Abstract
Reciprocal interactions between vascular endothelial cells and vascular mesenchymal cells are essential for angiogenesis. Here we show that the basic helix-loop-helix transcription factor, dHAND/Hand2, is expressed in the developing vascular mesenchyme and its derivative, vascular smooth muscle cells (VSMCs). Targeted deletion of the dHAND gene in mice revealed severe defects of embryonic and yolk sac vascular development by embryonic day 9.5. Vascular endothelial cells expressed most markers of differentiation. Vascular mesenchymal cells migrated appropriately but failed to make contact with vascular endothelial cells and did not differentiate into VSMCs. In a screen for genes whose expression was dependent upon dHAND (using subtractive hybridization comparing wild-type and dHAND-null hearts), the VEGF165 receptor, neuropilin-1, was found to be downregulated in dHAND mutants. These results suggest that dHAND is required for vascular development and regulates angiogenesis, possibly through a VEGF signaling pathway.
Introduction
Establishment of the mammalian vasculature is controlled by a complex process involving differentiation and assembly of multiple cell lineages. Rapid expansion of blood vessels is necessary for the tissue growth observed during embryonic development and tumor growth. Dysregulation of vascular development can result in excessive proliferation of vessels, as seen in hemangiomas, retinopathy of prematurity, and venous malformations (1). Because of the fundamental significance of vascular development to human biology and disease, there has been much interest in understanding the molecular mechanisms underlying the development and proliferation of vessels.
Anatomically, formation of blood vessels can be divided into 2 distinct steps. Vasculogenesis, the initial step, occurs when mesodermally derived angioblasts differentiate into vascular endothelial cells and assemble into vascular tubes (2). Subsequently, in a process known as angiogenesis, preexisting vessels sprout new branches that become remodeled into mature vessels (3). This step requires recruitment of vascular mesenchymal cells that encase endothelial tubes and differentiate into vascular smooth muscle cells (VSMCs), pericytes, and fibroblasts to promote the assembly and stabilization of the mature blood vessel wall (4). Reciprocal endothelial-mesenchymal interactions are essential for angiogenesis.
Recent studies have revealed several signaling pathways important during vasculogenesis and angiogenesis (1, 4). The secreted ligand angiopoietin-1 (Ang-1) and its receptor tyrosine kinase Tie2 (5–8), and ephrinB2 and its receptor EphB4 (9, 10), are critical for endothelial-mesenchymal interactions during angiogenesis. Within the heart, Ang-1 secretion from the endocardium is also necessary for trabeculation of the neighboring myocardium (8). TGF-β and its receptors play a role in vascular stabilization and remodeling and function through the Smad family of transcription factors (11–14). In contrast to these molecules that function mainly during angiogenesis, VEGF, a heparin-binding growth factor with high specificity for endothelial cells, is a central mediator of angiogenesis and vasculogenesis (4, 15). There are 5 isoforms produced from a single VEGF gene by alternative splicing. VEGF induces endothelial proliferation, promotes cell migration, and inhibits apoptosis (3). Inhibition of VEGF activity results in tumor regression, indicating that VEGF is necessary for the neovascularization associated with tumors (16). VEGF signals through its receptor tyrosine kinases, VEGFR-1/Flt-1 and VEGFR-2/KDR/Flk-1, which are expressed specifically on the surface of vascular endothelial cells and are required for both vasculogenesis and angiogenesis (17–20). Endothelial cells of VEGFR-1 mutant mouse embryos differentiate, but do not assemble properly into vessels (18), whereas VEGFR-2 mutants display more severe defects of vasculogenesis (17). Recently, another VEGF receptor, VEGFR-3, has been shown to be important for the remodeling and maturation of primary vascular networks into larger blood vessels (21). These data suggest that VEGF controls unique steps of vascular development through distinct receptors.
Neuropilin-1 is a specific receptor for the 165-amino acid form of VEGF (VEGF165) and is a potential regulator of VEGF-induced angiogenesis (22). Neuropilin-1 was first identified in the Xenopus tadpole nervous system (23) and found to play a role in semaphorin-mediated axonal chemorepellence (24). Neuropilin-1 is expressed in the endothelial and mesenchymal cells of the murine vasculature (25) and may mediate interactions between endothelial and mesenchymal cells. Mouse embryos overexpressing neuropilin-1 exhibit cardiovascular, peripheral nervous system (PNS) and limb defects (25). Mice deficient in neuropilin-1 have defects in PNS axonal guidance and die between embryonic day 10.5 and 12.5 from cardiovascular defects (26).
Despite recent progress in identification of signaling molecules and cell-surface receptors that regulate blood vessel formation, little is known currently about the nuclear transcriptional pathways that regulate or are regulated by the vascular signaling cascades. Tissue-specific members of the basic helix-loop-helix (bHLH) family of transcription factors are essential for development of many cell lineages (27–29), but have not been demonstrated to play a role in vascular development. The bHLH transcription factor dHAND (deciduum, heart, autonomic nervous system, neural crest-derived)/Hand2 is required for cardiac morphogenesis (30–32). Mice lacking dHAND have a single ventricle and aortic arch defects and die by embryonic day 11.0 (E11.0) from heart failure (31). Although the cardiac defects of dHAND mutants are well characterized as hypoplasia of the right ventricle segment and poor trabeculation of the remaining ventricle, the role of dHAND in development of the vasculature has not been examined previously. Here we demonstrate that dHAND is expressed in the developing vasculature and VSMCs and is required for normal vascular development. The dHAND-null endothelial cells differentiate but mesenchymal cells fail to differentiate into VSMCs. The vascular defects in dHAND-mutant embryos are similar to the angiogenic defects observed in VEGF and Flt1 mutants. In a screen to identify dHAND-dependent genes in the heart by suppressive subtractive hybridization, we found that expression of the VEGF165 receptor, neuropilin-1, was dHAND dependent in the heart and portion of the vasculature. These data indicate that dHAND plays a role in angiogenesis and may function through a VEGF signaling pathway.
