Abstract
We show a platform that merges a microfluidic chip with lensless imaging for CD4+ T-lymphocyte counting at resource-limited settings. To capture CD4+ T lymphocytes, anti-CD4 antibody was immobilized on a microfluidic chip. The captured cells were detected by a charge coupled device (CCD) sensor using lensless shadow imaging techniques. Gray scale shadow images of captured cells on the chip (24 mm × 4 mm × 50 μm) were enumerated in three seconds using an automatic cell counting software. The device achieved 70.2 ± 6.5% capture efficiency, 88.8 ± 5.4% capture specificity for CD4+ T-lymphocytes, 96 ± 1.6% CCD efficiency, and 83.5 ± 2.4% overall platform performance (n = 9 devices). This integrated platform has potential for point-of-care testing (POCT) to rapidly capture, image and count specific cell types from unprocessed whole blood.
I. Introduction
HIV remains the most serious infectious disease challenge to public health. As a result of inadequate access to HIV prevention and treatment, everyday, more than 6800 people contract HIV and more than 5700 people die from AIDS, globally[1]. In 2007, worldwide, 33.2 million people had HIV. 2.5 millions of these people were newly infected and 2.1 million died from AIDS. There is a lack of available monitoring technologies at resource limited settings. Blood cell isolation[2, 3] and enumeration methods are used to monitor progress of infectious diseases, such as HIV/AIDS[4]. Both CD4+ T-lymphocyte and viral load counts of patients have been used to monitor and initiate treatment of HIV disease using antiretroviral therapy (ART). ART is started for infected persons with CD4+ T-lymphocyte counts below 200 ~ 350 cells per microliter[4].The CD4+ T-lymphocyte count is performed currently 3 ~ 4 times a year in the developed world, and twice a year in the developing world using fluorescent activated cell count and sorting systems (FACS)[5].
In resource limited settings, current advanced technologies such as FACSCount (Becton Dickinson, CA, USA) face a significant challenge to monitor or count thousands of cells because of equipment costs ($27,000), reagent costs ($5 ~ 20), limited throughput (30 ~ 50 samples/day), need for an experienced operator, and maintenance costs[6]. There is a need for rapid diagnostic and monitoring systems that are simple-to-use, inexpensive, reliable, and disposable enhancing current monitoring methods. There have been ideas to create smaller flowcytometers targeting global health and point-of-care applications with limited functionality such as Guava EasyCD4 assay[7]. However, the equipment costs about $35,000. There is a need to lower these costs even further for developing countries. Simple microfluidic approaches merged with rapid detection and counting could provide new avenues in this field.
Microfluidic chip and detection platform for infectious diseases could impact current global health problems[8]. These technologies could provide ease of use and minimal sample preparation steps for point-of-care testing (POCT)[9, 10]. To use microfluidic approaches to count CD4+ T-lymphocytes from whole blood, three significant challenges need to be addressed: (1) Capture and isolation of CD4+ cells from whole blood with a microfluidic chip in a high throughput manner. (2) Detecting the captured cells rapidly. We choose to develop a lensless CD4 cell detection system which we address in this paper. (3) Merging the microfluidic chip with a wide field-of-view (FOV) lensless imaging technology, and (4) rapid automated counting of cells from captured images. As a solution to the first challenge, it was earlier demonstrated that CD4+ T lymphocytes can be captured selectively from whole blood using microfluidic channels by fluorescent labeling or label-free techniques[11-13]. CD4+ T-lymphocytes can be captured from whole blood either by mechanical filtering[13], or employing polydimethylsiloxane (PDMS) microfluidic devices with anti-CD4 antibody immobilization on channel surfaces[11]. These systems employ disposable microfluidic devices, but they require fluorescent labeling or long hours of counting under a microscope to determine CD4+ T-lymphocyte counts. Both approaches could be difficult to adopt for POCT at resource limited settings, since whole process needs to be performed in minutes rather than hours to be useful. This poses the following challenges to detect and quantify cells rapidly. As a first step to achieve the second challenge[14], cells free floating in microfluidic channels and placed between glass slides were detected by lensless cell shadow imaging. The key unresolved step that we demonstrate in this paper is to merge a properly fabricated microfluidic chip that can capture CD4 T cells from blood with a lensless imaging system that can detect captured cells in a channel. As a result, this platform provides a solution to capture/detect CD4+ T-lymphocytes from blood samples. The last challenge is to rapidly enumerate the detected cells captured on the whole chip, which is performed by an automatic cell counting software. In this paper, we attack these challenges that are keys to develop a label-free lensfree CD4 T cell counting platform targeting resource limited settings.
