Abstract
The first and the rate-limiting enzyme of heme biosynthesis is δ-aminolevulinate synthase (ALAS), which is localized in mitochondria. There are 2 tissue-specific isoforms of ALAS, erythroid-specific (ALAS-E) and nonspecific ALAS (ALAS-N). To identify possible mitochondrial factors that modulate ALAS-E function, we screened a human bone marrow cDNA library, using the mitochondrial form of human ALAS-E as a bait protein in the yeast 2-hybrid system. Our screening led to the isolation of the β subunit of human ATP-specific succinyl CoA synthetase (SCS-βA). Using transient expression and coimmunoprecipitation, we verified that mitochodrially expressed SCS-βA associates specifically with ALAS-E and not with ALAS-N. Furthermore, the ALAS-E mutants R411C and M426V associated with SCS-βA, but the D190V mutant did not. Because the D190V mutant was identified in a patient with pyridoxine-refractory X-linked sideroblastic anemia, our findings suggest that appropriate association of SCS-βA and ALAS-E promotes efficient use of succinyl CoA by ALAS-E or helps translocate ALAS-E into mitochondria.
Introduction
The first and the rate-limiting enzyme of the heme biosynthetic pathway is δ-aminolevulinate synthase (ALAS). The enzyme requires glycine and succinyl coenzyme A (CoA) as substrates, and pyridoxal 5′-phosphate as a cofactor (1, 2). There are 2 isoforms of ALAS in vertebrates, erythroid-specific (ALAS-E) and nonspecific ALAS (ALAS-N), which are encoded by separate genes (3). Human genes encoding ALAS-N (ALAS1) and ALAS-E (ALAS2) have been mapped to chromosomal regions 3p21.1 (4) and Xp11.21 (5, 6), respectively. ALAS-N is expressed ubiquitously in all tissues, whereas ALAS-E is expressed exclusively in bone marrow erythroid cells. ALAS-N undergoes marked induction when hepatic drug metabolism is enhanced, whereas ALAS-E induction occurs when uncommitted stem cells are induced to undergo erythroid cell differentiation (7). Thus ALAS-N induction is necessary for the synthesis of microsomal cytochrome P450 in the liver, whereas ALAS-E induction is required for the synthesis of ALA for hemoglobin formation in erythroid cells (8, 9). It has been shown that both ALAS isozymes are regulated at several levels, including transcription (10, 11), translation (12), translocation into mitochondria (13–15), and the catalytic activity of the enzyme (16). Additionally, each ALAS isozyme is regulated in a tissue-specific manner (17), suggesting that there is an exquisitely fine mode of control for ALAS synthesis in each tissue (18).
Sideroblastic anemia (SA) is a group of heterogeneous disorders that are characterized by hypochromic and microcytic anemia, with a marked increase in iron-laden erythroblasts (known as ringed sideroblasts) in the bone marrow (19, 20). Many patients with inherited SA are male; thus they are termed X-linked SA (XLSA). It has been demonstrated that patients with XLSA have decreased ALAS activity in bone marrow erythroblasts and a mutation in the ALAS2 gene (21–30), indicating that a mutation in ALAS2 underlies this disorder. Many patients with XLSA respond to pyridoxine treatment, suggesting that ALAS2 mutation may involve the pyridoxal 5′-phosphate–binding site in ALAS-E, whereas some patients show no response to pyridoxine treatment. For example, the D190V mutation identified in a patient with pyridoxine-refractory XLSA led to abnormal processing of the NH2-terminal presequence of ALAS-E, resulting in an unstable enzyme protein (30). These findings suggest that aberrant intramitochondrial processing of the presequence of ALAS-E may be involved in some cases of pyridoxine-refractory XLSA.
Succinyl CoA, 1 of the 2 substrates for ALAS reaction, is formed exclusively within mitochondria by succinyl CoA synthetase (SCS). The enzyme catalyzes phosphorylation of GDP and ADP (31). The enzyme also catalyzes a reverse reaction: the synthesis of succinyl CoA from succinate and CoA, which is used for ALA synthesis and activation of ketone bodies (32). There are 2 isoforms of SCS, the GTP-specific (G-SCS) and ATP-specific isoforms (A-SCS) (33). Recently, it has been shown that the 2 isoforms of SCS contain the identical α subunit, although they differ in their β subunits, indicating that the β subunit determines the specificity of nucleotide binding (33, 34).
Using the yeast 2-hybrid system, we examined possible association of human ALAS-E with a factor that may influence the level and activity of ALAS-E in mitochondria. Our findings demonstrated that there is specific association of human ALAS-E with the human β subunit of A-SCS (hSCS-βA). In addition, our findings suggest that this association may be an important process in normal mitochondria, and its failure may be responsible for some cases of pyridoxine-refractory XLSA.
Methods
Chemicals and enzymes.
All chemicals were obtained from Sigma Chemical Co. (St. Louis, Missouri, USA). Restriction enzymes and modifying enzymes were obtained from New England Biolabs Inc. (Beverly, Massachusetts, USA). DNA polymerase for PCR (Advantage 2 cDNA Polymerase Mix) was obtained from CLONTECH Laboratories Inc. (Palo Alto, California, USA).
Yeast 2-hybrid assay.
All vectors and yeast strains that were used for yeast 2-hybrid assays were available in the MATCHMAKER Two-Hybrid System 2 (CLONTECH Laboratories Inc.). This is a complete GAL4-based 2-hybrid system that provides a transcriptional assay for detecting specific protein-protein interactions in yeast. The pAS2-1 vector is used for expressing protein fused with the GAL4 DNA-binding domain, whereas pACT2 vector is used for expressing protein fused with the GAL4 DNA-activation domain. Because both pAS2-1 and pACT2 have a nutritional marker for selection (tryptophan for pAS2-1 and leucine for pACT2), we used a minimal synthetic dropout medium (SD medium) that lacked tryptophan and leucine for selection of transformants. Because the association of each protein activates the HIS3 reporter gene that has been integrated into the genome of the Y190 strain, all yeast 2-hybrid assays were performed using SD agar plates devoid of tryptophan, leucine and histidine, but supplemented with 40 mM of 3-amino-1,2,4-triazole (3AT) (SD/–Trp/–Leu/–His/+3AT).