Methods
Section and whole-mount in situ hybridization.
In situ hybridization to mouse embryo sections was performed with riboprobe labeled with 35S as described previously (30). Antisense riboprobe was synthesized with SP6 RNA polymerase (MAXIScript; Ambion Inc., Austin, Texas, USA) from dHAND full-length cDNA. Hybridization was performed on tissue sections of E9.5 and E10.5 mouse embryos. After hybridization, sections were washed and dipped in Kodak NTB-2 emulsion. Slides were exposed for 14 days and then developed in D19 (Eastman Kodak Co. Scientific Imaging Systems, New Haven, Connecticut, USA) for 3 minutes.
Whole-mount RNA in situ hybridizations for dHAND, Ang-1, COUP-TFII, SM22α, and neuropilin-1were performed using digoxigenin-labeled antisense riboprobes. In situ hybridizations were performed as described previously (31). Briefly, embryos were hybridized with digoxigenin-labeled riboprobes at 60°C for 18 hours. After a series of washes, embryos were incubated with alkaline phosphatase–conjugated anti-digoxigenin antibodies at room temperature for 1 hour. After another series of washes, embryos were incubated in a substrate color-reaction mixture (Roche Molecular Biochemicals, Indianapolis, Indiana, USA) for 10–12 hours in darkness. Color reaction was terminated by fixing embryos in 4% paraformaldehyde and 0.1% glutaraldehyde.
Breeding and genotyping of mice.
Mice heterozygous for the dHAND mutation were generated as described previously (31). To generate transgenic mice in a dHAND-null background, male mice heterozygous for the Tie2-lacZ transgene (33) and the SM22lacZ transgene (34) were mated to dHAND heterozygous females. Male offspring that were heterozygous for dHAND and the lacZ transgene were mated with dHAND heterozygous females to obtain embryos that expressed the lacZ transgene and were heterozygous or homozygous for the targeted dHAND allele. Mothers were sacrificed and their uteri dissected to harvest E8.0–10.5 embryos. Genomic DNA was prepared from tail biopsies or from embryonic yolk sacs and subjected to PCR and/or Southern blot for genotyping as described previously (31–34).
Immunochemistry.
Whole-mount immunochemistry was performed with mAb MEC13.3 (rat anti-mouse platelet-derived endothelial adhesion molecule; PharMingen, San Diego, California, USA), against mouse platelet endothelial cell adhesion molecule-1 (PECAM-1) (35). Embryos were fixed in 4% paraformaldehyde/PBS at 4°C overnight and dehydrated in methanol. Embryos were bleached in 5% hydrogen peroxide in methanol for 4–5 hours at room temperature, followed by rehydration and blocking in PBSMT (3% skim milk and 0.1% Triton-X-100 in PBS) at room temperature twice for 1 hour. Embryos were incubated in 10 μg/mL anti-mouse PECAM in PBSMT at 4°C overnight, washed with PBSMT at 4°C and secondary horseradish peroxidase–conjugated antibody in PBSMT at 4°C overnight. Embryos were again washed in PBSMT at 4°C and rinsed in PBST (0.1% Triton-X-100, PBS) at room temperature for 20 minutes. For signal detection, embryos were incubated in 0.3 mg/mL diaminobenzidine (Sigma Chemical Co., St. Louis, Missouri, USA) in PBST for 20 minutes, and hydrogen peroxide was added to a final concentration of 0.03% and incubated overnight. The staining reaction was terminated by rinsing the embryos in PBST. Embryos were postfixed in 4% paraformaldehyde/PBS at 4°C overnight. To better observe the vasculature of the embryos, embryos were dehydrated through graded steps into 100% methanol and cleared in a solution of benzyl benzoate/benzyl alcohol (2:1).
β- galactosidase staining.
Embryos were dissected in 4°C PBS and fixed in 2% paraformaldehyde/PBS with phenol red for 30 minutes on ice. After rinsing with PBS, embryos were incubated at room temperature overnight in 0.1% X-gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mM magnesium chloride, 0.002% NP-40, 0.01% sodium deoxycholate, PBS, pH 7.0. After staining, the embryos were rinsed in PBS and postfixed at 4°C overnight in 4% paraformaldehyde and 0.1% glutaraldehyde in PBS. Embryos were dehydrated and cleared in a solution of benzyl benzoate/benzyl alcohol (2:1).
Histology.
For histologic analysis, embryos were embedded in paraffin after fixation. Transverse and sagittal sections were made at 7-μm intervals throughout the embryos. Paraffin was cleared in xylene, and photographs of sections were taken without counterstaining to illustrate color reaction after either immunochemistry, β-gal reaction, or whole-mount in situ hybridization.
Electron microscopy.
Embryos were fixed in 2.5% glutaraldehyde in PBS at 4°C overnight and post-fixed with 1% osmium tetroxide for 1 hour. After washings, embryos were dehydrated in increasing concentrations of ethanol and infiltrated and embedded in LK-112 medium. Ultrathin transverse sections were cut with a diamond knife, mounted on 200-mesh grids, and stained with uranyl acetate and lead citrate. Sections were viewed with and photomicrographs obtained through an electron microscope.
RT-PCR.