Targeting the detection and counting challenges, there have been efforts to use electrodes integrated into microfluidic channels to indirectly quantify the number of cells using cell lysate electrical impedance[15] or using a local electric current change[16]. However, these methods may suffer from patient-to-patient variation, multiple wash steps, and low signal-to-noise levels. Instead of these indirect methods, recently direct cell detection methods through wide FOV have been developed such as lensless systems to image cells, (LUCAS: a lensless, ultra wide-field cell monitoring array platform based on shadow imaging)[14], to track Caenorhabditis elegans motion[17], and to detect antibody binding[18]. To have a platform system that achieves multiple functionalities of capture/detection/counting with whole blood comprises various additional integration difficulties such as, signal to noise ratio (SNR).
We focus on developing an integrated platform targeting point-of-care applications at resource-limited settings to count CD4+ T-lymphocytes from whole blood. We also present a method that creates microfluidic chips without using expensive photo-lithographical approaches. We demonstrate challenges and provide solutions to create an integrated platform that achieves sequentially; (i) selective rare cell capture on a microfluidic chip, (ii) detection of captured cells rapidly by a lensless CCD imaging platform, and (iii) automated cell counting to create an inexpensive system to enumerate CD4+ T-lymphocytes from blood samples.
We designed our chips as shown in fig. 1 and fabricated simply with a laser cutter instead of using expensive cleanroom equipment. Anti-CD4 antibody was immobilized on only one surface of the chip to capture the CD4 T lymphocytes. The microfluidic chip can be directly imaged with the CCD imaging platform and cells can be counted by automatic cell counting software. These steps take less than a minute. To image shadow patterns of captured cells with the CCD image sensor (KODAK, KAI-11002, Rochester, NY), the microchip was placed on the CCD surface. One gray color image of the entire channel surface was taken in one second. The sensor features more than 11 million square pixels (9 μm wide), across the active sensor array area, 37.25 mm × 25.70 mm, fig. 2. The large dimensions of the KODAK CCD chip allowed us to use commercially available microscope cover slides (24 mm × 35 mm × 0.10 mm). The white light, emitted by a halogen lamp (Micro-Lite, FL3000, Three Rivers, MA) with an annual light guide, passes through the PMMA cover and reaches the captured cells. A point white light source can be assumed as planar light source, if the light source is set up far from an object, i.e. a cell (fig 2a). Light intensity of a cell shadow image is determined by diffraction, which can be calculated by Rayleigh-Sommerfeld diffraction integral[14]. Fresnel number (N = πD2/4zλ) was used for determining whether our system was in diffraction (N > 1) or Fraunhofer region (N < 1). Our system operates in the Fraunhofer region (N ≈ 0.1) with the following conditions: cell diameter (D = 10 μm), distance (z = 1.4 mm), and wavelength of light source (λ = 400 ~ 700 nm, white light). Although we operate in this region, the shadow image can be recognized by a CCD sensor. Figure 2c shows the entire channel image captured by a CCD after the cell capture process. Enlarged figure shows that the dark rings are from diffracted light forming the shadow cell images. When the distance between the cell and CCD surface was increased, the ring diameter of shadow image was enlarged. This effect was observed until SNR between the shadow and background light intensity reached to the detection limit of CCD pixels. Higher z values for our system (1.4 mm) is more beneficial for low CCD resolution, since it exaggerates the shadow image to larger than the actual cell size. An automatic cell counting software can resolve a threshold signal level, which determines the boundaries between cell membranes and background[19]. The CCD images were analyzed to count cells and characterize a distribution of captured cells as a function of distance from the channel inlet using the public domain NIH Image program.
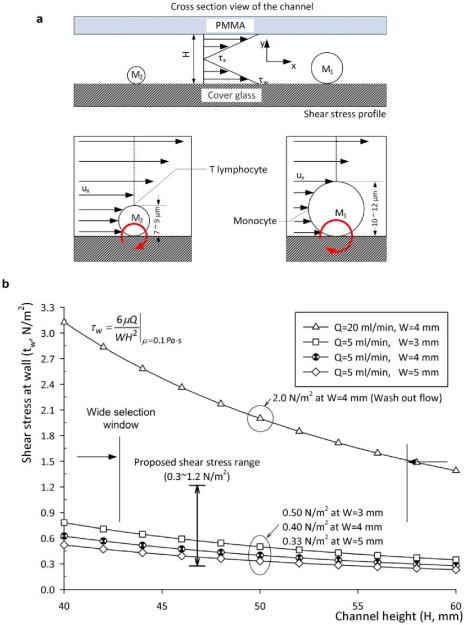
Fig. 1.