To construct an expression plasmid that encodes a bait protein as a fusion protein with GAL4 DNA-binding domain, human ALAS-E cDNA was digested with AflII, its sticky end was filled using Klenow DNA polymerase, and then it was digested with BamHI. Fragments were then isolated and subcloned into pAS2-1 expression vector. This was digested with NdeI, its sticky end was filled, and it was digested with BamHI. The junction sequence was confirmed by DNA sequencing. The resulting vector (pAS-AE) encoded a fusion protein consisting of GAL4 DNA-binding domain adjoined to human mature ALAS-E (amino acids 54–587). For yeast 2-hybrid screening assays, both pAS-AE and human bone marrow MATCHMAKER cDNA library (CLONTECH Laboratories Inc.) were cotransformed into the Y190 yeast host strain using the lithium acetate method. After initial selection of positive clones using SD/–Trp/–Leu/–His/+3AT plates, a secondary screening was performed by colony lifting followed by β-galactosidase assays.
To confirm specific protein-protein interactions, we prepared expression vectors for ALAS-E mutant cDNAs by subcloning amino acids 54–587 of each mutant’s cDNA into a filled-in NdeI site of the pAS2-1 vector. These expression vectors were termed pAS-TA, pAS-MO, and pAS-KK, for the D190V, R411C, and M426V mutants, respectively. To construct human ALAS-N expression vector, cDNA for mature human ALAS-N obtained from human placenta cDNA library (CLONTECH Laboratories Inc.) by PCR (using 5′ CATATGCAGATCAAAGAAACCCCTCCGGCC-3′, and 5′ GTCGACGCTAGCCTGAGCAGATACCAACTTG-3′ as primers) was subcloned into pGEM-T-Easy vector (Promega Corp., Madison, Wisconsin, USA). After confirmation of its sequence by DNA sequencing, cDNA clone was digested by NdeI. Then cDNA fragment was isolated and subcloned into the NdeI site of pAS2-1 (pAS-AN). Additionally, cDNA fragment was subcloned into the NdeI site of pGEM-Easy (self-ligated pGEM-T-Easy vector) and digested with EcoRI. Then cDNA fragment was isolated and subcloned into the EcoRI site of pACT2 (pACT-AN). The junction sequence of fusion proteins and the direction of the insert were confirmed by DNA sequencing and restriction analysis with appropriate enzymes.
Northern blot analysis for SCS subunit expression.
Human Multiple Tissue Northern Blot and Human Immune System Multiple Tissue Northern Blot II were purchased from CLONTECH Laboratories Inc. Human SCS-βA cDNA had been isolated by yeast 2-hybrid screening. Human SCS-βG cDNA (GenBank accession number AF058954) and human SCS-α cDNA (GenBank accession number AF104921) were obtained from human placenta cDNA library using PCR. Each cDNA — and human β-actin cDNA, which was used as control — was labeled with [32P]dCTP and used as a probe. Prehybridization, hybridization, and washing of these blots were performed according to the manufacturer’s protocol. The signals were detected by autoradiography.
Isolation of SCS-βA cDNA that encodes the full-length protein.
To obtain cDNA that encodes the full-length SCS-βA protein, Human Heart 5′ Stretch Plus cDNA library (CLONTECH Laboratories Inc.) was screened using SCS-βA cDNA, which had been isolated by yeast 2-hybrid screening. Additionally, we performed a 5′ rapid amplification of cDNA ends assay (5′ RACE), using a SMART PCR cDNA Amplification Kit (CLONTECH Laboratories Inc.) and human heart cDNA library to obtain sequence information upstream of the known end, at the 5′ end of human SCS-βA mRNA. All procedures were performed according to the manufacturer’s protocol.
Transient expression and coimmunoprecipitation analysis.
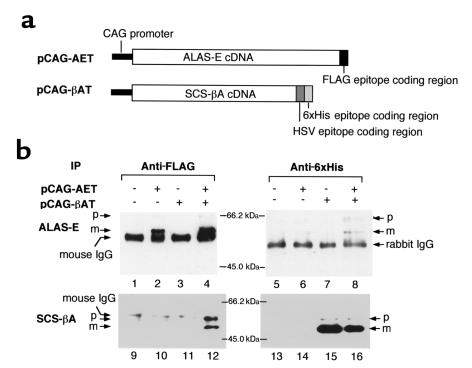
Chinese hamster ovary (CHO) cells were maintained in culture with DMEM supplemented with 10% FCS under 5% CO2, at 37°C. To construct eukaryotic expression vectors for expression of a FLAG epitope tagged with ALAS-E protein, SalI site was introduced by PCR into the 3′ end of the coding region of ALAS-E. After nucleotide sequence confirmation, cDNA fragment that had been obtained by digestion with XhoI and SalI was subcloned into the XhoI and SalI sites of pFLAG-CTS vector (Sigma Chemical Co.). The plasmid was digested with HindIII and XmnI, its sticky end was filled using Klenow DNA polymerase, and the cDNA fragment was isolated and subcloned into an EcoRV site of pGEM-Easy vector. Because this cDNA insert lacks its 5′ half, cDNA fragment of ALAS-E digested with NdeI and XhoI, subcloned into pGEM-Easy vector, was subcloned into the NdeI/XhoI site of the vector pGEM-AET. Next, pGEM-AET cDNA fragment digested with EcoRI was isolated and subcloned into the EcoRI site of the pCAG expression vector (35). The resulting plasmid (pCAG-AET) expressed ALAS-E protein as a FLAG-tagged protein at its COOH terminus (see Figure 3a). To construct expression vectors for HSV-6xHis epitope–tagged hSCS-βA protein, NdeI site was introduced into an expected first ATG (Figure 1, arrow 3) and into the 3′ end of the coding region of hSCS-βA by PCR. After confirmation of nucleotide sequences, the cDNA fragment obtained by digestion with NdeI and NheI was subcloned into the NdeI and NheI sites of pET-25b(+) vector (Novagen, Madison, Wisconsin, USA). Next, the plasmid was digested with XbaI and BlpI, its sticky end was filled using Klenow DNA polymerase, and cDNA fragment was isolated and subcloned into a blunt-ended EcoRI site of the pCAG expression vector. The resultant plasmid (pCAG-βAT) expressed hSCS-βA protein as an HSV-6xHis–tagged protein at its COOH terminus (see Figure 3a).
