FUT-1 (Fig. 7, A and B). FUT-1 only worked on hexosaminidase-processed substrates lacking non-reducing terminal GlcNAc, whereas FUT-6 did not have this restriction. The results also showed that there was no specific order of α1,3-fucosylation on the two core GlcNAcs.
FIGURE 7.
Four routes of modification of compound 10 to obtain trifucosylated core structures. Alkylamine-modified 10 was core α1,6-fucosylated (with FUT-8) and then modified by FUT-1, FUT-6, β-hexosaminidase, and GALT-1 in various serial reactions (A–D); reactions were monitored by MALDI-TOF MS. Quasimolecular ions in the mass spectra are [M + Na]+; transfer of fucose or galactose is indicated by respective gain of 146 or 162 mass units and removal of N-acetylglucosamine by loss of 203 mass units. Red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose.
Considering that many core fucose residues are also capped with galactose, it was also of interest to examine the point at which the core α1,6-fucose can be galactosylated, using the only proven nematode galactosyltransferase, C. elegans GALT-1 (30). Galactose was only transferred by GALT-1 to the core α1,6-fucose (Fig. 7, C and D); no further galactosylation appeared to occur on any of the α1,3-linked fucose residues. Furthermore, the action of GALT-1 was prevented by preincubation with either FUT-1 or FUT-6. The downstream α1,3-fucosylation by FUT-1 and FUT-6 on substrates carrying the GalFuc epitope followed pathways similar to those of the non-galactosylated glycans as described above.
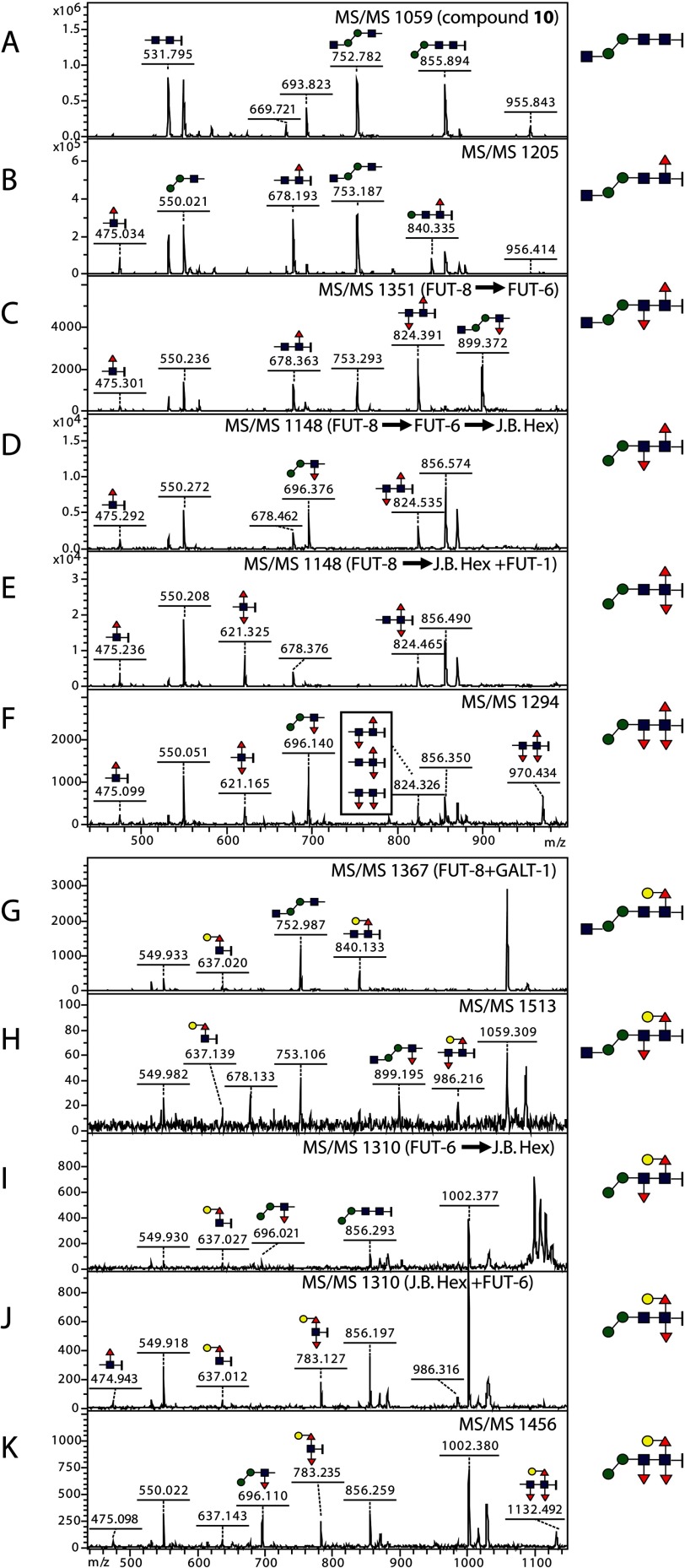
MS/MS of glycan products (Fig. 8) was then employed to examine the location of the transferred fucose residues. The core α1,6-fucose introduced by FUT-8 is always associated with the reducing terminal GlcNAc and the alkylamine linker; this results in the HexNAc1Fuc1-(CH2)5NH2 ion (m/z 475; Fig. 8, B–F) unless it is further modified with GALT-1 (G–K). Difucosylated compounds resulted from the action of FUT-8 and FUT-1 display diagnostic ions such as HexNAc1Fuc2-(CH2)5NH2 (m/z 621; Fig. 8, E and F) and, when galactosylated by GALT-1, Hex1HexNAc1Fuc2-(CH2)5NH2 (m/z 783; J and K). FUT-6-modified compounds possess either Hex2HexNAc1Fuc1 ion (m/z 696; Fig. 8, D, F, I, and K) or Hex2HexNAc2Fuc1 ion (m/z 899; C and H). The trifucosylated final products display the HexNAc2Fuc3-(CH2)5NH2 fragment (m/z 970; Fig. 8F) or its galactosylated form Hex1HexNAc2Fuc3-(CH2)5NH2 (m/z 1132; K).