Total RNA was purified from whole body, hearts, or yolk sacs of wild-type and dHAND-null embryos at E9.5, and adult mouse heart, aorta, and liver (Trizol; GIBCO BRL, Rockville, Maryland, USA) and A10 cell line (ATCC #CRL-1476; American Type Culture Collection, Rockville, Maryland, USA). After RT reaction, PCR analysis of several endothelial markers was performed using oligonucleotides specific for Flk-1, Tie-1, and Tie-2, described previously (17, 36), and numerous other vascular markers (primer sequences available upon request). PCR primers specific for dHAND (upper: 5′-TACCAGCTACATCGCCTACCT-3′; lower: 5′-TCACTGCTTGAGCTCCAGGG-3′) and neuropilin-1 (upper: 5′- TGAAAAATACCCCAACTG-3′; lower: 5′- CCTGAATGATGACACCTCTT-3′) were used under the following conditions: 94°C, 5 minutes; cycles of 94°C, 1 minute; 60°C, 1 minute; 72°C, 1 minute; and 72°C, 7 minutes. PCR products were analyzed after serial cycles in the linear range of amplification. RNA loading was controlled by amplification of the housekeeping gene, GAPDH. Negative controls were performed for each sample using non–reverse transcribed RNA.
Suppressive subtractive hybridization.
One microgram of total RNA, after DNase treatment, from pooled wild-type or dHAND-null E9.5 hearts was used for generation of cDNA by the SMART cDNA synthesis kit (CLONTECH Laboratories Inc., Palo Alto, California, USA). PCR-Select (CLONTECH) was used for subtractive hybridization as described by CLONTECH. Briefly, 2 pools of RsaI-digested wild-type cDNA were used as tester and ligated to unique adapters. RsaI-digested dHAND-null cDNA was used as a driver and was not ligated to any adapter sequences. Hybridization of the tester population with excess driver and subsequent amplification of subtracted species with the 2 unique adapters present on the tester cDNA resulted in a pool of PCR fragments theoretically present in wild-type, but not dHAND-null, cDNA. TA cloning (Invitrogen Corp., San Diego, California, USA) of the PCR product and subsequent DNA isolation and sequencing of selected clones was performed. Forty clones were initially sequenced. Basic local alignment search tool (BLAST) was used for searching sequence homology to DNA and expressed sequence tag (EST) databases.
Results
Vascular expression of dHAND.
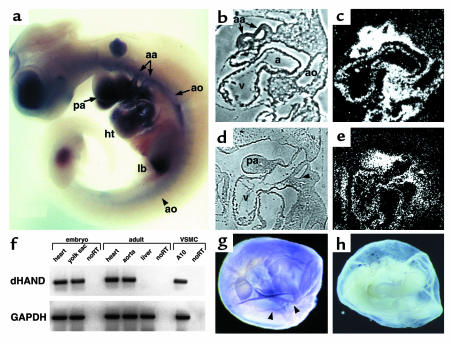
The expression of dHAND in the vasculature has not been examined in detail previously. Whole-mount in situ hybridization at E10.0 revealed dHAND expression in the aorta and aortic arch arteries in addition to the pharyngeal arches and heart (Figure 1a). Expression of dHAND was enhanced in the rostral, compared with the caudal, vasculature. Histologically, dHAND was expressed in the walls of the dorsal aorta and aortic arch arteries at E9.5 (Figure 1, b and c) and in the heart (endocardium and myocardium) and pharyngeal arches. The hybridization signal in the blood vessels was wider than the thickness of the endothelium, suggesting that the mesenchymal cells surrounding the vessels expressed dHAND. At E10.5, dHAND expression was localized to the vascular mesenchyme between the third and fourth aortic arch arteries (Figure 1, d and e), which later gives rise to VSMCs and to the mesenchyme of the pharyngeal arch. The vascular expression of dHAND was subsequently downregulated and became undetectable by in situ hybridization around E12.5. However, dHAND expression was detectable by RT-PCR in the adult mouse aorta (Figure 1f). Consistent with aortic expression of dHAND, we found that dHAND mRNA was present in a vascular cell line (A-10; ATCC #CRL-1476) that is derived from rat aortic smooth muscle cells (Figure 1f). In addition to expression in the embryo proper, dHAND was also detectable in the developing yolk sac. In situ hybridization detected dHAND expression along the yolk sac vessels during the process of remodeling at E9.5–10.0 (Figure 1, g and h). RT-PCR confirmed dHAND expression in the yolk sac at E9.5 (Figure 1f).
Figure 1.
Expression of dHAND in the developing vasculature. Whole-mount in situ hybridization revealed dHAND expression in the aortic arch arteries (aa) and aorta (ao) of an E10.0 embryo (a). Expression in the pharyngeal arches (pa), heart (ht), and limb bud (lb) was also seen. Sagittal section of an E9.5 embryo (b and c) showed expression in the aorta and aortic arch arteries. Sagittal section of an E10.5 embryo (d and e) demonstrated dHAND expression in the mesenchyme (m) of the third and fourth aortic arch arteries (arrowheads) and in the pharyngeal arch (pa) mesenchyme. b and d represent phase images of c and e, respectively. Expression of dHAND was also observed in the vascular smooth muscle cell line, A10, and in the adult heart and aorta by RT-PCR (f). RNA integrity was assayed by amplification of GAPDH. Yolk sac expression of dHAND was observed by RT-PCR in f and whole-mount in situ hybridization (arrowheads indicate vessels) (g). (h) Sense control.
Abnormal vascular development in dHAND mutants.