Figure 3.
Co-immunoprecipitation of ALAS-E and SCA-βA. (a) Schematic representation of FLAG-tagged ALAS-E expression vector and HSV-6xHis–tagged SCS-βA expression vector. (b) Coimmunoprecipitation analysis of ALAS-E and SCS-βA. Full-length human ALAS-E and human SCS-βA were expressed as FLAG epitope–tagged and HSV-6xHis epitope–tagged proteins, respectively. After immunoprecipitation with anti-FLAG monoclonal antibody (lanes 1–4 and 9–12) or anti-6xHis polyclonal antibody (lanes 5–8 and 13–16), the immune complex was examined by Western blot analysis, using as the primary antibody either anti-FLAG monoclonal antibody (lanes 1–8), anti-6xHis polyclonal antibody (lanes 9–12), or anti-HSV monoclonal antibody (lanes 13–16). p, precursor protein; m, mature protein; IP, antibody used for immunoprecipitation.
Figure 1.
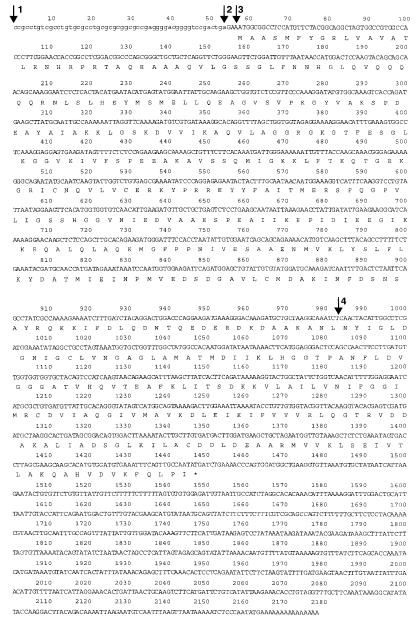
cDNA sequence of human SCS-βA (Accession AB035863). Arrow 1: 5′ end of cDNA that has been identified by 5′ RACE; arrow 2: 5′ end of the insert of pTriplEx-SCSβA; arrow 3:expected first ATG; arrow 4: 5′ end of cDNA in pGAD-SCSβA, pAS-SCSβA, and pACT-SCSβA.
These vectors were transfected into CHO cells using GenePORTER transfection reagent (Gene Therapy Systems Inc., San Diego, California, USA) according to the manufacturer’s protocol. Briefly, 10 μg of each plasmid and 50 μL of GenePORTER reagent was mixed in 5 mL of serum-free medium. This replaced the medium of CHO cells cultured on a dish 10-cm in diameter. Five hours after transfection, 5 mL of culture medium containing 20% FCS was added to the dish. Twelve hours later, the culture medium was replaced with fresh medium containing 10% FCS, and incubation was continued for another 24 hours. For immunoprecipitation, cells were washed twice with PBS, then harvested by scraping in PBS using a spatula. After centrifugation of the cell suspension, mitochondrial fractions were prepared as described previously (13). Each fraction was resuspended in cell-lysis buffer (20 mM HEPES at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 μg/mL leupeptin, 10 μg/mL chymostatin, 10 μg/mL pepstatin A, 10 μg/mL of antipain, 1 mM PMSF, and 1.8 μg/mL aprotinin) and immunoprecipitated with anti-FLAG M2 monoclonal antibody (Sigma Chemical Co.) or His-probe (H-15) polyclonal antibody (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA). Immunoprecipitates were analyzed by SDS-PAGE, followed by immunoblot analysis using anti-FLAG M2 monoclonal antibody, His-probe (H-15), or HSV-Tag monoclonal antibody (Novagen). Immunocomplexes were revealed by ECL (Amersham Life Sciences Inc., Arlington Heights, Illinois, USA).
Results
Yeast 2-hybrid screening.
We screened a human bone marrow cDNA library twice using 2 separate yeast 2-hybrid screening methods. The first screening, of 4 × 106 colonies by the standard method, yielded 1 positive clone. The second screening, of 7 × 106 colonies by the “His3 jump-start” method (the method of choice when a bait protein only weakly interacts with a partner), identified 2 positive clones. Sequence analysis of these clones and homology search using BLAST (36) revealed that all these clones encoded hSCS-βA (pGAD-SCSβA; Figure 1, arrow 4). We subcloned the insert of this clone into pAS2-1 (pAS-SCSβA) or pACT2 (pACT-SCSβA) for further analysis by 2-hybrid assays (Figure 1, arrow 4).
Northern blot analysis for hSCS.
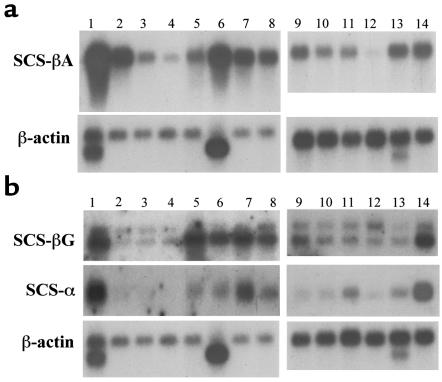
Levels of hSCS-βA mRNA were examined in various human tissues using Human Multiple Tissue Northern Blot (Figure 2a, lanes 1–8) and Human Immune System Multiple Tissue Northern Blot System II (Figure 2a, lanes 9–14) (CLONTECH Laboratories Inc.). The strongest signal was observed in the heart (lane 1), followed by skeletal muscle (lane 6), brain (lane 2), and kidney (lane 7, Figure 2a). This is consistent with the role of A-SCS in ATP production in these tissues, which are known to be highly dependent on oxidative metabolism (34). In hematopoietic systems (Figure 2a), a strong signal was observed in bone marrow (lane 13) and fetal liver (lane 14), followed by spleen (lane 9); only a weak signal was detected in peripheral blood leukocytes (lane 12). One size of mRNA (about 2,200 bases) was detected in all tissues. These findings suggest that hSCS-βA is expressed ubiquitously in various tissues, including those that are active in heme biosynthesis.