Schematic view of the mechanical cell filtration method and shear stress at the fluidic channel floor based on channel geometry and flow rate. (a) Cell selection method in a microfluidic channel and design parameters. The subscript 1 and 2 is used for monocytes and T-lymphocytes, respectively. Cell sizes are based on microvilli and ruffles are estimated from SEM images. The moment ratio between monocytes and T-lymphocytes is derived from: PxA1d1/PxA2d2 = (d1/d2)3 = 2.37, for 12 μm and 9 μm cell diameters, respectively. The area, A, and diameter, d represent cross sectional area and diameter of each cell. Px represents pressure at a distance x from the channel entrance. It has a uniform magnitude as a function of y. The height, H, is the fluidic channel height. Fluid velocity and shear stress as a function of x, and shear stress at wall are described by Ux, τx, and τw, respectively. (b) Calculated shear stress as a function of channel height for 5 μl/min and 20 μl/min volumetric flow rate. Shear stress range of 0.3 ~ 1.2 N/m2 was chosen to maximize cell attachment and 2 N/m2 to achieve mechanical filtration. The design parameters were 4 mm (W) × 50 μm (H) and flow rate, 5 μl/min and 20 μl/min.
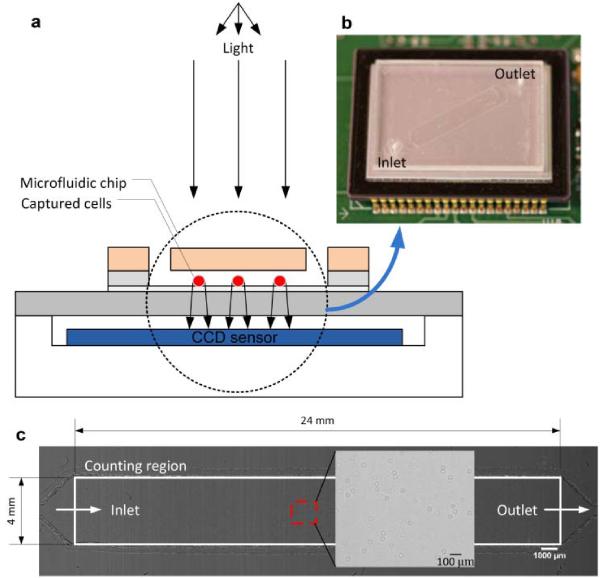
Fig. 2.
A schematic view of the CCD imaging platform: (a) CCD imaging platform to detect the captured cells. When light is incident on the captured cells, the cell membrane diffracts and transmits light. A shadow of the captured CD4+ T-lymphocytes generated by diffraction can be imaged by the CCD in one second. Image is obtained with the lens-less CCD imaging platform. (b) Picture of microfluidic chip and CCD imaging platform. Field of view of the CCD sensor is 35 mm × 25 mm. The entire microfluidic device can be imaged without alignment by simply placing the microfluidic channel on the sensor. (c) Image taken with the lens-less CCD imaging platform and magnified view at the microfluidic channel centre is shown. The magnified picture shows an image obtained by diffraction. Scale bar, 100 μm.
II. Results
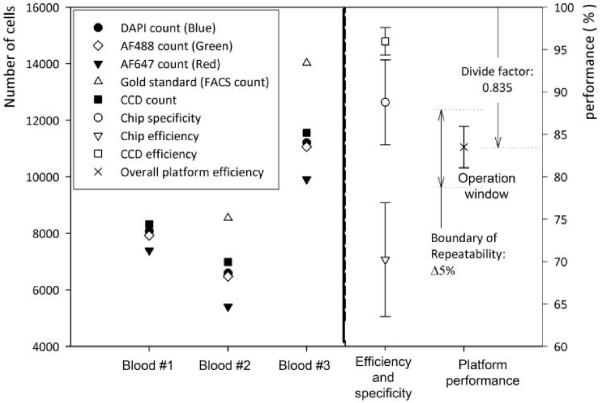
Figure 3 show microfluidic chip capture specificity, capture efficiency, CCD efficiency, and overall platform performance. The average of nine devices was used to evaluate the efficiency and specificity of the CCD imaging platform. The chip specificity and efficiency are related to the surface chemistry and shear based mechanical filtration methods. The average value of the three blood samples shows 88.8 ± 5% capture specificity for CD4+ cells and 70.2 ± 6.7% capture efficiency (n = 9 devices).