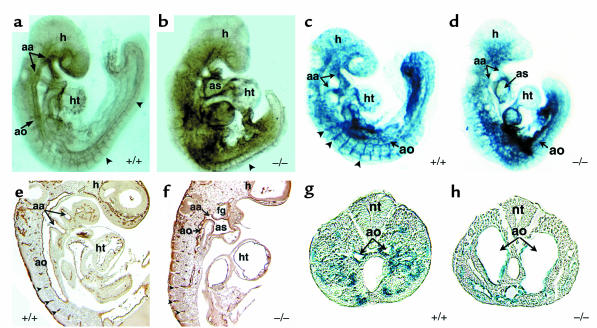
Mouse embryos homozygous-null for dHAND lack a right ventricle and have a poorly trabeculated left ventricle, resulting in death by E11.0 from heart failure (31). Although dHAND is also thought to play a role in the neural crest component of aortic arch development, we sought to determine its role during vasculogenesis and/or angiogenesis. Because heart failure might cause secondary vascular defects, all analyses of dHAND mutants were performed at E9.5, before any evidence of cardiac or growth failure. Unlike wild-type E9.5 embryos that had a grossly visible vascular pattern, dHAND mutants did not have an apparent vasculature, but rather had pooling of blood in extravascular regions of the embryo. Whole-mount immuno-chemical staining of E9.5 dHAND-null embryos using a mAb against the endothelial-specific marker PECAM-1 (Figure 2, a and b) revealed the presence of differentiated endothelial cells. However, the vasculature was grossly disorganized throughout the embryo with the more rostral vessels being more severely affected. Histological analysis of E9.5 mutant embryos showed that the aortic arch arteries were patent but the rostral aorta was poorly patterned, without an apparent lumen (Figure 2, e and f).
Figure 2.
Endothelial development in dHAND mutants. Whole-mount immunochemistry revealed that endothelial cells expressed PECAM-1 protein appropriately in wild-type (a) and mutant (b) E9.5 embryos, but displayed a disorganized pattern in dHAND mutants (b). The rostral portion of dHAND-null embryos was more severely affected than the caudal region, where the aorta (ao) and somitic arteries (arrowheads) were visible. Sagittal section of PECAM-1 antibody–stained wild-type (e) and mutant (f) embryos revealed disorganization of the dorsal aorta of dHAND-null embryos, where the aortic lumen was evident. Note patency of the aortic arch artery (aa) in the mutant (f). β-galactosidase activity in wild type (c) and mutant (d) embryos harboring lacZ under control of the Tie2 promoter revealed disorganization of lacZ expression in dHAND-null embryos compared with wild-type embryos. Transverse sections of wild-type (g) and mutant (h) embryos in the caudal region demonstrate the dilated nature of caudal vessels of dHAND-null embryos. ht, heart; as, aortic sac; h, head; nt, neural tube; fg, foregut.
To further examine endothelial development in dHAND mutants, transgenic mice containing a lacZ marker under control of the endothelial-specific Tie2 promoter (33) were crossed into the dHAND-null background. As observed with PECAM expression, Tie2 expression was intact, but vascular development was disrupted in the dHAND-null embryos at E9.5 (Figure 2, c and d). Interestingly, histologic analysis revealed that the vasculature was formed but dilated in the caudal portion of the embryo (Figure 2, g and h). The disorganization of the rostral vasculature and dilation of caudal vessels were similar to that seen in VEGF-deficient or MEF2C homozygous-null embryos, although the etiology of this observation in either model is unclear (19, 20, 37).
Because endothelial differentiation appeared relatively normal in dHAND mutants, we examined the expression of several endothelial markers, as well as other factors implicated in vasculogenesis and angiogenesis. By semiquantitative RT-PCR, transcripts of VEGF and its receptors Flk-1 and Flt-1, and Ang-1 and its receptors, Tie1 and Tie2, were detected at similar levels in wild-type and dHAND mutant embryos at E9.5 (Figure 3). Similarly, expression of the bHLH transcription factor, ARNT (38), and the transcription factor, MEF2C (37), both implicated in vascular development, were unaffected in dHAND mutants (Figure 3). These results suggest that the vascular defects of dHAND-null embryos do not result from altered expression of the factors examined, although we cannot eliminate the possibility of subtle changes in gene expression in specific regions of mutant embryos.
Figure 3.
Normal endothelial cell differentiation in dHAND-null embryos. Semiquantitative RT-PCR analysis of wild-type (+/+) and dHAND-null (–/–) embryos using primers specific to the endothelial cell markers Flk-1, Tie1, Tie2, Flt-1, and VEGF revealed equal levels of expression suggesting normal differentiation and quantity of endothelial cells in dHAND mutants. Similarly, the mesenchymal marker, Ang-1, was unaffected in dHAND mutants, as were the transcription factors MEF2C and ARNT. GAPDH expression indicates equal RNA loading.
Abnormal vascular remodeling in dHAND-null yolk sacs.
The vitelline circulation in the embryonic yolk sac represents the earliest circulatory system and is the first site of vasculogenesis and angiogenesis in the embryo. Normally, a honeycomb-like vascular plexus is evident by E8.5; subsequent remodeling of the vasculature (angiogenesis) results in defined vessels by E9.5 (Figure 4a). However, dHAND-null yolk sacs failed to form a normal vasculature at E9.5 (Figure 4b). Expression of lacZ under control of the Tie2 promoter revealed that endothelial cells were present in both wild-type and dHAND-null yolk sacs at E9.5 (Figure 4, c and d). In wild-type yolk sacs, lacZ expression demarcated the formation of large vitelline vessels and a fine network of smaller vessels (Figure 4c). In contrast, lacZ expression in yolk sacs lacking dHAND displayed a honeycomb-like plexus pattern of endothelial cells with no remodeling into a vascular network (Figure 4d). Histologic analyses revealed that although capillary-like vessels containing blood cells with an endothelial cell lining (Figure 4e) were seen in the wild-type yolk sac, no distinct blood vessels were evident in the dHAND-mutant yolk sac (Figure 4f). Large cavities containing blood cells and lined by endothelial cells (Figure 4f) were observed in dHAND mutants suggesting that vasculogenesis proceeds, but angiogenesis is blocked in the mutant yolk sac.
Figure 4.