Figure 2.
Expression of SCS-βA mRNA in various human tissues, examined by Northern blot. (a) mRNA levels of hSCS-βA. (b) mRNA levels of hSCS-βG and hSCS-α. Lane 1: heart; lane 2: brain; lane 3: placenta; lane 4: lung; lane 5: liver; lane 6: skeletal muscle; lane 7: kidney; lane 8: pancreas; lane 9: spleen; lane 10: lymph nodes; lane 11: thymus; lane 12: peripheral blood leukocytes; lane 13: bone marrow; lane 14: fetal liver.
In contrast to hSCS-βA, 2 different sizes of SCS-βG mRNA (2.5 kb and 3.4 kb) were detected in all tissues examined (Figure 2b), whereas just 1 size of mRNA was detected for SCS-α (Figure 2b). SCS-βG mRNA (Figure 2b) was strongly expressed in heart (lane 1), liver (lane 5), skeletal muscle (lane 6), kidney (lane 7), pancreas (lane 8), and fetal liver (lane 14), whereas SCS-α mRNA was expressed in heart (lane 1), kidney (lane 7), and fetal liver (lane 14). Both SCS-α mRNA and SCS-βG mRNA levels were relatively low in bone marrow (lane 13).
Screening of heart cDNA library and 5′ RACE experiment.
Because cDNA clones that had been isolated by yeast 2-hybrid screening were devoid of their 5′ ends, we screened for clones with longer lengths in a human heart cDNA library that expressed a high level of SCS-βA (Figure 2a, lane 1). Screening of 2 × 105 plaques yielded approximately 40 positive clones. A clone with the longest insert (pTriplEx-SCSβA; Figure 1, arrow 2) was then chosen for transient expression in CHO cells. Additionally, results of 5′ RACE experiments using human heart cDNAs provided information upstream (5′) of the known sequence of hSCS-βA mRNA (Figure 1, arrow 1). Although we did not identify the transcription start site of hSCS-βA, transient transfection followed by immune precipitation analysis revealed (as discussed below) that pTriplEx-SCSβA had in fact encoded the full-length hSCS-βA precursor.
Transient expression followed by coimmunoprecipitation analysis of ALAS-E and SCS-βA.
When pCAG-AET or pCAG-βAT was introduced into CHO cells, each protein was detected in mitochondria by immunoprecipitation with an antibody corresponding to each tagged epitope (Figure 3b, lanes 2, 4, 15, and 16). When pCAG-AET was expressed in CHO cells, 3 bands were detected in immune complex of anti-FLAG antibody (Figure 3b, lanes 2 and 4). A band with the smallest molecular weight (56 kDa) that was observed in all lanes (Figure 3b, lanes 1–4) represented mouse IgG (anti-FLAG antibody) that was used for immunoprecipitation. Estimated molecular sizes of the 2 other bands were 65 kDa and 60 kDa; these sizes were very similar to the calculated size of the precursor (65.7 kDa) and the mature form (60.6 kDa) of FLAG-tagged ALAS-E protein, respectively. When pCAG-βAT was expressed in CHO cells, 2 bands were detected in an immune complex produced by anti-6xHis polyclonal antibody (Figure 3, lanes 15 and 16). Estimated sizes of the 2 bands were 55 kDa and 50 kDa, slightly larger than the sizes of an expected precursor (51.9 kDa) and a mature (46.2 kDa) epitope-tagged SCS-βA protein. Because these bands were specifically detected only in pCAG-βAT–transfected cells, these bands should represent the precursor and the mature form of epitope-tagged SCS-βA protein. When both SCS-βA and ALAS-E cDNA were expressed, both proteins were detected in an immune complex from CHO cells, using an anti-FLAG antibody (Figure 3b, lanes 4 and 12) or anti-6xHis antibody (Figure 3b, lanes 8 and 16). In contrast, SCS-βA was not detected in an immune complex with anti-FLAG antibody from cells in which either SCS-βA (Figure 3b, lane 11) or ALAS-E (Figure 3b, lane 10) was expressed alone. ALAS-E was also not detectable in an immune complex with anti-6xHis antibody from cells in which either SCS-βA (Figure 3b, lane 7) or ALAS-E (Figure 3b, lane 6) was expressed alone. These findings conclusively indicate that there is specific binding of ALAS-E to SCS-βA.
Our findings also show that both the precursor and the mature form of both SCS-βA and ALAS-E were detectable in the immune complex with anti-FLAG (Figure 3, lane 12) or anti-6xHis antibody (Figure 3, lane 8). These findings suggest that ALAS-E associates with SCS-βA not only in the mitochondria, but also in the cytoplasm and at the outer membrane of mitochondria.
Additionally, it should be noted that 2 bands were detected for hSCS-βA in each immune complex with a corresponding antibody (Figure 3, lanes 12, 15, and 16). SCS is a nuclear-gene encoded mitochondrial enzyme, which is translated as a precursor (pre–SCS-βA) in the cytoplasm and then translocated into mitochondria. Similarly to ALAS-E, its presequence (the signal sequence for targeting into mitochondria) would be processed to yield a mature enzyme within mitochondria. Because the presequence is present at the NH2 terminus of the protein, the 2 detected bands should represent the precursor and the mature form of SCS-βA. Thus our findings are consistent with pTriplEx-SCSβA encoding the full-length precursor protein of hSCS-βA.
Interaction of hSCS-βA with ALAS isoforms.