FIGURE 8.
MS/MS of substrates and products elucidating the formation of trifucosylated glycans. Spectra A–F display the fragmentation patterns of the non-galactosylated glycans Hex2HexNAc2–3Fuc0–3-(CH2)5NH2 with m/z 1059, 1205, 1351, 1148, and 1294 (see Fig. 7, A and B), whereas spectra G–K display those of the galactosylated glycans Hex3HexNAc2–3Fuc0–3-(CH2)5NH2 with m/z 1367, 1513, 1310, and 1456 (see Fig. 7, C and D). All the ions annotated are [M + Na]+, and predicted structures of the key ions are shown in Consortium for Functional Glycomics format; the alkylamine linker, -(CH2)5NH2, is represented by a short vertical bar at the right of N-acetylglucosamine. 475, HexNAc1Fuc1-(CH2)5NH2; 621, HexNAc1Fuc2-(CH2)5NH2; 637, Hex1HexNAc1Fuc1-(CH2)5NH2; 678, HexNAc2Fuc1-(CH2)5NH2; 696, Hex2HexNAc1Fuc1; 783, Hex1HexNAc1Fuc2-(CH2)5NH2; 824, HexNAc2Fuc2-(CH2)5NH2; 899, Hex2HexNAc2Fuc1; 970, HexNAc2Fuc3-(CH2)5NH2; 1132, Hex1HexNAc2Fuc3-(CH2)5NH2. Red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose.
Formation of a Trifucosylated N-Glycan Core in Vitro
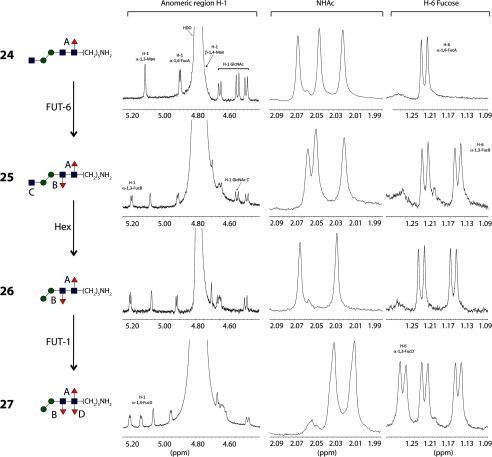
One of the biosynthetic pathways toward the formation of the trifucosylated N-glycan core (FUT-8 → FUT-6 → hexosaminidase → FUT-1; Fig. 9) was performed on a larger scale, starting with compound 10 (0.9 mg, 0.87 μmol). The sequential introduction of the fucose residues was easily monitored by following the NMR chemical shifts of the H-1 and H-6 protons of the fucose residues with different linkages (Fig. 9 and Table 2). Initially, the pentasaccharide 10 was treated with FUT-8 and GDP-Fuc to introduce the core α1,6-fucose, the fucosylated product 24 was purified, and 1H NMR was recorded. The H-1 of the fucose appeared as a doublet at δ 4.90 ppm with a coupling constant of JH-1,H-2 = 3.7 Hz. The characteristic doublet corresponding to H-6 of the newly introduced fucose appeared at δ 1.22 ppm. This compound was subjected to reaction with FUT-6 to yield compound 25, and the introduction of an α1,3-fucose in the distal GlcNAc could be demonstrated by the appearance of a doublet at δ 5.19 ppm with a coupling constant of JH-1,H-2 = 3.7 Hz, corresponding to H-1. Additionally, a doublet corresponding to H-6 appeared at δ 1.15 ppm. This difucosylated glycan 25 was treated with jack bean hexosaminidase. The removal of the terminal GlcNAc is demonstrated by the disappearance of a doublet at δ 4.54 ppm corresponding to H-1 and one singlet at δ 2.05 ppm corresponding to the acetyl group of this residue (glycan 26). Finally, the reaction with FUT-1 led to the formation of the trifucosylated core structure 27. The introduction of core α1,3-fucose was detected in the 1H NMR spectra by the appearance of a doublet at δ 5.21 ppm with a coupling constant of JH-1,H-2 = 4.0 Hz corresponding to H-1 and a doublet at δ 1.26 ppm for the H-6 of the fucose. The chemical shifts for this trifucosylated structure are in agreement with those reported previously for a similar compound prepared by chemical synthesis (34).