Fig. 3.
Efficiency of the microfluidic chip and the CCD imaging platform.
The CCD efficiency was obtained by the ratio of CCD count and all captured cells, (CCD efficiency = CCD count / blue stained cell count). It indicates CCD imaging efficiency based on signal to noise ratio of imaged cells. We observed that CCD efficiency is 96 ± 1.6%. This high efficiency shows that the shadow diffraction image gives a sufficiently high signal to noise ratio.
CD4+ cell capture in the channel can be performed in less than 10 minutes including all the steps. After the cell capture step, it takes less than 20 seconds to get the CD4+ cell counts using the CCD sensor system (one second to capture the whole image of cells in the microfluidic channel, three seconds to run the automated cell counting software). This time budget is based on our experimental data. The enumeration time may be further reduced by employing smaller sample volumes (e.g., 5 μl of whole blood) without sacrificing the CD4 counting accuracy. Such a rapid CD4 count allows high throughput at resource-limited settings, when compared to existing systems (e.g., magnetic beads: 5 ~ 10 test per day) and flow cytometry (30 ~ 50 test per day, including incubation times for fluorescent cell staining)[20].
Further, this overall platform performance was defined by the ratio of CCD image count and absolute number of target cells obtained from gold standard, i.e. flow cytometry, (overall platform performance = CCD image count / gold standard). ‘Overall platform performance’ is the key descriptor of this device, since the CCD count “all cells” would be clinically used to estimate the CD4+ cell count. The overall platform performance was 83.5 ± 2.44 %. The repeatable performance and small standard deviation, ± 2.44 %, allow correcting for the length dependent count bias (in this case we divide by 0.835). The corrected CD4+ cell count estimate is clinically acceptable (± 10 % overall count error)[6].
III. Conclusions
We presented a novel method to build a point-of-care device that is merged with lensless imaging for rapid cell counting, i.e. CD4 counts. The lensless CCD imaging platform merged with label-free cell capture has potential for resource-limited settings, since it eliminates the need for fluorescent imaging; it reduces the time for cell capture, imaging, and counting to a few minutes from hours. The integrated platform to merge the microchip with the CCD system was successful to capture, image and automatically count the CD4+ T-lymphocytes. This integrated system poses a promising future direction for point-of-care testing especially focusing on global health applications at resource limited settings.
Acknowledgment
Dr. Demirci would like to thank the Wallace H. Coulter Foundation for the Young Investigator in Bioengineering Award and CIMIT (Center for Integration of Medicine & Innovative Technology). The authors acknowledge NIH grant RO1 AI081534. Dr. Kuritzkes would like to acknowledge NIH grants RR016482 and AI060354 (Harvard University Center for AIDS Research).
References
- [1].WHO . 2007 UNAIDS annual report: Knowing your epidemic. Joint United Nations Programme on HIV/AIDS (UNAIDS); GENEVA: 2008. [Google Scholar]
- [2].Gascoyne P, Mahidol C, Ruchirawat M, Satayavivad J, Watcharasit P, Becker FF. Microsample preparation by dielectrophoresis: isolation of malaria. Lab Chip. 2002 May;2:70–5. doi: 10.1039/b110990c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shelby JP, White J, Ganesan K, Rathod PK, Chiu DT. A microfluidic model for single-cell capillary obstruction by Plasmodium falciparum-infected erythrocytes. Proceedings of the National Academy of Sciences. 2003 Dec 9;100:14618–22. doi: 10.1073/pnas.2433968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hammer SM, Eron JJ, Jr., Reiss P, Schooley RT, Thompson MA, Walmsley S, Cahn P, Fischl MA, Gatell JM, Hirsch MS, Jacobsen DM, Montaner JSG, Richman DD, Yeni PG, Volberding PA. Antiretroviral Treatment of Adult HIV Infection: 2008 Recommendations of the International AIDS Society-USA Panel. JAMA. 2008 Aug 6;300:555–570. doi: 10.1001/jama.300.5.555. 2008. [DOI] [PubMed] [Google Scholar]