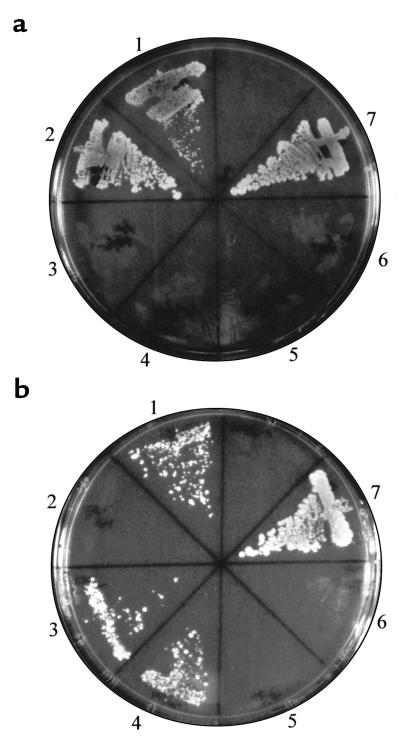
We then examined complex formation of ALAS isoforms with SCS-βA, using the yeast 2-hybrid system. Figure 4a shows the interaction of SCS-βA with ALAS-E (area 1). SCS-βA did not interact with the mature form of human ALAS-N (area 3). Similar findings were observed when expression vectors for fusion proteins were exchanged (Figure 4a, areas 2 and 4). When the Y190 yeast strain was transformed with pAS2-1 and pACT-SCSβA (Figure 4a, area 5), or pAS-SCSβA and pACT2 (Figure 4a, area 6), protein-protein interaction was not observed. These findings indicate that interaction of SCS-βA with ALAS is specific for ALAS-E and does not occur with ALAS-N.
Figure 4.
Association of ALAS with SCS-βA. (a) Association of ALAS-E or ALAS-N with SCS-βA. Y190 yeast strain was transfected by each vector, and selection was made using an SD/–Trp/–Leu/–His/+3AT plate. The pVA3-1 and pTD1-1 vectors are positive controls in the yeast 2-hybrid system. Areas represented by numbers: 1: pAS-AE and pACT–SCS-βA; 2: pAS–SCS-βA and pACT-AE; 3: pAS-AN and pACT–SCS-βA; 4: pAS–SCS-βA and pACT-AN; 5: pAS2-1 and pACT–SCS-βA; 6: pAS–SCS-βA and pACT2; 7: pVA3-1 and pTD1-1. (b) Association of SCS-βA with ALAS-E mutants from XLSA. Y190 yeast strain was transfected by each vector, and selection was made using an SD/–Trp/–Leu/–His/+3AT plate. The vectors pVA3-1 and pTD1-1 were used as positive controls. 1: pAS-AE and pACT–SCS-βA; 2: pAS-TA and pACT–SCS-βA; 3: pAS-MO and pACT–SCS-βA; 4: pAS-KK and pACT–SCS-βA; 5: pAS2-1 and pACT–SCS-βA; 6: pAS–SCS-βA and pACT2; 7: pVA3-1 and pTD1-1.
Interaction of hSCS-βA with mutant ALAS-E.
We also examined the interaction of SCS-βA with ALAS-E mutants that had been identified in patients with XLSA. Two mutants of ALAS-E, R411C and M426V, which had been identified in patients with pyridoxine-responsive XLSA (29, 30), were found to show similar interaction with SCS-βA (Figure 4b, areas 3 and 4), as did the wild-type ALAS-E (Figure 4b, area 1). However, a pyridoxine-refractory D190V mutant, which results in an unstable enzyme, failed to interact with SCS-βA (Figure 4b, area 2). Interaction of the wild-type and pyridoxine-responsive mutants of ALAS-E with SCS-βA, but not with a pyridoxine-refractory mutant, was also demonstrated when their expression vectors were exchanged (data not shown). Although the cDNA insert of SCS-βA used in Figure 4 is shorter than the one that encoded the mature SCS-βA, identical results were observed when we used the mature SCS-βA (amino acids 53–463, Figure 1) as a fusion protein in the yeast 2-hybrid assay (data not shown).
Discussion
It is well known that expression of ALAS-E is controlled at several levels, including transcription (37), translation (38), and importation into mitochondria (14, 15). For example, the promoter in human ALAS-E is known to contain several putative erythroid-specific cis-acting elements, such as GATA-1, CACCC, and CCAAT (7); these elements work together to promote erythroid-specific transcription (11). At the translational level, an iron-responsive element in the 5′ nontranslated region of ALAS-E mRNA controls its translation in response to iron concentration in cells (38, 39). Additionally, it is known that binding of heme to the heme-responsive motif of ALAS-E protein negatively regulates its importation into mitochondria (14, 15). Heme synthesized in mitochondria could potentially inhibit ALAS activity, though this mechanism appears to play a less significant role than other ALAS-inhibiting mechanisms (16). It is not known, however, whether the function and activity of ALAS-E can be modulated by other proteins in mitochondria with which it interacts.
In this study, we examined this question by screening for a protein that may associate with the mature form of ALAS-E, thereby influencing its function in mitochondria, by using the yeast 2-hybrid system. Our findings unequivocally demonstrate that the mature form of ALAS-E associates with SCS-βA in mitochondria. Because both ALAS-E and SCS are thought to be either loosely associated with the inner membrane or present in the matrix of mitochondria, SCS may supply succinyl CoA, 1 of the 2 ALAS-E substrates, in the form of an enzyme-enzyme complex. Both the precursor forms of ALAS-E and SCS-βA were detected in the immune complex with anti-6xHis (Figure 3b, lane 8) and anti-FLAG (Figure 3b, lane 12) antibodies, respectively. These findings suggest that these proteins may function as transporters for each other from cytosol to mitochondria or within mitochondria.