FIGURE 9.
Preparation of the trifucosylated compound 27. Shown is the synthetic pathway toward the formation of the trifucosylated core N-glycan 27 (left) and a comparison of the significant regions of the 1H NMR spectra of the fucosylated N-glycans. Chemical shifts corresponding to annotated residues A, B, C, and D are highlighted in the relevant parts of the spectra. The NHAc chemical shifts are those of the three (compounds 24 and 25) or two (26 and 27) GlcNAc residues, whereas those in the H-6 fucose region derive from the one (24), two (25 and 26), or three (27) fucose residues in each compound. Red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose.
TABLE 2.
Selected 1H NMR data for di- and trifucosylated compounds 24–27 (see also Fig. 9)
| Compound | Anomeric region | NHAc | H-6 Fuc |
|---|---|---|---|
| 24 | 5.11 (s, 1H, H-1α1–3Man), 4.90 (d, J = 3.7 Hz, 1H, H-1α1–6Fuc), 4.69 (s, 1H, H-1β1–4Man), 4.65 (d, J = 7.8 Hz, 1H, H-1GlcNAc), 4.54 (d, J = 8.3 Hz, 1H, H-1GlcNAc), 4.48 (d, J = 7.9 Hz, 1H, H-1GlcNAc) | 2.07 (s, 3H), 2.04 (s, 3H), 2.02 (s, 3H) | 1.22 (d, J = 6.6 Hz, 3H) |
| 25 | 5.19 (d, J = 3.7 Hz, 1H, H-1α1–3Fuc), 5.08 (s, 1H, H-1α1–3Man), 4.91 (d, J = 3.7 Hz, 1H, H-1α1–6Fuc), 4.70 (s, 1H, H-1β1–4Man), 4.67–4.62 (m, 2H, H-5Fuc, H-1GlcNAc), 4.54 (d, J = 8.5 Hz, 1H, H-1GlcNAc), 4.48 (d, J = 8.2 Hz, 1H, H-1GlcNAc) | 2.06 (s, 3H), 2.05 (s, 3H), 2.02 (s, 3H) | 1.22 (d, J = 6.6 Hz, 3H), 1.15 (d, J = 6.4 Hz, 3H) |
| 26 | 5.20 (d, J = 3.6 Hz, 1H, H-1α1–3Fuc), 5.07 (s, 1H, H-1α1–3Man), 4.92 (d, J = 3.8 Hz, 1H, H-1α1–6Fuc), 4.70 (s, 1H, H-1β1–4Man), 4.68–4.63 (m, 2H, H-5Fuc, H-1GlcNAc), 4.49 (d, J = 8.0 Hz, 1H, H-1GlcNAc) | 2.06 (s, 3H), 2.03 (s, 3H) | 1.22 (d, J = 6.5 Hz, 3H), 1.15 (d, J = 6.5 Hz, 3H) |
| 27 | 5.21 (d, J = 4.0 Hz, 1H, H-1α1–3Fuc), 5.14 (d, J = 3.5 Hz, 1H, H-1α1–3Fuc), 5.06 (s, 1H, H-1α1–3Man), 4.95 (d, J = 3.6 Hz, 1H, H-1α1–6Fuc), 4.69 (s, 1H, H-1β1–4Man), 4.68 − 4.62 (m, 3H, 2xH-5Fuc, H-1GlcNAc), 4.47 (d, J = 7.9 Hz, 1H, H-1GlcNAc) | 2.03 (s, 3H), 2.01 (s, 3H) | 1.26 (d, J = 6.5 Hz, 3H), 1.22 (d, J = 6.6 Hz, 3H), 1.14 (d, J = 6.5 Hz, 3H) |
DISCUSSION
Fucosyltransferase Substrate Screening
The substrate specificities of glycosyltransferases can be very subtle, and apparently small changes to glycan structures distant to the site of glycosylation can have an impact on whether a glycan is an acceptor or not; whether or not a protein-linked glycan is modified depends also on factors such as accessibility on the protein surface, glycosyltransferase expression levels, and the concentrations of the nucleotide sugar donors. The traditional view (35) was that for each glycosidic linkage, there is a specific enzyme (“one linkage-one enzyme”). However, it later became obvious that, in many circumstances, multiple enzymes can form the same linkage, or one enzyme may be able to form multiple related linkages. This scenario is shown by the activities of the six proven human α1,3-fucosyltransferases forming Lewis epitopes, one of which (Fuc-TIII) can form either α1,3 or α1,4 linkages, dependent on the substrate (36). In the past, screening of glycosyltransferase specificities was often unsystematic and reliant on the availability of natural sources of glycosylation precursors; generally, even for invertebrate enzymes, such as FUT-6, previously used substrates were those based on acceptors for mammalian enzymes, which led to misleading results.