- [5].WHO . Patient Monitoring Guidelines for HIV Care and Antiretroviral Therapy. WHO; GENEVA: 2008. [Google Scholar]
- [6].WHO . CD4+ T-Cell Enumeration Technologies; Technical Information. WHO; GENEVA: 2008. [Google Scholar]
- [7].Spacek LA, Shihab HM, Lutwama F, Summerton J, Mayanja H, Kamya M, Ronald A, Margolick JB, Nilles TL, Quinn TC. Evaluation of a low-cost method, the Guava EasyCD4 assay, to enumerate CD4-positive lymphocyte counts in HIV-infected patients in the United States and Uganda. Journal of acquired immune deficiency syndromes. 2006 Apr 15;41:607–10. doi: 10.1097/01.qai.0000214807.98465.a2. [DOI] [PubMed] [Google Scholar]
- [8].Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annual review of biomedical engineering. 2008;10:107–44. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- [9].Chin CD, Linder V, Sia SK. Lab-on-a-chip devices for global health: Past studies and future opportunities. Lab on a Chip. 2007;7:41–57. doi: 10.1039/b611455e. [DOI] [PubMed] [Google Scholar]
- [10].Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- [11].Cheng X, Irimia D, Dixon M, Sekine K, Demirci U, Zamir L, Tompkins RG, Rodriguez W, Toner M. A microfluidic device for practical label-free CD4+ T cell counting of HIV-infected subjects. Lab on a Chip. 2007;7:170–178. doi: 10.1039/b612966h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cheng X, Irimia D, Dixon M, Ziperstein JC, Demirci U, Zamir L, Tompkins RG, Toner M, Rodriguez WR. A microchip approach for practical label-free CD4+ T-cell counting of HIV-infected subjects in resource-poor settings. Journal of Acquired Immune Deficiency Syndromes. 2007 Jul 1;45:257–61. doi: 10.1097/QAI.0b013e3180500303. [DOI] [PubMed] [Google Scholar]
- [13].Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, Hsiang M, Peter T, Zavahir S, Thior I, Romanovicz D, Bernard B, Goodey AP, Walker BD, McDevitt JT. A Microchip CD4 Counting Method for HIV Monitoring in Resource-Poor Settings. PLoS Medicine. 2005 Jul 01;2:663–672. doi: 10.1371/journal.pmed.0020182. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ozcan A, Demirci U. Ultra wide-field lens-free monitoring of cells on-chip. Lab on a Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- [15].Cheng X, Liu Y.-s., Irimia D, Demirci U, Yang L, Zamir L, Rodriguez WR, Toner M, Bashir R. Cell detection and counting through cell lysate impedance spectroscopy in microfluidic devices. Lab on a Chip. 2007;7:746–755. doi: 10.1039/b705082h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Y-N, Kang Y, Xu D, Chon CH, Barnett L, Kalams SA, Li D, Li D. On-chip counting the number and the percentage of CD4+ T lymphocytes. Lab on a Chip. 2008;8:309–315. doi: 10.1039/b713932b. [DOI] [PubMed] [Google Scholar]
- [17].Cui X, Lee LM, Heng X, Zhong W, Sternberg PW, Psaltis D, Yang C. Lensless high-resolution on-chip optofluidic microscopes for Caenorhabditis elegans and cell imaging. Proceedings of the National Academy of Sciences. 2008;105:10670–10675. doi: 10.1073/pnas.0804612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ozkumur E, Needham JW, Bergstein DA, Gonzalez R, Cabodi M, Gershoni JM, Goldberg BB, Unlu MS. Label-free and dynamic detection of biomolecular interactions for high-throughput microarray applications. Proceedings of the National Academy of Sciences. 2008 Jun 10;105:7988–7992. doi: 10.1073/pnas.0711421105. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Seo S, Su T-W, Erlinger A, Ozcan A. Multi-color LUCAS: Lensfree On-chip Cytometry Using Tunable Monochromatic Illumination and Digital Noise Reduction. Cellular and Molecular Bioengineering. 2008;1:146–156. [Google Scholar]
- [20].Paltiel AD, Weinstein MC, Kimmel AD, Seage GR, III, Losina E, Zhang H, Freedberg KA, Walensky RP. Expanded Screening for HIV in the United States -- An Analysis of Cost-Effectiveness. New England Journal of Medicine. 2005 Feb 10;352:586–595. doi: 10.1056/NEJMsa042088. 2005. [DOI] [PubMed] [Google Scholar]