It has been suggested that A-SCS may participate in the Krebs cycle, whereas G-SCS may catalyze the reverse reaction to support heme synthesis (40). This suggestion was made based on changes in the ratio of G-SCS activity to A-SCS activity during experimentally induced diabetes mellitus and porphyria. The Km of A-SCS for succinate is also significantly higher than that of G-SCS (34). However, exact assessment of the ratio of A-SCS activity to G-SCS activity is difficult, because the apparent activity may arise from the other form of SCS and the recycling of endogenous nucleotides by nucleoside-diphosphate kinase (41). Thus, the precise mechanism of utilization of each isoform for specific metabolic reactions remains unknown. In bone marrow, SCS-βA, not SCS-βG, is the predominantly expressed form, as suggested by RT-PCR (33) and quantitatively demonstrated by Northern blot analysis (Figure 2, lane 13). Although SCS-βA interacts with ALAS-E, we could not detect interaction of either SCS-βG or SCS-α with either ALAS-E or ALAS-N in the yeast 2-hybrid system (data not shown). Therefore, in bone marrow erythroblasts, it should be A-SCS, rather than G-SCS, that contributes to heme synthesis, and it may do so by its specific association with ALAS-E, though the situation in other tissues remains less clear. It seems reasonable that A-SCS in erythroblasts may be involved in the direction of succinyl CoA synthesis through an SCS-βA/ALAS-E complex, which may allow A-SCS to overcome its high Km for succinate. Furthermore, association of ALAS-E with SCS-βA may also be involved in erythroid-specific expression of ALAS-E, because ALAS-N failed to associate with SCS-βA.
It should also be noted that pGAD-SCSβA (the original clone that was isolated by yeast 2-hybrid screening) encoded amino acids 310–463 of SCS-βA (indicated by arrow 4, Figure 1), and was used for subcloning into pAS-SCSβA and pACT-SCSβA, whereas the mature SCS-βA consists of 411 amino acids (53–463; Figure 1). The fact that both the shorter and the mature SCS-βA insert resulted in association with ALAS-E indicates that the domain for interaction resides in the last 153 amino acids of SCS-βA, i.e., in the last 3/8 of the mature form, or in the last 3/9 of the precursor form of SCS-βA.
Appropriate association of SCS-βA with ALAS-E also has an important implication in the pathogenesis of certain pyridoxine-refractory forms of XLSA. We have previously identified a D190V mutation of ALAS-E in a patient with pyridoxine-refractory XLSA that resulted in an unstable ALAS-E protein (30). Among approximately 23 mutations so far described in XLSA (42), D190V is the only mutant that has been shown to result in an unstable protein in vivo in the patient (30). When this mutant was expressed in vitro, the protein was of normal size for pre–ALAS-E. On the other hand, after translocation into mitochondria, the wild-type pre–ALAS-E was processed into a mature form, whereas the D190V mutant protein resulted in aberrant products with higher molecular weights than their normal mature forms. Because ALAS-E protein was markedly decreased in erythroblasts of the studied patient, this finding indicates that the aberrant products must have undergone extensive degradation (30). When we examined association of SCS-βA with ALAS-E, 2 pyridoxine-responsive mutants (R411C and M426V) were found to interact normally with SCS-βA, as did the wild type. The pyridoxine-refractory mutant (D190V) completely failed to associate with SCS-βA (Figure 4b). Because D190V mutation resulted in an unstable protein in vivo, it is possible that SCS-βA plays an important role in protecting ALAS-E from degradation in mitochondria. Such a failure of ALAS-E mutation to associate with SCS-βA may also accompany decreased access to succinyl CoA in bone marrow erythroblasts or reduced translocation into mitochondria. This possibility may be an important consideration in the case of pyridoxine-refractory XLSA that may result from unstable ALAS-E. Conversely, a mutation of SCS-βA that may disrupt its normal association with ALAS-E may lead to a pyridoxine-refractory SA.
When SCS-βA was expressed in CHO cells as an epitope-tagged protein, we were able to detect 2 bands by immunoprecipitation followed by Western blot analysis (Figure 3b, lanes 15 and 16). The sizes of the expressed epitope-tagged SCS-βA proteins (the precursor and the mature form) were slightly larger than the calculated molecular mass. Computing analysis of SCS-βA protein revealed that this protein has 2 sites for phosphorylation by protein kinase C, 1 site for phosphorylation by tyrosine kinase, and 1 site for N-glycosylation. Therefore it is quite possible that these modifications of the protein may increase the size of SCS-βA from its estimated molecular mass. SCS-βA is transcribed from its genomic DNA, synthesized on cytoplasmic ribosomes, and imported into mitochondria, after which its presequence is processed. Thus, the detected large and small bands represent the precursor and the mature form of SCS-βA, respectively. Because the precursor protein contained the presequence that permitted the importation of pre–SCS-βA into mitochondria, followed by proper processing of its presequence in mitochondria, this clone must encode the full-length SCS-βA protein. Additionally, cDNA sequences for the region including the first methionine of this clone are compatible with the consensus sequence of the translation start site (43). Although we did not confirm the transcription start site of SCS-βA, these findings are entirely consistent with the fact that this clone encodes a full-length SCS-βA protein.
In this report, we present novel evidence for the association of ALAS-E with SCS-βA. Although some protein complex formation has been previously suggested for enzymes in the heme biosynthetic pathway (44, 45), it has been difficult to unequivocally demonstrate a direct protein-protein interaction. In this respect, our study using yeast 2-hybrid screening is the first report of bona-fide protein-protein interaction of enzymes in the heme biosynthetic pathway. Yeast 2-hybrid assay is a powerful tool to use in examining specific protein-protein interactions and searching for a protein that can associate with the known protein of interest. Our studies have successfully demonstrated that this technique is useful for identifying interaction between 2 key proteins that are related to heme biosynthesis in mitochondria. Our strategy of using the mature sequence of a mitochondrial enzyme that is devoid of its presequence as the bait should be very useful in obtaining critical information about specific interaction with new proteins in mitochondria, and for investigation of the domains involved in such interactions.
Acknowledgments
We thank Norio Hayashi, Hiroyoshi Fujita, Masayuki Yamamoto, and George Drummond for their helpful discussion. We also thank Luba Garbaczewski and Sakiko Kusunoki for their excellent technical assistance in this work. This work was supported in part by United States Public Health Service Grant DK32890, the Chugai Fund for Molecular Hematology (S. Sassa), the Yamanouchi Molecular Medicine Research Fund, and Yamanouchi Foundation for Research on Metabolic Disorders (K. Furuyama).