However, the development of glycan arrays opens up new possibilities for examining these enzymes but to date has been (in terms of eukaryotic systems) limited to studying rather well characterized examples, such as mammalian fucosyl- and sialyltransferases or enzymes involved in plant cell wall biosynthesis (37–40). On the other hand, glycosyltransferases have been useful in synthesis of glycan libraries subsequently printed onto arrays (41).
Recently, some of us have developed a systematic array of N-glycans and N-glycan-like molecules on the basis of printing alkylamine-modified chemically synthesized oligosaccharides onto glass surfaces. These have been successfully appraised also using glycosyltransferases (a galactosyltransferase, a sialyltransferase, and two core fucosyltransferases) of known specificities, employing lectins and antibodies as detection reagents (6, 7, 42). A particular challenge, therefore, was to examine a glycosyltransferase from a model organism with an in vitro substrate specificity apparently not matching its role in vivo.
As summarized above, the α1,3-fucosyltransferase homologue FUT-6 from C. elegans can act as a Lewis-type enzyme in vitro, but a deletion in the fut-6 gene results in a loss of tetrafucosylated non-Lewis-type N-glycans in vivo; among the double fucosyltransferase mutants, no more than two fucose residues are present in the fut-6;fut-8 mutant. Although suggestive of a role for FUT-6 in N-glycan processing in vivo, our data resulting from on-chip fucosylation are the first to show its unique capacity to transfer fucose to the distal GlcNAc of N-glycans in vitro.
These data were then confirmed in assays with a variety of substrates in solution, followed by product characterization by MALDI-TOF MS/MS and NMR. The common element in both the N-glycan and Lewis-type acceptors for FUT-6 appears to be a Hexβ1,4GlcNAc unit, where the hexose can be either mannose or galactose. Interestingly, FUT-6 can transfer two fucoses to a galactosylated monoantennary N-glycan, one to the distal GlcNAc and one to the antenna (data not shown). However, only the transfer to the distal GlcNAc is meaningful in terms of the glycome of C. elegans, and, of those structures tested, only monoantennary N-glycans, lacking non-reducing terminal galactose as well as the α1,6-mannose of the trimannosyl core, are biologically significant substrates for FUT-6. The unusual specificity of this enzyme for such N-glycans correlates with the structures of the distally fucosylated monoantennary N-glycans observed in the hex-2;hex-3 mutant (17); our data also suggest a role for a Golgi-localized mannosidase activity removing the core α1,6-mannose during the biosynthesis of FUT-6-modified C. elegans N-glycans.
N-Glycan Core Modifications in Nematodes
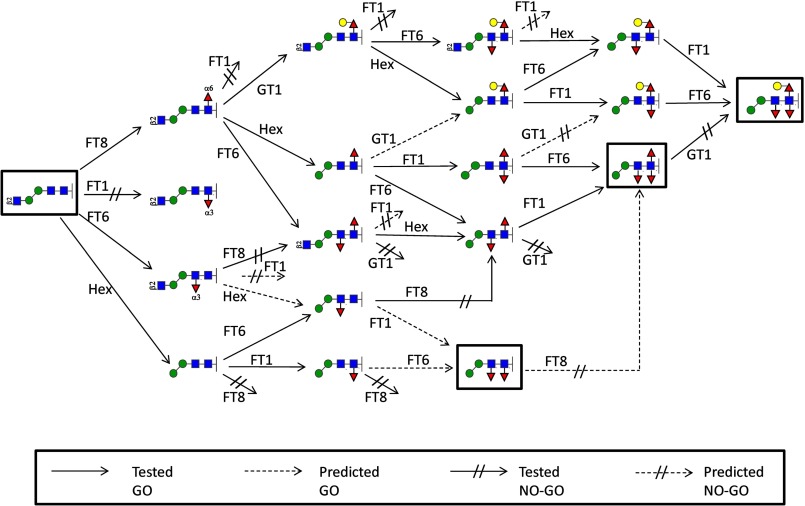
With the knowledge that FUT-6 can fucosylate the distal GlcNAc of the N-glycan core, it is easier to understand the enzymatic biosynthesis pathways of highly fucosylated N-glycan cores in C. elegans (13, 43). Thereby, the two fucosyltransferases (FUT-1 and FUT-8) are defined to solely fucosylate the proximal GlcNAc, whereas FUT-6 is the third core-modifying fucosyltransferase; thus, the order of fucosylation was of interest. Because the other α1,3-fucosyltransferase, FUT-1, can act on products of the core α1,6-fucosyltransferase FUT-8, but FUT-8 cannot act on FUT-1 products (31), the same rule might apply to FUT-6. Therefore, the order of fucosylation was tested on FUT-6 and FUT-8. Indeed, FUT-6-processed glycans (products of 1, 2, 8, and 10) cannot be modified by FUT-8 (data not shown), but FUT-8 products are acceptors for FUT-6. On the other hand, the action of GALT-1, which synthesizes the ligand for two galectins (the endogenous nematode LEC-6 and the fungal nematoxic CGL2), is inhibited when either distal or proximal α1,3-fucose is present. These reactions are summarized in Fig. 10; the substrate status of tested compounds is also shown in Table 3.