Yolk sac vascular defects in dHAND-null embryos. Unlike wild-type yolk sacs at E9.5 (a), dHAND-null yolk sacs at E9.5 display absence of visible vessels (b). Expression of lacZ under control of the Tie2 promoter in wild-type (c) and mutant (d) yolk sacs revealed a honeycomb-like vascular plexus in dHAND-mutants rather than the remodeled vessels present in the wild-type (e) and (f) represent histologic sections of c and d, respectively. Arrowheads indicate vessels; arrows indicate endothelial cells.
Vascular mesenchyme cells fail to differentiate into VSMCs in dHAND mutants.
Mesenchymal cells begin to invade the vasculature around E9.0 and later differentiate into VSMCs. Because dHAND was expressed in vascular mesenchymal cells and VSMCs, we examined the development of vascular mesenchymal cells in dHAND mutants. Ang-1 and COUP-TFII are expressed in mesenchymal cells and play a role in angiogenesis, possibly in a common molecular pathway (9, 39). Transcripts of these 2 genes were examined in dHAND-null embryos by RT-PCR and whole-mount in situ hybridization, and no difference was found between wild-type and dHAND-null embryos (Figure 3 and Figure 5, a and b; data not shown).
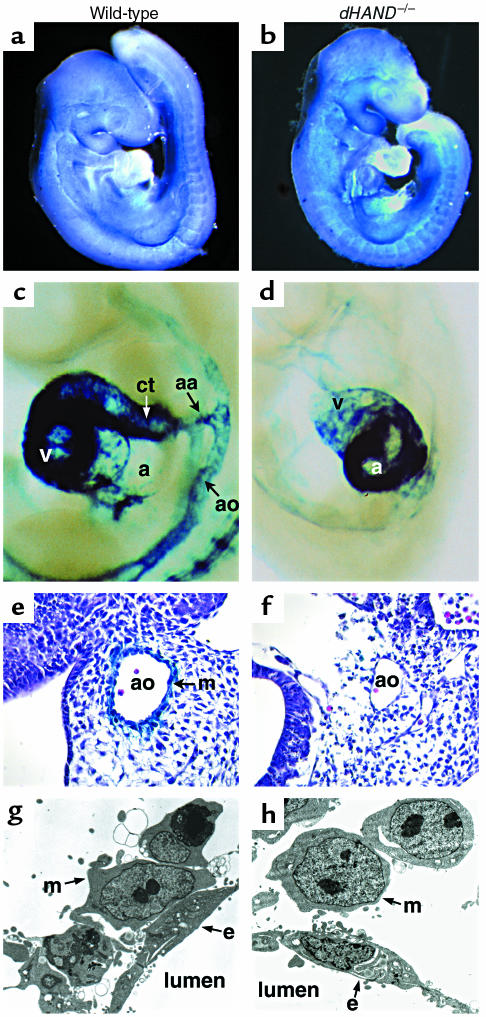
Figure 5.
Failure of vascular smooth muscle differentiation in dHAND mutants. Mesenchymal cells in wild-type (a) and dHAND mutants (b) expressed the mesenchymal marker, COUPTFII, throughout the embryo. Expression of lacZ under the control of the SM-22α promoter was seen in the aortic arch arteries (aa), aorta (ao), conotruncus (ct), and ventricle (v) of the heart of an E9.5 transgenic embryo (c). In the dHAND-null background, the SM-22-lacZ transgene expressed in the atrium (a) of the heart, but not in the vasculature (d) at E9.5. Embryos are shown in lateral views focusing on the cardiac and vascular areas. Histologic analysis of wild-type (e) and mutant (f) embryos revealed absence of lacZ expression in the cells surrounding the aorta in dHAND-null embryos. Electron microscopy of transverse sections through the rostral part of E9.5 wild-type (g) and dHAND-null (h) embryos revealed vascular mesenchymal (m) cells surrounding endothelial (e) cells. Wild-type mesenchymal cells developed cytoplasmic processes to contact endothelial cells, whereas dHAND-null mesenchymal cells remained rounded without contacting endothelial cells.
SM22α is an early marker for differentiated VSMCs and marks the developing vasculature only after vascular mesenchymal cells differentiate into VSMCs around E9.0 (40, 41). Unlike Ang-1 and COUP-TFII, SM22α was not detected in the dHAND-null vasculature by in situ hybridization (data not shown). To genetically confirm the absence of SM22α expression, transgenic mice containing a lacZ marker under control of a SM22α arterial VSMC-specific promoter (34) were crossed into the dHAND-null background. No expression of lacZ was detected in the vasculature of mice lacking dHAND at E9.5 or E10.5 (Figure 5, c and d). Ventricular expression of SM22α was not detected in dHAND mutants, although atrial expression was present (Figure 5, c and d). Histologic analysis confirmed absence of SM22α-expressing cells around the vasculature of mutants (Figure 5, e and f). These results suggest that mesenchymal cells were present in the absence of dHAND, but fail to differentiate into SM22α-expressing VSMCs.
To more accurately determine if vascular mesenchymal cells had migrated to the developing vasculature and to assess the relationship of vascular endothelial cells with mesenchymal cells, we performed electron microscopy of wild-type and dHAND-null E9.5 embryos. Transverse sections revealed that mesenchymal cells had migrated around the aorta of dHAND mutants, similar to wild-type (Figure 5, g and h). However, dHAND-null mesenchymal cells remained rounded without cytoplasmic processes that normally make contact with endothelial cells. Endothelial cells also began to appear abnormal at this stage, possibly secondary to insufficient interactions with the mesenchyme. Together, these observations suggest a role for dHAND in promoting vascular mesenchyme development and possibly endothelial-mesenchymal interactions.
Neuropilin-1 functions downstream of dHAND.