References
- 1.Whiting MJ, Granick S. δ-Aminolevulinic acid synthetase from chick embryo liver mitochondria. II. Immunochemical correlation between synthesis and activity in induction and repression. J Biol Chem. 1976;251:1347–1353. [PubMed] [Google Scholar]
- 2.Ferreira GC. Heme biosynthesis: biochemistry, molecular biology, and relationship to disease. J Bioenerg Biomembr. 1995;27:147–150. doi: 10.1007/BF02110029. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto M, et al. Isolation of recombinant cDNAs encoding chicken erythroid δ-aminolevulinate synthase. Proc Natl Acad Sci USA. 1985;82:3702–3706. doi: 10.1073/pnas.82.11.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutherland GR, et al. 5-Aminolevulinate synthase is at 3p21 and thus not the primary defect in X-linked sideroblastic anemia. Am J Hum Genet. 1988;43:331–335. [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter PD, Willard HF, Gorski JL, Bishop DF. Assignment of human erythroid δ-aminolevulinate synthase (ALAS2) to a distal subregion of band Xp11.21 by PCR analysis of somatic cell hybrids containing X;autosome translocations. Genomics. 1992;13:211–212. doi: 10.1016/0888-7543(92)90223-f. [DOI] [PubMed] [Google Scholar]
- 6.Cox TC, et al. Erythroid 5-aminolevulinate synthase is located on the X chromosome. Am J Hum Genet. 1990;46:107–111. [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto, M., Kim, L.-C., Nagai, T., Furuyama, K., and Engel, J.D. 1994. Structure and regulation of vertebrate δ-aminolevulinate synthase. In Regulation of heme protein synthesis. H. Fujita, editor. AlphaMed Press. Dayton, OH. 11–25.
- 8.Meguro K, Igarashi K, Yamamoto M, Fujita H, Sassa S. The role of the erythroid-specific delta-aminolevulinate synthase gene expression in erythroid heme synthesis. Blood. 1995;86:940–948. [PubMed] [Google Scholar]
- 9.Harigae H, et al. Deficient heme and globin synthesis in embryonic stem cells lacking the erythroid-specific 5-aminolevulinate synthase gene. Blood. 1998;91:798–805. [PubMed] [Google Scholar]
- 10.Yamamoto M, Hayashi N, Kikuchi G. Evidence for the transcriptional inhibition by heme of the synthesis of δ-aminolevulinate synthase in rat liver. Biochem Biophys Res Commun. 1982;105:985–990. doi: 10.1016/0006-291x(82)91067-1. [DOI] [PubMed] [Google Scholar]
- 11.Surinya KH, Cox TC, May BK. Transcriptional regulation of the human erythroid 5-aminolevulinate synthase gene. Identification of promoter elements and role of regulatory proteins. J Biol Chem. 1997;272:26585–26594. doi: 10.1074/jbc.272.42.26585. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto M, Hayashi N, Kikuchi G. Translational inhibition by heme of the synthesis of hepatic δ-aminolevulinate synthase in a cell-free system. Biochem Biophys Res Commun. 1983;115:225–231. doi: 10.1016/0006-291x(83)90993-2. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi K, Hayashi N, Kikuchi G. Translocation of δ-aminolevulinate synthase from the cytosol to the mitochondria and its regulation by hemin in the rat liver. J Biol Chem. 1980;255:1746–1751. [PubMed] [Google Scholar]
- 14.Lathrop JT, Timko MP. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science. 1993;259:522–525. doi: 10.1126/science.8424176. [DOI] [PubMed] [Google Scholar]
- 15.Munakata H, Furuyama K, Hayashi N. The mitochondrial transfer of the non-specific and the erythroid-specific 5-aminolevulinate synthase and the heme regulatory motif. J Biochem (Tokyo) 1996;68:792. (Abstr.) [Google Scholar]
- 16.Wolfson SJ, Bartczak A, Bloomer JR. Effect of endogenous heme generation on δ-aminolevulinic acid synthase activity in rat liver mitochondria. J Biol Chem. 1979;254:3543–3546. [PubMed] [Google Scholar]
- 17.Sassa S, Nagai T. The role of heme in gene expression. Int J Hematol. 1996;63:167–178. doi: 10.1016/0925-5710(96)00449-5. [DOI] [PubMed] [Google Scholar]
- 18.Kappas, A., Sassa, S., Galbraith, R.A., and Nordmann, Y. 1995. The porphyrias. In The metabolic and molecular basis of inherited disease. C.R. Scriver, A.L. Beaudet, W.S. Sly, and D. Valle, editors. McGraw-Hill Inc. New York, NY. 2103–2159.
- 19.Beutler, E. 1995. Hereditary and acquired sideroblastic anemia. In Williams hematology. E. Beutler, M.A. Lichtman, B.S. Coller, and T.J. Kipps, editors. McGraw-Hill Inc. New York, NY. 747–750.
- 20.Bottomley, S.S. 1993. Sideroblastic anemia. In Wintrobe’s clinical hematology. G.R. Le, T.C. Bithell, J. Foster, J.W. Athens, and J.N. Lukens, editors. Lea & Febiger. Philadelphia, PA. 852–871.