FIGURE 10.
Predicted biosynthetic routes for core N-glycan modifications in nematodes. Tested biosynthetic routes (solid lines) involved in the formation of the core chitobiose modifications are summarized based on our experimental data, including reactions of pyridylaminated sugars, dabsylated glycopeptides, and chemically synthesized compounds; predicted GO or NO-GO reactions (broken lines) are judged on substrate specificities of relevant glycoenzymes; short broken lines with a double bar are “dead ends.” FT, fucosyltransferase; Hex, jack bean hexosaminidase; GT1, galactosyltransferase GALT-1. Red triangles, fucose; yellow circles, galactose; blue squares, N-acetylglucosamine; green circles, mannose.
TABLE 3.
Substrate status of N-glycans
Proven substrates and non-substrates for recombinant C. elegans FUT-6 are shown using the nomenclature of the Consortium for Functional Glycomics. The boxed region surrounding one antenna of one N-glycan indicates the region modified by the Lewis-type activity; the other substrates are modified on their core regions.
However, fucosylation of the core is not the end of the story. FUT-6-processed substrates are then available for the action of a putative α1,2-galactosyltransferase-generating α-galactosylated fucose (Galα1,2Fucα1,3) epitope (17), which differs from the proximal GlcNAc-linked GalFuc (Galβ1,4Fucα1,6) epitope found in C. elegans as well as in planaria or mollusks. It is possible that an appropriate mix of approaches (analysis of mutants and use of substrates first identified by array screening) will be necessary for the identification of further glycosyltransferases required for the modification of glycans in C. elegans and other “lower” model or parasitic organisms.
Indeed, the postulated core modification pathways are not just applicable to the model organism C. elegans but also take place in parasitic nematodes, such as A. suum, H. contortus, and O. dentatum (17, 18). Therefore, based on glycomic data, we presume that the distal fucosylation reaction performed by FUT-6 is specific to a subset of nematodes. However, trifucosylation has not been detected to date in a number of other nematode species, including Trichinella spiralis (44) and Onchocerca volvulus (45); this appears to correlate with the phylogeny of nematode fucosyltransferases because obvious FUT-6 orthologues are lacking in the trichinellid and filarial species (data not shown).
Perspectives
Parasites have a high impact on quality of life as well as on agricultural productivity; on the other hand, because the immune systems evolved while being subject to the selection pressure of helminth infections, the absence of parasites is possibly associated with the huge increase in allergies and autoimmunity (46). Indeed, whereas glycosylation may play a role in the efficacy of vaccination against nematodes in farm animals (47), nematode glycans have been implicated in the effects of these organisms on mammalian immune systems (48) and may be relevant to the effects of controversial therapies against autoimmune diseases, in which patients ingest nematode eggs (49). It is of interest that trifucosylated glycans, similar to those prepared during the present study in vitro, are present on the H11 glycoprotein of H. contortus, a known vaccine antigen candidate (18). Thus, the definition of parasite glycan modification pathways and the utilization of the relevant enzymes may not only aid the identification of vaccine targets or the preparation of recombinant vaccine antigens with a more natural glycosylation pattern but also facilitate the production of immunomodulatory factors; furthermore, synthesis of trifucosylated glycan structures required for definition of the natural specificity of endogenous carbohydrate-binding proteins in nematodes or of nematoxic lectins becomes feasible. Indeed, because a fut-1;fut-6 double mutant is completely resistant to nematoxic CCL2, whereas fut-1 and fut-6 single mutants are either only partially or not resistant (20, 21), the FUT-6-mediated modification of nematode N-glycans is (either on its own or in combination with other residues) an interesting target for anthelminthic agents.
Acknowledgments
We thank Hermann Agis, Birgit Antlinger, and Lukas Sobczak for performing initial tests on FUT-6 and Markus Künzler (ETH Zürich) for the kind gift of recombinant nematode GALT-1 and recombinant CCL2. We also thank Raquel Pazos (CICbiomaGUNE) for help with microarray preparation.