To gain insight into potential mechanisms through which dHAND might function, we sought to identify dHAND-dependent genes by subtraction cloning. Subtraction cloning was performed by suppressive-subtractive hybridization in which E9.5 dHAND-null heart cDNA was subtracted from wild-type E9.5 heart cDNA, resulting in isolation of genes that were expressed in wild-type, but not dHAND-mutant, hearts (Figure 6a). Whereas numerous genes were identified and will be described elsewhere, one of the dHAND-dependent factors represented neuropilin-1, the VEGF165 receptor (22). Downregulation of neuropilin-1 in dHAND-mutant hearts was confirmed by semiquantitative RT-PCR (Figure 6b).
Figure 6.
Neuropilin-1 is downregulated in dHAND mutants. Neuropilin-1 was isolated as a dHAND-dependent gene by subtractive hybridization between dHAND-mutant and wild-type hearts (a). Quantitative RT-PCR of neuropilin-1 in wild-type (+/+) and dHAND-null (–/–) hearts confirmed downregulation of neuropilin-1 in dHAND-mutant heart and yolk sac RNA (b). Cycles of PCR amplification are shown. RNA loading was controlled by assaying GAPDH expression. Whole-mount in situ hybridization demonstrated neuropilin-1 expression in the heart (ht) and aorta (arrowheads) of an E9.0 embryo in right lateral view (c). At E9.5 (d), neuropilin-1 expression, viewed from the left lateral side, was seen in the aorta and aortic arch arteries (arrowheads), heart, pharyngeal arch (pa), limb bud (lb), and septum transversum (st). In dHAND mutants, neuropilin-1 was downregulated in the rostral aorta, pharyngeal arches, and heart (e). Expression in the caudal aorta (arrowhead), limb bud, and septum transversum was intact. Transverse sections through the rostral part of embryos in d and e revealed neuropilin-1 expression in the endothelial (e) and mesenchymal (m) cells of the wild-type aorta (f), but no expression of neuropilin-1 in the dHAND-null aorta (g). h, head.
The expression pattern of neuropilin-1 in early embryonic development has not been described previously. In addition to its expression in the heart, neuropilin-1 mRNA was found to be most prominent in the aorta, aortic arch arteries, pharyngeal arches, and limb bud of wild-type embryos by whole-mount in situ hybridization (Figure 6, c and d). The expression of neuropilin-1 described previously in the PNS and placenta and that described here is very similar to that of dHAND expression. Because neuropilin-1 was downregulated in the hearts of dHAND mutants, we examined the spatial regulation of neuropilin-1 expression in dHAND-null embryos. Neuropilin-1 was downregulated in the ventricle of the heart, as expected (Figure 6e). Interestingly, neuropilin-1 mRNA was severely downregulated in the rostral aorta and pharyngeal arches but was still detectable in the caudal aorta, correlating with the expression pattern of dHAND. Expression in the limb buds and septum transversum was normal in dHAND mutants (Figure 6e), providing internal controls for RNA integrity within the embryo. Histologic analysis also demonstrated downregulation of neuropilin-1 expression in the rostral dHAND-null vasculature (Figure 6, f and g). Neuropilin-1 expression was also seen in the yolk sac of E9.5 wild-type embryos (Figure 6b). Transcripts of neuropilin-1 were downregulated in the dHAND-null yolk sac by semiquantitative RT-PCR (Figure 6b).
Discussion
The results in this study demonstrate that dHAND is expressed in the developing vasculature and VSMCs and plays a role in vascular development. Whereas endothelial cell differentiation and vasculogenesis were relatively unaffected in dHAND mutants, endothelial cell patterning was disrupted. Defects in angiogenesis were observed with failure of VSMC differentiation. Finally, by subtraction cloning, the VEGF165 receptor, neuropilin-1, was found to be downregulated in dHAND mutants, suggesting one potential mechanism through which dHAND may function in cardiac and vascular development.
Reciprocal interactions between vascular endothelial cells and mesenchymal cells are essential for angiogenesis and maintenance of the vasculature. Based on a model of endothelial-mesenchymal interaction proposed by Folkman and D’Amore, vascular mesenchymal cells are recruited to endothelial cells through Ang-1-Tie2 signaling, then differentiate into VSMCs or pericytes, possibly being induced by TGF-β signaling (1). In dHAND mutants, numerous markers of endothelial cell development were normally expressed and the mesenchymal cells surrounded endothelial tubes, suggesting that the step of mesenchymal recruitment is intact. This observation is consistent with the results of marker analysis showing that Ang-1, Tie2, and COUP-TFII, which is a putative upstream regulator of the Ang-1-Tie2 signaling pathway (39), are expressed normally in dHAND-null embryos. However, in the absence of dHAND, VSMC differentiation was arrested based on the lack of expression of SM22α. The failure of vascular mesenchymal cells to express the VSMC marker SM-22α could be a result of inadequate interactions with endothelial cells or could be a direct effect of dHAND transcriptional regulation. A proximal CArG box in the SM-22α promoter is essential for smooth muscle–specific gene expression, but no binding sites for bHLH proteins (E-boxes) are required for SM-22α transcription (42). Therefore, SM-22α is unlikely to be a direct target of dHAND, but more likely serves as a marker for abnormal vascular smooth muscle differentiation.
TGF-β signaling pathways are also important for angiogenesis. Mouse embryos lacking endoglin, a TGF-β–binding protein expressed on the surface of endothelial cells, have vascular defects similar to those described here. Specifically, endoglin-null embryos have poor VSMC development and arrested endothelial remodeling, whereas vasculogenesis was normal (43). Targeted deletion of a number of other genes involved in the TGF-β signaling pathways have defects in remodeling of the yolk sac vasculature (11–14). It will be interesting to determine if dHAND-related pathways and TGF-β signaling converge during vascular development.