- 21.Cotter PD, Baumann M, Bishop DF. Enzymatic defect in “X-linked” sideroblastic anemia: molecular evidence for erythroid delta-aminolevulinate synthase deficiency. Proc Natl Acad Sci USA. 1992;89:4028–4032. doi: 10.1073/pnas.89.9.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox TC, et al. X-linked pyridoxine-responsive sideroblastic anemia due to a Thr388-to-Ser substitution in erythroid 5-aminolevulinate synthase. N Engl J Med. 1994;330:675–679. doi: 10.1056/NEJM199403103301004. [DOI] [PubMed] [Google Scholar]
- 23.Cotter PD, Rucknagel DL, Bishop DF. X-linked sideroblastic anemia: identification of the mutation in the erythroid-specific δ-aminolevulinate synthase gene (ALAS2) in the original family described by Cooley. Blood. 1994;84:3915–3924. [PubMed] [Google Scholar]
- 24.Cotter PD, et al. Late-onset X-linked sideroblastic anemia. Missense mutations in the erythroid delta-aminolevulinate synthase (ALAS2) gene in two pyridoxine-responsive patients initially diagnosed with acquired refractory anemia and ringed sideroblasts. J Clin Invest. 1995;96:2090–2096. doi: 10.1172/JCI118258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prades E, et al. A new mutation of the ALAS2 gene in a large family with X-linked sideroblastic anemia. Hum Genet. 1995;95:424–428. doi: 10.1007/BF00208968. [DOI] [PubMed] [Google Scholar]
- 26.Edgar AJ, Losowsky MS, Noble JS, Wickramasinghe SN. Identification of an arginine452 to histidine substitution in the erythroid 5-aminolaevulinate synthetase gene in a large pedigree with X-linked hereditary sideroblastic anaemia. Eur J Haematol. 1997;58:1–4. doi: 10.1111/j.1600-0609.1997.tb01402.x. [DOI] [PubMed] [Google Scholar]
- 27.Edgar AJ, Wickramasinghe SN. Hereditary sideroblastic anaemia due to a mutation in exon 10 of the erythroid 5-aminolaevulinate synthase gene. Br J Haematol. 1998;100:389–392. doi: 10.1046/j.1365-2141.1998.00569.x. [DOI] [PubMed] [Google Scholar]
- 28.Edgar AJ, Vidyatilake HM, Wickramasinghe SN. X-linked sideroblastic anaemia due to a mutation in the erythroid 5-aminolaevulinate synthase gene leading to an arginine170 to leucine substitution. Eur J Haematol. 1998;61:55–58. doi: 10.1111/j.1600-0609.1998.tb01061.x. [DOI] [PubMed] [Google Scholar]
- 29.Furuyama K, et al. R411C mutation of the ALAS2 gene encodes a pyridoxine-responsive enzyme with low activity. Br J Haematol. 1998;103:839–841. doi: 10.1046/j.1365-2141.1998.01050.x. [DOI] [PubMed] [Google Scholar]
- 30.Furuyama K, et al. Pyridoxine refractory X-linked sideroblastic anemia caused by a point mutation of the erythroid-specific 5-aminolevulinate synthase gene. Blood. 1997;90:822–830. [PubMed] [Google Scholar]
- 31.Nishimura JS. Succinyl-CoA synthetase structure-function relationships and other considerations. Adv Enzymol Relat Areas Mol Biol. 1986;58:141–172. doi: 10.1002/9780470123041.ch4. [DOI] [PubMed] [Google Scholar]
- 32.Ottaway JH, McClellan JA, Saunderson CL. Succinic thiokinase and metabolic control. Int J Biochem. 1981;13:401–410. doi: 10.1016/0020-711x(81)90111-7. [DOI] [PubMed] [Google Scholar]
- 33.Johnson JD, Mehus JG, Tews K, Milavetz BI, Lambeth DO. Genetic evidence for the expression of ATP- and GTP-specific succinyl CoA synthetases in multicellular eucaryotes. J Biol Chem. 1998;273:27580–27586. doi: 10.1074/jbc.273.42.27580. [DOI] [PubMed] [Google Scholar]
- 34.Johnson JD, Muhonen WW, Lambeth DO. Characterization of the ATP- and GTP-specific succinyl CoA synthetases in pigeon. The enzymes incorporate the same alpha-subunit. J Biol Chem. 1998;273:27572–27579. doi: 10.1074/jbc.273.42.27573. [DOI] [PubMed] [Google Scholar]
- 35.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Fujita H, Yamamoto M, Yamagami T, Hayashi N, Sassa S. Erythroleukemia differentiation. Distinctive responses of the erythroid-specific and the nonspecific δ-aminolevulinate synthase mRNA. J Biol Chem. 1991;266:17494–17502. [PubMed] [Google Scholar]
- 38.Melefors O, et al. Translational control of 5-aminolevulinate synthase mRNA by iron-responsive elements in erythroid cells. J Biol Chem. 1993;268:5974–5978. [PubMed] [Google Scholar]
- 39.Cox TC, Bawden MJ, Martin A, May BK. Human erythroid 5-aminolevulinate synthase: promoter analysis and identification of an iron-responsive element in the mRNA. EMBO J. 1991;10:1891–1902. doi: 10.1002/j.1460-2075.1991.tb07715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins TM, Weitzman PD. Distinct physiological roles of animal succinate thiokinases. Association of guanine nucleotide-linked succinate thiokinase with ketone body utilization. FEBS Lett. 1986;205:215–218. doi: 10.1016/0014-5793(86)80900-0. [DOI] [PubMed] [Google Scholar]
- 41.Kadrmas EF, Ray PD, Lambeth DO. Apparent ATP-linked succinate thiokinase activity and its relation to nucleoside diphosphate kinase in mitochondrial matrix preparations from rabbit. Biochim Biophys Acta. 1991;1074:339–346. doi: 10.1016/0304-4165(91)90083-s. [DOI] [PubMed] [Google Scholar]
- 42.May A, Bishop DF. The molecular biology and pyridoxine responsiveness of X-linked sideroblastic anaemia. Haematologica. 1998;83:56–70. [PubMed] [Google Scholar]
- 43.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higuchi M, Bogorad L. The purification and properties of uroporphyrinogen I synthase and uroporphyrinogen III cosynthase. Interactions between the enzymes. Ann NY Acad Sci. 1975;244:401–418. doi: 10.1111/j.1749-6632.1975.tb41545.x. [DOI] [PubMed] [Google Scholar]
- 45.Ponka P. Tissue-specific regulation of iron metabolism and heme synthesis: distinct control mechanisms in erythroid cells. Blood. 1997;89:1–25. [PubMed] [Google Scholar]