This work was supported in part by Austrian Fonds zur Förderung der Wissenschaftlichen Forschung Grants P23922 (to I. B. H. W.) and P21946 (to K. P.), by Spanish Ministerio de Ciencia e Innovación Projects CTQ2008-04444/BQA and CTQ2011-27874, by the Government of the Basque Country (Etortek grant), and by the European Commission (Euroglycanarrays Marie Curie ITN, PITN-GA-2008-215536).
- GalFuc
- galactose linked β1,4 to core α1,6-fucose
- LDNF
- fucosylated LacdiNAc (Gal(NAc)β1,4(Fucα1,3)GlcNAc)
- AAL
- A. aurantia lectin
- RCA
- R. communis agglutinin.
REFERENCES
- 1. Park S., Gildersleeve J. C., Blixt O., Shin I. (2013) Carbohydrate microarrays. Chem. Soc. Rev. 42, 4310–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agard N. J., Bertozzi C. R. (2009) Chemical approaches to perturb, profile, and perceive glycans. Acc. Chem. Res. 42, 788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ban L., Mrksich M. (2008) On-chip synthesis and label-free assays of oligosaccharide arrays. Angew. Chem. Int. Ed. Engl. 47, 3396–3399 [DOI] [PubMed] [Google Scholar]
- 4. Laurent N., Voglmeir J., Wright A., Blackburn J., Pham N. T., Wong S. C., Gaskell S. J., Flitsch S. L. (2008) Enzymatic glycosylation of peptide arrays on gold surfaces. Chembiochem 9, 883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Voglmeir J., Sardzík R., Weissenborn M. J., Flitsch S. L. (2010) Enzymatic glycosylations on arrays. Omics 14, 437–444 [DOI] [PubMed] [Google Scholar]
- 6. Sanchez-Ruiz A., Serna S., Ruiz N., Martin-Lomas M., Reichardt N. C. (2011) MALDI-TOF mass spectrometric analysis of enzyme activity and lectin trapping on an array of N-glycans. Angew. Chem. Int. Ed. Engl. 50, 1801–1804 [DOI] [PubMed] [Google Scholar]
- 7. Serna S., Yan S., Martin-Lomas M., Wilson I. B., Reichardt N. C. (2011) Fucosyltransferases as synthetic tools. Glycan array based substrate selection and core fucosylation of synthetic N-glycans. J. Am. Chem. Soc. 133, 16495–16502 [DOI] [PubMed] [Google Scholar]
- 8. Ban L., Pettit N., Li L., Stuparu A. D., Cai L., Chen W., Guan W., Han W., Wang P. G., Mrksich M. (2012) Discovery of glycosyltransferases using carbohydrate arrays and mass spectrometry. Nat. Chem. Biol. 8, 769–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rillahan C. D., Paulson J. C. (2011) Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 80, 797–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blaxter M. (2011) Nematodes. The worm and its relatives. PLoS Biol. 9, e1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tundup S., Srivastava L., Harn D. A., Jr. (2012) Polarization of host immune responses by helminth-expressed glycans. Ann. N.Y. Acad. Sci. 1253, E1–E13 [DOI] [PubMed] [Google Scholar]
- 12. Hein W. R., Harrison G. B. (2005) Vaccines against veterinary helminths. Vet. Parasitol. 132, 217–222 [DOI] [PubMed] [Google Scholar]
- 13. Hanneman A. J., Rosa J. C., Ashline D., Reinhold V. (2006) Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology 16, 874–890 [DOI] [PubMed] [Google Scholar]
- 14. Paschinger K., Gutternigg M., Rendić D., Wilson I. B. H. (2008) The N-glycosylation pattern of Caenorhabditis elegans. Carbohydr. Res. 343, 2041–2049 [DOI] [PubMed] [Google Scholar]
- 15. Paschinger K., Rendić D., Wilson I. B. H. (2009) Revealing the anti-HRP epitope in Drosophila and Caenorhabditis. Glycoconj. J. 26, 385–395 [DOI] [PubMed] [Google Scholar]
- 16. Schiller B., Hykollari A., Yan S., Paschinger K., Wilson I. B. H. (2012) Complicated N-linked glycans in simple organisms. Biol. Chem. 393, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan S., Bleuler-Martinez S., Plaza D. F., Künzler M., Aebi M., Joachim A., Razzazi-Fazeli E., Jantsch V., Geyer R., Wilson I. B. H., Paschinger K. (2012) Galactosylated fucose epitopes in nematodes. Increased expression in a Caenorhabditis mutant associated with altered lectin sensitivity and occurrence in parasitic species. J. Biol. Chem. 287, 28276–28290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haslam S. M., Coles G. C., Munn E. A., Smith T. S., Smith H. F., Morris H. R., Dell A. (1996) Haemonchus contortus glycoproteins contain N-linked oligosaccharides with novel highly fucosylated core structures. J. Biol. Chem. 271, 30561–30570 [DOI] [PubMed] [Google Scholar]