Similar to the endothelial-mesenchymal interaction in angiogenesis, reciprocal interactions between the endocardium and myocardium are essential for cardiogenesis. Development of myocardial trabeculations is dependent upon secreted signals from the endocardium, many of which are common to the vasculature, including VEGF, Ang-1, and their respective receptors. The phenotype of dHAND-null embryos was similar to VEGF-heterozygous embryos (19), not only in the vasculature, but also in the myocardium where trabeculations failed to form. An unbiased search for genes downstream of dHAND in the heart resulted in cloning of neuropilin-1 and demonstration of neuropilin-1 downregulation in the hearts of dHAND mutants. Further evidence of neuropilin-1 downregulation in the yolk sac and other portions of the dHAND-null vasculature suggests a possible link between VEGF signaling and dHAND function in the heart and vessels. Neuropilin-1 potentiates binding of VEGF165 with Flk-1, and mice lacking neuropilin-1 die from cardiovascular defects between E10.5 and E12.5, similar to dHAND. Thus, it is tempting to speculate that regulation of neuropilin-1 is important for dHAND’s role in not only cardiac but also vascular development.
Although our data indicate that vascular defects in dHAND-null embryos may be partly mediated by dysregulation of neuropilin-1, deficiencies in many yet unknown genes downstream of dHAND likely contribute to the phenotype. In addition, we cannot formally eliminate the possibility that a subtle hemodynamic change from the cardiac defect might contribute to some part of the dHAND-null phenotype, although all our analyses were performed before any signs of cardiac failure was apparent. Hemodynamic forces (shear stress of blood flow) are important for vascular maintenance (44), but its role in early vascular development remains unclear. Classic experiments in chick embryos suggest that hemodynamic forces may not be essential for initial development of the vasculature in vivo (45). Further studies are required to elucidate how gene defects and hemodynamic changes, respectively, could contribute to defects of early vascular development.
Although little is known about transcriptional regulation of vascular development, several recent reports have described mouse mutants harboring vascular defects overlapping those described here (37–39, 46, 47). A member of the myocyte enhancer factor-2 family of transcription factors, MEF2C, is required for VSMC differentiation and vascular morphogenesis (37). Embryos lacking MEF2C or dHAND have overlapping phenotypes in the heart (48) and vasculature (37). The MyoD family of bHLH proteins and the MEF2 family of transcription factors interact with one another and function synergistically during skeletal myogenesis (49). Because dHAND, rather than MyoD family members, is expressed in smooth and cardiac muscle, it is possible that dHAND functions together with MEF2 factors during cardiovascular development. The phenotypes after genetic alteration of dHAND and MEF2C in mice is consistent with such a model warranting further studies of direct interaction among these proteins.
The studies here showed that an unbiased screen for potential mediators of dHAND function in the cardiovascular system may have provided potential insight into the mechanism through which dHAND might support cardiovascular development. Subtraction of dHAND-null heart from wild-type heart cDNA provided a tractable approach to screening for many genes in molecular pathways regulated by dHAND (50). This type of screen selects genes that may be one or several molecular steps downstream. Taking advantage of this approach, we found neuropilin-1 as one of many genes downstream of dHAND. This could be a powerful method for functional genomics and can be used for any genes that are targeted in mice. Whether neuropilin-1 is a direct or indirect target for dHAND transcriptional activation remains to be determined and awaits identification and analysis of the neuropilin-1 promoter. Given the important role of VEGF signaling pathways in development and disease, it will be interesting to determine how neuropilin-1 and other dHAND-dependent genes are involved in vascular maintenance during embryogenesis, tumorigenesis, and other pathologic conditions.
After submission of this manuscript, a detailed analysis of the vascular defects in neuropilin-1 mutant embryos was reported (Kawasaki, T., et al. 1999. A requirement of neuropilin-1 in embryonic vessel formation. Development. 126:4895–4902). The vascular abnormalities overlapped those described here and those observed in endothelin-1 mutants where dHAND is downregulated.
Acknowledgments
The authors thank T. Sato for use of Tie2-lacZ transgenic mice and helpful discussions, X. Chao for technical assistance, S. Tsai for the COUPTFII probe, Michael Bennett for electron microscopy, and S. Johnson for manuscript preparation and graphics. This work was supported by grants from the NIH/NHLBI (R01 HL5781-01 and R01 HLDE62591-01) and March of Dimes to D. Srivastava.
References
- 1.Folkman J, D’Amore PA. Blood vessel formation: what is its molecular basis? Cell. 1996;87:1153–1155. doi: 10.1016/s0092-8674(00)81810-3. [DOI] [PubMed] [Google Scholar]
- 2.Risau W, Flamme I. Vasculogenesis. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 3.Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D. Signaling vascular morphogenesis and maintenance. Science. 1997;277:48–50. doi: 10.1126/science.277.5322.48. [DOI] [PubMed] [Google Scholar]
- 5.Dumont DJ, et al. Dominant-negative and targeted null mutations in the endothelial receptor tyrosine kinase, tek, reveal a critical in vasculogenesis of the embryo. Genes Dev. 1994;8:1897–1909. doi: 10.1101/gad.8.16.1897. [DOI] [PubMed] [Google Scholar]
- 6.Sato TN, et al. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature. 1995;376:70–74. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 7.Davis S, et al. Isolation of angiopoietien-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell. 1996;87:1161–1169. doi: 10.1016/s0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 8.Suri C, et al. Requisite role of angiopoietin-1, a ligand for the Tie2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang HU, Chen Z, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- 10.Adams RH, et al. Roles of ephrinB ligands and EphB receptors in cardiovascular development: demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev. 1999;13:295–306. doi: 10.1101/gad.13.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson MC, et al. Defective haematopoiesis and vasculogenesis in transforming growth factor-beta 1 knock out mice. Development. 1995;121:1845–1854. doi: 10.1242/dev.121.6.1845. [DOI] [PubMed] [Google Scholar]
- 12.Oshima M, Oshima H, Taketo MM. TGF-beta receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev Biol. 1996;179:297–302. doi: 10.1006/dbio.1996.0259. [DOI] [PubMed] [Google Scholar]
- 13.Chang H, et al. Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development. 1999;126:1631–1642. doi: 10.1242/dev.126.8.1631. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, et al. Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development. 1999;126:1571–1580. doi: 10.1242/dev.126.8.1571. [DOI] [PubMed] [Google Scholar]
- 15.Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999;13:9–22. [PubMed] [Google Scholar]
- 16.Kim KJ, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–844. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 17.Shalaby F, et al. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 18.Fong GH, Rossant J, Gertsenstein M, Breitman ML. Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature. 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 19.Carmeliet P, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 20.Ferrara N, et al. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 21.Dumont DJ, et al. Cardiovascular failure in mouse embryos deficient in VEGF receptor-3. Science. 1998;282:946–949. doi: 10.1126/science.282.5390.946. [DOI] [PubMed] [Google Scholar]
- 22.Soker S, Takashima S, Miao HQ, Neufeld G, Klagsburn M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell. 1998;92:735–745. doi: 10.1016/s0092-8674(00)81402-6. [DOI] [PubMed] [Google Scholar]
- 23.Takagi S, Tsuj T, Amagai T, Takamatsu T, Fujisawa H. Specific cell surface labels in the visual centers of xenopus laevis tadpole identified using monoclonal antibodies. Dev Biol. 1987;122:90–100. doi: 10.1016/0012-1606(87)90335-6. [DOI] [PubMed] [Google Scholar]
- 24.He Z, Tessier-Lavigne M. Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell. 1997;90:739–751. doi: 10.1016/s0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- 25.Kitsukawa T, Shimono A, Kawakami A, Kondoh H, Fujisawa H. Overexpression of a membrane protein, neuropilin, in chimeric mice causes anomalies in the cardiovascular system, nervous system and limbs. Development. 1995;121:4309–4318. doi: 10.1242/dev.121.12.4309. [DOI] [PubMed] [Google Scholar]
- 26.Kitsukawa T, et al. Neuropilin-semaphorin III/D-mediated chemorepulsive signals play a crucial role in peripheral nerve projection in mice. Neuron. 1997;19:995–1005. doi: 10.1016/s0896-6273(00)80392-x. [DOI] [PubMed] [Google Scholar]
- 27.Jan YN, Jan LY. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- 28.Olson EN, Klein WH. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiogenesis. Science. 1995;70:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava D, et al. Regulation of cardiac mesoderm and neural crest by the bHLH protein, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 32.Thomas T, Yamagishi H, Overbeek PA, Olson EN, Srivastava D. The bHLH factors, dHAND and eHAND, specify pulmonary and systemic cardiac ventricles independent of left-right sidedness. Dev Biol. 1998;196:228–236. doi: 10.1006/dbio.1998.8849. [DOI] [PubMed] [Google Scholar]
- 33.Schlaeger TM, Qin Y, Fujiwara Y, Magram J, Sato TN. Vascular endothelial cell lineage-specific promoter in transgenic mice. Development. 1995;121:1089–1098. doi: 10.1242/dev.121.4.1089. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Miano JM, Mercer B, Olson EN. Expression of the SM22 promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol. 1996;132:1–11. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldwin HS, et al. Platelet endothelial cell adhesion molecule-1 (PECAM-1/CD31): alternatively spliced, functionally distinct isoforms expressed during mammalian cardiovascular development. Development. 1994;120:2539–2553. doi: 10.1242/dev.120.9.2539. [DOI] [PubMed] [Google Scholar]
- 36.Iwama A, et al. Molecular cloning and characterization of mouse TIE and TEK receptor tyrosine kinase genes and their expression in hematopoietic stem cells. Biochem Biophys Res Commun. 1993;195:301–309. doi: 10.1006/bbrc.1993.2045. [DOI] [PubMed] [Google Scholar]
- 37.Lin Q, et al. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- 38.Maltepe E, Schmidt JV, Baunoch D, Bradfield CA, Simon CM. Abnormal angiogenesis and response to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature. 1997;386:403–407. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- 39.Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solway J, et al. Structure and expression of a smooth muscle cell-specific gene, SM22 alpha. J Biol Chem. 1995;270:13460–13469. doi: 10.1074/jbc.270.22.13460. [DOI] [PubMed] [Google Scholar]
- 41.Li L, Miano JM, Cserjesi P, Olson EN. SM22 alpha, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ Res. 1996;78:188–195. doi: 10.1161/01.res.78.2.188. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Liu Z-C, Mercer M, Overbeek P, Olson EN. Distinct serum response factor-mediated regulatory networks governing SM22 transcription in smooth, skeletal and cardiac muscle cells. Dev Biol. 1997;187:311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 43.Li DY, et al. Defective angiogenesis in mice lacking endoglin. Science. 1999;284:1534–1537. doi: 10.1126/science.284.5419.1534. [DOI] [PubMed] [Google Scholar]
- 44.Papadaki M, Eskin SG. Effects of fluid shear stress on gene regulation of vascular cells. Biotechnol Prog. 1997;13:209–221. doi: 10.1021/bp970029f. [DOI] [PubMed] [Google Scholar]
- 45.Chapman WB. The effect of the heart-beat upon the development of the vascular system in the chick. Am J Anat. 1918;23:175–203. [Google Scholar]
- 46.Kuo CT, et al. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 48.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 50.Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D. A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science. 1999;283:1158–1161. doi: 10.1126/science.283.5405.1158. [DOI] [PubMed] [Google Scholar]