- 19. Takeuchi T., Hayama K., Hirabayashi J., Kasai K. (2008) Caenorhabditis elegans N-glycans containing a Gal-Fuc disaccharide unit linked to the innermost GlcNAc residue are recognized by C. elegans galectin LEC-6. Glycobiology 18, 882–890 [DOI] [PubMed] [Google Scholar]
- 20. Schubert M., Bleuler-Martinez S., Butschi A., Wälti M. A., Egloff P., Stutz K., Yan S., Collot M., Mallet J. M., Wilson I. B. H., Hengartner M. O., Aebi M., Allain F. H., Künzler M. (2012) Plasticity of the β-trefoil protein fold in the recognition and control of invertebrate predators and parasites by a fungal defence system. PLoS Pathog. 8, e1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Butschi A., Titz A., Wälti M. A., Olieric V., Paschinger K., Nöbauer K., Guo X., Seeberger P. H., Wilson I. B., Aebi M., Hengartner M. O., Künzler M. (2010) Caenorhabditis elegans N-glycan core β-galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLOS Pathog. 6, e1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wälti M. A., Walser P. J., Thore S., Grünler A., Bednar M., Künzler M., Aebi M. (2008) Structural basis for chitotetraose coordination by CGL3, a novel galectin-related protein from Coprinopsis cinerea. J. Mol. Biol. 379, 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DeBose-Boyd R. A., Nyame A. K., Cummings R. D. (1998) Molecular cloning and characterization of an α1,3-fucosyltransferase, CEFT-1, from Caenorhabditis elegans. Glycobiology 8, 905–917 [DOI] [PubMed] [Google Scholar]
- 24. Nguyen K., van Die I., Grundahl K. M., Kawar Z. S., Cummings R. D. (2007) Molecular cloning and characterization of the Caenorhabditis elegans α1,3-fucosyltransferase family. Glycobiology 17, 586–599 [DOI] [PubMed] [Google Scholar]
- 25. Nyame A. K., Debose-Boyd R., Long T. D., Tsang V. C., Cummings R. D. (1998) Expression of Lex antigen in Schistosoma japonicum and S. haematobium and immune responses to Lex in infected animals. Lack of Lex expression in other trematodes and nematodes. Glycobiology 8, 615–624 [DOI] [PubMed] [Google Scholar]
- 26. Haslam S. M., Coles G. C., Morris H. R., Dell A. (2000) Structural characterisation of the N-glycans of Dictyocaulus viviparus. Discovery of the Lewisx structure in a nematode. Glycobiology 10, 223–229 [DOI] [PubMed] [Google Scholar]
- 27. Aranzamendi C., Tefsen B., Jansen M., Chiumiento L., Bruschi F., Kortbeek T., Smith D. F., Cummings R. D., Pinelli E., Van Die I. (2011) Glycan microarray profiling of parasite infection sera identifies the LDNF glycan as a potential antigen for serodiagnosis of trichinellosis. Exp. Parasitol. 129, 221–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paschinger K., Rendic D., Lochnit G., Jantsch V., Wilson I. B. (2004) Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J. Biol. Chem. 279, 49588–49598 [DOI] [PubMed] [Google Scholar]
- 29. Longmore G. D., Schachter H. (1982) Control of glycoprotein synthesis. VI. Product identification and substrate specificity studies of the GDP-l-fucose:2-acetamido-2-deoxy-β-d-glucoside (Fuc to Asn-linked GlcNAc) 6-α-l-fucosyltransferase in a Golgi-rich fraction form porcine liver. Carbohydr. Res. 100, 365–392 [DOI] [PubMed] [Google Scholar]
- 30. Titz A., Butschi A., Henrissat B., Fan Y. Y., Hennet T., Razzazi-Fazeli E., Hengartner M. O., Wilson I. B. H., Künzler M., Aebi M. (2009) Molecular basis for galactosylation of core fucose residues in invertebrates. Identification of Caenorhabditis elegans N-glycan core α1,6-fucoside β1,4-galactosyltransferase GALT-1 as a member of a novel glycosyltransferase family. J. Biol. Chem. 284, 36223–36233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paschinger K., Staudacher E., Stemmer U., Fabini G., Wilson I. B. H. (2005) Fucosyltransferase substrate specificity and the order of fucosylation in invertebrates. Glycobiology 15, 463–474 [DOI] [PubMed] [Google Scholar]
- 32. Rendić D., Klaudiny J., Stemmer U., Schmidt J., Paschinger K., Wilson I. B. H. (2007) Towards abolition of immunogenic structures in insect cells. Characterization of a honey-bee (Apis mellifera) multi-gene family reveals both an allergy-related core α1,3-fucosyltransferase and the first insect Lewis-histo-blood-group-related antigen-synthesizing enzyme. Biochem. J. 402, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iskratsch T., Braun A., Paschinger K., Wilson I. B. H. (2009) Specificity analysis of lectins and antibodies using remodeled glycoproteins. Anal. Biochem. 386, 133–146 [DOI] [PubMed] [Google Scholar]
- 34. Soderman P., Larsson E. A., Widmalm G. (2002) Synthesis of the trifucosylated N-linked hexasaccharide of a glycoprotein from Haemonchus contortus. Eur. J. Org. Chem. 2002, 1614–1618 [Google Scholar]
- 35. Hagopian A., Eylar E. H. (1968) Glycoprotein biosynthesis. Studies on the receptor specificity of the polypeptidyl:N-acetylgalactosaminyl transferase from bovine submaxillary glands. Arch. Biochem. Biophys. 128, 422–433 [DOI] [PubMed] [Google Scholar]
- 36. Weston B. W., Nair R. P., Larsen R. D., Lowe J. B. (1992) Isolation of a novel human α(1,3)fucosyltransferase gene and molecular comparison to the human Lewis blood group α(1,3/1,4)fucosyltransferase gene. Syntenic, homologous, nonallelic genes encoding enzymes with distinct acceptor substrate specificities. J. Biol. Chem. 267, 4152–4160 [PubMed] [Google Scholar]
- 37. Fazio F., Bryan M. C., Blixt O., Paulson J. C., Wong C. H. (2002) Synthesis of sugar arrays in microtiter plate. J. Am. Chem. Soc. 124, 14397–14402 [DOI] [PubMed] [Google Scholar]
- 38. Blixt O., Allin K., Bohorov O., Liu X., Andersson-Sand H., Hoffmann J., Razi N. (2008) Glycan microarrays for screening sialyltransferase specificities. Glycoconj. J. 25, 59–68 [DOI] [PubMed] [Google Scholar]
- 39. Shipp M., Nadella R., Gao H., Farkas V., Sigrist H., Faik A. (2008) Glyco-array technology for efficient monitoring of plant cell wall glycosyltransferase activities. Glycoconj. J. 25, 49–58 [DOI] [PubMed] [Google Scholar]
- 40. Šardzik R., Green A. P., Laurent N., Both P., Fontana C., Voglmeir J., Weissenborn M. J., Haddoub R., Grassi P., Haslam S. M., Widmalm G., Flitsch S. L. (2012) Chemoenzymatic synthesis of O-mannosylpeptides in solution and on solid phase. J. Am. Chem. Soc. 134, 4521–4524 [DOI] [PubMed] [Google Scholar]
- 41. Blixt O., Razi N. (2006) Chemoenzymatic synthesis of glycan libraries. Methods Enzymol. 415, 137–153 [DOI] [PubMed] [Google Scholar]
- 42. Serna S., Etxebarria J., Ruiz N., Martin-Lomas M., Reichardt N. C. (2010) Construction of N-glycan microarrays by using modular synthesis and on-chip nanoscale enzymatic glycosylation. Chemistry 16, 13163–13175 [DOI] [PubMed] [Google Scholar]
- 43. Struwe W. B., Reinhold V. N. (2012) The conserved oligomeric golgi (COG) complex is required for fucosylation of N-Glycans in C. elegans. Glycobiology 22, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morelle W., Haslam S. M., Olivier V., Appleton J. A., Morris H. R., Dell A. (2000) Phosphorylcholine-containing N-glycans of Trichinella spiralis. Identification of multiantennary lacdiNAc structures. Glycobiology 10, 941–950 [DOI] [PubMed] [Google Scholar]
- 45. Haslam S. M., Houston K. M., Harnett W., Reason A. J., Morris H. R., Dell A. (1999) Structural studies of N-glycans of filarial parasites. Conservation of phosphorylcholine-substituted glycans among species and discovery of novel chito-oligomers. J. Biol. Chem. 274, 20953–20960 [DOI] [PubMed] [Google Scholar]
- 46. Maizels R. M., Yazdanbakhsh M. (2003) Immune regulation by helminth parasites. Cellular and molecular mechanisms. Nat. Rev. Immunol. 3, 733–744 [DOI] [PubMed] [Google Scholar]
- 47. Ellis S. E., Newlands G. F., Nisbet A. J., Matthews J. B. (2012) Phage-display library biopanning as a novel approach to identifying nematode vaccine antigens. Parasite Immunol. 34, 285–295 [DOI] [PubMed] [Google Scholar]
- 48. Tawill S., Le Goff L., Ali F., Blaxter M., Allen J. E. (2004) Both free-living and parasitic nematodes induce a characteristic Th2 response that is dependent on the presence of intact glycans. Infect. Immun. 72, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Summers R. W., Elliott D. E., Qadir K., Urban J. F., Jr., Thompson R., Weinstock J. V. (2003) Trichuris suis seems to be safe and possibly effective in the treatment of inflammatory bowel disease. Am. J. Gastroenterol. 98, 2034–2041 [DOI] [PubMed] [Google Scholar]